Abstract
Engineering the gut microbiota to produce specific beneficial metabolites represents an important new potential strategy for treating chronic diseases. Our previous studies with bacteria engineered to produce N-acyl-phosphatidylethanolamines (NAPEs), the immediate precursors of the lipid satiety factors N-acyl-ethanolamides (NAEs), found that colonization of these bacteria inhibited development of obesity in C57BL/6J mice fed a high fat diet. Individual NAE species differ in their bioactivities. Intriguingly, colonization by our engineered bacteria resulted in increased hepatic N-stearoyl-ethanolamide (C18:0NAE) levels despite the apparent inability of these bacteria to biosynthesize its precursor N-stearoyl-phosphatidylethanolamine (C18:0NAPE) in vitro. We therefore sought to identify the factors that allowed C18:0NAPE biosynthesis by the engineered bacteria after colonization of the intestinal tract. We found that the species of NAPE biosynthesized by engineered bacteria depends on the species of dietary fatty acids available in the intestine, suggesting a simple method to fine tune the therapeutic effects of modified microbiota.
Keywords: N-acyl-phosphatidylethanolamines, N-acyl-ethanolamides, N-acyltransferases, fatty acids, gut microbiota, engineered bacteria
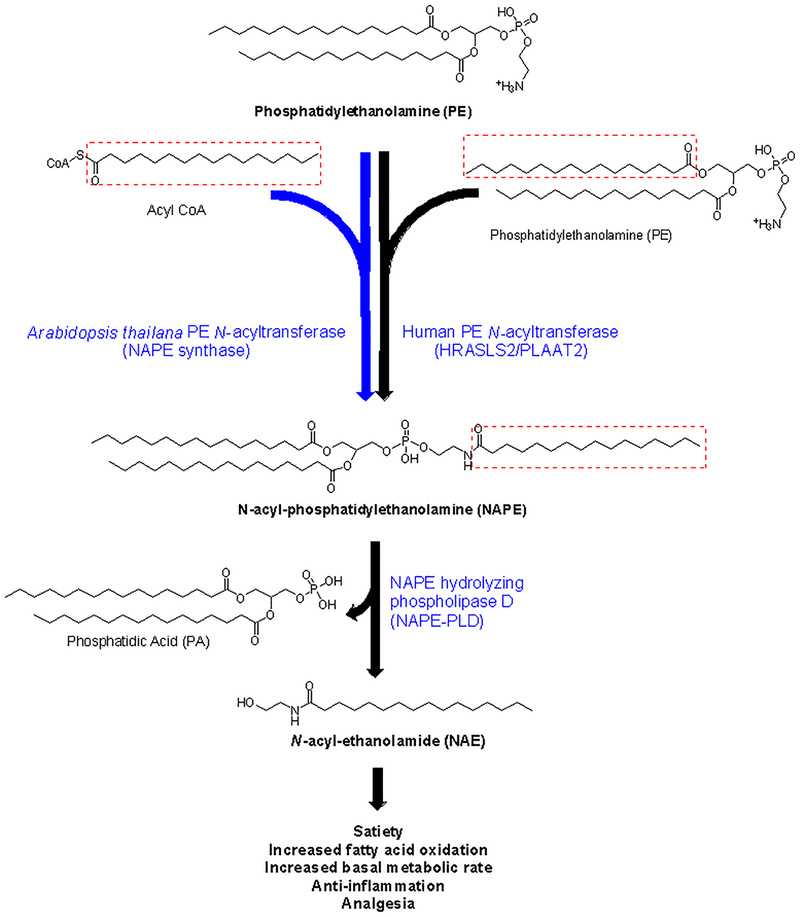
In recent years, it has become apparent that the gut microbiota may exert profound effects on host physiology including energy balance and fat accumulation. This paradigm suggests that incorporating microbes engineered to generate specific beneficial metabolites into an at-risk individual’s microbiota may be useful in treating obesity associated diseases. For example, the enterocytes of obese individuals fail to biosynthesize sufficient amounts of N-acyl-ethanolamides (NAEs) (Figure 1), lipids that act as satiety factors, in response to food intake1–3. We have shown that incorporating bacteria genetically engineered to biosynthesize N-acyl-phosphatidylethanolamines (NAPEs), the immediate precursor for NAE biosynthesis, into the intestinal microbiota has the potential to protect against development of obesity so long as there is sufficient active NAPE-hydrolyzing phospholipase D (NAPE-PLD) present in either the host or engineered bacteria to convert the NAPE to NAE4, 5.
Figure 1.
Biosynthesis and bioactivities of N-acyl-phosphatidylethanolamines (NAPEs) and N-acyl-ethanolamides (NAEs). N-palmitoyl-phosphatidylethanolamine (C16:0NAPE) is shown for illustration, but NAPE with a variety of N-acyl chain species can be produced by the two NAPE acyltransferases used for heterologous expression in E. coli in these studies. The human NAPE acyltransferase transfers an acyl group from the sn-1 group of phosphatidylethanolamine (PE) or phosphatidylcholine to a second PE molecule whereas the A. thaliana NAPE acyltransferase uses acyl CoA as the acyl donor to generate NAPEs. NAPE-PLD converts NAPEs to NAEs. Individual NAEs exert differing bioactivities depending on their N-acyl chain species.
The effectiveness of such incorporated bacteria depends on their ability to sustainably produce adequate amounts of the desired metabolite (e.g. NAPE / NAEs) in the complex environment present in the intestinal tract. Unfortunately, little is known about the factors that control metabolite production by engineered bacteria once incorporated into the gut microbiota. Factors that differ in the intestinal tract versus conditions typically used for in vitro expression by bacteria include temperature and the availability of amino acids, fatty acids, and other components needed for biosynthesis. Biosynthesis of desired products may also be reduced if the expressed enzyme catalyzes undesired alternative products in the presence of competing substrates arising from the diet or other intestinal bacteria. Such competing substrates become particularly important if the bioactivity of their products oppose those of the desired product. In the case of bacteria engineered to produce NAPEs, the specific N-acyl chain incorporated into the NAPE matters because individual NAE species have differing bioactivities. For instance, N-arachidonyl-ethanolamide (C20:4NAE, also known as anandamide) markedly increases appetite while N-palmitoyl-ethanolamide (C16:0NAE) or N-oleoyl-ethanolamide (C18:1NAE) induce satiation and suppress appetite. So the therapeutic value of incorporating bacteria designed to increase NAPE / NAE levels depends on which species of NAE these bacteria generate in vivo.
