Abstract
The United States Environmental Protection Agency's (EPA)1 2012 Recreational Water Quality Criteria included an Enterococcus spp. quantitative polymerase chain reaction (qPCR) method as a supplemental indicator-method. In 2012, performance of qPCR for beach monitoring remained limited, specifically with addressing interference. A systematic literature search of peer-reviewed publications was conducted to identify where Enterococcus spp. and E. coli qPCR methods have been applied in ambient waters. In the present study, we evaluated interference rates, contributing factors resulting in increased interference in these methods, and method improvements that reduced interference. Information on qPCR methods of interest and interference controls were reported in 16 papers for Enterococcus spp. and 13 papers for E. coli. Of the Enterococcus spp. qPCR methods assessed in this effort, the lowest frequencies of interference were reported in samples using Method 1609. Low frequencies of sample interference were also reported EPA's modified E. coli qPCR method, which incorporates the same reagents and interference controls as Method 1609. The literature indicates that more work is needed to demonstrate the utility of E. coli qPCR for widespread beach monitoring purposes, whereas more broad use of Method 1609 for Enterococcus spp. is appropriate when the required and suggested controls are employed.
Keywords: Recreational water, E. coli, Enterococcus
Graphical Abstract
1. Introduction
Quantitative polymerase chain reaction (qPCR) methodology offers the advantage of providing rapid detection results (2–6 h), allowing beach managers to make same-day decisions to protect recreators (Griffith and Weisberg, 2011; U.S. EPA, 2018). In contrast, water quality results for traditional culturable indicator methods are not available until 24–48 h after sampling (Haugland et al., 2016). When using qPCR-based enumeration methods in place of culturable indicator methods, sample interference should be assessed. Interference is defined as any process that results in lower quantitative estimates than expected or actual values (Haugland et al., 2012). For these methods, interference occurs when substances in the test sample inhibit polymerase function (i.e., PCR inhibition) or cause the DNA to be lost or unavailable for amplification interference (i.e., poor recovery of amplifiable target gene sequences) (Haugland et al., 2012). This interference can result in false negative results of the sample. Examples of substances causing interference include humic acids, coral sands, calcium, and certain types of clay particles; however, there are likely many other unidentified substances that can also contribute to qPCR interference (Goyer and Dandle, 2012; Kirs, 2016; Opel et al., 2009; Shanks et al., 2016).
In the 2012 Recreational Water Quality Criteria (RWQC), the United StatesEnvironmental Protection Agency (EPA) proposed qPCR Method 1611 to detect and quantify Enterococcus spp. in ambient water on a site-specific basis (U.S. EPA, 2012a). EPA provided qPCR Method 1611 for states’ consideration and possible use following demonstration of the method for beach monitoring purposes as part of the EPA National Epidemiological and Environmental Assessment of Recreational Water (NEEAR) studies (Wade et al., 2006, Wade et al., 2008, Wade et al., 2010). EPA’s Enterococcus spp. qPCR Method A, precursor of the subsequently published EPA Method 1611, was significantly associated with gastrointestinal (GI) illness in the human-impacted EPA NEEAR studies (Wade et al., 2006, Wade et al., 2008, Wade et al., 2010). However, at the time of the publication of the RWQC in 2012, EPA still had limited experience with the method’s performance across a broad range of environmental conditions. Users were cautioned to be aware of the potential for qPCR interference in various waterbodies, which may vary on a site-specific basis, and were encouraged to conduct a site-specific analysis of the method’s performance prior to use in a beach notification program or adoption of water quality standards based on the method.
Over time, several method adaptations, as reflected by the EPA’s migration from Method A to Method 1611 to Method 1609, have been created to better estimate and control sample interferences (U.S. Environmental Protection Agency (US-EPA), 2010a, U.S. Environmental Protection Agency (US-EPA), 2012b, U.S. Environmental Protection Agency (US-EPA), 2013b). When publishing qPCR results, authors have been encouraged to use controls for the identification of and to address the potential for sample interference (Bustin et al., 2009).
The objectives of this work are to better understand and identify: 1) where Enterococcus spp. and E. coli qPCR methods have been applied since 2010 (the time at which information gathering in support of the 2012 RWQC stopped); 2) the rate of interference when using molecular methods in those waterbodies; 3) method improvements that have reduced interference; and 4) method or water matrix attributes (e.g., turbidity) and dynamics of fecal contamination that may continue to contribute to poor method performance or increased interference.
2. Materials and methods
2.1. Systematic literature search and screening
A systematic literature search of the peer-reviewed literature for publications reporting qPCR monitoring data in recreational water in PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and Web of Science was performed. The search included keywords relating to specific indicator organisms of interest (i.e., Enterococcus spp. and E. coli), qPCR methodology, inhibition, and source water. The full set of literature search terms is provided in Table S1 in the Supplemental Material. The searched methods were not limited to publish EPA methods. The literature search was limited to English language peer-reviewed citations published between 2010 and March 2017. In support of the development of the 2012 RWQC, EPA evaluated published demonstrations on the application of qPCR in ambient water for water quality monitoring purposes (U.S. EPA, 2018). The cut-off date for this previous evaluation was 2010. Thus, this literature search focused on identifying studies published after previous research conducted in support of the 2012 RWQC.
Abstracts were subject to a primary screen for relevance and included papers using Enterococcus spp. qPCR and/or E. coli qPCR. Following the abstract screening, the full text of articles passing scope was reviewed to determine if ambient water samples were analyzed. Samples spiked with the target organism were determined to be not relevant. Studies also needed to provide information on the occurrence and/or evaluation of inhibition to be included in the review. Relevant studies were then reviewed to obtain specific information related to study location, sampling time, waterbody type, analytical method(s) applied, how interference was controlled, contamination source(s) and dynamics (e.g., wet-weather), water quality results, percent of samples inhibited, limit of quantitation, and percent recovery.
2.2. qPCR method improvements
We read the full text of articles that passed the primary screen and identified information on method improvements and the use of any qPCR interference controls applied to reduce interference described by the study authors. Common qPCR interference controls include: sample processing control (SPC); internal amplification control (IAC); dilution; ratio spiked test matrix/spiked control matrix; and calculation using delta-delta cycle threshold (Table 1).
Table 1.
