Abstract
Demyelination occurs following many neurological insults, most notably in multiple sclerosis (MS). Therapeutics that promote remyelination could slow the neurological decline associated with chronic demyelination; however, in vivo testing of candidate small molecule drugs and signaling cascades known to impact myelination is expensive and labor intensive. Here, we describe the development of a novel zebrafish line which uses the putative promoter of Myelin Protein Zero (mpz), a major structural protein in myelin, to drive expression of Enhanced Green Fluorescent Protein (mEGFP) specifically in the processes and nascent internodes of myelinating glia. We observe that changes in fluorescence intensity in Tg(mpz:mEGFP) larvae are a reliable surrogate for changes in myelin membrane production per se in live larvae following bath application of drugs. These changes in fluorescence are strongly predictive of changes in myelin specific mRNAs [mpz, 36K and myelin basic protein (mbp)] and protein production (Mbp). Finally, we observe that certain drugs alter nascent internode number and length, impacting the overall amount of myelin membrane synthesized and number of axons myelinated without significantly changing the number of myelinating oligodendrocytes. These studies demonstrate that the Tg(mpz:mEGFP) reporter line responds effectively to positive and negative small molecule regulators of myelination, and could be useful for identifying candidate drugs that specifically target myelin membrane production in vivo. Combined with high throughput cell-based screening of large chemical libraries and automated imaging systems, this transgenic line should be useful for rapid large scale whole animal screening to identify novel myelinating small molecule compounds in vivo.
Keywords: Zebrafish, Myelin protein zero, Oligodendrocytes, Myelination, In vivo
Graphical Abstract
INTRODUCTION
Proper neurologic function arises from the efficient transmission of electrical and chemical signals between individual neurons organized into functional networks. Complex nervous systems have evolved a highly specialized structure, myelin, which is the thick multilayered sheath that surrounds large caliber axons. Myelin is produced by oligodendrocytes in the central nervous system (CNS) and Schwann cells in the peripheral nervous system (PNS). In addition to the role of myelin in promoting rapid energy-efficient electrical conduction, a growing body of research suggests that myelin supports the overall health of the nervous system by supplying metabolic support in the form of lactate to axons (Funfschilling et al., 2012; Y. Lee et al., 2012), which extend long distances and are often isolated from neuronal cell bodies. Similarly, loss of critical myelin proteins such as proteolipid protein (PLP) or 2’,3’, cyclic nucleotide 3’ phosphodiesterase (CNP) alters myelin, leading to axon swelling and degeneration in animals as they age (Anderson et al., 1998; Griffiths et al., 1998; Lappe-Siefke et al., 2003; Rasband et al., 2005; Uschkureit, Sporkel, Stracke, Büssow, & Stoffel, 2000), which suggests that the long term health of many neurons is dependent on intact myelin.
Demyelination, or loss of the myelin sheath, is a common feature in a variety of neurological diseases (reviewed in (Chang et al., 2012; Trapp & Nave, 2008), and additionally often occurs following traumatic injury such as stroke (Aboul-Enein et al., 2003; Goldenberg-Cohen et al., 2005) or spinal cord injury (Almad, Sahinkaya, & McTigue, 2011; Smith & Jeffery, 2006). Perhaps the best described demyelinating disorder is multiple sclerosis (MS), where recurring immune attacks target myelin sheaths and/or oligodendrocytes, resulting in focal loss of myelin with resulting neurological deficits (for extensive reviews see (Fox, 2004; R. J. Franklin, 2002; Trapp & Nave, 2008). Disease severity is often related to the endogenous ability of resident oligodendrocyte progenitor cells (OPCs) to replace damaged myelin and repair neurological dysfunction (reviewed in (Fancy et al., 2010; R. J. Franklin & Ffrench-Constant, 2008). Currently, MS therapeutics target the immune component of the disease by blocking immune cell entry into the CNS, e.g.,Tysabri (Stüve & Bennett, 2007) or dampening of the inflammatory immune response e.g., copaxone (Kala, Miravalle, & Vollmer, 2011), fingolimod [Gilenya (Groves, Kihara, & Chun, 2013)] or dimethyl fumarate [Tecfidera (Scannevin et al., 2012)]. While these therapeutics, by reducing the immune component of the disease, have been effective in slowing disease progression, they are not designed to repair the existing damage in the CNS. Thus, there remains a great need to identify and optimize small molecule drugs that enhance oligodendrocyte development and endogenous repair mechanisms to replace myelin and protect vulnerable axons, particularly in actively demyelinated plaques.
High throughput small molecule screens to identify candidate pro-myelinating drugs have been developed using isolated oligodendrocyte lineage cells from rodent or human pluripotent stem-cell derived culture systems (Lariosa-Willingham et al., 2016; Peppard et al., 2015; Usher et al., 2010). However, these systems lack the complexity of cell-cell interactions found in intact nervous systems and provide limited information on potential therapeutic efficacy in vivo or systemic effects of drug delivery in whole animals. Because it remains time-consuming and expensive to assay myelination in vivo in rodents, zebrafish (Danio rario) have been identified as a useful model system for drug discovery screens. A large body of research shows that molecules and canonical signaling cascades driving myelination are conserved in the fish (for extensive reviews see (Tim Czopka & Lyons, 2011; Monk & Talbot, 2009; M. A. Preston & Macklin, 2014); that fish respond to drugs in a comparable manner to other animals (MacRae & Peterson, 2015); and that detailed cellular responses to bath application of drugs are readily quantifiable e.g., OPC migration, process extension and internode growth and stability, reviewed in (C. E. Buckley, A. Marguerie, W. K. Alderton, & R. J. M. Franklin, 2010; D’Rozario, Monk, & Petersen, 2017; M. A. Preston & Macklin, 2014). Overall, these data support using zebrafish larvae as a rapid, cost-efficient model system for screening candidate small molecules in an intact animal.
Given that zebrafish embryos are transparent and develop ex utero, and that rapid myelination of the spinal cord occurs during the first week of life, the current model was designed to rapidly assay oligodendrocyte morphology and myelin membrane production in live zebrafish larvae. The use of fluorescent markers for detailed quantification of oligodendrocyte behavior in response to small molecule drug exposure in real time has been well documented (see Buckley, Marguerie et al. 2010, Preston and Macklin 2014, D’Rozario, Monk et al. 2017) and can be used to provide insight into potential mechanisms of drug action. Tracking fluorophore accumulation in myelin-producing cells allows us to use fluorescence intensity as a surrogate for myelin gene promoter activity per se in lieu of more labor intensive approaches such as quantitative polymerase chain reaction (qPCR) of myelin gene transcripts, western blot or electron microscopy analysis. Furthermore, quantifying membrane-tethered fluorophore is an excellent proxy for myelin membrane production. Several transgenic lines that label oligodendrocyte lineage cells are available (see (M. A. Preston & Macklin, 2014). However, in these lines, transgene expression is also seen in non-oligodendrocyte lineage cell populations such as neurons in Tg(olig2:EGFP) (H.-C. Park, Mehta, Richardson, & Appel, 2002) or Tg(nkx2.2:mEGFP) (Kucenas, Snell, & Appel, 2008) or in neural crest-derived PNS progenitors in Tg(sox10:mRFP) reporter fish (Takada, Kucenas, & Appel, 2010), which complicates quantification of fluorescent intensity as a proxy for myelin gene expression in the spinal cord or in whole animals.
As our goal was to quantify myelin membrane production in the CNS, where remyelination is less efficient than seen in the PNS, we generated a novel fluorescent transgenic zebrafish line that specifically labels the processes of myelin producing cells. This transgenic zebrafish line was developed using the putative promoter for myelin protein zero (mpz), the zebrafish homolog of Protein Zero (P0), a major structural protein in myelin (Bai, Sun, Stolz, & Burton, 2011; Brosamle & Halpern, 2002). The mpz promoter was selected due to prior reports that suggested that mpz transcripts and Mpz protein expression is confined to the CNS in developing zebrafish (Brosamle & Halpern, 2002; C. E. Buckley, A. Marguerie, W. K. Alderton, & R. J. Franklin, 2010). This contrasts with other myelin promoters such as myelin basic protein (mbp) or proteolipid protein (plp), which are highly expressed in both the CNS and PNS in zebrafish (Brosamle and Halpern 2002) or 36K, which despite being an excellent marker for maturing CNS oligodendrocytes, is not conserved in all mammalian species (Morris et al., 2004). In these studies, we find that the mpz promoter drives expression of membrane-tethered Enhanced Green Fluorescent Protein (mEGFP) in the processes of oligodendrocyte lineage cells, allowing visualization of nascent myelin internodes in the CNS of live animals. Live confocal imaging of mpz transgenic embryos [e.g. Tg(mpz:mEGFP)] was used to quantitatively assay myelin membrane production in vivo in response to bath application of therapeutically relevant small molecules. Changes in fluorescent reporter response (EGFP intensity) were then compared to changes in gene expression using qPCR for myelin specific transcripts or by immunohistochemistry (IHC), western blot analysis for myelin basic protein (Mbp in zebrafish) expression and electron microscopic analysis of myelin production in the spinal cord of developing larvae. These studies demonstrate that mpz-driven mEGFP expression is a reliable surrogate for myelin production in developing zebrafish larvae and is useful for rapidly determining the pro-myelinating efficacy of small molecules in an in vivo environment. This model should complement cell culture-based screening platforms for discovery of candidate pro-myelinating small molecules.
METHODS
Isolation of putative 12.5kb mpz promoter:
Using recombination-mediated genetic engineering, or ‘Recombineering’ (P. Liu, Jenkins, & Copeland, 2003), the mpz promoter was isolated from a bacterial artificial chromosome (BAC) by gap repair using published protocols (Kitazono, 2009). Briefly, a query search of the Ensembl Genome Browser (www.ensembl.org) was used to identify and download a 20kb upstream sequence upstream (5’ −20kb-0 bp) of the zebrafish mpz-001 transcript start site. This sequence was used to search the UCSC genome browser (Zv9) and paired BAC ends were visualized to identify candidate BACs likely to contain mpz promoter sequence (5’ upstream of start codon). BAC CH73–42C9 (BacPAC Resources; http://bacpac.chori.org) was used to generate BAC plasmid DNA by midi prep (PureLink™ HiPure Plasmid Filter Kit, Invitrogen). RT-PCR confirmed the predicted mpz promoter sequence (3 distinct 2kb amplicons, data not shown). 500bp homology arms flanking a 12.5kb region immediately upstream of the predicted start ATG of mpz-001 (−12.5kb-12kb and −500bp-0bp) were joined with a ~25bp Fsel cassette using Fusion PCR and subcloned into p5E-MCS vector using XhoI and BamHI restriction sites (added during generation of 12.5kb homology cassette). Primers used for mpz homology arm isolation: Left arm (12.5kb-11.5kb upstream start ATG): Forward TCTTGTTTCCTCAAGGTAAATAAAC, Reverse TGGAGTAGGAGTGAAGAATTGGAAG, Right arm (−500bp to 0): Forward CTCTTTCCCTGTCAAACAGATGTTC, Reverse GATCTCTCTCCGATCTCCCCGCAGG). The resulting p5E-12H-mpz12.5kb targeting plasmid was linearized, dephosphorylated and electroporated into the recombination competent bacterial strain SW-102 containing BAC CH73–42C9 and heat shocked in induce ‘GAP repair’ and recovery ((Kitazono, 2009; P. Liu et al., 2003) of a Gateway compatible vector containing the putative mpz promoter (p5E-mpz12.5kb). Clones were screened by colony PCR and restriction digest, then sequenced (UC Denver Genomics Core), and a BLAST-n search was used to confirm retrieval of putative mpz promoter regions (data not shown).
Following isolation of the mpz promoter, Gateway Tol2 methodology (Kwan et al., 2007) was used to generate multiple mpz promoter driven fluorescent reporter constructs, pEXPRESS-mpz12.5kb:EGFP.pA.cmcl2:mCherry.tol2, pEXPRESS-mpz12.5kb:EGFP.CAAX.pA.cmcl2:mCherry.tol2 and pEXPRESS-mpz12.5kb:mCherry.CAAX.pA.CG2.tol2 lines, which expressed cytosolic EGFP, membrane tethered EGFP or mCherry; respectively. 1–5 nl of each construct (50μg/μl) was then injected into 1–2 cell wild type (AB or TL background) zebrafish embryos along with 25ng of capped Tol2 transposase mRNA to promote integration of the mpz reporter construct into the zebrafish genome. Live confocal imaging of injected embryos was used to confirm transient labeling of mature oligodendrocytes in vivo in the embryonic spinal cord (data not shown). Expressing F0 fish were raised to adulthood and screened for germ line transmission. Positive F1 embryos were raised as stable transgenic mpz reporter lines. Embryos/larvae (F2) from multiple F1 fluorescent reporter lines were screened for potential use in fluorescent quantification of myelination in vivo, e.g., using cytosolic or membrane tethered EGFP or mCherry. Several individual mpz12.5kb F2 founders were then raised to adulthood. Their embryos (F3) were screened for strong expression of mEGFP or mCherry in the spinal cord and brain and the highest expressing embryos were raised to adulthood (F3 generation). A Tg(mpz:Gal4-VP16;cmcl2:EGFP) line was also generated using available Gateway entry clones (Halpern et al., 2008). Founder lines were identified by green heart marker expression (cmcl2:EGFP), and out crossed with AB or TLs to produce a stable Tg(mpz:Gal4-VP16;cmcl2:EGFP) line. This line was also crossed with a Tg(UAS:Kaede) line (Bruce Appel, University of Colorado School of Medicine; unpublished), to drive Kaede (Ando, Hama, Yamamoto-Hino, Mizuno, & Miyawaki, 2002) expression in mpz-expressing cells.
