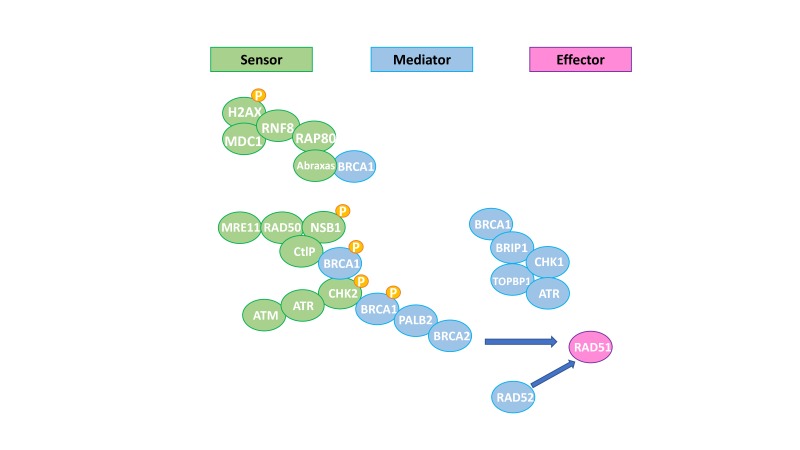
Mechanisms that protect DNA are crucial to preserve the genome integrity from injuries produced by environmental agents or those spontaneously generated during DNA metabolism.1 DNA single-strand breaks (SSBs) are repaired by mismatch repair (MMR), base excision repair (BER) and nucleotide excision repair (NER).1 Non-homologous end joining (NHEJ) or homologous recombination repair (HRR) processes double-strand breaks (DSBs).1 While NHEJ solves DSBs in a potentially inaccurate way, HRR is an error-free pathway that restores the genomic sequence of the broken DNA ends by using the sister chromatid as template for repair.1 Several proteins are involved in the HRR pathway: some of them act as sensors of DSBs such as γH2AX, ATM and ATR, leading to the activation of signal mediator proteins (ie, BRCA1, BRCA2 and PALB2). The final event of the HRR pathway is the loading of a small nuclear protein called RAD51 onto single-stranded DNA, where it promotes strand invasion and replication fork stabilisation (figure 1).2 Tumours with HRR deficiency (HRD) were described for the first time in cancers that harbour germline mutations of the tumour suppressors BRCA1 and BRCA2 (BRCA1/2).3 Nonetheless, genetic and epigenetic events can also result in inactivation of other HRR components, leading to HRD in sporadic cancers.3
Figure 1.
Homologous recombination pathway. Adapted from De Picciotto et al.50
HRD is harboured by approximately 13% and 15% of ovarian and triple negative breast cancers (TNBC), respectively, and it is attributable to germline BRCA1/2 (gBRCA1/2) mutations.4 5 Furthermore, 50% and 40% of ovarian and TNBC, respectively, are characterised by harbouring HRD in the absence of gBRCA1/2 mutations.4 6 Also, 10%–12% of advanced prostate cancer harbour germline or somatic BRCA2 inactivation and up to 25% contain a DNA repair defect.7 As HRR is required for the repair of DSBs generated during DNA interstrand cross-link (ICL) resolution, HRR-deficient tumours are sensitive to ICL-generating platinum chemotherapy.3 8 Moreover, BRCA1/2-mutant cells are sensitive to PARP inhibitors (PARPi), a new class of drugs that block SSB repair, favouring accumulation of DSB that HRR-deficient cells cannot repair.9 PARPi also trap PARP onto DNA causing replication stress that is toxic in these cells.9 Several PARPi have been approved for the treatment of ovarian and breast cancers.10–14 EMA approved olaparib and rucaparib as maintenance treatments for platinum-sensitive ovarian cancer with germline/somatic BRCA1/2-mutation; and niraparib was labelled as maintenance treatment for patients who are in response to platinum-based chemotherapy.10 12 15 Olaparib and talazoparib have been approved for patients with advanced breast cancer and a gBRCA1/2 mutations who have previously been treated with chemotherapy.13–15 The current clinical challenge is the identification of BRCA1/2 wild type (WT) patients who harbour alterations in the HRR pathway and share molecular features of BRCA1/2-mutated tumours (the so-called ‘BRCAness’ phenotype) and who may also benefit from similar therapeutic approaches.16