Common qPCR interference controls.
| Interference controls |
Abbreviation | Application | Common types | References |
|---|---|---|---|---|
| Sample processing control | SPC | A non-target DNA sequence used to estimate recovery efficiency. The control involves spiking a known quantity of non-target DNA into the sample prior to processing. | Salmon testes DNA (i.e., Sketa 22) | U.S. EPA, 2012b;U.S. EPA, 2013b |
| Internal amplification control | IAC | A non-target DNA sequence added to the reaction mix prior to the qPCR reaction. If the non-target DNA does not amplify as expected, a problem with the qPCR reaction is indicated (e.g., DNA polymerase inhibition). | IAC5 (refers the specific plasmid DNA primer template sequence used as part of this assay) | U.S. EPA, 2012b;U.S. EPA, 2013b |
| Dilution | Dilution | Dilution of the sample can result in dilution of other compounds that interfere with DNA amplification. Different dilutions can be compared (i.e., serial dilutions). | 5× 25× |
Cao et al., 2012 |
| Ratio spiked test matrix/spiked control matrix | STM/SCM | The recovery of target DNA sequences (gene copies) from target organisms spiked into the water samples (STM) can be compared to the recovery of DNA from spiked target organisms in control samples (SCM). The STM/SCM ratio can provide an additional measure of interference caused by inhibitors in the water matrix. | Not applicable | Haugland et al., 2016 |
| Addition of higher salmon DNA concentrations to samples during extraction | Not applicable | Demonstrated at one tropical site to reduce interference due to DNA loss during sample extraction. | 25× increase in salmon DNA concentration | Haugland et al., 2012 |
| Calculation using delta-delta cycle threshold | ΔΔCt | A method to estimateEnterococcus spp. in a water sample, accounting for recovery and partial inhibition. The ΔΔCt is calculated from the ΔCt (Enterococcus assay Ct – Sketa SPC assay Ct value) for the water sample and for the calibrator/positive control sample and then subtracting the calibrator/positive control ΔCt from the water sample ΔCt. | Not applicable | U.S. EPA, 2010a;U.S. EPA, 2012b;U.S. EPA, 2013b |
When available, we also reported the percentage of sample interference. The percentages of sample interference were either reported directly by the study’s author in the paper, or derived by calculating the percentage based on the total number of samples and the number of samples for which interference was reported. In some cases, the percentage of sample interference could not be identified in the paper (or was not reported for both dilution measures), and was labeled in Table 3and Table 4 as “Not reported.”
Table 3.
Summary of interference rates for Enterococcus spp. qPCR methods.
| Reference | Water type | Location (number of sites) |
Fecal source | Number of samples undiluted (percent interference) |
Number of samples diluted 5× (percent interference) |
Strategies to test interferencea |
|---|---|---|---|---|---|---|
| EPA Method 1609 (Environmental Master Mix) | ||||||
| Dorevitch et al., 2017 | Freshwater | IL (9) | Waste water, non-point source | 1256 (1.1) | 540 (0.37) | SPC (Sketa 22) (Ct 3) |
| Haugland et al., 2016 | Marine | FL, CA, NC (9) | Not reported | 241 (10) | 356 (4) | SPC (Sketa 22) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Haugland et al., 2016 | Freshwater | WI, OH, FL (13) | Not reported | 491 (11) | 419 (3) | SPC (Sketa 22) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Sivaganesan et al., 2014 | Freshwater | OH, KY, IN, PA, IA (7) | Non-point source, spiked samples, waste water, animal and human waste | 221 (5) | 221 (3) | SPC (Sketa 22) (Ct 3) IAC (IAC5) (Ct 1.5) |
| Haugland et al., 2012 | Freshwater | OH, KY (5) | Non-point source, spiked samples, waste water, animal and human waste | 268c (0) | 268c (0.7) | SPC (Sketa 22) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | 133 (0) | Not reported | SPC (Sketa 2)d (Ct 3)b |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | 133 (11) | 133 (0) | IAC (IAC5) (Ct 1.7)b |
| EPA Method 1611 (Universal Master Mix) | ||||||
| Haugland et al., 2016 | Marine | FL, CA, NC (9) | Not reported | 240c (>40)e | 359 (7) | SPC (Sketa 22) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Haugland et al., 2016 | Freshwater | WI, OH, FL (13) | Not reported | 490c (>40)e | 419 (6) | SPC (Sketa 22) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Sivaganesan et al., 2014 | Freshwater | OH, KY, IN, PA, IA (7) | Non-point source, spiked samples, waste water, animal and human waste | 221 (53) | 221 (11) | SPC (Sketa 22) (Ct 3) IAC (IAC5) (Ct 1.5) |
| Cao et al., 2013 | Marine | CA (9c) | Spiked samples, waste water, human waste | 12 (0) | Not reported | SPC (Sketa 22) (Ct threshold not indicated) |
| Converse et al., 2012a | Freshwater | WI (1) | Animal waste | 80 (0) | Not reported | SPC (Sketa 22) (Ct 3) |
| Converse et al., 2012b | Marine | CA (3) | Non-point source | 1200 (7) | Not reported | SPC (Sketa 22) (Ct 3) |
| Haugland et al., 2012 | Freshwater | OH, KY (5) | Non-point source, spiked samples, waste water, animal and human waste | 268c (18) | 268c (1.5) | SPC (Sketa 22) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Haugland et al., 2012 | Marine | PR (6c) | Not reported | Not reported | 684 (32)b | SPC (Sketa 22) (Ct 3) IAC (IAC5) (Ct 1.5) |
| EPA Method A | ||||||
| Raith et al., 2014 | Marine | CA (9) | Not reported | Not reported | 306 (5)a | SPC (Sketa 2) (Ct 3)f |
| Zimmer-Faust et al., 2014 | Marine, freshwater | CA, Mexico (18) | Non-point source, animal waste, waste water, spiked samples | 82 (0) | Not reported | SPC |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | 133 (7) | 133 (1) | SPC (Sketa 2) (Ct 3)b |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | 133 (42) | 133 (7) | IAC (IAC5) (Ct 1.7)b |
| Haugland et al., 2012 | Freshwater | OH, KY (5) | Non-point source, spiked samples, waste water, animal and human waste | 268c (30) | 268c (7) | SPC (Sketa 2) (Ct 3)b IAC (IAC5) (Ct 1.5) |
| Haugland et al., 2012 | Marine | PR (6c) | Not reported | Not reported | 895 (36)b | SPC (Sketa 2) (Ct 3) IAC (Ct 1.5) |
| Haugland et al., 2012 | Freshwater | AZ, CA, GA, HI, IA, IN, LA, MD, MN, NC, NJ, NY, WA, WI (27) | Not reported | Not reported | 108 (7) | SPC (Sketa 2) (Ct 3) |
| Sauer et al., 2011 | Freshwater | WI | Non-point source, waste water | 214 (<1) | Not reported | IAC |
| Abdelzaher et al., 2010 | Marine | FL (1) | Non-point source, human waste | 12 (0) | Not reported | SPC |
| Scorpion | ||||||
| Raith et al., 2014 | Marine | CA (9) | Not reported | Not reported | 306 (5)g | SPC (Ct > 1.7) |
| Cao et al., 2013 | Marine | CA (9c) | Spiked samples, waste water, human waste | Not reported | 12 (0) | SPC (Sketa 22)d |
| Converse et al., 2012b | Marine | CA (3) | Non-point source | 1200 (16) | Not reported | SPC (Ct 1.6) |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | 133 (42) | 133 (4) | SPC (Sketa 2) (Ct 3)b |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | 133 (56) | 133 (18) | IAC (Ct 1.7)b |
| Noble et al., 2010 | Marine, freshwater | CA | Urban runoff, spiked samples, waste water | 238 (<5) | Not reported | SPC (Lactococcus) (Ct 1.5) IAC (Ct 1.5) |
| Other | ||||||
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | TaqFast method: 133 (9) | TaqFast method: 133 (1) | SPC (Sketa 2) (Ct 3)b |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | TaqFast method: 133 (53) | TaqFast method: 133 (8) | IAC (IAC5) (Ct 1.7)b |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | Taq Fastfast method: 133 (23) | Taq Fastfast method: 133 (2) | SPC (Sketa 2) (Ct 3)b |
| Cao et al., 2012 | Marine, freshwater | CA, IL (52) | Non-point source, human waste | Taq Fastfast method: 133 (90) | Taq Fastfast method: 42 (37) | IAC (IAC5) (Ct 1.7)b |
| Bergeron et al., 2011 | Marine | Spain, France | Spiked samples, waste water, human waste | 85 (0) | Not reported | qPCR control |
| Santiago-Rodriguez et al., 2012h | Freshwater | PR | Non-point source, animal waste | 130 (0) | 130 (0)i | Not reported |
| Wang et al., 2016h | Freshwater, marine | CA (8) | Not reported | 24 (0) | Not reported | compare to digital PCR results |
Some studies did not specify an interference cut-off value; only one Ct value is provided.