We had previously found that transformation of a commensal bacteria, E. coli Nissle 1917 (EcN), with a plasmid for heterologous expression of NAPE synthase, the PE N-acyltransferase of A. thaliana, resulted in production of significant amounts of N-hexadecaenoyl-PE (C16:1NAPE), along with lesser amounts of N-palmitoyl-PE (C16:0NAPE), N-methylenehexadecanoyl-PE (C17cycNAPE) and N-octadecaenoyl-PE (C18:1NAPE) when these bacteria (pNAPE-EcN) were cultured in conditions typically used for bacterial expression of recombinant protein (Luria-Bertani media at 28°C)5. These bacteria appeared to lack the ability to produce other NAPE species such as N- stearoyl-PE (C18:0NAPE) or N-arachidonyl-PE (C20:4NAPE), even though the purified enzyme can produce these species. Administration of pNAPE-EcN in the drinking water of wild-type C57BL/6J mice significantly increased liver NAE levels and protected the mice against diet induced obesity5. Surprisingly, the major species of NAEs that were increased in the liver by this treatment were C16:0NAE and C18:0NAE, suggesting that once incorporated into the intestinal microbiota, pNAPE-EcN gained the ability to produce C18:0NAPE and lost the ability to produce C16:1NAPE and C17cycNAPE5. Because individual NAPE/NAE species have differing bioactivities, it is critical to identify and appropriately control the factors that determine the species and extent of NAPE produced in the intestinal tract before attempting to utilize these engineered bacteria in human trials.
We therefore examined the effect of culture conditions and media components on the amount and species of NAPEs produced. We also examined whether different NAPE species were produced by heterologous expression of a mammalian PE N-acyltransferase (which uses the sn-1 O-acyl chains of phospholipids as the acyl donor6, 7) in E. coli compared to heterologous expression of NAPE synthase, the A. thaliana PE N-acyltransferase (which uses Acyl CoA as the acyl donor8), that we had used previously. We found that there was a significant effect of amino acid supplementation and temperature on the species of NAPE generated; however, the most important determinant appeared to be the presence of exogenous fatty acids. Saturated and monounsaturated fatty acids significantly increased formation of their respective NAPE species, but exogenous arachidonic acid or docosahexaenoic acid had no effect. Surprisingly, the type of PE N-acyltransferase used for heterologous expression had relatively little effect on extent of NAPE production or the NAPE species generated. Using fecal NAPEs as a surrogate measure for NAPE biosynthesis by engineered bacteria in the intestinal lumen, we found that administration engineered EcN initially produced C16:1NAPE and C7cycNAPE after administration, but after colonization of mice fed high fat diet based on lard, they primarily produced C16:0NAPE, C18:0NAPE, and C18:1NAPE.
Results and Discussion
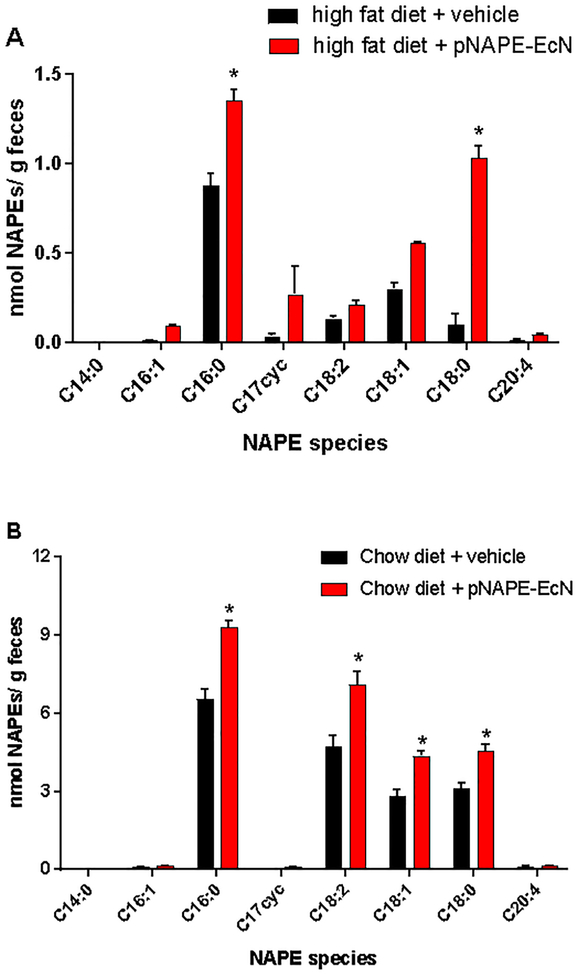
We previously found that C18:0NAE increased in the liver of mice administered E. coli Nissle 1917 (EcN) heterologously expressing A. thaliana NAPE synthase (pNAPE-EcN) in their drinking water for 8 weeks, despite the lack of C18:0NAPE synthesis when pNAPE-EcN were grown in Luria-Bertani (LB) media. This result suggested that when pNAPE-EcN colonized the intestinal tract, it significantly altered the species of NAPE it produced. To determine if pNAPE-EcN did indeed change their NAPE profile after colonization, we administered pNAPE-EcN for two weeks in the drinking water of C57BL/6J mice fed a high-fat diet (60%kcal fat), and then measured fecal NAPE species one week after ending active administration of the engineered bacteria. We found that fecal NAPE levels were markedly higher in the mice that had received pNAPE-EcN compared to vehicle and that C16:0NAPE and C18:0NAPE were the predominant species (Figure 2A). No C16:1NAPE was detected in feces under these conditions. These results support the notion that the increases in hepatic C18:0NAE levels were indeed due to a shift to C18:0NAPE biosynthesis by pNAPE-EcN after colonization.