Different approaches are currently being investigated to identify BRCA1/2 WT tumours that can benefit from DNA-damaging agents and PARPi based on the presence of HRD, that is, (1) scores capturing large genomic aberrations, so-called ‘genomic scars’, (2) analysis of mutational signatures or (3) point mutations identified in HRR genes using DNA sequencing panels.9 17–19 In BRCA1/2-mutant cells, chromosomal spreads reveal increased gross chromosomal rearrangements. This led to the development of assays to evaluate the ‘genomic scars’ caused by the loss of HRR function using SNP array data.3 Two commercial genomic scar assays have been tested to identify tumours with HRD in clinical trials. The ‘myChoice HRD’ assay by Myriad tests for the presence of loss of heterozygosity (LOH), telomeric allelic imbalance (TAI), and large-scale state transitions (LST) across the genome.17 The readout of this assay is presented as an ‘HRD score’: a tumour with an HRD score ≥42 is labelled as HRD-positive. The ‘FoundationFocus CDx BRCALOH’ is designed to detect the presence of mutations in the BRCA1/2 genes and the percentage of the genome affected by LOH in DNA from tumour tissue samples of patients with ovarian cancer.20 According to the FoundationFocus test, tumours are categorised as LOH-high if score is ≥16. On the other hand, mutational signatures are characteristic patterns left on the cancer genome by each mutational process: for example, HRD has been associated with the ‘signature 3’ described by Alexandrov et al.21–23 ‘Signature 3’ is also able to accurately classify missense BRCA1/2 mutations with known functional implications and it is associated with silencing of RAD51C and BRCA1 by promoter methylation.24
So, what is the current clinical evidence around the use of genomics scars to quantify HRD and its impact on treatment decision-making? The main open question is whether genomic scars are predictive biomarkers of response to platinum salts or PARPi, beyond BRCA1/2 mutation.
In advanced ovarian cancer, the ARIEL2 study demonstrated the efficacy of the PARPi rucaparib as monotherapy in gBRCA1/2 mutated and/or LOH-high relapsed, platinum-sensitive ovarian cancer, and the ARIEL3 trial demonstrated the benefit of rucaparib as maintenance therapy in platinum-sensitive recurrent patients who responded to platinum, regardless of the LOH status (table 1).12 20 The NOVA trial investigated the role of the PARPi niraparib as maintenance therapy in platinum-sensitive ovarian cancer and showed that patients with BRCA1/2 mutations or HRD-positive according to myChoice assay benefited from PARPi.11 Nevertheless, niraparib also improved PFS in BRCA1/2 WT patients with an HRD-negative test, although the magnitude of the benefit was smaller compared to BRCA1/2-mutated or HRD-positive patients (table 1).11 From these trials, one may conclude that in the platinum-sensitive population the HRD-genomic scars provide information regarding the magnitude of the clinical benefit, given the high probability of response after platinum sensitivity. Indeed, the magnitude of the benefit is higher among those who are HRD-positive or BRCA1/2-mutated. Most importantly, in the ovarian population, further investigations are needed to verify if an HRD test may be useful to select platinum-resistant tumours that may benefit from PARPi or to identify long responders to PARPi and/or platinum salts.
Table 1.
Efficacy of PARPi according to HRD status in ovarian cancer
| Clinical trial | Drug | Study population | HRD role |
| ARIEL-212 | Rucaparib, monotherapy | Relapsed, platinum-sensitive ovarian cancer | Higher efficacy in gBRCA1/2-mutated and/or LOH-high compared with LOH-low tumours. Not powered to show a difference between LOH-high and LOH-low tumours |
| ARIEL-320 | Rucaparib, maintenance therapy | Relapsed, platinum-sensitive ovarian cancer | Efficacy regardless of LOH-status. Magnitude of the benefit dependent on LOH |
| NOVA-trial11 | Niraparib, maintenance therapy | Relapsed, platinum-sensitive ovarian cancer | Efficacy regardless of HRD-status. Magnitude of the benefit dependent on HRD |
HRD, homologous recombination repair deficiency; LOH, loss of heterozygosity; PARPi, PARP inhibitors.