Zebrafish Husbandry and Live Imaging
All animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Colorado School of Medicine. Embryos were raised at 28.5°C in egg water or embryo medium (EM) and staged according to hours post fertilization (hpf), days post fertilization (dpf), and morphological criteria (Kimmel, Ballard, Kimmel, Ullmann, & Schilling, 1995). At the end of experiments, larvae were sacrificed with an overdose of >2% tricaine (Ethyl 3-aminobenzoate methanesulfonate salt [MESAB]; Sigma) in EM or processed for further analysis (see below). For live imaging, 2–6dpf larvae were embedded in 1% low-melt agarose containing 0.1% tricaine. Live confocal images were acquired on a Leica SP5 laser scanning microscope (Leica Microsystems GmbH, Wetzlar, Germany) using Leica Application Suite Advanced Fluorescence (LAS AF) Software. For quantification of mEGFP reporter intensity, 20X confocal z-stack images (lateral view, 2μm step size) of equivalent truck regions (identified by dorsal fin in bright field, segments 8–16) were collected using identical gain and off set values. mEGFP intensity was calculated on 3D reconstructions of each confocal z-stack using the surfaces tracing plugin (IMARIS imaging processing software) to calculate a ‘max intensity surface’ for each larvae [n=4–6 larvae per experiment (N)]. To control for potential variation in transgene expression between individual founders, only larvae from single founder outcrosses were included in each experiment. Data from N=6 total experiments were pooled and mEGFP intensity values compared between DMSO control and drug treated groups. Significance was calculated using 1-way ANOVA statistical analysis software (GraphPad Prism software).
Drug Treatments
Small molecule drugs were purchased from Tocris or Sigma Aldrich, dissolved in DMSO following manufacturer’s recommendations, stored at −20oC or −80oC and thawed fresh for each experiment. Prior to drug treatment, larvae were sorted for EGFP expression, dechorinated and placed in 6-well plates at a maximum of 25 per well. Larvae were treated with EM + 1% DMSO (control) or 1% DMSO + drug EM+drug (or DMSO). EM+drug (or DMSO) was refreshed every 24hrs during treatment period, which spanned 2–6dpf depending on the specific developmental time frame tested, e.g 2–3dpf for OPC specification and migration, 2–4dpf for initiation of myelination, 3–6dpf for ongoing myelination and 2–6dpf or 2–8dpf for electron microscopy analysis. At the end of each experiment, larvae were collected for live imaging, snap frozen for qPCR or Western blot analysis, or fixed in 4% PFA for immunohistochemistry or in 2% glutaraldehyde and 4% paraformaldehyde for EM (see below).
qPCR
For each experimental condition, 15–20 embryos were anesthetized 0.1% tricaine, and snap-frozen with liquid nitrogen. Samples were stored in 300 μl Trizol Reagent (Thermo Fisher) at −80oC. RNA was isolated using Direct-zol™ RNA miniprep Kit plus DNase treatment (Zymo Research). After isolation, 100 ng of mRNA was reverse transcribed using iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad). RT-qPCR was performed using Taqman Universal PCR Master Mix (Thermo Fisher) on a StepOnePlus realtime PCR Machine (Applied Biosystems). Amplification proceeded as follows: 50oC for 2 min, 95oC for 10 min, and then cycling 40 times at 95oC for 15 sec and 50oC for 1 min. Taqman probes (Thermo Fisher Scientific) were used for mRNA analysis including mpz (Dr03131915_m1), 36k (flj13639; Dr034338676_m1), and reference genes gapdh (Dr03436842_m1), and rpl13 (Dr03101114_g1). Commercially available probes for mbp were not available, therefore a probe against mbpa-201 (ENSDARG00000036186, jh71mbpa) spanning exons 1 and 2, and shared by all mbpa transcript isoforms, was designed through Eurofins (Longmont, CO) using the following sequences: Forward: GTTCTTCGGAGGAGACAAGAAGAG, Reverese: GTCTCTGTGGAGAGGAGGATAGATGA. All experiments followed the MIQE Guidelines for qPCR (SA Bustin 2009).
mpz in situ hybridization
To generate an mpz antisense probe for in situ hybridization (ISH), the sequences of known protein coding transcripts for mpz-001 (ENSDART00000109251), mpz-003 (ENSDART00000132682), mpz-004 (ENSDART00000136818) and mpz-201 (ENSDART00000056372) were downloaded from the Ensembl genome browser (Zv9, www.ensembl.org) and aligned using tools available on the T-COFFEE Multiple Sequence Alignment Server (http://tcoffee.crg.cat). A 454bp amplicon shared by all transcripts was amplified using KAPA HiFi PCR Kit (Kapa Biosystems) and the following primers: Forward: TTCTCCAGAA GTGTCCTTTACATGGCATTA & Reverse: AAAAGAAGGATCACAGCAAAAACAG), and was then subcloned into pGEM-T easy vector system (Promega). DIG-labeled sense and antisense probes against mpz were synthesized following manufacturer’s recommendations (Roche Applied Biosciences). Whole-mount ISH in 2–6dpf wild type (AB), Tg(mpz:mEGFP) or Tg(olig2:EGFP) larvae was performed as previously described (Mathews & Appel, 2016). Probe hybridization was visualized using BM purple (Roche) or Fluorescent-ISH (FISH) using a TSA® Plus Cyanine 3 (Cy3) detection kit (Perkin-Elmer). For FISH/IHC, larvae were incubated in ms αEGFP antibody (Living Colors, JL-8, Clontech Laboratories) following FISH as described in (Seredick, Van Ryswyk, Hutchinson, & Eisen, 2012). Following probe development, larvae were either cryosectioned and imaged with a QImaging Retiga EXi color CCD camera mounted on a compound microscope and imported into Adobe Photoshop or whole-mounted in agarose and imaged by confocal microscopy as described above.
Immunohistochemistry (IHC)
Following drug treatments, 6dpf Tg(mpz:mEGFP) larvae were fixed 4%PFA/1X PBS overnight, washed 3× in 1X PBS, frozen in 1.5% agar/5% sucrose cryoprotectant, then sectioned at 15–20μM. Sections were preblocked in 1%BSA/1% heat inactivated Sheep Serum/ 1X PBS + 0.2% Triton X, and incubated overnight in antibody specific for zebrafish Mbp (1:500, gift from Bruce Appel) and msEGFP (JL-8, 1:1000, see above). Sections were incubated with fluorophore-conjugated secondary antibodies (1:1000, MolecularProbes), after which they were mounted in Fluoromount and imaged by confocal microscopy, as described above, using identical gain settings.
Western Blot
For each experimental condition, 20–25 larvae per treatment condition per experiment (N) were anesthetized in 0.1% tricaine in EM, collected, snap frozen in liquid nitrogen and stored at −80°C until processing. To collect protein, larvae were thawed on ice, then homogenized in 1% Sodium Dodecyl Sulfate (SDS) in RIPA Buffer plus cOmplete™ mini protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail set II (Calbiochem) using a 0.1 ml mortar and pestle (Electron Microscopy Sciences). Lysates were then sonicated and centrifuged at max speed, to collect the supernatant. Protein concentrations were quantified using a Pierce BCA Protein Assay Kit (Thermo Scientific). Samples were diluted in RIPA buffer + Trident 6× Laemmli SDS (GeneTex) + 15% 2-Mercaptoethanol (Sigma), heated at 95oC for 5 minutes, chilled on ice and centrifuged at max speed at 4 °C for 10 minutes. Protein (50 ug) was loaded into 4–20% Mini-Protean TGX Precast Gels (Biorad) in 1× Running Buffer (BioRad) and electrophoresed then transferred to PVDF-FL Immobilon membranes (Millipore). Blots were blocked in 5% BSA/TBST, probed with zebrafish-specific Mbp antibody (1:1000, as above, from Bruce Appel) and alpha-Tubulin (1:5000, Cell Signaling), and visualized using LiCor compatible infrared-secondary antibodies (1:20,000, LiCor). Band intensity (Mbp/alpha-Tubulin) was calculated using Image Studio software. Relative band intensity (drug/vehicle treated) was compiled from N=4 experiments, and significance calculated using paired t-tests using statistical analysis software (GraphPad Prism).
Internode quantification
For mosaic labeling of individual oligodendrocytes, we microinjected of 1–5nl of pEXPR-mbp:mEGFP.cmcl2:mCherry.tol2 (25ng/ul) + capped tol2 mRNA (1ng/ul) into 1–2 cell AB or Tg(sox10:TagRFP-T) embryos. Injected embryos were treated from 2–4dpf in EM+1% DMSO or EM+drug as described above. Confocal stack images (0.3μM step size, 3× sampling) of individual oligodendrocytes in the dorsal spinal cord were collected. To match regions to be quantified and to control for potential differences in growth rate along the rostral caudal axis of the spinal cord, images were collected during a maximum 3 hour window from segments along the yolk extension (identified by dorsal fin, approximately segments 8–12). At least 50 individual cells per treatment group were imaged and data were pooled from multiple experiments. Internode length, process number per cell, and total internode length (sum of the length of all internodes per individual cell) were calculated using 3D reconstruction and filament tracing software (IMARIS imaging processing software). Statistical significance was calculated between DMSO and treatment groups using paired t-tests in statistical analysis software (GraphPad Prism).
Electron Microscopy Analysis
For electron microscopy, all steps were assisted by the use of a PELCO Biowave Pro tissue processor (Ted Pella). Zebrafish were first fixed in 2% glutaraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) (adapted from Czopka and Lyons, 2011 PMID:21951525). Following primary fixation, the specimens were rinsed in 0.1 M sodium cacodylate buffer and post-fixed in a reduced osmium mixture consisting of 2% osmium tetroxide and 0.1 M imidazole (pH 7.5). Specimens were rinsed in distilled water and then stained en bloc with 8% uranyl acetate. Dehydration was carried out through a graded series of ethanol (50%, 70%, 95%, 100%) and acetone (100%) before the samples were finally infiltrated and embedded in EMbed 812 (Electron Microscopy Sciences) and cured for 48 hours at 60oC in an oven. The zebrafish were trimmed to the same location on the rostral-caudal axis and ultrathin sections (65 nm) were cut and mounted on Formvar-coated slot grids and viewed at 80 kV on a FEI Tecnai G2 transmission electron microscope (Hillsboro, OR) with an AMT side-mount digital camera (Woburn, MA). Electron Microscopy data was collected from just above the yolk sac extension, (~somites #12–14) Transverse sections of the spinal cord for each larvae were captured, with focus on the regions containing dorsal and ventral myelinated axons. Images were blinded and number of myelinated axons quantified using Fiji then summed for each individual larva. The total number of ventral and dorsal myelinated axons per fish were summed then averaged across treatment groups (Total larvae analyzed per group were as follows: 2–8dpf; DMSO n=16, GC1 n=15 and 2–6dpf; DMSO n=12, rapamycin n=11) and compared using unpaired t-tests in Graphpad.