Figure 2.
Administration of pNAPE-EcN results in increased fecal C18:0 NAPE levels. Feces were collected from mice after ending eight-week treatment with pNAPE-EcN or from untreated mice receiving 0.125% gelatin in standard drinking water during this same period (n=5 mice per group, mean±SEM). Fecal NAPEs were measured after extraction by LC/MS. A. Fecal NAPE levels of mice fed a high fat diet based on lard. B. Fecal NAPE levels of mice fed low fat chow diet based on plant components. For both, 2-way ANOVA p<0.001 for both treatment and species. *p<0.05 compared to vehicle by Sidak’s multiple comparison test.
Because high fat diets alter several aspects of intestinal biology that might affect bacterial responses, we performed similar studies in mice fed a standard low fat chow diet. Once again, fecal NAPE levels were increased in mice that received pNAPE-EcN, with significant increases in C16:0NAPE, C18:0NAPE, C18:1NAPE, and C18:2NAPE, but only trivial amounts of C16:1NAPE (Figure 2B). These results confirm that pNAPE-EcN undergoes a significant shift in the species of NAPE biosynthesized after administration to mice, suggesting that factors present in the intestinal tract but not in our standard culture conditions altered the NAPE species biosynthesized. We therefore sought to determine how individual factors relevant to the intestinal environment affected the profile NAPE of species produced.
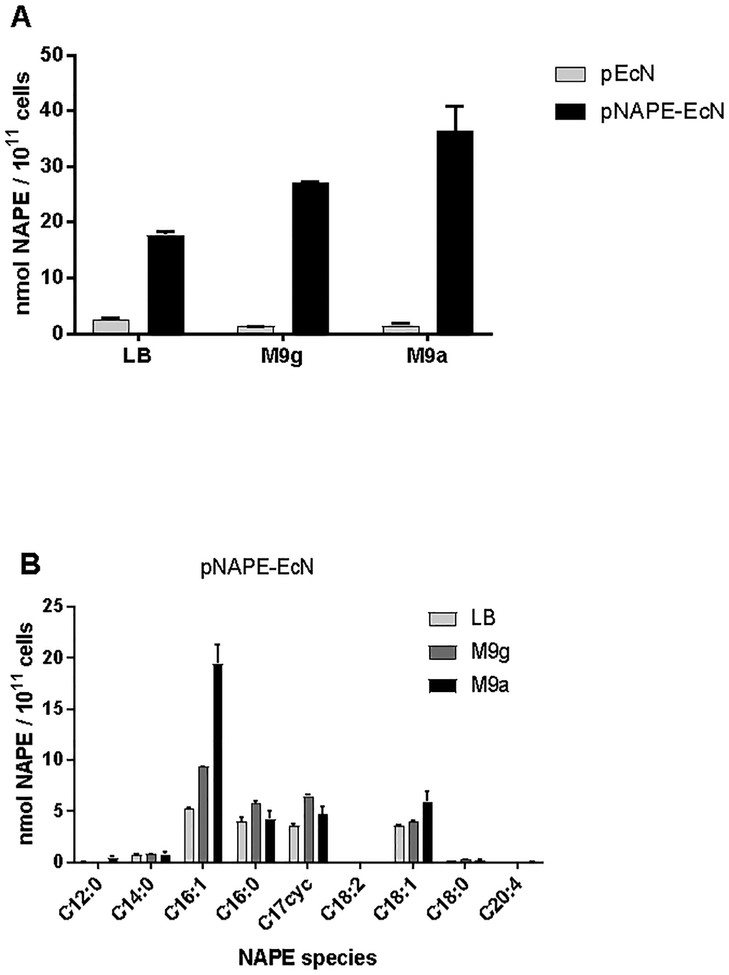
Rigorous determination of the factors that control NAPE biosynthesis requires completely defined media, but standard LB media contains undefined component mixtures such as yeast extract and tryptone. We therefore assessed the effect of culturing pNAPE-EcN in a minimal salt media (M9) supplemented with either glucose (M9g) or amino acids (M9a) as a carbon source under aerobic conditions. Control EcN transformed only with empty expression plasmid lacking the A. thaliana NAPE synthase gene (pEcN) grew well in all three media and, as expected, produced very little NAPE (Figure 3A). In contrast, pNAPE-EcN produced significant amounts of NAPE in all three of the media, with M9a media generating the highest levels of NAPE per cell (Figure 3A). In all three media, C16:1NAPE was the predominant NAPE species formed. Cultivation in M9a media resulted in a more marked difference in C16:1NAPE biosynthesis relative to other NAPE species (Figure 3B). C18:0NAPE or C20:4NAPE were not produced in any significant amount by pNAPE-EcN or pEcN in any of the three media conditions. Our finding that C16:1NAPE and C18:1NAPE levels are markedly lower in M9g compared to M9a media is consistent with previous reports that limiting available nitrogen reduces the relative abundance of C16:1 and C18:1 O-acyl chains in E. coli phospholipids9.
Figure 3.
NAPE biosynthesis by pEcN or pNAPE-EcN in minimal media with different carbon sources. NAPEs were measured by LC/MS in bacteria grown overnight in LB, M9+glucose (M9g) or M9+aminoacids (M9a) media then induced in same media with IPTG for 18 h. A. Effect of media on total NAPE production by pEcN and pNAPE-EcN. B. Effect of media on profile of NAPE species produced by pNAPE-EcN.