Other interference controls evaluated (dilution and/or STM/SCM).
Interference rates shown are based on SPC assay only, IAC assay results were generally in agreement when available.
Deviated from Method 1609 by using Sketa2 rather than Sketa22 SPC assay.
Average interference rate was not reported separately for marine and freshwater in undiluted samples. However, separate rates are reported for 5× diluted samples.
The SPC control was evaluated using Ct shift acceptance thresholds of 3.0 and 1.7. When using the 1.7 Ct acceptance threshold, 22% interference was found.
Composite of undiluted and 5× dilution results. 5× dilutions analyzed only for undiluted samples that failed Sketa2 assay acceptance criterion.
qPCR conducted with 1× TaqMan Universal PCR Master Mix and the Entero1 23S rRNA gene assay.
10-fold dilution.
Table 4.
Summary of interference rates for E. coli qPCR methods.
| Reference | Water type |
Location (number of sites) |
Fecal source | Number of samples undiluted (percent interference) |
Number of samples 5× diluted (percent interference) |
Strategies to test interferencea |
|---|---|---|---|---|---|---|
| EPA Method C [EC23S857]b | ||||||
| Chern et al., 2011 | Marine, freshwater | MA, PR (12) | Non-point source | 25 (0)c | Not reported | SPC (Sketa 2) (Ct > 3) |
| Peed et al., 2011 | Freshwater | OH (9) | Non-point source, waste water | 215 (2.1) | Not reported | IAC (CowM2)c(Ct 35.1 ± 1.8) |
| Molina et al., 2014 | Marine | SC, FL (5) | Non-point source | 471d (7) | Not reported | IAC (CowM2)c(Ct 33.8 ± 1.6) |
| Scorpion | ||||||
| Krometis et al., 2013 | Freshwater | NC (4) | Non-point source | 94 (31) | 29 (20)e | SPC (Sketa 2) (Ct > 1.5) |
| Painter et al., 2013 | Freshwater | TX (1) | Human and animal waste | Not reported | 102 (19)f | IAC |
| Converse et al., 2012a | Freshwater | WI (1) | Animal waste | 80 (0) | Not reported | SPC (Sketa 22) (Ct 3) |
| Noble et al., 2010 | Marine, freshwater | CA (6) | Spiked sample, waste water, urban runoff | 226 (<5) | Not reported | Ct shift (> 1.5) |
| Other | ||||||
| Cloutier and McLellan, 2017 | Freshwater | WI (6) | Not reported | 124 | Not reported | Not reported |
| Byappanahalli et al., 2015 | Freshwater | IN (1) | Not reported | 5d (0) | Not reported | SPC |
| Walker et al., 2013 | Marine, freshwater | PR, Trinidad (44) | Non-point source, waste water | 210 (0) | Not reported | IAC (Ct <2) |
| Zhang et al., 2012 | Freshwater | MO (1) | Spiked sample, non-point source | 10d (8) | Not reported | IAC |
| Bergeron et al., 2011 | Marine | Spain, France (3) | Spiked sample, waste water | 80 | 0 | Ct shift (24.5 ± 0.5 cycles) |
| Sauer et al., 2011 | Freshwater | WI (4) | Non-point source, waste water | 220 (<1) | Not reported | IAC |
Some studies did not specify an interference cut-off value. Thus, only one Ct value is provided.
Current draft method calls for the use of salmon DNA SPC with Sketa22 assay, IAC5 plasmid and assay for inhibition control, 56 degrees Celsius annealing temperature for thermal cycling, and EMM reagent. Some of these provisions were not followed in the reported studies.
IAC using CowM2 plasmid DNA.
Estimated.
10× dilution.
DNA not diluted but cleaned using post-extraction technique (Chelex®100 and solubilization/binding buffer QX1).
3. Results
3.1. Literature screening and review
The literature search returned 337 unique results, of which 54 were relevant based on the abstract screening (Fig. 1). An additional 13 studies were identified through other sources (e.g., cited in another paper and identified via a hand search). The full-text of these 67 studies was reviewed. Upon review, some studies were determined to be out of scope for reasons such as failure to evaluate organisms of interest to this effort or the use of spiked samples, non-ambient water, or only non-molecular based methods. Some studies categorized as in scope evaluated multiple organisms and/or methods of interest. A total of 32 studies included Enterococcus qPCR and 22 included E. coli qPCR (Fig. 1).
Fig. 1.
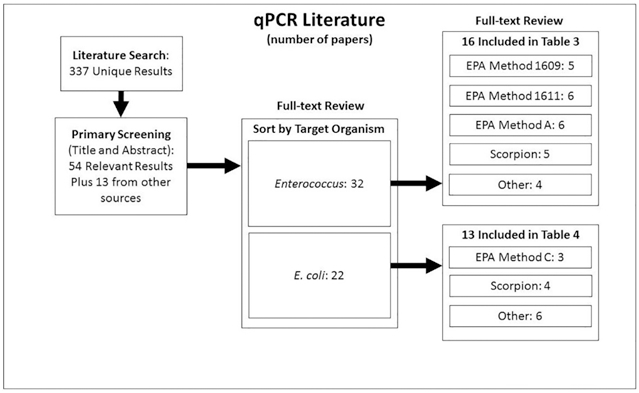
Overview of the literature review process. Some studies included in the full-text review reported multiple organisms and/or multiple qPCR methods. Others were excluded as they were found to be out of scope for this effort.