In TNBC, several trials have investigated if HRD-genomic scars predict response to DNA-damaging agents or to the addition of carboplatin to standard chemotherapy beyond BRCA1/2 mutation (table 2). In the neoadjuvant setting, Telli et al retrospectively assessed the predictive value of the ‘myChoice HRD’ assay in three single-arm trials testing platinum-based therapy.25 Patients who were HRD-positive had a higher probability to achieve a complete pathological response or minimal residual disease (RCB 0-I) after platinum chemotherapy, even among BRCA1/2 WT tumours.25 The GeparSixto trial evaluated the benefit of the addition of carboplatin to anthracycline/taxane-based neoadjuvant chemotherapy in TNBC and analysed the predictive and prognostic value of testing for HRD by the composite biomarker including germline/somatic BRCA1/2 mutations and the ‘myChoice’ assay.6 Among all patients with TNBC, addition of carboplatin resulted in a marked increment in pCR rates in HRD-positive tumours (from 33.9 to 63.5%, P=0.001), and in HRD-negative tumours (from 20 to 29.6%, P=0.399). However, according to the test of interaction, HRD did not predict for carboplatin benefit in this study. Of note, the control arm in this trial lacked cyclophosphamide, which might have overestimated the carboplatin benefit. Other observations regarding the prognostic value of HRD genomic scars will require a powered study to demonstrate the improvement in disease-free survival (DFS) or overall survival (OS). Litton et al have recently showed the efficacy of PARPi talazoparib in the neoadjuvant setting in patients with BRCA1/2 mutations. In this setting, an HRD test could be useful to identify patients with BRCA1/2 WT who can also benefit from PARPi.26 Finally, in the adjuvant setting, Sharma et al evaluated the predictive role of the ‘myChoice HRD’ in TNBC to predict outcome of adjuvant anthracycline and cyclophosphamide regimen.27 The study showed a better DFS in patients with high HRD, even beyond gBRCA1/2 status.
Table 2.
Efficacy of platinums or DNA-damaging chemotherapy according to HRD status in breast cancer
| Clinical trial | Drug | Study population | HRD role |
| PrECOG 0105 Cisplatin-1 trial Cisplatin-2 trial51 |
Platinum salts | Neoadjuvant TNBC | Patients who were HRD-positive had higher complete pathological response |
| Gepar-Sixto trial6 | Carboplatin | Neoadjuvant TNBC | Patients who were HRD-positive had a better prognosis compared with HRD-negative. No robust conclusions regarding the predictive role of HRD for addition of carboplatin |
| SWOG S9313 trial27 | Doxorubicin and cyclophosphamide | Adjuvant TNBC | Patients who were HRD-positive had a better DFS, even beyond gBRCA1/2 status |
| TBCRC009 trial28 | Platinum salts | Advanced, first or second line TNBC | Higher HRD scores were reported in responding patients, independent of BRCA1/2 mutational status. |
| TNT trial29 | Carboplatin | Advanced, first line TNBC | ORR did not correlate with HRD-score of the primary tumours. |
HRD, homologous recombination repair deficiency; ORR, overall response rate; TNBC, triple negative breast cancers.
In metastatic TNBC, Isakoff et al conducted a phase II trial aimed to investigate the predictive role of genomic scars to platinum salts. Higher HRD scores were reported in responding patients, independent of BRCA1/2 mutational status.28 However, the predictive role of this HRD test was not confirmed in the TNT trial, a randomised phase III trial comparing the efficacy of first-line carboplatin versus docetaxel in patients with advanced TNBC.29 According to the preplanned biomarker analysis, carboplatin resulted in higher overall response rates (ORR) among patients harbouring a gBRCA1/2 mutation, but not in subjects with other profiles associated with HRR dysfunction such as high HRD-score, BRCA1 methylation, or BRCA1 mRNA-low, mostly evaluated in the primary tumours.29 These results could be partially explained by the fact that genomic scars tested in the primary tumour may have lower prediction power for response in the advanced setting because metastatic tumours may have restored the HRR function and become resistant to platinum. As in the GeparSixto trial, HRD-positive tumours were more likely to respond to any chemotherapy regimens compared with the HRD-negative ones. Several open questions may raise from the previous statements: first, that no data are available comparing the HRD status in early and advanced breast cancer, and second, that further studies are required to dissect the role of recovering the HRR function in predicting resistance to PARPi and platinum salts.30 Furthermore, despite the OlympiAD and EMBRACA trials demonstrated the efficacy of PARPi in BRCA1/2-mutated metastatic breast cancer,13 14 still a relevant proportion of patients did not respond. Further research is needed to investigate if an HRD test would help to refine the subgroup more likely to benefit.