RESULTS
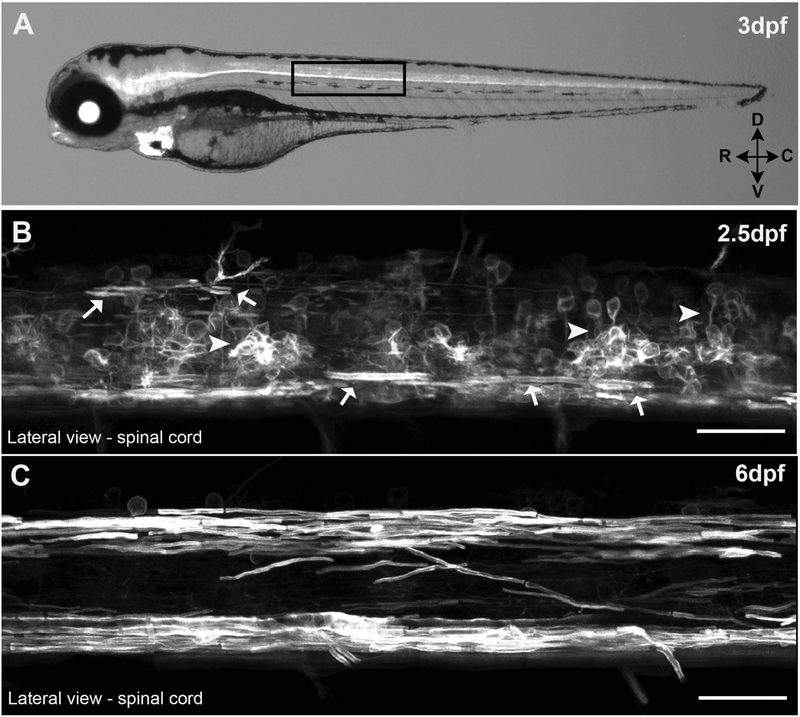
Mpz, the zebrafish homolog of mammalian Protein zero (P0), has previously been reported to be expressed exclusively in the CNS, where it is important for the intermembrane compaction of myelin in the fish (Bai et al., 2011; Brosamle & Halpern, 2002), but it has not been reported to be expressed in the PNS during early development (Brosamle & Halpern, 2002; C. E. Buckley et al., 2010). Genomic databases (Zfin and Ensembl) indicated a single copy of the mpz gene in the zebrafish genome located on chromosome 2. Sequence queries using the UCSC genome browser identified several Bacterial Artificial Chromosomes (BACs) that likely contained the full upstream cis promoter of the mpz gene. Since no suitable restriction sites for isolating a significant portion (>2kb) of the putative promoter for mpz were available, we used recombination based genetic engineering, or ‘Recombineering’ (P. Liu et al., 2003) to isolate a 12.5kb fragment of the BAC located immediately upstream of start codon of mpz-001, the most abundant mpz splice isoform present in zebrafish myelin (Bai et al., 2011). Once this 12.5kb putative mpz promoter sequence was isolated, Gateway modular cloning was used to generate several mpz-driven expression vectors to test the efficacy of multiple fluorescent reporters (e.g. membrane tethered vs cytosolic EGFP, and mCherry; data not shown) to label oligodendrocyte lineage cells. These clones were used to generate a series of mpz transgenic lines, some optimized for cell body stain/quantification and others optimized for visualizing oligodendrocyte membrane production and nascent internode morphology in live animals. The major line described here, Tg(mpz:mEGPF) (Figure 1A–C), strongly expressed a membrane-tethered variant of EGFP (CAAX; prenylated EGFP, hereafter referred to as ‘mEGFP’) in oligodendrocyte lineage cells (Figure 1B) and myelinating oligodendrocytes (Figure 1C), which are readily visible from 2.5dpf on in the spinal cord of developing larvae.
Figure 1: Tg(mpz:mEGFP) fish.
(A) 3 day post fertilization (dpf) larvae expressing membrane tethered EGFP under the control of a 12.5kb mpz promoter fragment. High magnification confocal z-stack images of the lateral spinal cord show some myelinating oligodendrocytes (arrows) at 2.5dpf (B) and robustly myelinated spinal tracks by 6 dpf (C) in live larvae. (scale bar = 25μM)
Transient activation of the 12.5kb mpz reporter was observed in nascent neural tube, beginning at ~16–18hpf, presumably labeling a subset of neural progenitor cells (data not shown). This early promoter activation is similar to previous reports (Schweitzer, Becker, Becker, & Schachner, 2003) and to the transient activation seen in other myelin protein promoters such as PLP in the developing spinal cord (Harlow, Saul, Culp, Vesely, & Macklin, 2014). Similarly, in some founder lines, mEGFP expression was observed in the digestive track of developing larvae (data not shown), consistent with reports of P0 promoter activation in the gut of mice during development (M. J. Lee et al., 2001). mEGFP expression also occurred in the developing lens of the eye (data not shown), consistent with published in situ hybridization data for mpz transcripts (Thisse, B., Thisse, C. 2004, ZFIN Direct Data Submission [http://zfin.org]) and low expression of mEGFP was observed in the developing heart, as we found, using an isolated myelin basic protein (mbp) promoter fragment (Almeida, Czopka, Ffrench-Constant, & Lyons, 2011; Hines, Ravanelli, Schwindt, Scott, & Appel, 2015; Jung et al., 2010) (unpublished observations). Since no purified zebrafish-specific Mpz antibodies were available for immunohistochemistry, we could not determine whether mpz promoter activity outside the CNS, or in non-oligodendrocyte lineage cells, resulted in protein expression.
By 2.5dpf, cells with the morphology of OPCs and myelinating oligodendrocytes (i.e., producing nascent internodes in the dorsal spinal cord and elongating sheaths surrounding ventral Mauthner axons) were clearly visible in the spinal cord (Figure 1B, arrows). At 2.5dpf some non-oligodendrocyte specific expression of mEGFP was also seen in some interneurons/neural progenitors likely due to persistent mEGFP fluorophore expression produced during the transient activation of the promoter earlier in neural tube development described above. However, by 3dpf, this non-oligodendrocyte specific expression in the spinal cord was largely reduced, and replaced by the intense mEGFP expression from expanding oligodendrocyte processes and nascent internodes, which were clearly seen in the 6dpf spinal cord by live confocal imaging (Figure 1C). Furthermore, any residual low level non-oligodendrocyte specific mEGFP expression in the spinal cord from the early ~16–18hr promoter activation required a very high gain setting to visualize from 4dpf on and was quite dim in comparison to the intense expression seen in myelinating membranes (see supplemental Figure 1). Following preliminary characterization of the mpz:mEGFP reporter, multiple F1 Tg(mpz:mEGFP) founder lines were screened for the strongest oligodendrocyte specific expression (and the least non-myelin specific expression), then propagated by outcrossing to at least the F3 generation.
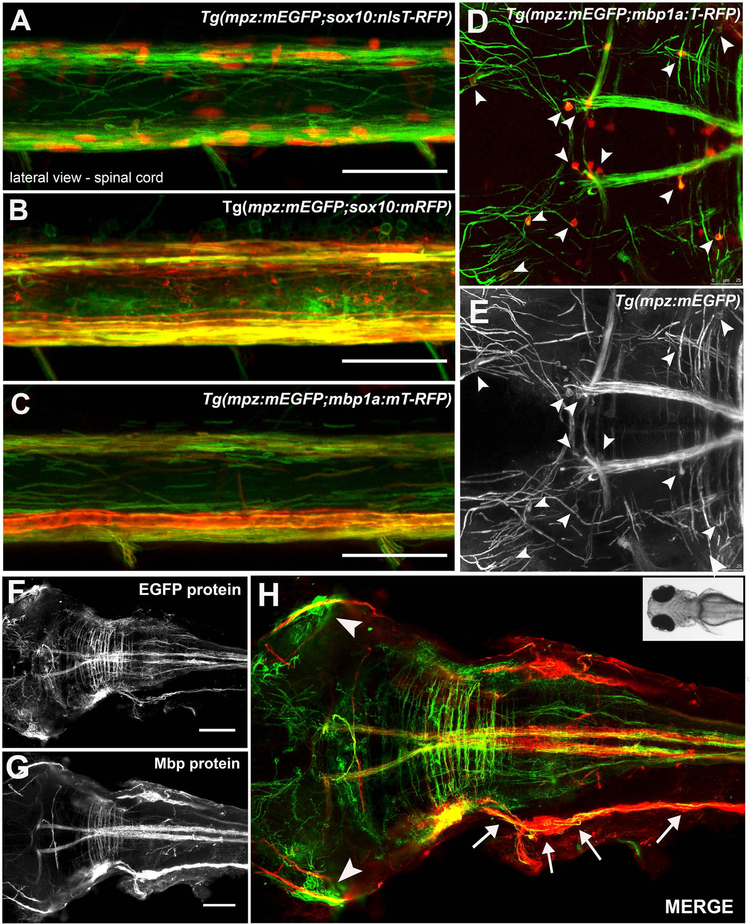
To compare specific labeling of myelinating glia by the mpz:mEGFP reporter with other well characterized oligodendrocyte lineage promoters, the Tg(mpz:mEGFP) line was crossed with multiple lines generated using previously characterized promoters (Hines et al., 2015; Takada et al., 2010) such as Tg(sox10:nls-T-RFP) (Figure 2A), Tg(sox10:mRFP) (Figure 2B) and Tg(mbp1a:mT-RFP) or Tg(mbp1a:T-RFP) (Figure 2C, G,H). Lateral spinal cords in 4–6dpf larvae were imaged to confirm the co-expression of mEGFP with Sox10-driven nls-T-RFP (Figure 2A) or mbp1a- driven membrane tethered T-RFP (mT-RFP; Figure 2B,C), or cytosolic T-RFP (Figure D,E and supplemental Figure 2) in cells that have the classic morphology and localization of myelinating oligodendrocytes in zebrafish spinal cord. To visualize the specificity of the mpz reporter for myelinating glia in the entire CNS, we also bred the mpz:mEGFP and mbp:T-RFP reporters into the Casper background, which lacks all pigment cells (White et al., 2008), allowing better visualization of the developing CNS, especially in the brain, from 3–6dpf (see supplemental Figure 2). Live imaging confirmed that there was consistent co-expression of the mpz membrane reporter (green cell membranes; Figure 2D,E arrowheads) seen at low level surrounding the mbp cytoplasmic reporter (red cell bodies; Figure 2D arrowheads) by 4–6dpf in the CNS of Tg(mpz:mEGFP × mbp1a:T-RFP) larvae. Similarly, whole-mount fluorescent in situ hybridization (F-ISH) with a DIG-labeled antisense probe that recognizes a 454bp region shared by all mpz transcripts was combined with IHC for EGFP protein in 6dpf Tg(olig2vu:EGFP) or at 6dpf Tg(mpz:mEGFP) larvae (supplemental Figure 3). Double positive EGFP+/mpz+ cells were observed in the spinal cord and hindbrain, consistent with myelinating oligodendrocytes. Finally, we performed whole-mount IHC in 6dpf Tg(mpz:mEGFP) larvae using an antibody specific for zebrafish Mbp protein, and observed strong co-localization of Mbp protein with the mEGFP reporter in the brain, spinal cord, lateral line, and cranial nerves of larvae (Figure 2F-H), consistent with known sites of myelination in the fish. Altogether, these data support that the mpz:mEGFP reporter, strongly and specifically labels myelinating cells, including oligodendrocytes and to some extent Schwann cells, in the developing zebrafish nervous system.
Figure 2: The mpz:mEGFP reporter co-localized with known oligodendrocyte markers.
mpz reporter expression (green) co-labeled oligodendrocyte lineage cells in the spinal cord at 6dpf in Tg(sox10:T-RFPnls+) (red nuclei, A), Tg(sox10:mRFP+) (red membranes, B) and Tg(mbp1a:mRFP) (red membranes, C) transgenic lines, imaged laterally by live confocal microscopy. Similarly, T-RFP positive cell bodies (arrowheads, D) also were faintly mEGFP positive (arrowheads D, E) within mEGFP-positive myelinated tracks in the brain of a 6dpf Tg(mpz:mEGFP;mbp1a:T-RFP)casper larvae, imaged dorsally by live confocal microscopy. Whole mount immmunohistochemistry for EGFP protein (green F,H) and Mbp protein (G,H red) was consistent with mpz reporter expression specifically in myelinating glia in the brain, spinal cord, lateral lines (arrows) and cranial nerves (arrowheads) of 6dpf Tg(mpz:mEGFP) larvae, although MBP expression was much stronger in lateral line than mpz reporter expression, consistent with low expression of mpz in Schwann cells at this stage. (scale bar 50μM, A-C; 100μM, D-F, 25μM, A-C).
Although previous reports indicated that expression of mpz transcripts is CNS specific in developing larvae from 2–10dp (Brosamle & Halpern, 2002; Clare E Buckley et al., 2010; Schweitzer et al., 2003), as noted above, we observed weak expression of mEGFP in myelinating Schwann cells in the lateral line and cranial nerves of developing larvae from 4–6dpf (Figure 2F,H). mpz transcripts have been seen in the PNS of juveniles and adults (Schweitzer et al., 2003), although at significantly lower levels than mbp transcripts or Mbp protein, and our data showing high Mbp protein expression with weak mpz:mEGFP expression in the lateral line at 6dpf (Figure 2G,H) are consistent with that. Given that the mbp1a promoter is also specifically expressed in myelinating oligodendrocytes, we also compared the intensity of several membrane reporter lines made using the mbp1a promoter. Our mpz:mEGFP reporter was consistently expressed earlier than the mbp:T-RFP reporter, with nascent myelinated tracks readily visible at ~3dpf in the spinal cord and hindbrain (see Supplemental Figure 2). The mpz:mEGFP reporter was also consistently brighter than the mbp1a:mT-RFP or mbp1a:mEGFP reporters in the spinal cord (Supplemental Figure 4) during our drug treatments (e.g 2–4dpf or 3–6dpf) and strongly labeled oligodendrocyte membranes in both the dorsal and ventral spinal cord while only dimly labeling Schwann cells in the PNS (Supplemental Figure 4). There was significant variation in the membrane reporters made using the mbp1a promoter, where they were either highly expressed in the PNS and only dimly expressed in the CNS (mbp1a:mTRFP, supplemental Figure 4) or most intensely expressed around large ventral Mauthner axons and only weakly expressed in oligodendrocytes in the dorsal spinal cord (mbp1a:mEGFP, supplemental Figure 4).