3.2. Advancements in Enterococcus spp. qPCR methods
3.2.1. EPA methods
EPA developed Method A for the detection and enumeration of Enterococcus spp. using qPCR. EPA Method A was successfully applied to EPA’s NEEAR study (Haugland et al., 2005; Wade et al., 2006, Wade et al., 2008, Wade et al., 2010). The freshwater sites in the Great Lakes and four temperate marine beaches demonstrated minimal to no interference, but the tropical marine beach site samples from Puerto Rico exhibited significant interference (Haugland et al., 2012; U.S. EPA, 2010b). As a result, EPA’s Method A was updated and published as EPA Method 1611. Updates included two interference controls: 1) a requirement of the SPC assay to use Sketa 22 (a more robust version of the original Sketa 2 assay used to detect the Salmon DNA SPC); and 2) a recommendation for using the IAC assay. As in EPA Method A, the method employed a reagent called Universal Master Mix (UMM) (TaqMan; Applied Biosystems, Foster City, CA) (U.S. EPA, 2012b). However, even with these updates, EPA Method 1611 still was found to result in high levels of interference (>10%) in inland freshwater samples, unless samples were diluted five-fold or more (Haugland et al., 2012, Haugland et al., 2016; Sivaganesan et al., 2014). Dilution is a standard methodological approach to lessen interference in water samples.
To further address the potential for interference, EPA developed and published EPA Method 1609 (U.S. EPA, 2013b). Method 1609 uses a newer reagent called Environmental Master Mix (EMM) (TaqMan; Applied Biosystems, Foster City, CA) and has produced results with lower levels of interference in undiluted samples) (Cao et al., 2012; Haugland et al., 2012, Haugland et al., 2014, Haugland et al., 2016). The EMM provides a more sophisticated chemistry than the previously developed UMM to amplify and analyze complementary DNA and DNA targets in water samples with known inhibitory substances (TaqMan; Applied Biosystems, Foster City, CA). Like EPA Method 1611, EPA Method 1609 requires the SPC interference control assay using Sketa 22 and recommends the IAC assay.
3.2.2. Non-EPA methods
Most other qPCR methods for measuring Enterococcus spp. in ambient water have been applied by a single research laboratory. The exception is the Scorpion-based qPCR assay from Noble and colleagues (Noble et al., 2010). The Scorpion qPCR technology uses a different master mix (OmniMix, Cepheid, Inc., Sunnyvale, CA) and processing controls (Smartbeads, Cepheid Inc., Sunnyvale, CA), and was designed to be faster than other qPCR chemistries.
Table 2 summarizes analytical details related to reducing interference and the strategies for controlling for interference in both EPA and non-EPA qPCR methods. The Scorpion-based method was included in Table 2 because multiple papers evaluated the method, published by Noble and colleagues, in ambient waters (Noble et al., 2010).
Table 2.
Enterococcus spp. qPCR methods.
| Method | Analytical permutations | Performance/interference evaluation analyses | References | |||
|---|---|---|---|---|---|---|
| Master mix | Recommended sample extract dilution |
SPC: acceptance range | IAC: acceptance range | Target sequence copy or cell equivalent spike recovery: acceptance range |
||
| EPA Method 1609 | Environmental Master Mix | Undiluted (5× diluted optional) | Sketa 22: Test sample Ct within 3 units of calibrator samples (mandatory in method) | IAC5: Test sample Ct within 1.5 units of negative control samples (recommended in method) | Target sequence copy: 50–200%a | Haugland et al., 2016; U.S. EPA, 2013b |
| EPA Method 1611 | Universal Master Mix | 5× diluted | Sketa 22: Test sample Ct within 3 units of calibrator samples (mandatory in method) | IAC5: Test sample Ct within 1.5 units of negative control samples (recommended in method) | Target sequence copy: 50–200%a | Haugland et al., 2012; U.S. EPA, 2012b |
| EPA Method A | Universal Master Mix | 5× or 25× diluted | Sketa 2b: Test sample Ct within 3 units of uninhibited reference samples | Not evaluated | Cell equivalent: Acceptance ranges defined by study resultsc; Freshwater: detect – 333% Marine Water: detect – 1123% |
U.S. EPA, 2010a |
| Scorpion method | OmniMix | 10× diluted, if needed | Lactococcus(SmartBeads): 1.5 Ct shift | Enterococcus internal control template, Lactococcus internal control template (SmartBeads): 1.5 Ct shift | Not evaluated | Noble et al., 2010 |
Recovery ratio of spiked test matrix (filters and retentates from collected water samples spiked with Enterococcus spp. cells) to spiked control matrix (clean filters spiked with Enterococcus spp. cells).
Sketa 22 was also evaluated.
Recovery ratio of estimated qPCR cell equivalents in spiked test matrix to estimated CFU in the spikes. Spiking done with 550 CFU Bioballs™ (bioMérieux).
3.3. Recent application of Enterococcus spp. qPCR methods to ambient waters (2010–2017)
Table 3 summarizes results from the 16 papers that include information on the selected Enterococcus spp. qPCR methods.
In a national study focusing primarily on potentially problematic sites, including both coastal fresh and marine waters and inland freshwaters, EPA Method 1609 showed an average qPCR interference rate of 10% (range 0–22%) and 11% (range 0–24%) in undiluted samples from nine and 12 individual temperate marine and freshwater sites, respectively, based on the SPC and IAC controls (Haugland et al., 2016). Average interference rates from other studies were lower (Table 3). A five-fold dilution of the water sample extracts from the national study reduced the average interference rates to 4% and 3% for temperate marine and freshwaters, respectively; and reduced the interference rates to acceptable frequencies (<10%) at most sites (9/9 marine and 10/12 freshwater) (Haugland et al., 2016; U.S. EPA, 2013a).
In contrast, EPA Method 1611 exhibited a much higher average interference rate in undiluted samples, ranging from 18 to 53%, in studies of corresponding temperate marine and freshwater sites. A five-fold dilution of the water sample extracts again significantly reduced the interference rate in both freshwaters and marine waters to acceptable frequencies (<10%) at most sites studied. For EPA’s qPCR Method A, the interference rate is significantly higher when using Sketa 2, as compared to using Sketa 22 in Method 1611 for analyses of inland freshwater samples (Table 3).
Only one of the studies shown in Table 3 addressed the potential reason for interference in the water samples tested (Haugland et al., 2012). Haugland and colleagues explored the causes of the discrepancy in criterion failure rates for control assays among samples collected from the Ohio River and Boquerón Bay (Haugland et al., 2012). Authors noted the predominance of polymerase inhibitory compounds (i.e., calcium, iron, iron containing compounds, and tannic acid) may have affected the amplification results of both the IAC and SPC in the Ohio River study, whereas the presence of DNA binding compounds (i.e., humic acid and melanin) may have affected the SPC assay results in the Boquerón Bay study (Haugland et al., 2012). Kinzelman and co-authors speculated that changes in environmental conditions (e.g., turbidity) due to runoff from land during precipitation events could have been a factor for interference in that particular study (Kinzelman et al., 2011). Additionally, Wang and colleagues spiked qPCR reactions with organic (humic acid, 5 ng/μL) and inorganic (calcium, 2.0 mM) matter to test their inhibitory effects on PCR reactions (Wang et al., 2016). The study found that small concentrations of both caused significant inhibition. Too few studies provided adequate information on fecal source dynamics to draw any meaningful conclusions on how fecal sources might impact the likelihood of interference (Table 3).