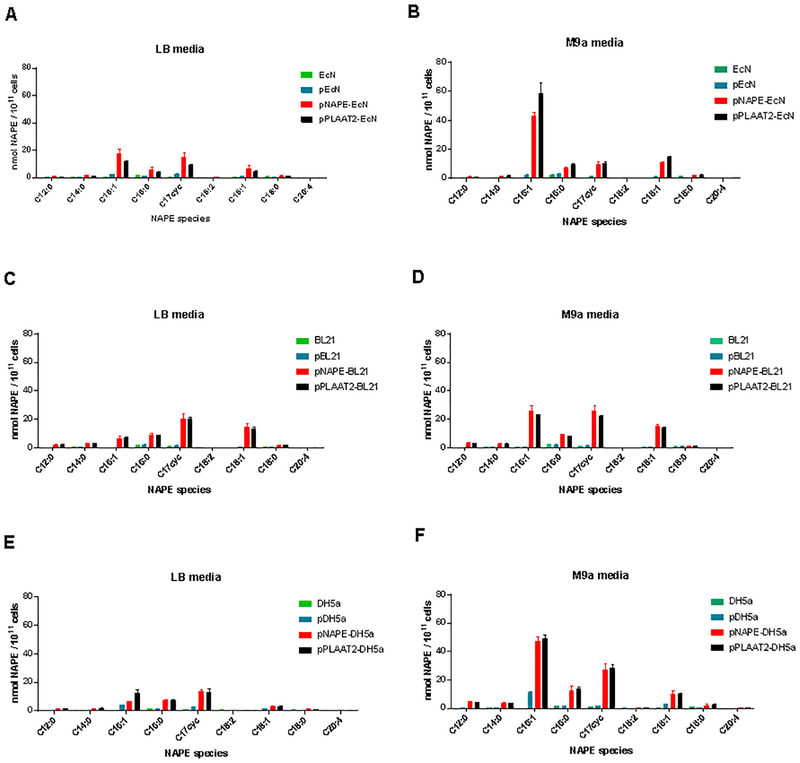
While NAPE synthase, the PE N-acyltransferase of A. thaliana, utilizes acyl CoAs as the acyl donor for NAPE synthesis, the human calcium-independent PE N-acyltransferase Phospholipase A/acyltransferase-2 (PLAAT2 also known as HRASLS-2), utilizes the sn-1 O-acyl chain from a second phospholipid (either phosphatidylcholine or phosphatidylethanolamine) as the acyl donor for NAPE synthesis (Figure 1). Given this difference in mechanism for NAPE synthesis, we hypothesized that heterologous expression of PLAAT2 in EcN would produce a NAPE profile that markedly differed from that of pNAPE-EcN. We therefore generated an expression plasmid for human PLAAT2 (Supplemental Figure 1), and transformed EcN with this expression plasmid (pPLAAT2-EcN). We then cultured these engineered bacteria in LB or M9a media. For comparison, we cultured pEcN and pNAPE-EcN in these same media. Surprisingly, pPLAAT2-EcN generated very similar NAPE profiles as pNAPE-EcN when cultivated in the same media (Figure 4A and B), and once again the profile produced in M9a differed from that of LB media.
Figure 4.
Comparison of NAPE profiles generated by transformation of three E. coli strains with human NAPE acyltransferase PLAAT2 or the Arabidopsis thaliana NAPE acyltransferase in minimal media. A. Engineered EcN in LB media. B. Engineered EcN in M9a media. C. Engineered B21 in LB media. D. Engineered B21 in M9a media. E. Engineered DH5a in LB media. F. Engineered DH5a in M9 media.
To assess whether the similarity in NAPE profile for the two enzymes was characteristic of their expression in E. coli more generally, we also transformed the two PE N-acyltransferases into the common lab strains E. coli BSL21(de3) and E. coli DH5a and cultivated each of these bacterial strains in either LB or M9a media. While we found that individual strains of E. coli markedly differed in terms of their NAPE profile in each media under aerobic conditions, once again heterologous expression of human PLAAT2 gave very similar NAPE profile as heterologous expression of A. thaliana NAPE synthase (Figure 4C–F). Of interest, E. coli BL21(de3) expressing either PE N-acyltransferase generated significantly more C17cycNAPE and C18:1NAPE than when these enzymes were expressed in EcN, but cultivation in M9a media still shifted the NAPE profile to significantly more C16:1NAPE synthesis than found for cultivation in LB media. Heterologous expression of the two enzymes in E. coli DH5a gave results intermediate between the two other E. coli strains, with similar C16:1NAPE synthesis but more C17cycNAPE than for EcN. The similarity of NAPE profile generated by the two different PE N-acyltransferases suggest that their two acyl donor pools (acyl CoAs and O-acyl chains of phospholipids) are in direct equilibrium. Previous studies have shown that acyl CoA can be converted to the acyl ACP derivative for use in phospholipid synthesis10, 11 and that the O-acyl chains of phospholipids can undergo hydrolysis and conversion to their acyl CoA derivatives12, providing a mechanism for this acyl donor pool equilibrium.