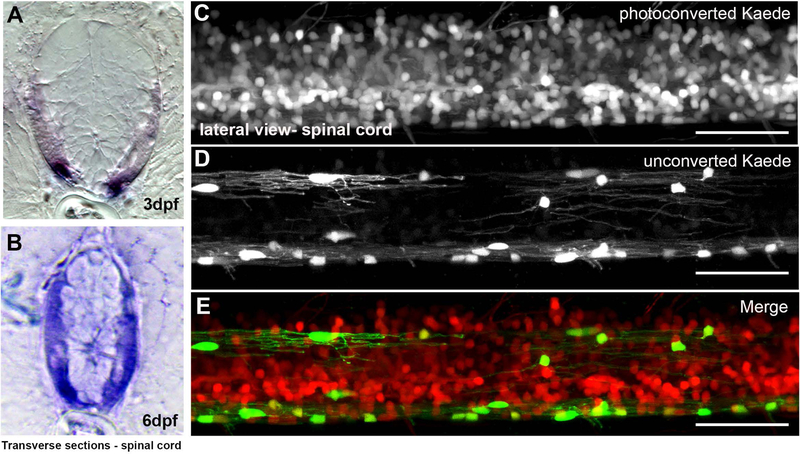
Given that our goal was to use mpz-driven mEGFP accumulation as a surrogate for CNS myelination, we wanted to confirm that our putative mpz promoter was expressed specifically in myelinating oligodendrocytes in the spinal cord after 2.5dpf (see Figure 1B) and that it does not continue to be expressed in neural progenitors after its transient activation at ~16–18hrs. First, ISH of spinal cord sections (Figure 3) showed that mpz transcripts were highly expressed in myelinated tracts between 3dpf (Figure 3A) and 6dpf (Figure 3B). To look more closely at when our mpz transgene was actively expressed, we photoconverted Kaede (Hatta, Tsujii, & Omura, 2006; Leung & Holt, 2008) to follow the temporal activation of the mpz promoter during spinal cord development. Here, 2.5–3dpf (60–72hrs) Tg(mpz:Gal4-VP16;UAS:Kaede) larvae were anesthetized, mounted in agarose and exposed to UV light for approximately 3 minutes to irreversibly convert Kaede protein from green to red, which can be stable for several days (Sattarzadeh, Saberianfar, Zipfel, Menassa, & Hanson, 2015). To precisely determine when mpz promoter activity becomes confined to oligodendrocytes, we photoconverted Kaede every 2hrs (e.g., photoconversion at 60hrs, 62hrs, 64hrs, ect.). After confirming total conversion of Kaede to red by confocal imaging (data not shown), embryos were removed from agarose, returned to the incubator and allowed to develop normally. At 6dpf, UV exposed larvae expressed residual red Kaede protein (from the photoconversion at 2.5–3dpf) in many cells including some oligodendrocyte lineage cells and other neural progenitors in spinal cord (produced during transient activation of the mpz promoter at ~16–18hrs; see Figure 1B). However, newly produced green Kaede protein, i.e., non-photoconverted Kaede protein, was observed only in cells with both the morphology and localization of myelinating oligodendrocytes in the spinal cord (Figure 3C–E), and in the hindbrain and cranial nerves outside the CNS (supplemental Figure 5). Based on these data, it is clear that the mpz promoter is active only in myelin-producing cells after ~2.8dpf (66hrs). Therefore, changes in EGFP fluorescence in the mpz:mEGFP reporter line after 2.8dpf are directly related to changes in mpz gene activation specifically in myelinating glia.
Figure 3: The mpz reporter was restricted to myelinating cells in the spinal cord of developing zebrafish.
In situ hybridization for mpz transcripts (purple) at 3dpf (A) and 6dpf (B) in transverse spinal cord sections showed that mpz expression increased in the lateral spinal cord of developing embryos during nascent myelination in larvae. In order to assess when mpz promoter activity was specific to oligodendrocyte lineage cells, the whole spinal cord of Tg(mpz:Gal4;UAS:Kaede) larvae was photoconverted at 2.5dpf and larvae allowed to develop until 6dpf. Presence of photoconverted, stabilized Kaede protein in the spinal cord of Tg(mpz:Gal4;UAS:Kaede) larvae at 6dpf showed that the mpz reporter had been active before 2.5dpf in numerous spinal cord neural progenitors including oligodendrocyte lineage cells (C, E, red cells; converted Kaede). However, the mpz reporter was active only in myelinating oligodendrocytes after 2.5dpf (D, E, green cells; unconverted Kaede). (scale bar = 50μM)
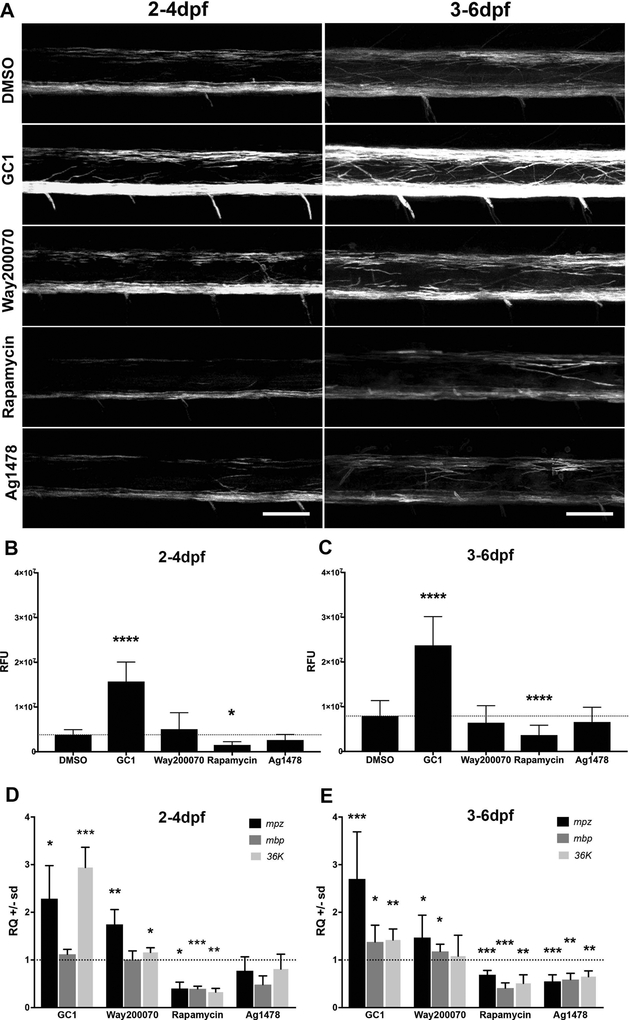
Next, to test the efficacy of the mpz:mEGFP reporter line as a model to assay changes in myelination in vivo, larvae were bath exposed to several small molecule drugs previously shown to impact signaling cascades important for oligodendrocyte development including 10nM GC1, a thyroid receptor (TRβ) agonist (Baxi et al., 2014), 5–10μM Way200070, an estrogen receptor (ERβ) agonist (Khalaj, Hasselmann, Augello, Moore, & Tiwari-Woodruff, 2016), 20mM rapamycin, an inhibitor of mTOR signaling (Abraham & Wiederrecht, 1996) and 1.5μM Ag1478, an Epidermal Growth Factor Receptor (EGFR) antagonist (Pruvot, Cure, Djiotsa, Voncken, & Muller, 2014) that reduces ERK1/2 signaling. Developmental analysis of untreated 1–6dpf embryos by qPCR showed that significant upregulation of myelin-specific gene transcripts (mpz, mbp, 36K) began between 2–3dpf and increased steadily through 6dpf (Supplemental Figure 6), consistent with previous characterization of the temporal specification and differentiation of myelinating glia in the fish (reviewed in Preston & Macklin, 2015). To target distinct stages of oligodendrocyte development, Tg(mpz:mEGFP) larvae were bath treated from 2–4dpf (initiation of myelination) or from 3–6dpf (ongoing myelination) (Figure 4A) and images of equivalent spinal cord segments were collected from live larvae by confocal microscopy. Total spinal cord EGFP intensity (Relative Fluorescent Units, RFUs) of 3D reconstructions; Figure 4B–C) of drug-treated larvae were compared to vehicle-treated larvae (1% DMSO). Imaging intensity was optimized for the strong signal intensity in oligodendrocyte processes, thereby minimizing any residual non-oligodendrocyte specific mEGFP expression in the spinal cord (see supplemental Figure 1). During initiation of myelination (2–4dpf), significant increases in relative EGFP intensity were observed in GC1-treated fish but not in Way200070-treated larvae, relative to DMSO treated controls (Figure 4B). Conversely, EGFP intensity was significantly decreased in rapamycin-treated but not in Ag1478-treated (Figure 4B).
Figure 4: The mpz reporter responded to bath application of small molecule drugs.
Tg(mpz:mEGFP) larvae were incubated in embryo medium with vehicle, 10nM GC1, 10μM WAY200070, 20μM Rapamycin, or 1.5μM AG1478 in 1% DMSO during critical periods of oligodendrocyte lineage progression including initiation of myelination (2–4dpf, A-left panels) or active myelination (3–6dpf, A-right panels). Larvae were anesthetized with tricaine, mounted in low-melt agarose and live imaged using confocal microscopy (A). (B-C) Representative max projections of 3D reconstructions using Imaris image processing software were used to quantify total changes in fluorescence intensity (relative fluorescence units [RFUs]) and compared to vehicle treated controls using equivalent regions of the lateral spinal cord. (D-E) qPCR analysis of mpz, mbp and 36K mRNAs following drug treatments in age-matched larvae was consistent with the quantified changes in EGFP intensity measurements (RFUs). (scale bar = 50μM; statistical analysis: 1-way Anova, p<0.05* <0.01** <0.001*** <0.0001****)
qPCR analysis for common myelin mRNAs showed relatively comparable responses to the drugs (Figure 4D–E). mpz, mbp and 36K transcripts, which are the most well characterized myelin-specific transcripts in zebrafish (Q. Bai, M. Sun, D. B. Stolz, & E. A. Burton, 2011b; C. E. Buckley et al., 2010; Morris et al., 2004) were quantified, and the changes in EGFP intensity generally mirrored changes in mpz mRNA expression as assayed by qPCR (Relative changes [RQ] for GC1: 2.29 fold increase, p<0.0166; Way200070: 1.75 fold increase, p<0.0079; rapamycin: 0.40 fold vs DMSO, p<0.0182 and Ag1478: 0.77 fold decrease, p<0.402; Figure 4D). 36k mRNA levels were generally regulated more consistently with mpz mRNA levels and the mpz:mEGFP reporter response. Most myelin mRNAS were particularly inhibited by treatment with rapamycin, which is unsurprising given the known impact of mTOR signaling during myelination. There were, however, some distinctions in RNA accumulation relative to the mpz:mEGFP reporter responses. The most striking difference is the lack of responsiveness of mbp RNAs to some of the treatments. For example, there is a small, non-significant, increase in RFU quantification at 2–4dpf treatment with Way200070, with statistically significant increases in mpz and 36K mRNA for those samples, but no change in mbp mRNA. This may reflect altered regulation of mbp mRNA transcription/stabilization, which is not unexpected, as mbp mRNA is tightly regulated for transportation out into the accumulating myelin membranes for local translation (for a detailed review see (Muller, Bauer, Schafer, & White, 2013). The differences in mRNA accumulation relative to mpz:mEGFP detection may also reflect increased impact of drugs on cells in brain and hindbrain, which are not included in the imaging of individual fish in spinal cord.