Overall, EPA Enterococcus qPCR (Method 1609) resulted in lower frequencies of interference in analyzed samples, as compared to other methods (EPA Method A, Method 1611, and the Scorpion-based method). Use of the EMM and, when necessary, sample dilution addressed interference at the nine marine and 23 (of 25) freshwater sites in 10 states investigated in EPA studies (Haugland et al., 2012, Haugland et al., 2016; Sivaganesan et al., 2014).
3.4. Advancements in E. coli qPCR methods
EPA has developed a draft qPCR method for E. coli (referred to as draft Method C) (U.S. EPA, 2018). This method was first described by Chern and colleagues and uses EC23S857 primers (Chern et al., 2011). A total of three studies were identified in the literature that referred to the use of EC23S857 primers in their methodology, including Chern and colleagues (Chern et al., 2011). Modifications to improve the method have been made by several study authors. In addition to using Sketa 22 for an SPC, Peed and colleagues and Molina and colleagues used the CowM2 plasmid as an IAC, which was originally developed by EPA researchers for bovine-specific microbial source tracking (Molina et al., 2014; Peed et al., 2011; Shanks et al., 2008). The method also employs the EMM, which minimizes interference. Additionally, the current EPA draft Method C calls for the use of salmon DNA SPC with Sketa22 assay, the IAC5 plasmid and assay for inhibition control, and 56 degrees Celsius annealing temperature for thermal cycling.
Over the past few years, other researchers have developed qPCR methods for E. coliand tested those methods in ambient waters, using a variety of available primers and probes specific to E. coli (Table 4). These methods have not been directly compared to EPA’s E. coli qPCR method in ambient waters, and thus differences in performance are unclear.
Table 4 summarizes results from the 13 papers that included information on E. coliqPCR methods, including EPA draft Method C. The 13 studies all illustrate low rates of interference (<10%) (Table 4), and overall EPA draft Method C has similar performance characteristics as EPA Method 1609. However, the number of sites and samples reported using EPA draft Method C is significantly less than those reported using Enterococcus spp. qPCR methods. Additionally, there are no peer-reviewed demonstrations of its use for routine monitoring at this time.
3.5. General advancements in molecular methods
With additional development, other molecular-based enumeration tools offer promise for microbial monitoring purposes. Digital PCR (dPCR), for example, is an emerging technology for determining the quantity of target DNA sequences in a sample. While traditional qPCR involves measuring DNA products in a single tube after each qPCR cycle, dPCR partitions the sample into thousands to millions of smaller reactions that are examined individually for binary endpoint results (presence/absence). The DNA density is then estimated from the fraction of positives using Poisson statistics. The dPCR methodology offers several key potential advantages over qPCR, including the elimination of standard curves, thus reducing the labor and materials associated with regularly running batch standards and the biases associated with calibration model variability (Wang et al., 2016). However, it is important to note that a positive standard control is still recommended by dPCR experts (Huggett et al., 2013). As a result, practitioners will still need to create and maintain a standard reference material as a positive control for routine testing. The dPCR methodology also offers improved repeatability (i.e., the precision of an assay among replicates of the same sample over a short period of time) and reproducibility (i.e., the consistency in results among operators, runs, or laboratories), resulting in the detection of a 1.25-fold difference in the DNA template (qPCR can typically only detect a two-fold difference) (Cao et al., 2016a). Additional advantages may include decreased interference rates due to sample partitioning, an increased tolerance for PCR inhibitor concentrations, and a superior multiplexing ability (Cao et al., 2016a, Cao et al., 2016b; Wang et al., 2016).
There are also several potential limitations of dPCR, as compared to qPCR. First, given this is a new technology, there would likely be costs associated with implementing it in a laboratory and obtaining the necessary instrumentation and supplies (Huggett et al., 2013). Secondly, the detectable range is smaller for dPCR, and currently the upper limit of quantitation of dPCR is four orders of magnitude lower than that of qPCR. Thus, sample dilution is required when measuring high concentrations of DNA targets, like those potentially found in sewage spills (Cao et al., 2016b). Additionally, Poisson statistics require uniformity in the partitions for accurate endpoint results. Viscous DNA, due to high concentrations or long templates, can result in uneven distributions, biasing the partitions and leading to potentially inaccurate results. If double-stranded DNA is denatured into single strands, the template is effectively increased because single-strands can occupy different partitions, which could lead to up to a two-fold overestimation by dPCR (Cao et al., 2016b).
4. Discussion
We reviewed the abstracts of 337 unique peer-reviewed published studies and identified a total of 67 studies that reported water quality monitoring data collected using Enterococcus spp. qPCR (i.e., EPA Method 1609, EPA Method 1611, Scorpion-based method, and other methods) and/or E. coli spp. qPCR (EPA draft Method C, Scorpion-based method, and other methods).
The use of Enterococcus spp. qPCR and the effects of sample interference were more frequently reported in the literature than E. coli qPCR. Of the available Enterococcusspp. qPCR methods, EPA Method 1609 had the fewest number of samples with interference (as compared to EPA Method A, Method 1611, and the Scorpion-based method) when the proper controls were in place (Haugland et al., 2014, Haugland et al., 2016).
Several researchers have identified environmental sources of sample interference and proposed approaches for its mitigation. There is some indication that coral sands, silt, and humic and tannic acids contribute to sample interference during qPCR reactions (Goyer and Dandle, 2012; Kirs, 2016; Opel et al., 2009; Shanks et al., 2016). However, the contributions of these compounds vary on a site-specific basis. For example, coral sands present in Hawaii’s tropical waters have been observed to contribute to high levels of sample interference (40–70%) (Kirs, 2016). Coral sands are believed to interfere by adsorbing DNA during the rapid DNA extraction process making them unavailable for PCR amplification (Kirs, 2016). A similar phenomenon was observed for kaolinite clay particles, which have properties similar to those of coral sands (Shanks et al., 2016). The presence of silt in samples can decrease method performance and the efficacy of the DNA extraction process (Goyer and Dandle, 2012; Kirs, 2016). High silt levels are characteristic of samples taken from waterbodies containing mud or influenced by stormwater and high-energy waves. Humic and tannic acids are produced by trees in deciduous forests found in areas on the eastern coast of the United States. Humic acid contributes to sample interference by binding to DNA and limiting available template, while tannic acid binds to DNA and inhibits polymerase function (Opel et al., 2009).