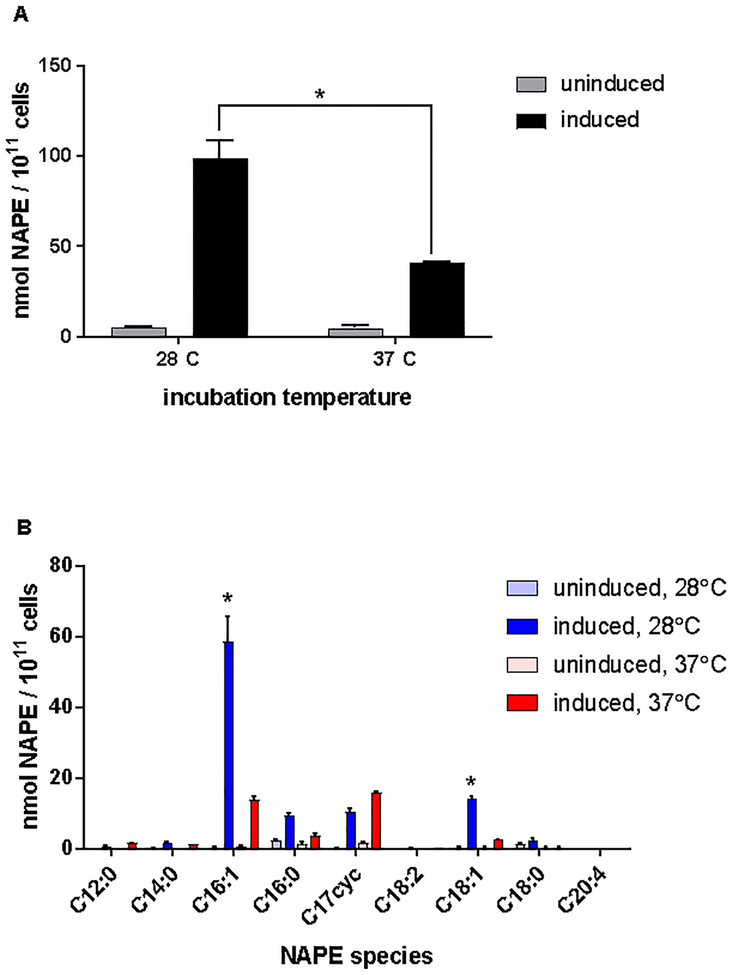
Culture temperature have previously been shown to alter the species of O-acyl chains present in the phospholipids of E. coli9. While the temperature within the intestinal tract of mice is approximately 37°C, we have typically prepared pNAPE-EcN for administration by induction at 28°C, as we had previously found this temperature gave better NAPE expression than induction at 37°C5. To determine the effect of temperature on NAPE expression by pPLAAT-EcN, we induced pPLAAT-EcN with and without IPTG before cultivation at 28°C and 37°C overnight in M9a media under aerobic conditions. Induction with IPTG increased NAPE biosynthesis by pPLAAT2-EcN 18-fold at 28°C, but only 8-fold at 37°C (Figure 5A). Furthermore, the species of NAPE significantly differed at the two temperatures, with C16:1NAPE being predominant when cultivation occurred at 28°C, but not at 37°C (Figure 5B). Levels of C16:0NAPE and C18:1NAPE were also reduced at 37°C, while levels of C17cycNAPE were increased. These findings are in keeping with previous reports that increasing cultivation temperature increases the relative abundance of saturated fatty acids while decreasing levels of monounsaturated fatty acids9,
Figure 5.
Effect of incubation temperature on NAPE biosynthesis by pPLAAT2-EcN. pPLAAT2-EcN were grown in M9a media, induced with IPTG and then incubated overnight at either 37°C or 28°C. NAPEs were measured by LC/MS. A. Total NAPEs. B. Profile of NAPE species produced. * p<0.05 compared to 37°C.
LB and M9a media contain negligible amounts of fatty acids, so the only species of fatty acids available as acyl donors for NAPE biosynthesis when engineered E. coli are cultured in these media are those endogenously synthesized by E. coli themselves. The vast majority of all fatty acids in E.coli are esterified as the O-acyl chains of phosphatidylethanolamine and previous reports examining E. coli cultivated at 30°C in LB media showed that the major O-acyl chain species are C18:1 (30% total fatty acids), C16:0 (29%), C16:1 (23%), C14:0 (4.1%) and C17cyclopropane (3.4%), with no significant amounts of C18:0, C18:2, or C20:4 detected9. Because none of our previously tested culture conditions gave rise to C18:0NAPE biosynthesis, and both esterified and unesterified fatty acids would be present in the intestinal tract after a meal13, we hypothesized that utilization of exogenous fatty acids for NAPE biosynthesis accounted for the altered NAPE biosynthesis profile.
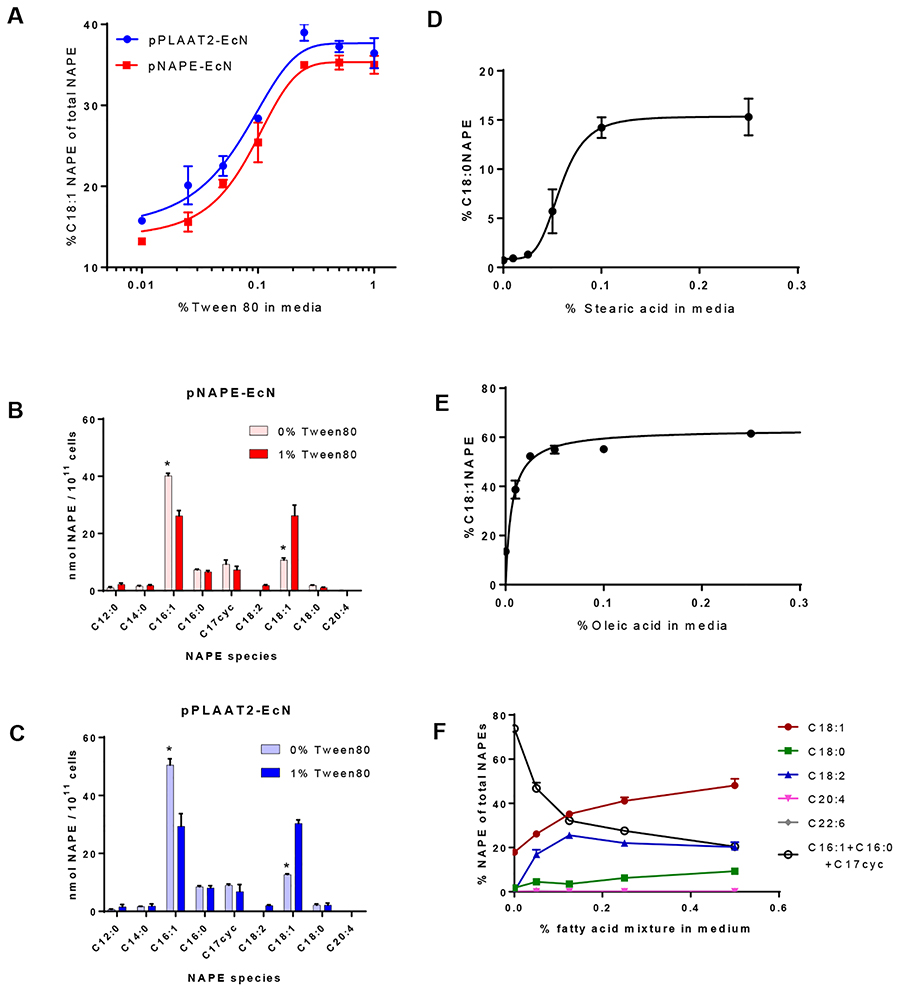
To determine if making exogenous esterified fatty acids available to the engineered bacteria could alter their profile of NAPE species, we supplemented M9a media with increasing concentrations of Tween 80, a water soluble source of esterified oleic acid (C18:1n-9 fatty acid). We found a concentration dependent increase in the extent of C18:1NAPE biosynthesis when Tween 80 was added to the culture media of pNAPE-EcN or pPLAAT2-EcN (Figure 6A). We note that our LC/MS analytical method does not distinguish between C18:1NAPE where the N-acyl chain is C18:1n-9 (which would result from incorporation of exogenously added oleic acid) from C18:1n-7 (which would result from incorporation of endogenously synthesized cis-vaccenic acid). Thus our measurement of C18:1NAPE includes both species of C18:1NAPE. C18:1NAPE represented ~15% of the total NAPE produced in the absence of Tween 80, but represented ~35% of total NAPE for Tween 80 concentration ≥0.25%. Absolute levels of total NAPEs produced were similar across the various concentrations of added Tween 80, so that C18:1NAPE biosynthesis appeared to progressively displace C16:1NAPE biosynthesis as Tween 80 concentration increased in both pNAPE-EcN (Figure 6B) and pPLAAT2-EcN (Figure 6C). Above 0.25%Tween80, no further enrichment occurred suggesting that uptake of Tween80 (or oleic acid released from Tween 80) was saturated, which would prevent further enrichment of C18:1 into the acyl CoA pool and then the phospholipid O-acyl chain pool. Given the similarity of the results in pPLAAT2-EcN and pNAPE-EcN, we chose to focus our remaining studies on pPLAAT2-EcN as use of the human PE N-acyltransferase would be preferred over A. thaliana PE N-acyltransferase for eventual human trials of these engineered bacteria.
Figure 6.