To test if our drugs were also able to modulate on-going (active) myelination, in addition to the initiation of myelination, we treated fish from 3–6dpf, when dorsally migrating OPCs have already migrated to their target axons and begun generating myelinating processes. Here, we found that changes in EGFP intensity largely mirrored the changes in EGFP intensity seen in the 2–4dpf treatments (Figure 4B). EGFP intensity was significantly increased in GC1-treated larvae and but not in Way200070-treated larvae (Figure 4B), and decreases in EGFP intensity in rapamycin-treated but not in Ag1478-treated larvae (Figure 4B) were also observed, which were similar to the 2–4dpf treatment groups. These faithfully represented changes in increases (GC1 2.7 fold change; p<0.00028) or decreases (rapamycin 0.69 fold change; p<0.02441) in mpz mRNA expression assayed by qPCR (Figure 4E, Supplemental Table 1). In general, the mRNA responses of larvae treated from 2–4dpf were more consistent with the RFU data than samples treated from 3–6dpf. This perhaps suggests RNA responses may be more sensitive to current regulation, while EGFP signal (as assayed by mEGFP accumulation) is more stable and captures functional changes in cell morphology from early changes in RNA signal. Nevertheless, again, rapamycin had an inhibitory effect on both RFU data representing membrane accumulation and RNA data representing accumulated RNAs (Figure 4E, supplemental Table 1).
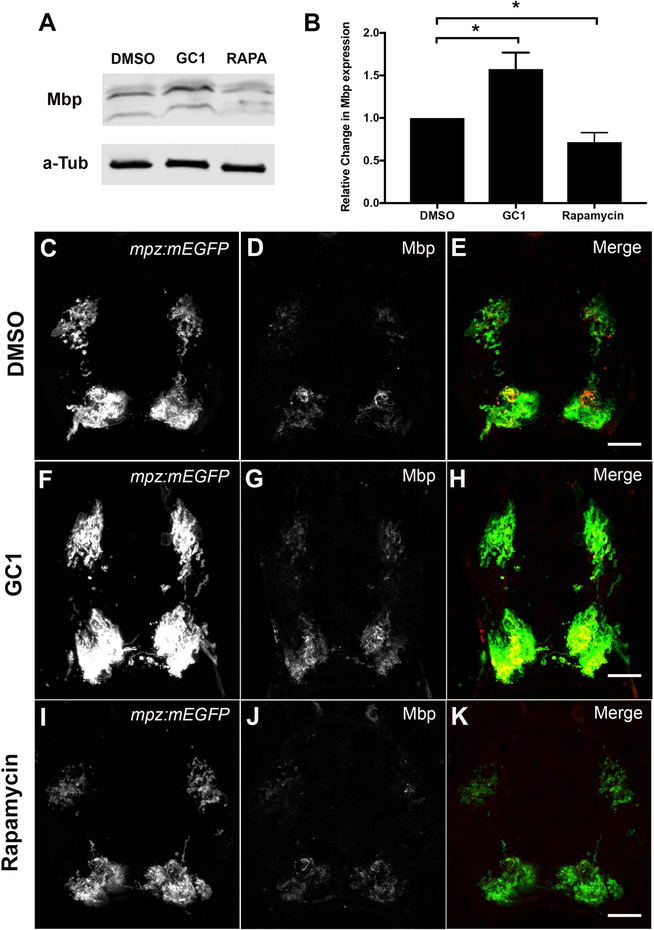
We next asked if the observed changes in mpz promoter activation or inhibition and mRNA levels following drug treatments (EGFP reporter and qPCR) also resulted in significant changes in myelin protein production in zebrafish larvae. Here, we focused on the two drugs with the most significant impact on membrane fluorescence across treatment periods (2–4dpf and 3–6dpf): GC1 as positive regulator of myelin transcription and rapamycin as a negative regulator of myelin transcriptional programs. To quantitatively assay changes in Mbp protein levels, we performed western blot analysis on larvae treated from 2–4dpf with DMSO, GC1 (10nM) or rapamycin (20μM). Consistent with our EGFP intensity and qPCR analysis, GC1 increased Mbp protein expression by ~60% (1.58 +/− 0.097 fold increase, p<0.0157) (Figure 5A–B), while rapamycin decreased Mbp protein expression by ~30% (0.72 +/− 0.055 fold decrease, p<0.0238) (Figure 5A–B). To confirm changes in Mbp protein specifically in the spinal cord, which is first detectable by IHC at 6dpf, we also performed IHC with antibodies against Mbp and EGFP on transverse sections of 6dpf Tg(mpz:mEGFP) larvae treated from 3–6dpf with DMSO (1%, Figure 5C–E), GC1 (10nM; Figure 5F–H) or rapamycin (20μM, Figure 5I–K), showing protein expression changes comparable to the mRNA changes.
Figure 5: Western blot analysis and immunohistochemistry for Mbp protein expression correlated with mpz reporter activity in drug-treated larvae.
(A) Representative western blot of Mbp protein expression in 2–4dpf DMSO, GC1 (10nM), or rapamycin (20μM) treated larvae. (B) Quantification of western blots showed that GC1 treatment significantly increased Mbp expression relative to DMSO treated controls (alpha tubulin loading control), while rapamycin significantly decreased Mbp protein expression in developing larvae. Immunohistochemistry of transverse spinal cord sections of 6dpf Tg(mpz:mEGFP) larvae treated with DMSO (C-E), GC1 (F-H) or rapamycin (I-K) showed changes in Mmb and EGFP expression in myelinated tracts in response to drug treatments. (Scale bar = 10μM; statistical analysis: 1-way Anova, p<0.05* <0.01** <0.001*** <0.0001****)
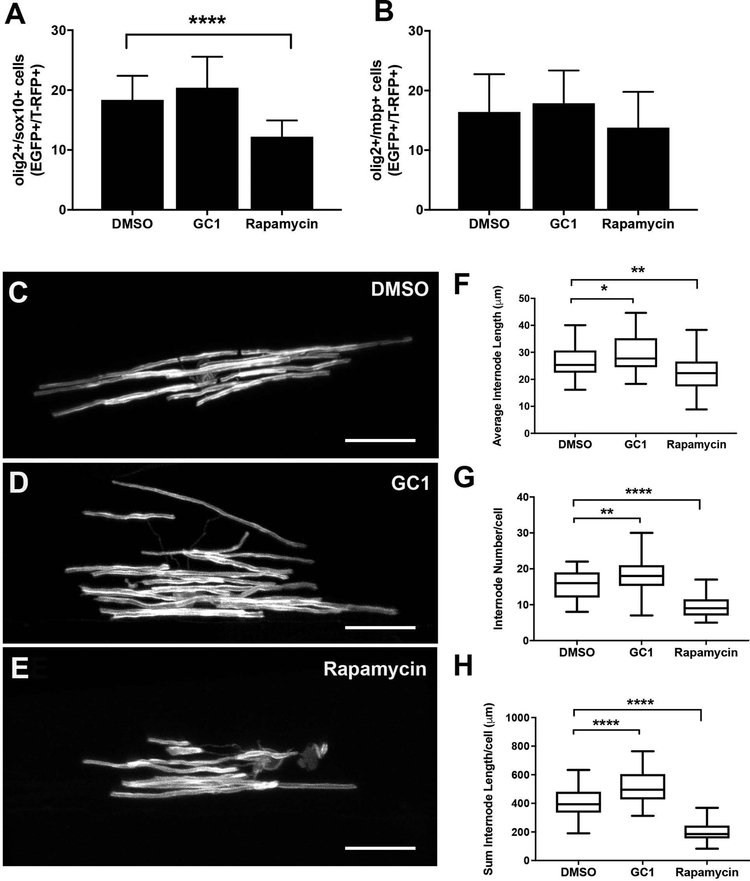
In order to better understand potential cellular mechanisms that might contribute to the observed alterations in myelin RNA/protein expression, we repeated our 2–4dpf drug treatments in oligodendrocyte lineage reporter lines that label cell bodies (Hines et al., 2015; H.-C. Park et al., 2002; Takada et al., 2010). First, we counted EGFP+/RFP+ cells in dorsal spinal cord of 2–4dpf drug treated Tg(olig2:EGFP;sox10:T-RFP) or Tg(olig2:EGFP;mbp:T-RFP) larvae. GC1 treated embryos had no change to the total number of olig2+/sox10+ cells (20.4 vs 18.4 double positive cells, p>0.1782) or olig2+/mbp+ cells (17.8 vs 16.4, p>0.495) compared to control larvae (Figure 6, A-B). In contrast, rapamycin treated larvae had a significant decrease in the number of olig2+/sox10+ (12.2 vs 18.4, p>0.0001) but no changes in total number of olig2+/mbp+ cells (13.8 vs 16.4 p>0.171) compared to control larvae (Figure 6, A-B). Taken together, these data indicate that GC1 had little impact on the total number of oligodendrocyte lineage cells or their differentiation into myelinating oligodendrocytes, although mbp reporter expression (as assayed by T-RFP intensity) appeared to slightly increase in GC1 treated embryos (Supplemental Figure 7) consistent with increases in mbp mRNA concentration (see Figure 4).
Figure 6: Morphological changes in individual oligodendrocytes following drug treatment.
Average number of dorsal oligodendrocyte lineage cells (olig2+/sox10+) was quantified from 3D reconstructions of live images of the lateral spinal cord of Tg(olig2:EGFP; sox10:T-RFP) larvae (A) or myelinating oligodendrocytes (olig2+/mbp+) from Tg(olig2:EGFP; mbp1a:T-RFP) larvae (B) following 10nM GC1 or 20μM rapamycin treatment (2–4dpf). (C-E) Representative images of individual scatter labeled oligodendrocytes and quantitative analysis of internode length (F), number of internodes (G) and total internode length generated per cell (H) following 10nM GC1 or 20μM rapamycin treatment (2–4dpf). (scale bar =25μM, statistical analysis: 1-way Anova, p<0.05* <0.01** <0.001*** <0.0001****)
In comparison, exposure to rapamycin reduced the number of total oligodendrocyte lineage cells in the dorsal spinal cord, but did not prevent the available OPCs from differentiating into myelinating oligodendrocytes as assayed by mbp promoter activation. However, the intensity of mbp reporter expression (e.g. T-RFP intensity) was also slightly downregulated (see Supplemental Figure 7), suggesting reduced promoter activity, which is consistent with the reduction in mbp mRNA concentration (see Figure 4), and with the known inhibition of active myelination by rapamycin.
Given that normal numbers of total number of oligodendrocyte lineage cells in our GC1-treated larvae, we reasoned that the changes in mEGFP intensity and myelin transcripts and proteins might result from changes in myelin membrane production per individual cell. Mosaic analysis allowed quantification of the morphology of individual oligodendrocytes. Here, 1–2-cell AB embryos were injected with a construct encoding a 2.5kb mbp1a promoter fragment driving expression of mEGFP (pEXPRESS-mbp1a2.5kb:mEGFP.cmcl2-RFP.tol2), which scatter-labelled individual mbp-expressing oligodendrocytes in the spinal cord of zebrafish larvae. Injected embryos were then treated from 2–4dpf with either GC1 or rapamycin, and live imaged to assay changes in the morphology of single cells (Figure 6C–E). We measured three separate parameters of oligodendrocyte morphology including average length of nascent internodes, average number of internodes per cell, and the total internode production per cell (sum of the length of the internodes produced per individual cell; Figure 6F–H). Consistent with changes in EGFP intensity, qPCR, and western analysis, we found that GC1 treatment significantly increased both the length (29.43μM vs 26.4μM compared to DMSO, p<0.026; Figure 6F) and number (18.2 vs 15.7 compared to DMSO, p<0.0026; Figure 6G) of nascent internodes of individual oligodendrocytes. Conversely, rapamycin treatment reduced both the length (22.26μM vs 15.69μM, p< 0.0028; Figure 6F) and number (9.6 vs 15.7 compared to DMS0, p<0.0001; Figure 6G) of nascent internodes. Furthermore, these changes in individual cell morphology resulted in highly significant changes in total amount of internode myelin produced per cell, presumably resulting from changes in the myelinating capacity of each oligodendrocyte in response to drug treatment. Here, GC1 increased the total length of internodes produced by each cell an average of ~20% (512.6μM vs 409.9μM, p<0.0001; Figure 6H), while rapamycin decreased the total length of internodes produced by ~50% (205.5μM vs 409.9μM, p<0.0001; Figure 6H).