In prostate cancer, the interest in developing DNA damaging and PARPi-based therapeutic strategies arises from the enrichment for DNA damage response gene mutations among cases with advanced disease. The TOPARP-A trial evaluated the antitumor activity of olaparib in advanced prostate cancer, identifying a strong association between the presence of certain DNA repair defects and response to olaparib.31 Preliminary data from the TRITON2 study, evaluating rucaparib, confirmed the high response rate to PARPi of BRCA1/2-deficient prostate cancers.32 Nevertheless, the predictive value of other defects in DNA repair genes such as ATM, FANCA or CHEK2 remains yet to be validated in randomised studies. None of the HRD score tests has been validated yet in prostate cancer clinical trials. Of note, in two recent studies, the prevalence of LOH-high signatures, based on the FoundationOne assay, among BRCA2-mutated prostate cancer was lower than for the BRCA1/2-mutated ovarian cancer setting.33 34
A current limitation of the genomic scar assays is the impossibility to capture tumour evolution processes, such as a restoration of the HRR function in response to therapy-selective pressure. As an alternative, it could be useful to incorporate functional biomarkers based on dynamic assays that assess the activity of a repair pathway. A crucial step of HRR is mediated by the RAD51 protein.1 In vivo and in vitro studies supported the highly sensitive and specific predictive power of lack of nuclear RAD51 foci to PARPi response.30 35–37 One limitation is that the RAD51 assay may fail to identify ATM-mutated tumours that can benefit from PARPi.31 38–40 There are several other tests currently used in research to identify tumours with a similar biological behaviour as the BRCA1/2-mutated ones. Those tests take into account not only the copy number changes but also methylation or gene expression profiles.41–43 Even if preclinical and retrospective data suggest that these tests are predictive of high dose alkylating chemotherapy and PARPi response, the lack of prospective validation and concerns regarding their large-scale feasibility seem to be major issues for their clinical application.43–49
We can summarise that genomic scars associated with HRR defects in ovarian cancer identify patients who obtain maximum benefit from PARPi maintenance after platinum response. In early breast cancer, HRD-genomic scars have shown a high correlation to DNA-damaging chemotherapy response, and a definitive word to predict the benefit of adding carboplatin is warranted. In prostate cancer, recent data suggest that HRR gene-mutated tumours, including gBRCA2, are sensitive to PARPi. Nonetheless, it is of concern that HRD is observed less-frequently than in ovarian cancers.
As future perspectives, research is needed to confirm if clinical implementation of HRD tests might be useful to identify patients with platinum-resistant ovarian cancer who may benefit from PARPi. Likewise, some BRCA1/2 WT, HRD-positive breast cancers could respond to PARPi. Also, the prognostic role of HRD should be further investigated with ad hoc trials in order to recognise patients with early breast cancer candidates for a targeted strategy. Prospective comparison between HRD-genomic scars and functional dynamic tests such as the RAD51 assay is encouraged.
Footnotes
Contributors: All the authors substantially contributed to the conception of the work. BP drafted the work and all the authors revised it critically for important intellectual content. All the authors finally approved the version published.
Funding: BP was supported by ESMO with the aid of a grant from Roche. JM is supported by a Prostate Cancer Foundation Young Investigator Award.
Disclaimer: Any views, opinions, findings, conclusions or recommendations expressed in this material are those solely of the authors and do not necessarily reflect those of ESMO or Roche.
Competing interests: VS declares a non‐commercial research agreement with AstraZeneca and Tesaro. JB has participated in Advisory Board for Clovis, Tesaro, Medivation and AstraZeneca. JM has participated in Advisory Board for AstraZeneca and Janssen Speakers bureau for Sanofi, Astellas.
Patient consent for publication: Not required.
Provenance and peer review: Commissioned; externally peer reviewed.