Much like the variability in the presence of environmental sources of interference, the efficacy of approaches to mitigate these sources of interference varies on a site-specific basis. Although Haugland and colleagues reported the efficacy of sample dilution and the use of the EMM in addressing interference in inland freshwater samples, other researchers have observed less favorable outcomes when applying these approaches in coastal marine water samples (Cao et al., 2012; Haugland et al., 2012, Haugland et al., 2016). While reducing the concentration of inhibitors, dilution can also potentially reduce the target concentration to levels below the detection limit and decrease method sensitivity (Cao et al., 2012).
Other important considerations for qPCR methods include sample preparation and extraction. Improper sample preparation can introduce materials, including compounds known or suspected to contribute to sample interference, which can create high variability among qPCR results (Bustin et al., 2009). Utilizing the correct extraction method and following its protocol is also important as technologies and protocols vary between methods (Bustin et al., 2009). Different nucleic acidextraction methods may impact the levels of interference observed in these studies as there can be variability in the binding of DNA to extraction columns in the presence of environmental sources of interference (Guo et al., 2009). Thus, the concentration of extracted template could vary when different extraction methods are employed (Bustin et al., 2009).
Overall, EPA Method 1609 has a more robust performance, with no sample dilution required in most instances, and a lower overall interference rate, as compared to other EPA methods (Draft Method A, EPA Method 1611). Sample dilution and use of the EMM addressed inhibition at the nine marine and 23 of the 25 potentially problematic freshwater sites in 10 states comprehensively investigated by the EPA since 2010 (Haugland et al., 2012, Haugland et al., 2016). Based on these results, broad use of EPA Method 1609 is appropriate, when the required and suggested controls are employed. The exception is when coral sands are known to be present in the water. Use of the EMM, the Sketa 22 SPC assay, and optional use of the IAC assay both reduces interference and identifies whether interference was observed in the qPCR sample (U.S. EPA, 2018).
Based on the available literature, more work is needed to demonstrate that E. coliqPCR is also ready for use for routine water quality monitoring. Although low rates of interference (<10%) have been reported, the number of samples and sample sites were much smaller than those included in studies of Enterococcus spp. qPCR. Studies with larger sample sizes and more sampling sites are needed to determine the characteristics of a sampling site where the use of E. coli qPCR is suitable for monitoring purposes. A peer-reviewed demonstration of the use of E. coli qPCR EPA draft Method C (the method first described by Chern and colleagues) is needed to determine if this method specifically is suitable for use (Chern et al., 2011; U.S. EPA, 2018). Additionally, a direct comparison between EPA’s E. coli qPCR method and other E. coli qPCR methods that use a variety of available primers and probes specific to E. coli is needed to determine the advantages and disadvantages of each method in ambient waters. These analyses should include consideration of the efficacy of available strategies on reducing the rate of interference, such as dilution.
Although our search was limited to literature published from 2010 to 2017, we acknowledge previous analyses and monitoring efforts that utilized qPCR to assess water quality. One relevant study not described in this effort due to its publication outside of the years included in this literature search strategy but acknowledged in both EPA’s 2012 RWQC and 2017 Five-Year Review of the 2012 Recreational Water Quality Criteria (U.S. Environmental Protection Agency (US-EPA), 2012a, U.S. Environmental Protection Agency (US-EPA), 2018) is Lavender and Kinzelman’s (2009) evaluation of E. coli qPCR against culturable methods at Wisconsin beaches. The authors reported that all five monitored sites demonstrated some potential for interference from ambient DNA present in the waterbody (Lavender and Kinzelman, 2009). Overall, this study demonstrated the utility of E. coli qPCR on a site-specific basis (U.S. EPA, 2018). In addition, EPA published water quality monitoring data in their National Rivers and Streams Assessment 2008–2009 and used qPCR to measure Enterococcus spp. (U.S. EPA, 2016). However, these analyses and assessments generally did not address sample interference. Prior to 2010, products to address sample interference in qPCR methodologies, such as the EMM and UMM, were limited as this technology was still emerging. Our literature search was targeted to include years in which published studies were more likely to include consideration of sample interference and the utilization of advanced technology to reduce the sample interferences, and to supplement the previous work done in EPA’s 2012 RWQC to evaluate the use of qPCR for beach monitoring purposes.
Finally, it should be noted that there are few publications to-date that have evaluated E. coli qPCR methods in ambient waters (Cao et al., 2015, Cao et al., 2016a, Cao et al., 2016b; Wang et al., 2016). Thus, the method is not broadly recommended for routine monitoring, at this time.
5. Conclusions
This systematic literature search provides insight on the progress made in demonstrating the utility of qPCR methodologies for quantifying and enumerating Enterococcus spp. and E. coli in ambient waters. Numerous studies demonstrating the utility of qPCR methods for the detection and enumeration of Enterococcus spp. were identified, while only limited data are available for E. coli. In the studies identified in this review, low levels of qPCR interference were reported in samples collected from both coastal marine waters and coastal and inland freshwaters for Enterococcus spp. and E. coli, respectively. One exception is the application of these qPCR methodologies in tropical, marine waters in Hawaii (Kirs, 2016). Thus, at this time, the use of E. coli qPCR for routine water quality monitoring should be considered on a site-specific basis as more work is done to demonstrate its utility whereas the use of EPA Method 1609 for the detection of Enterococcus is appropriate when the required and suggested controls are employed.
The following is the supplementary data related to this article.
Supplementary Material
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The research described in this article was funded by the EPA Office of Water, Office of Science and Technology under contract #EP-C-11–005 to ICF, LLC. This work has been subject to formal Agency review. The views expressed in this article are those of the authors and do not necessarily reflect the views or policies of the Environmental Protection Agency, and no official endorsement should be inferred. We are grateful to Kevin Oshima, Jamie Strong, Shari Barash, and Elizabeth Behl for their invaluable review and technical edit of our draft manuscript. The authors declare no competing financial interest.