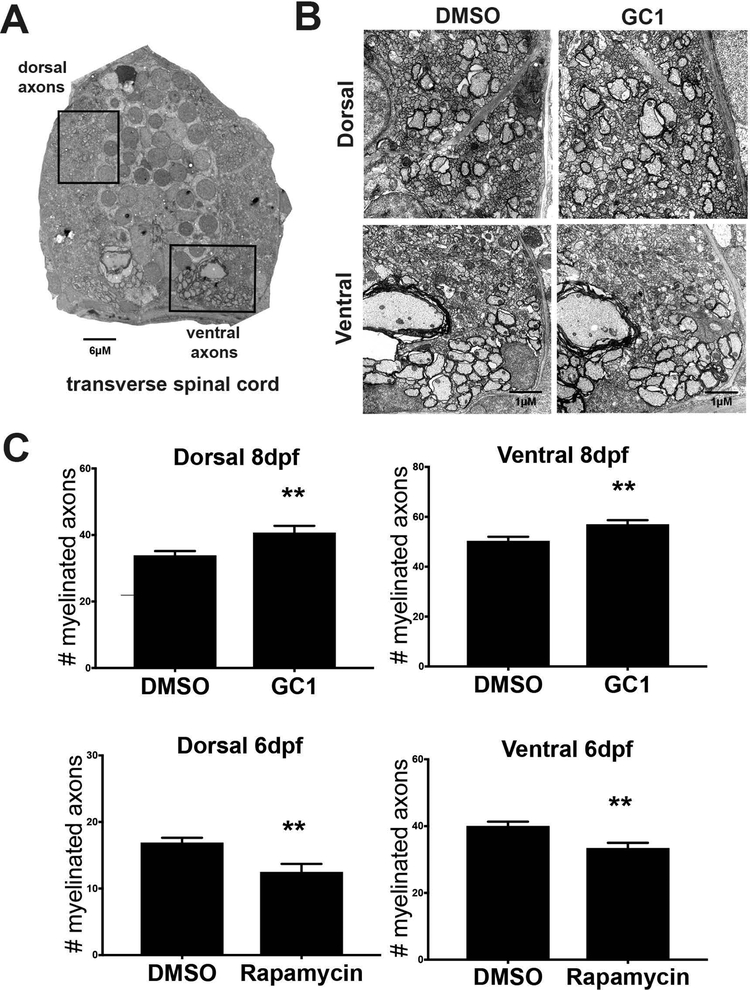
Finally, to confirm that quantifiable changes in the mpz:mEGFP reporter, myelin transcript expression, protein expression and individual cellular morphology are predictive of changes in myelin at the ultrastructure level, we performed electron microscopic analysis on the spinal cords of DMSO and drug treated larvae. We found that myelin structure can be highly variable in the fish at 4dpf and highly variable between larvae (data not shown) so we focused on 8dpf after compaction of myelin begins the larval zebrafish (reviewed Preston & Macklin, 2015). Here, we bath treated embryos from 2–8dpf with GC1 (10μM) or rapamycin (20μM) and processed equivalent regions of the spinal cord for electron microscopy as those imaged for mEGFP quantification (see Figure 4). Total number of myelinated axons were counted in both the ventral and dorsal spinal cord (Figure 7A, boxed regions). GC1 treatment increased the total number of myelinated axons at 8dpf as compared to vehicle treated controls in both the ventral (57.0 v 50.4; p<0.0074; Figure 7B–C) and dorsal (33.9 v 40.7; p<0.0071; Figure 7:B-C) spinal cord. Unfortunately, rapamycin treatment from 2–8dpf was lethal to the larvae, likely due to systemic suppression of mTOR signaling in the developing embryos. Therefore, the maximum treatment window possible for analysis of the impact of rapamycin was 2–6dpf. Consistent with the decreases in mEGFP+ intensity, myelin transcript expression and Mbp protein, rapamycin treatment decreased the total number of myelinated axons as compared to vehicle treated controls in both the ventral (40.1 vs 33.4, p<0.0042; Figure 7: C) and dorsal (16.9 vs 12.5, p<0.0057; Figure 7:C) spinal cord.
Figure 7: Electron micrograph analysis of myelinated axons following drug treatment.
Transverse spinal cord sections were collected from wildtype larvae following drug treatment with 10nM GC1 (2–8dpf) or 20μM rapamycin (2–6dpf). A representative image (A) shows dorsal and ventral regions of the spinal cord containing myelinated axons. Boxed areas were used for detailed analysis (scale bar=6μM). Myelinated axons were imaged at high magnification (B, image shown is from 8dpf, scale bar=1μM) and quantified (C). Electron micrograph analysis showed that GC1 treatment increased the number of myelinated axons in both the dorsal and ventral spinal cord of fish treated from 2–8dpf, while rapamycin treatment 2–6dpf decreased the number of myelinated axons in both the dorsal and ventral spinal cord. (statistical analysis: unpaired t-test, p<0.05* <0.01** <0.001*** <0.0001****)
DISCUSSION
Recovery of function following MS attacks depends both on reducing the inflammatory/immune attack on myelin and the local inflammation, then efficient replacement of damaged myelin (extensively reviewed in (Fancy et al., 2010; R. J. Franklin & Ffrench-Constant, 2008). OPCs are clearly recruited to demyelinated lesions but they fail to mature into myelinating oligodendrocytes (see (R. J. M. Franklin & Blakemore, 1997; M. Preston & Sherman, 2011; Sherman & Back, 2008). Furthermore, this remyelination failure becomes more pronounced over time in patients (R. J. Franklin, Zhao, & Sim, 2002; Ibanez et al., 2003). Therefore, identifying small molecule therapies that specifically promote oligodendrocyte differentiation and myelin production is important for the repair process, with the goal of slowing the degeneration of demyelinated axons (Bjartmar & Trapp, 2001). High throughput small molecule screens have identified several promising drugs from FDA approved libraries (Eleuteri et al., 2017; Mei et al., 2014; Porcu et al., 2015). For example, two separate high throughput screens of FDA approved libraries using cultured OPCs independently identified Clemastine, an antihistamine that enhances oligodendrocyte differentiation and myelination in rodents (Li, He, Fan, & Sun, 2015; J. Liu et al., 2016; Mei et al., 2014). However, many of the promising hits from such cell screens have failed to show efficacy in vivo or still need to be tested rigorously using in vivo rodent models. Given the cost and time of testing candidate molecules in whole animals, in vivo testing represents a major bottleneck in the drug discovery pipeline, especially for translating in vitro findings into the pre-clinical validation needed to rapidly advance therapeutically relevant candidate drugs into clinical trials.
The data presented here demonstrate that mpz promoter-driven mEGFP accumulation/fluorescence intensity can be used to rapidly and non-invasively follow myelin membrane production in the spinal cords of developing zebrafish larvae. Thus our mpz:mEGFP reporter line is a quick and rapid in vivo measure of the relative impact of small molecule drugs on myelin gene activation and cellular morphology. We have shown that Tg(mpz:mEGFP) reporter fish responds to both positive (GC1) and negative (rapamycin) small molecule regulators of signaling cascades known to influence myelination in other species such as rodents and humans. Importantly, we found that the change in mEGFP accumulation in response to bath application of drugs is strongly correlated with activation of myelin specific genes, and is predictive of changes in myelin-specific protein production (Mbp) and changes in the total numbers of myelinated axons. Thus, it is a useful proxy for altered myelination in vivo in general. We used several well characterized transgenic lines including Tg(olig2:EGFP) and Tg(sox10:T-RFP) (Blasky, Pan, Moens, & Appel, 2014; H. C. Park, Shin, Roberts, & Appel, 2007; Takada et al., 2010); as well as, several reporter lines made with a previously isolated regulatory sequence for mbp1a (Almeida et al., 2011; Jung et al., 2010; Ravanelli & Appel, 2015), to more closely assess the responses of oligodendrocyte lineage cells to GC1 and rapamycin to better understand the specific impact of each drug on myelination. We found that following GC1 treatment, the number of myelinated axons in the spinal cord increased without changing the number of oligodendrocyte lineage cells. By contrast, following rapamycin treatment, despite reducing the overall number of oligodendrocyte lineage cells (olig2+/sox10+), oligodendrocytes (olig2+/mbp+) were still able to initiate myelination, as assayed by activation of the mbp promoter (e.g mbp1a:T-RFP). However, these oligodendrocytes expressed significantly lower levels of both the mpz:mEGFP and mbp1a:T-RFP reporter (Supplemental Figure 7), and larvae had reduced levels of myelin specific mRNAs and Mbp protein.
To better understand the cellular mechanisms underlying these changes, we used single cell labeling of myelinating oligodendrocytes (mosaic labeling of mbp1a:mEGFP+ cells in the dorsal spinal cord) which allowed us to quantify the number and length of internodes, i.e., the overall myelin membrane production by an individual cell. Oligodendrocytes in GC1-treated embryos produced more nascent internodes, which were increased in length, while oligodendrocytes in rapamycin-treated embryos had fewer nascent internodes that were shorter in length. Electron microscopy demonstrated that these observed changes in individual cellular morphology correlated with significant changes in the total number of myelinated axons, which were increased in response to the positive regulator (GC1) and decreased in response to the negative regulator (rapamycin). Altogether, these data support the hypothesis that the total amount of myelin generated by individual cells can be modulated by small molecule drug treatment, and that these small changes in individual cell morphology can account for significant changes in total myelin membrane produced on a global level. As in other zebrafish studies (Almeida et al., 2011; T. Czopka, Ffrench-Constant, & Lyons, 2013), these findings indicate that the myelinating potential of an individual oligodendrocyte is highly plastic, responding to both intrinsic and extrinsic cues, and thus it is a prime target for therapeutic manipulation.
In validating our model, we tested several drugs that have been identified as targeting potentially therapeutically relevant signaling cascades for demyelinating injuries and diseases. We found that two, GC1 and rapamycin, significantly impacted mpz promoter activation (mEGFP expression) and importantly, modulated myelinating process generation and membrane production (accumulation of mEGFP in expanding membranes). We also observed changes in the spinal cord at the ultrastructure level in response to GC1 and rapamycin treatment, with a ~20% change in the total number of axons selected for myelination. Longer treatment periods may enhance these changes over time, although significant functional recovery has been shown with as little as 25% recovery of compact myelin (Readhead et al., 1987). Interestingly, we found that other drugs, such as the ERβ agonist Way200070 and the EGF Receptor antagonist Ag1478, were able to alter myelin gene activation (in particular 36K or mpz RNA transcripts) but did not result in significant changes in membrane production. As noted above, this could result from the fact that qPCR analysis uses whole larvae and thus captures changes in transcriptional activation of myelin genes in the brain and the PNS in addition to the spinal cord where our confocal imaging was done. Alternatively, these differences could reflect acute changes in transcriptional activation that do not always reflect or result in changes in cellular morphology, e.g. increases or decreases in myelin membrane production.
While our model will clearly not replace in vivo myelination/remyelination models in rodents, it could significantly increase the speed and decrease the cost of the preliminary in vivo characterization of hundreds of candidate drugs, and be useful in identifying the most promising (and therapeutically relevant molecules) to move up the pipeline. For example, we found that GC1 robustly and reliably increased mEGFP intensity (and myelin gene/protein expression) at a nanomolar concentration whether it was delivered in single or multiple doses (data not shown), or whether treatment occurred during initiation (2–4dpf) or on-going (3–6dpf) myelination. Consistent with these observations in the fish, GC1 has been shown to strongly promote myelination in rodents and human stem cell-derived oligodendrocyte lineage cells (Baxi et al., 2014) and is currently being tested for its therapeutic potential. These results also highlight that pharmacologic modulation of complicated signaling cascades that drive myelination, such as those studied here, also need to be tested in an in vivo context for efficacy. For example, recent work from our lab and others has shown that mTOR signaling, a fundamental driver of myelination that impacts progenitor differentiation and membrane synthesis, may be vastly more complex than previously appreciated. Here, in the PNS, mTOR activity is initially elevated in progenitor cells, but must be downregulated to allow Schwann cells to begin differentiation. Next, mTOR signaling must then be upregulated to promote active myelination programs (Figlia, Norrmen, Pereira, Gerber, & Suter, 2017). We found that rapamycin treatment unexpectedly did not block OPC differentiation into mbp+ oligodendrocytes but strongly impacted the morphology of individual cells and reduced their myelinating capacity. As such, zebrafish studies using narrow treatment windows and time lapse imaging should provide important insight into how complicated signaling cascades can be specifically modulated to control oligodendrocyte lineage progression to drive myelination in real time.
An ideal screen for molecules that specifically target CNS myelination would use a reporter that is expressed strongly and specifically in oligodendrocytes but not in other cell types. The mpz reporter presented here is highly and specifically expressed in myelinating glia after ~2.8dpf. However, there is brief transient activation of the promoter in neural progenitors at ~16–18hpf, which is consistent with previous in situ hybridization analysis for mpz RNA (Schweitzer et al., 2003). This early promoter activation results in a faint but persistent expression of EGFP in non-oligodendrocyte cells during our treatment timeframes, likely due to the prolonged stability of EGFP protein (Progatzky, Dallman, & Lo Celso, 2013). However, 3D imaging demonstrated no change in this non-oligodendrocyte signal in response to drug treatments, consistent with EGFP stability rather than ongoing regulation in non-myelinating cells. Given the distinct localization and morphology of myelinating oligodendrocytes, it is easy to identify any false positive hits due to non-specific expression in other cells. In contrast to earlier reports for mpz expression (Brosamle & Halpern, 2002;(Schweitzer et al., 2003), we found that our transgene is weakly expressed in PNS Schwann cells during our treatment window (2–6dpf). However, we found that mpz:mEGFP expression in the PNS was quite low in comparison to CNS expression, and significantly lower than other myelin specific proteins such as myelin basic protein, which is readily detectable in the lateral line a from ~3dpf by in situ hybridization (Brosamle & Halpern, 2002) and by ~5–6dpf when staining for Mbp protein in our studies. Therefore, we believe our mpz:mEGFP reporter is useful for identifying drugs that enhance CNS myelination specifically.