References
- 1.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: important differences with common interests. Nat Rev Cancer 2012;12:372 10.1038/nrc3181-c3 [DOI] [Google Scholar]
- 2.Murai J, Huang S-yinN, Das BB, et al. . Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 2012;72:5588–99. 10.1158/0008-5472.CAN-12-2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoppe MM, Sundar R, Tan DSP, et al. . Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst 2018;110:704–13. 10.1093/jnci/djy085 [DOI] [PubMed] [Google Scholar]
- 4.Bell D, Berchuck A, Birrer M, et al. . Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akashi-Tanaka S, Watanabe C, Takamaru T, et al. . BRCAness predicts resistance to taxane-containing regimens in triple negative breast cancer during neoadjuvant chemotherapy. Clin Breast Cancer 2015;15:80–5. 10.1016/j.clbc.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 6.Loibl S, Weber KE, Timms KM, et al. . Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response-final results from GeparSixto. Ann Oncol 2018;29:2341–7. 10.1093/annonc/mdy460 [DOI] [PubMed] [Google Scholar]
- 7.Robinson D, Van Allen EM, Wu Y-M, et al. . Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. 10.1016/j.cell.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heeke AL, Baker T, Lynce F, et al. . Prevalence of homologous recombination deficiency among all tumor types. JCO 2017;35(15_suppl):1502 10.1200/JCO.2017.35.15_suppl.1502 [DOI] [Google Scholar]
- 9.O'Connor MJ. Targeting the DNA damage response in cancer. Mol Cell 2015;60:547–60. 10.1016/j.molcel.2015.10.040 [DOI] [PubMed] [Google Scholar]
- 10.Ledermann JA, Harter P, Gourley C, et al. . Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol 2016;17:1579–89. 10.1016/S1470-2045(16)30376-X [DOI] [PubMed] [Google Scholar]
- 11.Mirza MR, Monk BJ, Herrstedt J, et al. . Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med 2016;375:2154–64. 10.1056/NEJMoa1611310 [DOI] [PubMed] [Google Scholar]
- 12.Swisher EM, Lin KK, Oza AM, et al. . Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol 2017;18:75–87. 10.1016/S1470-2045(16)30559-9 [DOI] [PubMed] [Google Scholar]
- 13.Robson M, Im S-A, Senkus E, et al. . Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377:523–33. 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 14.Litton JK, Rugo HS, Ettl J, et al. . Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med 2018;379:753–63. 10.1056/NEJMoa1802905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Medicines Agency Available: https://www.ema.europa.eu/medicines/human/
- 16.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110–20. 10.1038/nrc.2015.21 [DOI] [PubMed] [Google Scholar]
- 17.Watkins JA, Irshad S, Grigoriadis A, et al. . Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res 2014;16 10.1186/bcr3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgson DR, Dougherty B, Lai Z, et al. . 435 candidate biomarkers of PARP inhibitor sensitivity in ovarian cancer beyond the BRCA genes. European Journal of Cancer 2015;51 10.1016/S0959-8049(16)30269-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abkevich V, Timms KM, Hennessy BT, et al. . Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012;107:1776–82. 10.1038/bjc.2012.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman RL, Oza AM, Lorusso D, et al. . Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet 2017;390:1949–61. 10.1016/S0140-6736(17)32440-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 2014;15:585–98. 10.1038/nrg3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. . Signatures of mutational processes in human cancer. Nature 2013;500:415–21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng G, Chun-Jen Lin C, Mo W, et al. . Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun 2014;5 10.1038/ncomms4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polak P, Kim J, Braunstein LZ, et al. . A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet 2017;49:1476–86. 10.1038/ng.3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telli ML, Timms KM, Reid J, et al. . Homologous recombination deficiency (HRD) score predicts response to Platinum-Containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res 2016;22:3764–73. 10.1158/1078-0432.CCR-15-2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litton JK, Scoggins M, Hess KR, et al. . Neoadjuvant talazoparib (TALA) for operable breast cancer patients with a BRCA mutation (BRCA+). JCO 2018;36(15_suppl):508 10.1200/JCO.2018.36.15_suppl.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma P, Barlow WE, Godwin AK, et al. . Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313). Ann Oncol 2018;29:654–60. 10.1093/annonc/mdx821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isakoff SJ, Mayer EL, He L, et al. . TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. JCO 2015;33:1902–9. 10.1200/JCO.2014.57.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tutt A, Tovey H, Cheang MCU, et al. . Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TnT trial. Nat Med 2018;24:628–37. 10.1038/s41591-018-0009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz C, Castroviejo-Bermejo M, Gutiérrez-Enríquez S, et al. . RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol 2018;29:1203–10. 10.1093/annonc/mdy099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mateo J, Carreira S, Sandhu S, et al. . DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med Overseas Ed 2015;373:1697–708. 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abida W, Bryce AH, Vogelzang NJ, et al. . 793PDPreliminary results from TRITON2: a phase II study of rucaparib in patients (PTS) with metastatic castration-resistant prostate cancer (mCRPC) associated with homologous recombination repair (HRR) gene alterations. Ann Oncol 2018;29(suppl_8). 10.1093/annonc/mdy284.002 [DOI] [Google Scholar]