References
- Abdelzaher AM, Wright ME, Ortega C, Solo-Gabriele HM, Miller G, Elmir S, Newman X, Shih P, Bonilla JA, Bonilla TD, et al. , 2010. Presence of pathogens and indicator microbes at a non-point source subtropical recreational marine beach. Appl. Environ. Microbiol. 76 (3), 724–732. 10.1128/AEM.02127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron P, Oujati H, Catalan Cuenca V, Huguet Mestre JM, Courtois S, 2011. Rapid monitoring of Escherichia coli and Enterococcus spp. in bathing water using reverse transcription-quantitative PCR. Int. J. Hyg. Environ. Health 214 (6), 478–484. 10.1016/j.ijheh.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. , 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4),611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Byappanahalli MN, Nevers MB, Whitman RL, Ishii S, 2015. Application of amicrofluidic quantitative polymerase chain reaction technique to monitor bacterial pathogens in beach water and complex environmental matrices. Environ. Sci. Technol. Lett. 2 (12), 347–351. 10.1021/acs.estlett.5b00251. [DOI] [Google Scholar]
- Cao Y, Griffith JF, Dorevitch S, Weisberg SB, 2012. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J. Appl. Microbiol. 113 (1),66–75. 10.1111/j.1365-2672.2012.05305.x. [DOI] [PubMed] [Google Scholar]
- Cao Y, Sivaganesan M, Kinzelman J, Blackwood AD, Noble RT, Haugland RA, Griffith JF, Weisberg SB, 2013. Effect of platform, reference material, and quantification model on enumeration of Enterococcus by quantitative PCR methods. Water Res. 47 (1), 233–241. 10.1016/j.watres.2012.09.056.739 [DOI] [PubMed] [Google Scholar]
- Nappier SP et al. Science of the Total Environment 671 (2019) 732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Raith MR, Griffith JF, 2015. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res. 70, 337–349. 10.1016/j.watres.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Cao Y, Griffith JF, Weisberg SB, 2016a. The next-generation PCR-based quantification method for ambient waters: digital PCR. Methods Mol. Biol. 1452, 113–130. 10.1007/978-1-4939-3774-5_7. [DOI] [PubMed] [Google Scholar]
- Cao Y, Raith MR, Griffith JF, 2016b. A duplex digital PCR assay for simultaneous quantification of the Enterococcus spp. and the human fecal-associated HF183 marker in waters. J. Vis. Exp. (109) 10.3791/53611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern EC, Siefring S, Paar J, Doolittle M, Haugland RA, 2011. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol. 52 (3), 298–306. 10.1111/j.1472-765X.2010.03001.x. [DOI] [PubMed] [Google Scholar]
- Cloutier DD, McLellan SL, 2017. Distribution and differential survival of traditional and alternative indicators of fecal pollution at freshwater beaches. Appl. Environ. Microbiol. 83 (4). 10.1128/aem.02881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse RR, Kinzelman JL, Sams EA, Hudgens E, Dufour AP, Ryu H, Santo-Domingo JW, Kelty CA, Shanks OC, Siefring SD, et al. , 2012a. Dramatic improvements in beach water quality following gull removal. Environ. Sci. Technol. 46 (18),10206–10213. 10.1021/es302306b. [DOI] [PubMed] [Google Scholar]
- Converse RR, Griffith JF, Noble RT, Haugland RA, Schiff KC, Weisberg SB, 2012bCorrelation between quantitative PCR and culture-based methods for measuring Enterococcus spp. over various temporal scales at three California marine beaches. Appl.Environ. Microbiol. 78 (4), 1237–1242. 10.1128/AEM.07136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorevitch S, Shrestha A, DeFlorio-Barker S, Breitenbach C, Heimler I, 2017. Monitor-ing urban beaches with qPCR vs. culture measures of fecal indicator bacteria: implications for public notification. Environ. Health 16 (1), 45. 10.1186/s12940-017-0256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer C, Dandle C. 2012. Quantification of microorganisms targeting conserved genes in complex environmental samples using quantitative polymerase chain reaction In: Quantitative Real-time PCR in Applied Microbiology. Norfolk, UK: Caister AcademicPress. [Google Scholar]
- Griffith JF, Weisberg SB, 2011. Challenges in implementing new technology for beach water quality monitoring: lessons from a California demonstration project. Mar. Technol. Soc. J. 45 (2), 65–73. 10.4031/MTSJ.45.2.13. [DOI] [Google Scholar]
- Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH, 2009. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 9 (4), 261–265. 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP, 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39(4), 559–568. 10.1016/j.watres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Siefring S, Lavender J, Varma M, 2012. Influences of sample interference and interference controls on quantification of enterococci fecal indicator bacteria in surface water samples by the qPCR method. Water Res. 46 (18), 5989–6001. 10.1016/j.watres.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Siefring SD, Varma M, Dufour AP, Brenner KP, Wade TJ, Sams E,Cochran S, Braun S, Sivaganensan M, 2014. Standardization of enterococci density estimates by EPA qPCR methods and comparison of beach action value exceedances in river waters with culture methods. J. Microbiol. Methods 105, 59–66. 10.1016/j.mimet.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Siefring S, Varma M, Oshima KH, Sivaganesan M, Cao Y, Raith M,Griffith J, Weisberg SB, Noble RT, et al. , 2016. Multi-laboratory survey of qPCR enterococci analysis method performance in U.S. coastal and inland surface waters.J. Microbiol. Methods 123, 114–125. 10.1016/j.mimet.2016.01.017. [DOI] [PubMed] [Google Scholar]
- Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, et al. , 2013. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 59 (6),892–902. 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- Kinzelman J, Bushon RN, Dorevitch S, Noble R, 2011Comparative Evaluation of Molecular and Culture Methods for Fecal Indicator Bacteria for Use in Inland Recreational Waters. WERF. p. PATH7R09. [Google Scholar]
- Kirs M, 2016Application of Rapid qPCR-based Tests for Enterococci (Method 1611) in Hawaiian Coastal Waters. U.S. EPA Recreational Waters Conference, New Orleans, LA. [Google Scholar]
- Krometis LA, Noble RT, Characklis GW, Denene Blackwood A, Sobsey MD, 2013. Assessment of E. coli partitioning behavior via both culture-based and qPCR methods. Water Sci. Technol. 68 (6), 1359–1369. 10.2166/wst.2013.363. [DOI] [PubMed] [Google Scholar]
- Lavender JS, Kinzelman JL, 2009. A cross comparison of QPCR to agar-based or defined substrate test methods for the determination of Escherichia coli and enterococci inmunicipal water quality monitoring programs. Water Res. 43 (19), 4967–4979. 10.1016/j.watres.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Molina M, Hunter S, Cyterski M, Peed LA, Kelty CA, Sivaganesan M, Mooney T, Prieto L, Shanks OC, 2014. Factors affecting the presence of human-associated and fecal indicator real-time quantitative PCR genetic markers in urban-impacted recreational beaches. Water Res. 64, 196–208. 10.1016/j.watres.2014.06.036. [DOI] [PubMed] [Google Scholar]