Early et al. have also recently reported a fluorescence-based myelination screening model in zebrafish using mbp regulatory sequences (Early et al., 2018), which is expressed only in CNS and PNS myelinating glia, and does not appear to be transiently activated in neural progenitor cells like the mpz promoter described here. While there was robust expression of our mbp reporters following injection during mosaic-labeling experiments (unpublished observations), unfortunately the transgenic reporter lines we generated using this mbp promoter were quite dim compared to our mpz reporter. This presented technical difficulties for manual sorting and imaging, but more importantly, our mbp-driven reporter lines were also highly variable between individual founders. We observed that the mbp transgene was either strongly expressed in Schwann cells in the lateral line, but only weakly in the CNS, or more strongly expressed in oligodendrocytes myelinating Mauthner axons and ventral spinal cord axons but only weakly expressed in dorsal oligodendrocytes. This was a particular problem during the earliest treatment periods (2–4dpf) when myelination of the dorsal spinal cord is just beginning. These differences in transgene expression could be due to factors such as the relatively small size (2.5kb) of the mbp regulatory elements used versus the ~12.5kb mpz promoter we used, or possibly to the insertion site of our transgene reporters, which can strongly impact reporter strength, even in stable lines (Walker et al., 2012). Another possibility is that the isolated mbp regulatory sequences are more susceptible to silencing or genome regulation than the mpz regulatory sequences. To minimize some of this promoter and positional variability, transgenic animals are often generated using modified BACs, which contain more endogenous upstream and intronic regulatory elements. However, this requires modified BACs for each reporter line (ie EGFP, mEGFP, RFP, etc.) and they often show reduced efficiency for germline transmission as compared to Tol2 based integration (Bussmann & Schulte-Merker, 2011). As such, we used a combination of the two methods to isolate the largest upstream regulatory element possible from a BAC for Tol2 transgenesis (see methods for more details). Alternatively, a non-technical and potentially very important explanation for the differences in these reporters may be due to differences in the timing of each myelin promoter activation during development. Here, the mpz:mEGFP reporter was strongly activated in myelinating oligodendrocytes beginning at ~3dpf, while the mbp reporters including mbp1a:T-RFP, mbp1a:mT-RFP and mbp1a:mEGFP were challenging to detect manually prior to ~5dpf. Thus, the early and strong activation of the mpz:mEGFP reporter allowed us to assay the effects of drugs on the differentiation and internode production beginning at ~3–4dpf, when the first wave of myelinating oligodendrocytes begins wrapping axons in the dorsal spinal cord.
Despite the relatively narrow temporal window studied here (2–8dpf), this model targets oligodendrocyte lineage progression through several key developmental stages, including proliferation and migration of OPCs, followed by target recognition, axonal wrapping and nascent myelin compaction. Our imaging was confined to a distinct region of the zebrafish trunk to capture and quantify only spinal cord myelination that occurs before pigmentation obscures the CNS. The scale of our screen was also limited to a small number of compounds due to temporal limitations of manual handling of larvae and labor intensive confocal imaging. In the future, efficient screening of 1000’s of compounds for large scale screens will require specialized equipment for automated larvae handling and imaging scripts such as developed by the Lyons lab (Early et al., 2018). It is also interesting to note that one itineration of the Early et al. (2018) screen used a cytoplasmic marker for labeling oligodendrocytes to identify changes specifically in oligodendrocyte cell number as a parameter for identifying candidate molecules (Early et al., 2018). In contrast, our membrane tethered reporter measures not only promoter activity, but also membrane production, which may be independent of changes in oligodendrocyte cell numbers, e.g., such as following GC1 or rapamycin treatment where the total number of mbp+ oligodendrocytes was unchanged. Given that remyelination failure in MS lesions may result from local disruption of differentiation programs in individual cells (reviewed in (Harlow & Macklin, 2014; M. Preston & Sherman, 2011; Sherman & Back, 2008), identification of drugs that specifically target the plasticity of the remaining/recruited oligodendrocytes by targeting differentiation programs or membrane production may be particularly important for enhancing remyelination (Wilson, Scolding, & Raine, 2006). Altogether, the use of multiple myelin reporters, such as a cytoplasmic mbp reporter for cell counts and a mpz membrane-tethered reporter to assay internode production, could provide a more comprehensive picture of the therapeutic potential of candidate small molecules identified via in vivo screening.
Supplementary Material
Main Points:
Remyelination therapeutics need to be tested in vivo for efficacy.
Fluorescent zebrafish can be used to follow myelin membrane production in live larvae.
Drug screening in whole larvae predicts drug efficacy.
Acknowledgements
We would like to thank current and past members of Bruce Appel’s laboratory for technical assistance and helpful theoretical discussions, as well as the generous gifts of a zebrafish-specific Mbp antibody, the Tg(sox10-T-RFP) line, and plasmids containing the mbp1a(2.5kb) and sox10(7.2kb) zebrafish promoters for Gateway cloning. We would like to thank Steve Seredick for technical advice in BAC ‘recombineering’ and Tanya Brown and Hannah Hathaway for manuscript draft review and critiques. We would also like to thank the University of Colorado Anschutz Medical Campus Zebrafish Core Facility and Staff for zebrafish care and technical assistance. Finally, our graphical abstract was generated using a modified cartoon zebrafish originally generated by Lizzy Griffiths. The authors declare no conflict of interests or financial interests in the original work presented here. This work was generously supported by The National Multiple Sclerosis Society (NMSS) project grant (RG 5334-A-12) and NINDS (NS078386) to WBM and an NMSS Postdoctoral Fellowship (FG 2078-A-1) to MAP.
Literature Cited
- Aboul-Enein F, Rauschka H, Kornek B, Stadelmann C, Stefferl A, Bruck W, … Lassmann H (2003). Preferential loss of myelin-associated glycoprotein reflects hypoxia-like white matter damage in stroke and inflammatory brain diseases. J Neuropathol Exp Neurol, 62(1), 25–33. [DOI] [PubMed] [Google Scholar]
- Abraham RT, & Wiederrecht GJ (1996). Immunopharmacology of rapamycin. Annu Rev Immunol, 14, 483–510. doi: 10.1146/annurev.immunol.14.1.483 [DOI] [PubMed] [Google Scholar]
- Almad A, Sahinkaya FR, & McTigue DM (2011). Oligodendrocyte fate after spinal cord injury. Neurotherapeutics, 8(2), 262–273. doi: 10.1007/s13311-011-0033-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RG, Czopka T, Ffrench-Constant C, & Lyons DA (2011). Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development, 138(20), 4443–4450. doi: 10.1242/dev.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Schneider A, Barrie JA, Klugmann M, McCulloch MC, Kirkham D, … Griffiths IR (1998). Late-onset neurodegeneration in mice with increased dosage of the proteolipid protein gene. The Journal of comparative neurology, 394(4), 506–519. [DOI] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, & Miyawaki A (2002). An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A, 99(20), 12651–12656. doi: 10.1073/pnas.202320599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Sun M, Stolz DB, & Burton EA (2011). Major isoform of zebrafish P0 is a 23.5 kDa myelin glycoprotein expressed in selected white matter tracts of the central nervous system. J. Comp Neurol, 519(8), 1580–1596. doi: 10.1002/cne.22587 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxi EG, Schott JT, Fairchild AN, Kirby LA, Karani R, Uapinyoying P, … Calabresi PA (2014). A selective thyroid hormone beta receptor agonist enhances human and rodent oligodendrocyte differentiation. Glia, 62(9), 1513–1529. doi: 10.1002/glia.22697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjartmar C, & Trapp BD (2001). Axonal and neuronal degeneration in multiple sclerosis: mechanisms and functional consequences. Curr.Opin.Neurol, 14(3), 271–278. [DOI] [PubMed] [Google Scholar]
- Blasky AJ, Pan L, Moens CB, & Appel B (2014). Pard3 regulates contact between neural crest cells and the timing of Schwann cell differentiation but is not essential for neural crest migration or myelination. Dev Dyn, 243(12), 1511–1523. doi: 10.1002/dvdy.24172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosamle C, & Halpern ME (2002). Characterization of myelination in the developing zebrafish. Glia, 39(1), 47–57. doi: 10.1002/glia.10088 [DOI] [PubMed] [Google Scholar]
- Buckley CE, Marguerie A, Alderton WK, & Franklin RJ (2010). Temporal dynamics of myelination in the zebrafish spinal cord. Glia, 58(7), 802–812. doi: 10.1002/glia.20964 [DOI] [PubMed] [Google Scholar]
- Buckley CE, Marguerie A, Alderton WK, & Franklin RJM (2010). Temporal dynamics of myelination in the zebrafish spinal cord. Glia, 58(7), NA–NA. doi: 10.1002/glia.20964 [DOI] [PubMed] [Google Scholar]
- Bussmann J, & Schulte-Merker S (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development, 138(19), 4327–4332. doi: 10.1242/dev.068080 [DOI] [PubMed] [Google Scholar]
- Chang A, Staugaitis SM, Dutta R, Batt CE, Easley KE, Chomyk AM, … Trapp BD (2012). Cortical remyelination: A new target for repair therapies in multiple sclerosis. Annals of Neurology, 72(6), 918–926. doi: 10.1002/ana.23693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T, Ffrench-Constant C, & Lyons DA (2013). Individual oligodendrocytes have only a few hours in which to generate new myelin sheaths in vivo. Dev Cell, 25(6), 599–609. doi: 10.1016/j.devcel.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czopka T, & Lyons DA (2011). Dissecting Mechanisms of Myelinated Axon Formation Using Zebrafish. In (Vol. 105, pp. 25–62): Elsevier. [DOI] [PubMed] [Google Scholar]
- D’Rozario M, Monk KR, & Petersen SC (2017). Analysis of myelinated axon formation in zebrafish. Methods Cell Biol, 138, 383–414. doi: 10.1016/bs.mcb.2016.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early JJ, Cole KL, Williamson JM, Swire M, Kamadurai H, Muskavitch M, & Lyons DA (2018). An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. Elife, 7. doi: 10.7554/eLife.35136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleuteri C, Olla S, Veroni C, Umeton R, Mechelli R, Romano S, … Agresti C (2017). A staged screening of registered drugs highlights remyelinating drug candidates for clinical trials. Sci Rep, 7, 45780. doi: 10.1038/srep45780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy SP, Kotter MR, Harrington EP, Huang JK, Zhao C, Rowitch DH, & Franklin RJ (2010). Overcoming remyelination failure in multiple sclerosis and other myelin disorders. Exp.Neurol, 225(1), 18–23. doi:S0014–4886(09)00513–5[pii]; 10.1016/j.expneurol.2009.12.020 [doi] [DOI] [PubMed] [Google Scholar]
- Figlia G, Norrmen C, Pereira JA, Gerber D, & Suter U (2017). Dual function of the PI3K-Akt-mTORC1 axis in myelination of the peripheral nervous system. Elife, 6. doi: 10.7554/eLife.29241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EJ (2004). Immunopathology of multiple sclerosis. Neurology, 63(12 Suppl 6), S3–7. [DOI] [PubMed] [Google Scholar]
- Franklin RJ (2002). Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci, 3(9), 705–714. doi: 10.1038/nrn917 [DOI] [PubMed] [Google Scholar]
- Franklin RJ, & Ffrench-Constant C (2008). Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci, 9(11), 839–855. doi: 10.1038/nrn2480 [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Zhao C, & Sim FJ (2002). Ageing and CNS remyelination. Neuroreport, 13(7), 923–928. [DOI] [PubMed] [Google Scholar]
- Franklin RJM, & Blakemore WF (1997). To what extent is oligodendrocyte progenitor migration a limiting factor in the remyelination of multiple sclerosis lesions? Multiple sclerosis (Houndmills, Basingstoke, England), 3(2), 84–87. doi: 10.1177/135245859700300205 [DOI] [PubMed] [Google Scholar]