Exogenous fatty acids alter the species of NAPE biosynthesized by pNAPE-EcN and pPLAAT2-EcN. A. Effect of Tween 80 (esterified C18:1n-9) on the amount of C18:1NAPE (as %total NAPEs) biosynthesized by pNAPE-EcN and pPLAAT2-EcN grown in M9a. B. Effect of Tween 80 on NAPE profile for pNAPE-EcN. * p<0.05 Sidak’s multiple comparison test for individual NAPE species. C. Effect of Tween 80 on NAPE profile for pPLAAT2-EcN. * p<0.05 Sidak’s multiple comparison test for individual NAPE species. D. Effects of stearic acid addition on % of total NAPE biosynthesized by pPLAAT2-EcN that are C18:0NAPE. E. Effects of oleic acid addition on % of total NAPE biosynthesized by pPLAAT2-EcN that are C18:1NAPE. F. Effects of adding a mixture of five free fatty acids (C18:0, C18:1, C18:2, C20:4, C22:6) on the profile of NAPE species biosynthesized by pPLAAT2-EcN The contribution of the three most prominent NAPE species produced in the absence of the fatty acid mixture (C16:1NAPE, C16:0NAPE, and C17cycNAPE) were summed for comparison.
One initial impetus for determining the factors that controlled the species of NAPE produced by our engineered bacteria was our finding that levels of C18:0NAE were markedly increased in the liver of mice administered the engineered bacteria. We therefore sought to determine if exogenously adding stearic acid resulted in significant formation of C18:0NAPE. In the absence of added stearic acid, <1% of total NAPE was C18:0NAPE, while supplementation of M9a media with at least 0.1% stearic acid resulted in approximately 15% of total NAPE being C18:0NAPE (Figure 6D). Similar levels of total NAPEs were synthesized when M9a media was supplemented with 0 to 0.25% unesterified stearic acid, so that addition of unesterified stearic acid did not exert any toxic effects at these concentrations.
Although these results clearly indicated that exogenous stearic acid could be used for C18:0NAPE biosynthesis, the total enrichment for C18:0NAPE was less than the enrichment for C18:1NAPE that we had found with Tween80 supplementation. We hypothesized that EcN had a more limited capacity to take up unesterified fatty acid than fatty acids esterified in water soluble forms (i.e. Tween). We therefore determined to what extent unesterified oleic acid was utilized by pPLAAT-EcN when supplemented into M9a media. To our surprise, we found that supplementation of M9a media with unesterified oleic acid resulted in even greater enrichment of C18:1NAPE than did its supplementation with Tween 80 (Figure 6E). Supplementation with at least 0.025% oleic acid resulted in about 55% of total NAPE being C18:1NAPE. This result clearly demonstrated that uptake and utilization of unesterified fatty acids for NAPE synthesis was robust, but suggested that the extent of uptake and/or utilization of individual fatty acids might substantially differ.
Because oleic acid contains a double bond that stearic acid lacks, we next determined the effect of the number of double bonds present in exogenous unesterified fatty acids on their utilization for NAPE synthesis. To do this, we supplemented M9a media with a fatty acid mixture containing equimolar amounts of stearic acid (C18:0), oleic acid (C18:1n-9), linoleic acid (C18:2n-6), arachidonic acid (C20:4n-6), and docosahexaenoic acid (C22:6n-3). All five of these fatty acids are typically found in human diets, although their relative abundance varies markedly. Even at the highest concentration of added fatty acids (0.5% fatty acid mixture), less than 0.5% of total NAPEs were either C20:4NAPE or C22:6NAPE (Figure 6F). In contrast, levels of both C18:1NAPE and C18:2NAPE were significantly increased by addition of the fatty acid mixture, with C18:2NAPE reaching maximal levels of 26% of total NAPE when media included at least 0.1% fatty acid mixture, and C18:1NAPE reaching 48% of total NAPE when media included 0.5% fatty acid mixture. C18:0 appeared to poorly compete with C18:1 and C18:2, as even at the highest concentration of added fatty acids C18:0NAPE represented only 9% of total NAPEs. Our results suggest that when exogenous fatty acids are available, they are the predominant source of acyl donors for NAPE biosynthesis, but that when various exogenous fatty acids compete for utilization, the number of double bonds present is not the primary determinant of their utilization.