- 33.Chowdhury S, McDermott R, Piulats JM, et al. . 795PDGenomic profiling of circulating tumour DNA (ctDNA) and tumour tissue for the evaluation of rucaparib in metastatic castration-resistant prostate cancer (mCRPC). Ann Oncol 2018;29(suppl_8). 10.1093/annonc/mdy284.004 [DOI] [Google Scholar]
- 34.Agarwal N, Sokol ES, Lara P, et al. . 51OPan-cancer assessment of BRCA1/2 genomic alterations (GAS) by comprehensive genomic profiling (CGP) of tissue and circulating tumor DNA (ctDNA). Ann Oncol 2018;29(suppl_8). 10.1093/annonc/mdy269.049 [DOI] [Google Scholar]
- 35.Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, et al. . A Rad51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med 2018;10:e9172 10.15252/emmm.201809172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naipal KAT, Verkaik NS, Ameziane N. Functional ex vivo assay to select homologous recombination − deficient breast tumors for PARP inhibitor treatment functional ex vivo assay to select homologous recombination – de FI cient breast tumors for PARP, 2014: 4816–26. [DOI] [PubMed] [Google Scholar]
- 37.Graeser M, Mccarthy A, Lord CJ. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodall J, Mateo J, Yuan W, et al. . Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 2017;7:1006–17. 10.1158/2159-8290.CD-17-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakr A, Oing C, Köcher S, et al. . Involvement of ATM in homologous recombination after end resection and Rad51 nucleofilament formation. Nucleic Acids Res 2015;43:3154–66. 10.1093/nar/gkv160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe N, Turner NC, Lord CJ, et al. . Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res 2006;66:8109–15. 10.1158/0008-5472.CAN-06-0140 [DOI] [PubMed] [Google Scholar]
- 41.Lips EH, Laddach N, Savola SP, et al. . Quantitative copy number analysis by multiplex ligation-dependent probe amplification (MLPA) of BRCA1-associated breast cancer regions identifies BRCAness. Breast Cancer Res 2011;13 10.1186/bcr3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schouten PC, Grigoriadis A, Kuilman T, et al. . Robust BRCA1-like classification of copy number profiles of samples repeated across different datasets and platforms. Mol Oncol 2015;9:1274–86. 10.1016/j.molonc.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konstantinopoulos PA, Spentzos D, Karlan BY, et al. . Gene Expression Profile of BRCA ness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. JCO 2010;28:3555–61. 10.1200/JCO.2009.27.5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schouten PC, Marmé F, Aulmann S, et al. . Breast cancers with a BRCA1-like DNA copy number profile recur less often than expected after high-dose alkylating chemotherapy. Clin Cancer Res 2015;21:763–70. 10.1158/1078-0432.CCR-14-1894 [DOI] [PubMed] [Google Scholar]
- 45.Vollebergh MA, Lips EH, Nederlof PM, et al. . An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann Oncol 2011;22:1561–70. 10.1093/annonc/mdq624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehmann BD, Bauer JA, Chen X, et al. . Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67. 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuda H, Baggerly KA, Wang Y, et al. . Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res 2013;19:5533–40. 10.1158/1078-0432.CCR-13-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Severson TM, Peeters J, Majewski I, et al. . BRCA1-like signature in triple negative breast cancer: molecular and clinical characterization reveals subgroups with therapeutic potential. Mol Oncol 2015;9:1528–38. 10.1016/j.molonc.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Severson TM, Wolf DM, Yau C, et al. . The BRCA1ness signature is associated significantly with response to PARP inhibitor treatment versus control in the I-SPY 2 randomized neoadjuvant setting. Breast Cancer Res 2017;19 10.1186/s13058-017-0861-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.De Picciotto N, Cacheux W, Roth A, et al. . Ovarian cancer: Status of homologous recombination pathway as a predictor of drug response. Crit Rev Oncol Hematol 2016;101:50–9. 10.1016/j.critrevonc.2016.02.014 [DOI] [PubMed] [Google Scholar]
- 51.Telli ML, Jensen KC, Vinayak S, et al. . Phase II Study of Gemcitabine, Carboplatin, and Iniparib As Neoadjuvant Therapy for Triple-Negative and BRCA1 / 2 Mutation–Associated Breast Cancer With Assessment of a Tumor-Based Measure of Genomic Instability: PrECOG 0105. JCO 2015;33:1895–901. 10.1200/JCO.2014.57.0085 [DOI] [PMC free article] [PubMed] [Google Scholar]