- Funfschilling U, Supplie LM, Mahad D, Boretius S, Saab AS, Edgar J, … Nave KA (2012). Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature, 485(7399), 517–521. doi: 10.1038/nature11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg-Cohen N, Guo Y, Margolis F, Cohen Y, Miller NR, & Bernstein SL (2005). Oligodendrocyte dysfunction after induction of experimental anterior optic nerve ischemia. Investigative ophthalmology & visual science, 46(8), 2716–2725. doi: 10.1167/iovs.04-0547 [DOI] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson TJ, Yool D, Thomson CE, Schwab MH, … Nave K-AA (1998). Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science (New York, NY), 280(5369), 1610–1613. [DOI] [PubMed] [Google Scholar]
- Groves A, Kihara Y, & Chun J (2013). Fingolimod: Direct CNS effects of sphingosine 1-phosphate (S1P) receptor modulation and implications in multiple sclerosis therapy. Journal of the neurological sciences. doi: 10.1016/j.jns.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, & Leach SD (2008). Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish, 5(2), 97–110. doi: 10.1089/zeb.2008.0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow DE, & Macklin WB (2014). Inhibitors of myelination: ECM changes, CSPGs and PTPs. Exp Neurol, 251, 39–46. doi: 10.1016/j.expneurol.2013.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow DE, Saul KE, Culp CM, Vesely EM, & Macklin WB (2014). Expression of proteolipid protein gene in spinal cord stem cells and early oligodendrocyte progenitor cells is dispensable for normal cell migration and myelination. J Neurosci, 34(4), 1333–1343. doi: 10.1523/JNEUROSCI.2477-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, & Omura T (2006). Cell tracking using a photoconvertible fluorescent protein. Nat Protoc, 1(2), 960–967. doi: 10.1038/nprot.2006.96 [DOI] [PubMed] [Google Scholar]
- Hines JH, Ravanelli AM, Schwindt R, Scott EK, & Appel B (2015). Neuronal activity biases axon selection for myelination in vivo. Nature Publishing Group. doi: 10.1038/nn.3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez C, Shields SA, El-Etr M, Leonelli E, Magnaghi V, Li WW, … Franklin RJ (2003). Steroids and the reversal of age-associated changes in myelination and remyelination. Prog Neurobiol, 71(1), 49–56. [DOI] [PubMed] [Google Scholar]
- Jung SH, Kim S, Chung AY, Kim HT, So JH, Ryu J, … Kim CH (2010). Visualization of myelination in GFP-transgenic zebrafish. Dev Dyn, 239(2), 592–597. doi: 10.1002/dvdy.22166 [DOI] [PubMed] [Google Scholar]
- Kala M, Miravalle A, & Vollmer TL (2011). Recent insights into the mechanism of action of glatiramer acetate. Journal of Neuroimmunology, 235(1–2), 9–17. doi: 10.1016/j.jneuroim.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Khalaj AJ, Hasselmann J, Augello C, Moore S, & Tiwari-Woodruff SK (2016). Nudging oligodendrocyte intrinsic signaling to remyelinate and repair: Estrogen receptor ligand effects. J Steroid Biochem Mol Biol, 160, 43–52. doi: 10.1016/j.jsbmb.2016.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, & Schilling TF (1995). Stages of embryonic development of the zebrafish. Dev Dyn, 203(3), 253–310. doi: 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kitazono AA (2009). Improved gap-repair cloning method that uses oligonucleotides to target cognate sequences. Yeast, 26(9), 497–505. doi: 10.1002/yea.1680 [DOI] [PubMed] [Google Scholar]
- Kucenas S, Snell H, & Appel B (2008). nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol, 4(2), 71–81. doi:S1740925X09990123 [pii]; 10.1017/S1740925X09990123[doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, … Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn, 236(11), 3088–3099. doi: 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, … Nave K-A (2003). Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nature genetics, 33(3), 366–374. doi: 10.1038/ng1095 [DOI] [PubMed] [Google Scholar]
- Lariosa-Willingham KD, Rosler ES, Tung JS, Dugas JC, Collins TL, & Leonoudakis D (2016). A high throughput drug screening assay to identify compounds that promote oligodendrocyte differentiation using acutely dissociated and purified oligodendrocyte precursor cells. BMC Res Notes, 9(1), 419. doi: 10.1186/s13104-016-2220-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Calle E, Brennan A, Ahmed S, Sviderskaya E, Jessen KR, & Mirsky R (2001). In early development of the rat mRNA for the major myelin protein P(0) is expressed in nonsensory areas of the embryonic inner ear, notochord, enteric nervous system, and olfactory ensheathing cells. Dev Dyn, 222(1), 40–51. doi: 10.1002/dvdy.1165 [DOI] [PubMed] [Google Scholar]
- Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, … Rothstein JD (2012). Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature, 487(7408), 443–448. doi: 10.1038/nature11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, & Holt CE (2008). Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nat Protoc, 3(8), 1318–1327. doi: 10.1038/nprot.2008.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He Y, Fan S, & Sun B (2015). Clemastine rescues behavioral changes and enhances remyelination in the cuprizone mouse model of demyelination. Neurosci Bull, 31(5), 617–625. doi: 10.1007/s12264-015-1555-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dupree JL, Gacias M, Frawley R, Sikder T, Naik P, & Casaccia P (2016). Clemastine Enhances Myelination in the Prefrontal Cortex and Rescues Behavioral Changes in Socially Isolated Mice. J Neurosci, 36(3), 957–962. doi: 10.1523/JNEUROSCI.3608-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, & Copeland NG (2003). A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res, 13(3), 476–484. doi: 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae CA, & Peterson RT (2015). Zebrafish as tools for drug discovery. Nat Rev Drug Discov, 14(10), 721–731. doi: 10.1038/nrd4627 [DOI] [PubMed] [Google Scholar]
- Mathews ES, & Appel B (2016). Cholesterol Biosynthesis Supports Myelin Gene Expression and Axon Ensheathment through Modulation of P13K/Akt/mTor Signaling. J Neurosci, 36(29), 7628–7639. doi: 10.1523/JNEUROSCI.0726-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Fancy SPJ, Shen YA, Niu J, Zhao C, Presley B, … Chan JR (2014). Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med, 20(8), 954–960. doi: 10.1038/nm.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk KR, & Talbot WS (2009). Genetic dissection of myelinated axons in zebrafish. Curr.Opin.Neurobiol, 19(5), 486–490. doi:S0959–4388(09)00110-X [pii]; 10.1016/j.conb.2009.08.006 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JK, Willard BB, Yin X, Jeserich G, Kinter M, & Trapp BD (2004). The 36K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia, 45(4), 378–391. doi: 10.1002/glia.10338 [DOI] [PubMed] [Google Scholar]
- Muller C, Bauer NM, Schafer I, & White R (2013). Making myelin basic protein -from mRNA transport to localized translation. Front Cell Neurosci, 7, 169. doi: 10.3389/fncel.2013.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-C, Mehta A, Richardson JS, & Appel B (2002). olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Developmental biology, 248(2), 356–368. [DOI] [PubMed] [Google Scholar]
- Park HC, Shin J, Roberts RK, & Appel B (2007). An olig2 reporter gene marks oligodendrocyte precursors in the postembryonic spinal cord of zebrafish. Dev Dyn, 236(12), 3402–3407. doi: 10.1002/dvdy.21365 [DOI] [PubMed] [Google Scholar]
- Peppard JV, Rugg CA, Smicker MA, Powers E, Harnish E, Prisco J, … Chandross KJ (2015). High-content phenotypic screening and triaging strategy to identify small molecules driving oligodendrocyte progenitor cell differentiation. J Biomol Screen, 20(3), 382–390. doi: 10.1177/1087057114559490 [DOI] [PubMed] [Google Scholar]
- Porcu G, Serone E, De Nardis V, Di Giandomenico D, Lucisano G, Scardapane M, … Ragnini-Wilson A (2015). Clobetasol and Halcinonide Act as Smoothened Agonists to Promote Myelin Gene Expression and RxRgamma Receptor Activation. PLoS One, 10(12), e0144550. doi: 10.1371/journal.pone.0144550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston M, & Sherman LS (2011). Neural stem cell niches: roles for the hyaluronan-based extracellular matrix. Front Biosci (Schol Ed), 3, 1165–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, & Macklin WB (2014). Zebrafish as a model to investigate CNS myelination. Glia. doi: 10.1002/glia.22755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progatzky F, Dallman MJ, & Lo Celso C (2013). From seeing to believing: labelling strategies for in vivo cell-tracking experiments. Interface Focus, 3(3), 20130001. doi: 10.1098/rsfs.2013.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruvot B, Cure Y, Djiotsa J, Voncken A, & Muller M (2014). Developmental defects in zebrafish for classification of EGF pathway inhibitors. Toxicol Appl Pharmacol, 274(2), 339–349. doi: 10.1016/j.taap.2013.11.006 [DOI] [PubMed] [Google Scholar]
- Rasband MN, Tayler J, Kaga Y, Yang Y, Lappe-Siefke C, Nave KA, & Bansal R (2005). CNP is required for maintenance of axon-glia interactions at nodes of Ranvier in the CNS. Glia, 50(1), 86–90. doi: 10.1002/glia.20165 [DOI] [PubMed] [Google Scholar]
- Ravanelli AM, & Appel B (2015). Motor neurons and oligodendrocytes arise from distinct cell lineages by progenitor recruitment. Genes Dev, 29(23), 2504–2515. doi: 10.1101/gad.271312.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Readhead C, Popko B, Takahashi N, Shine HD, Saavedra RA, Sidman RL, & Hood L (1987). Expression of a myelin basic protein gene in transgenic shiverer mice: correction of the dysmyelinating phenotype. Cell, 48(4), 703–712. [DOI] [PubMed] [Google Scholar]
- Sattarzadeh A, Saberianfar R, Zipfel WR, Menassa R, & Hanson MR (2015). Green to red photoconversion of GFP for protein tracking in vivo. Sci Rep, 5, 11771. doi: 10.1038/srep11771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannevin RH, Chollate S, Jung MY, Shackett M, Patel H, Bista P, … Rhodes KJ (2012). Fumarates promote cytoprotection of central nervous system cells against oxidative stress via the nuclear factor (erythroid-derived 2)-like 2 pathway. J Pharmacol Exp Ther, 341(1), 274–284. doi: 10.1124/jpet.111.190132 [DOI] [PubMed] [Google Scholar]
- Schweitzer J, Becker T, Becker CG, & Schachner M (2003). Expression of protein zero is increased in lesioned axon pathways in the central nervous system of adult zebrafish. Glia, 41(3), 301–317. doi: 10.1002/glia.10192 [DOI] [PubMed] [Google Scholar]
- Seredick SD, Van Ryswyk L, Hutchinson SA, & Eisen JS (2012). Zebrafish Mnx proteins specify one motoneuron subtype and suppress acquisition of interneuron characteristics. Neural Dev, 7, 35. doi: 10.1186/1749-8104-7-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman LS, & Back SA (2008). A ‘GAG’ reflex prevents repair of the damaged CNS. Trends Neurosci, 31(1), 44–52. doi: 10.1016/j.tins.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Smith PM, & Jeffery ND (2006). Histological and ultrastructural analysis of white matter damage after naturally-occurring spinal cord injury. Brain pathology (Zurich, Switzerland), 16(2), 99–109. doi: 10.1111/j.1750-3639.2006.00001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüve O, & Bennett JL (2007). Pharmacological Properties, Toxicology and Scientific Rationale for the use of Natalizumab (Tysabri) in Inflammatory Diseases. CNS Drug Reviews, 13(1), 79–95. doi: 10.1111/j.1527-3458.2007.00003.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Kucenas S, & Appel B (2010). Sox10 is necessary for oligodendrocyte survival following axon wrapping. Glia, 58(8), 996–1006. doi: 10.1002/glia.20981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, & Nave K-A (2008). Multiple Sclerosis: An Immune or Neurodegenerative Disorder? Annual Review of Neuroscience, 31(1), 247–269. doi: 10.1146/annurev.neuro.30.051606.094313 [DOI] [PubMed] [Google Scholar]
- Uschkureit T, Sporkel O, Stracke J, Büssow H, & Stoffel W (2000). Early onset of axonal degeneration in double (plp−/−mag−/−) and hypomyelinosis in triple (plp−/−mbp−/−mag−/−) mutant mice. The Journal of Neuroscience, 20(14), 5225–5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher LC, Johnstone A, Erturk A, Hu Y, Strikis D, Wanner IB, … Bixby JL (2010). A chemical screen identifies novel compounds that overcome glial-mediated inhibition of neuronal regeneration. J Neurosci, 30(13), 4693–4706. doi: 10.1523/JNEUROSCI.0302-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SL, Ariga J, Mathias JR, Coothankandaswamy V, Xie X, Distel M, … Mumm JS (2012). Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLoS One, 7(1), e29916. doi: 10.1371/journal.pone.0029916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, … Zon LI (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell, 2(2), 183–189. doi: 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HC, Scolding NJ, & Raine CS (2006). Co-expression of PDGF alpha receptor and NG2 by oligodendrocyte precursors in human CNS and multiple sclerosis lesions. J Neuroimmunol, 176(1–2), 162–173. doi: 10.1016/j.jneuroim.2006.04.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.