The inability of pPLAAT2-EcN to biosynthesize C20:4NAPE most likely results from the inability of E. coli to take up and incorporate arachidonic acid into O-acyl chains of its phospholipids. Exogenous fatty acid are taken up in E. coli by vectorial transport14, 15, with the outer membrane protein FadL facilitating movement of the fatty acid through the two leaflets of the outer membrane into the acidic environment of the periplasm. Protonation of the fatty acid anion in the periplasm allows it to cross the two leaflets of the inner membrane, a process further facilitated by conversion of the fatty acid to its acyl CoA derivative by FadD. Loss of either FadL or FadD blocks uptake of exogenous long-chain fatty acids15. Previous studies found that FadL had the highest binding capacity for C16:0 fatty acid, followed by C18:1 and had poor binding capacity for C14:0 or shorter fatty acids16. FadD has been shown to utilize various saturated fatty acids from C8 to C18, with C12:0 having the highest Vmax17, 18. To the best of our knowledge, no studies with C18:2, C20:4, or C22:6 fatty acid have been reported for either FadL or FadD. Based on our findings, we would predict that C18:2 is an effective substrate for both FadL and FadD, but that C20:4 and C22:6 fatty acids are not.
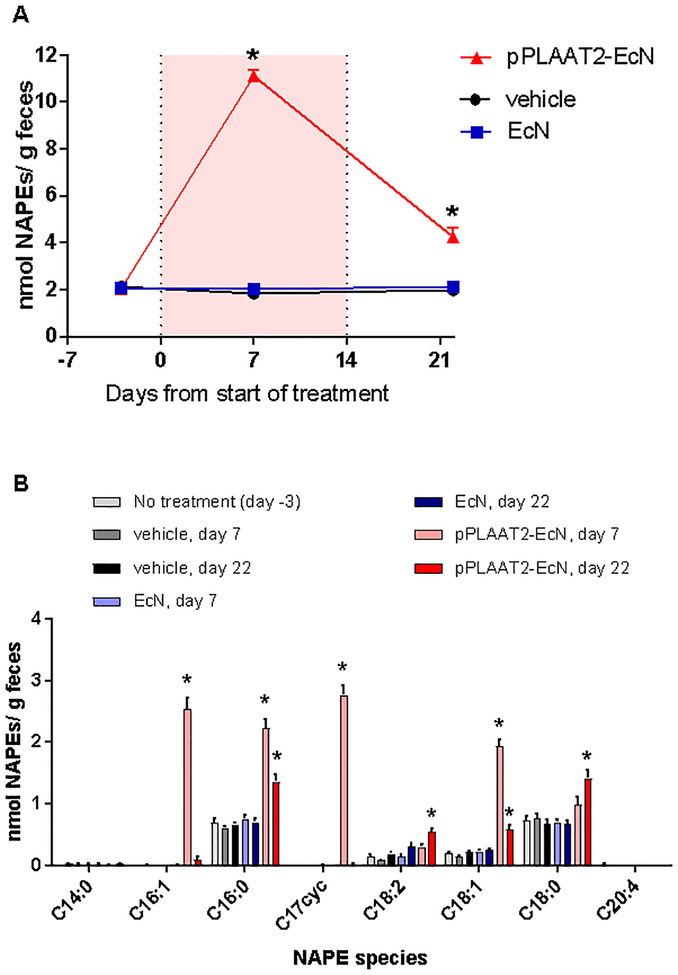
Based on these results, we hypothesized that the profile of NAPE species generated by pPLAAT2-EcN in the intestinal tract would undergo a change after colonization because they would be able to utilize exogenous fatty acids present in the lumen of intestinal tract from digested diets. For these studies, we fed C57BL/6J mice a high fat diet (60%kcal) based on lard that contains primarily C16:0, C18:0, and C18:1 fatty acids. We administered pPLAAT2-EcN at 5×109cfu/ml in the drinking water of C57BL/6J mice for two weeks (experimental day 0 to day 14), and measured NAPE levels in feces collected prior to administration (experimental day -3), one week after the start of bacterial administration (experimental day 7), and one week after the end of bacterial administration (experimental day 22). As controls, fecal NAPEs were also measured in mice receiving drinking water with either vehicle only or control EcN without plasmid (EcN).
Treatment with vehicle only or control EcN did not significantly alter fecal NAPE levels compared to levels prior to treatment (Figure 7A). In contrast, NAPE levels were significantly higher in feces collected during bacterial administration (day 7) and after bacterial administration (day 22) from mice treated with pPLAAT2-EcN. In feces collected during day 7 of pPLAAT2-EcN administration, the predominant species of NAPEs that were increased over the controls were C16:1NAPE and C17cycNAPE, followed by C18:1NAPE and then C16:0NAPE (Figure 7B). In feces collected 8 days after cessation of pPLAAT2-EcN administration (day 22), the predominant species that differed from baseline were C16:0NAPE and C18:0NAPE, followed by C18:1NAPE and C18:2NAPE. These findings are consistent with the notion that in the initial administration period, the intestinal NAPE profile reflects the NAPE species generated by induction in M9a media (i.e. predominantly C16:1NAPE and C17cycNAPE), but that after colonization this profile subsequently shifts to reflect the fatty acid composition of fatty acids in the diet fed to the animals (i.e. predominantly C16:0NAPE, C18:0NAPE, and C18:1NAPE). We note that when the matching time point and diet is compared, the fecal NAPE profile of mice treated with pPLAAT2-EcN (Figure 7B) is very similar to the profile of mice treated with pNAPE-EcN (Figure 2A). This would be expected from the results of the in vitro studies showing that the two strains generated similar NAPE profiles. We also note that mice treated with pNAPE-EcN but fed a chow diet have relatively high levels of C18:2NAPE, consistent with the utilization of the relatively high levels of C18:2 fatty acid reported for this diet.