- Noble RT, Blackwood AD, Griffith JF, McGee CD, Weisberg SB, 2010. Comparison ofrapid quantitative PCR-based and conventional culture-based methods for enumeration of Enterococcus spp. and Escherichia coli in recreational waters. Appl. Environ. Microbiol. 76 (22), 7437–7443. 10.1128/AEM.00651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opel KL, Chung D, McCord BR, 2009. A study of PCR inhibition mechanisms using real time PCR. J. Forensic Sci. 55 (1), 25–33. 10.1111/j.1556-4029.2009.01245.x. [DOI] [PubMed] [Google Scholar]
- Painter SM, Pfau RS, Brady JA, McFarland AM, 2013. Quantitative assessment of Naegleria fowleri and Escherichia coli concentrations within a Texas reservoir. J. Water Health 11 (2), 346–357. 10.2166/wh.2013.162. [DOI] [PubMed] [Google Scholar]
- Peed LA, Nietch CT, Kelty CA, Meckes M, Mooney T, Sivaganesan M, Shanks OC,2011. Combining land use information and small stream sampling with PCR-based methods for better characterization of diffuse sources of human fecal pollution. Environ. Sci. Technol. 45 (13), 5652–5659. 10.1021/es2003167. [DOI] [PubMed] [Google Scholar]
- Raith MR, Ebentier DL, Cao Y, Griffith JF, Weisberg SB, 2014. Factors affecting the relationship between quantitative polymerase chain reaction (qPCR) and culture-based enumeration of Enterococcus in environmental waters. J. Appl. Microbiol.116 (3), 737–746. 10.1111/jam.12383. [DOI] [PubMed] [Google Scholar]
- Santiago-Rodriguez TM, Tremblay RL, Toledo-Hernandez C, Gonzalez-Nieves JE, Ryu H, Santo Domingo JW, Toranzos GA, 2012. Microbial quality of tropical inland waters and effects of rainfall events. Appl. Environ. Microbiol. 78 (15), 5160–5169. 10.1128/aem.07773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer EP, Vandewalle JL, Bootsma MJ, McLellan SL, 2011. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res. 45 (14),4081–4091. 10.1016/j.watres.2011.04.049. [DOI] [PubMed] [Google Scholar]
- Shanks OC, Atikovic E, Blackwood AD, Lu J, Noble RT, Domingo JS, Seifring S,Sivaganesan M, Haugland RA, 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 74 (3),745–752. 10.1128/AEM.01843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Oshiro R, Haugland RA, Madi T, Brooks L, Field KG,Sivaganesan M, 2016. Data acceptance criteria for standardized human-associated fecal source identification quantitative real-time PCR methods. Appl. Environ. Microbiol. 82 (9), 2773–2782. 10.1128/AEM.03661-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaganesan M, Siefring S, Varma M, Haugland RA, 2014. Comparison of Enterococcusquantitative polymerase chain reaction analysis results from Midwest U.S. river samples using EPA Method 1611 and Method 1609 PCR reagents. J. Microbiol. Methods 101, 9–17. 10.1016/j.mimet.2014.03.004. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (US-EPA), 2010aMethod A: Enterococci in Waterby TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay. [Google Scholar]
- U.S. EPA, Officeof Water. EPA/821/R-10/004. U.S. Environmental Protection Agency (US-EPA) 2010b. Report on 2009 National Epidemiologic and Environmental Assessment of Recreational Water Epidemiology Studies (NEEAR 2010 - Surfside & Boquerón). EPA/600/R-10/168. https://archive.epa.gov/nheerl/neear/web/pdf/report2009v5_508comp.pdf [Accessed: January 10, 2018]. [Google Scholar]
- U.S. Environmental Protection Agency (US-EPA). 2012a. Recreational Water Quality Criteria. U.S. EPA, Office of Water; EPA/820/F-12/058.https://www.epa.gov/wqc/2012-recreational-water-quality-criteria [Accessed: January 10, 2018]. [Google Scholar]
- U.S. Environmental Protection Agency (US-EPA). 2012b. Method 1611: Enterococci in Water by TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay. [Google Scholar]
- U.S.EPA, Office of Water. EPA/821/R-12/008.https://www.epa.gov/sites/production/files/2015-08/documents/method_1611_2012.pdf [Accessed: January 10, 2018].
- U.S. Environmental Protection Agency (US-EPA). 2013a. Acceptability of the EPA qPCRTest at Your Beach. U.S. EPA, Office of Water; EPA/820/R-13/012. https://www.epa.gov/sites/production/files/2015-09/documents/acceptability-of-epa-qpcr-test-at-your-beach_dec-2013.pdf [Accessed: January 10, 2018]. [Google Scholar]
- U.S. Environmental Protection Agency (US-EPA). 2013b. Method 1609: Enterococci in Water by TaqMan® Quantitative Polymerase Chain Reaction (qPCR) With Internal Amplification Control (IAC) Assay. EPA/820/R-13/005. https://nepis.epa.gov/Exe/ZyPDF.cgi/P100MYZZ.PDF?Dockey=P100MYZZ.PDF [Accessed: January 10, 2018].
- U.S. Environmental Protection Agency (US-EPA). 2016. National Rivers and Streams Assessment 2008–2009: A collaborative survey. U.S. EPA, Office of Water and Office ofResearch and Development; EPA/841/R-16/007. http://www.epa.gov/national-aquatic-resource-surveys/nrsa [Accessed: March 30, 2018]. [Google Scholar]
- U.S. Environmental Protection Agency (US-EPA). 2018. 2017 Five-year Review of the2012 Recreational Water Quality Criteria. EPA/823/R-18/001. https://www.epa.gov/wqc/five-year-review-2012-recreational-water-quality-criteria [Accessed: July 5,2018].
- Wade TJ, Calderon RL, Sams E, Beach M, Brenner KP, Williams AH, Dufour AP,2006. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Perspect. 114 (1),24–28. 10.1289/ehp.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade TJ, Calderon RL, Brenner KP, Sams E, Beach M, Haugland R, Wymer L,Dufour AP, 2008. High sensitivity of children to swimming-associated gastrointestinal illness: results using a rapid assay of recreational water quality. Epidemiology. 19(3), 375–383. 10.1097/EDE.0b013e318169cc87. [DOI] [PubMed] [Google Scholar]
- Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, Wymer L, Rankin CC, Love D, Li Q, et al. , 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health 9, 66 10.1186/1476-069X-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TJ, Bachoon DS, Otero E, Ramsubhag A, 2013. Detection of verotoxin producing Escherichia coli in marine environments of the Caribbean. Mar. Pollut. Bull. 76(1–2), 406–410. 10.1016/j.marpolbul.2013.08.019. [DOI] [PubMed] [Google Scholar]
- Wang D, Yamahara KM, Cao Y, Boehm AB, 2016. Absolute quantification of Enterococcal 23S rRNA gene using digital PCR. Environ. Sci. Technol. 50 (7), 3399–3408. 10.1021/acs.est.5b05747. [DOI] [PubMed] [Google Scholar]
- Zhang YY, Riley LK, Lin MS, Hu ZQ, 2012. Determination of low-density Escherichiacoli and Helicobacter pylori suspensions in water. Water Res. 46 (7), 2140–2148. 10.1016/j.watres.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Zimmer-Faust AG, Thulsiraj V, Ferguson D, Jay JA, 2014. Performance and specificity of the covalently linked immunomagnetic separation-ATP method for rapid detection and enumeration of enterococci in coastal environments. Appl. Environ. Microbiol. 80(9), 2705–2714. 10.1128/aem.04096-13.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