Figure 7.
Effects of pPLAAT2-EcN administration and colonization on fecal NAPE levels and profile in mice feeding on high fat diet. A. Time course of total fecal NAPE levels for each treatment B. Effect of time and treatment on NAPE profile. 2way ANOVA p<0.0001 for species and treatment. *P<0.05 Dunnet post-hoc test compared to sample collected prior to treatment (no treatment).
Our finding that fecal NAPE levels from mice previously treated with pPLAAT2-EcN through experimental day 14 continue to be elevated at day 22 is consistent with the notion that colonized pPLAAT2-EcN continue to biosynthesize NAPE via PLAAT2 expression. How induction of PLAAT2 expression continues in the intestinal tract during these eight days is unclear. Basal PLAAT2 gene expression from our plasmid is repressed by the binding of LacI to the lac operator sequence near the T7 promoter (Supplemental Figure 1), so that binding of an inducer to LacI, which leads to the release of LacI from the lac operator, is required to induce robust PLAAT2 expression and NAPE biosynthesis (Figure 4). Although we add isopropyl β-D-1-thiogalactopyranoside (IPTG), a synthetic ligand for LacI, to pPLAAT2-EcN cultures to induce PLAAT2 expression prior to administration, subsequent centrifugation of pPLAAT2-EcN followed by removal of supernatant and resuspension of pPLAAT2-EcN in the drinking water with 0.125% gelatin likely removes almost all of this IPTG. It is therefore unlikely that sufficient residual IPTG remains in the intestinal tract seven days after last bacterial administration (day 22) to induce PLAAT2 expression. It seems more likely that native ligands for LacI derived from the diet such as β-galactosyl glycerol or related β-galactosides19, 20 are the primary inducers of PLAAT2 expression in colonized pPLAAT2-EcN.
In summary, our studies demonstrate that the remarkable switch in the NAPE species found when engineered bacteria are administered to mice results from their utilization of exogenous dietary fatty acids. This suggests that the NAPE profile of individuals treated with pPLAAT2-EcN or pNAPE-EcN could be optimized simply by changing the fatty acid profile of their diet. Individual NAEs differ in their efficacy for various therapeutic bioactivities, so that the desired NAPE species will vary depending on the particular clinical indication that these engineered bacteria are used to treat. Our previous studies using pNAPE-EcN focused on inhibiting obesity by reducing food intake and increasing basal metabolic rate. In this regard, enhancing C18:1NAPE production by enriching diets with oleic acid may be beneficial, as others have reported that C18:1NAE is the most potent NAE at reducing food intake in rodents21, 22. Altering the fatty acid profile of a high fat diet to enrich for C18:1n-9 (or for n-3 fatty acids) has already been utilized to reduce the adverse effects of the high fat diet on bacterial infection23, so such enrichment with C18:1n-9 could provide dual benefits in pPLAAT2-EcN treated individuals. Further testing to determine the optimal NAPE profile for obesity treatment is needed; however, as one group has reported that C18:0NAE is most potent at inhibiting very short term food intake after overnight fasting24. One reassuring implication of our results is that it should not be necessary to deplete diets of arachidonic acid (which is abundant in the diet of many Western countries) to achieve anti-obesity effects because pPLAAT2-EcN does not utilized arachidonic acid to form C20:4NAPE, which would lead to increased C20:4NAE levels and enhanced feeding drive.
For treating clinical conditions besides obesity, enhancing levels of other NAPE species besides C18:1NAPE may be beneficial. For instance, other beneficial effects of NAEs include analgesia and reduced inflammation. In regards to treatment of chronic pain, enhancing C16:0NAPE biosynthesis may be most beneficial as a meta-analysis of data from twelve clinical studies demonstrated significant effect of C16:0NAE administration on neuroinflammation and chronic pain25. Again, the optimal NAPE profile for chronic pain treatment would need to be characterized as C18:1NAE26 and C18:0NAE27 have also been reported to exert anti-inflammatory and analgesic effects.
Experimental Procedure
Materials:
Magnesium sulfate, triethylammonium acetate buffer, Tween 80, and methylamine solution (40 wt. % in water) were purchased from Sigma-Aldrich (St. Louis, MO). Organic solvents including methanol, chloroform, ethanol, 1-butanol and acetonitrile were all HPLC grade purchased from Thermo Fisher Scientific (Waltham, MA). Calcium chloride dihydrate, ammonium chloride, sodium phosphate dibasic anhydrous, potassium phosphate monobasic, and sodium chloride were also purchased from Thermo Fisher Scientific (Waltham, MA). Casamino acids (vitamin-free) from ACROS organics. A total of five fatty acids were used in this study: stearic acid and linoleic acid (Matreya LLC, Pleasant Gap, PA), oleic acid, arachidonic acid and docosahexadecanoic acid (Cayman chemical, Ann Arbor, MI). Ampicillin sodium salt and isopropyl-β-D-thiogalactoside (IPTG) were obtained from Research Products International (RPI; Mt. Prospect, IL). Sep-Pak silica cartridges were purchased from the Waters Corporation (Milford, MA). Kinetex 2.6μ C18 100Å column 50×2.1 mm was purchased from Phenomenex (Torrance, CA).
Bacterial strains preparation:
E. coli Nissle 1917 (EcN) was obtained as a gift from ArdeyPharm, GmbH. The pQE-80L-At expression plasmid (vector for Arabidopsis thaliana N-acyltransferase) and its insertion into EcN have been previously described5. The plasmid for human NAPE-acyltransferase HRASLS2 / PLAAT2 (pPLAAT2) was synthesized on our behalf by Vectorbuilder (Santa Clara, CA). This was done by insertion into pET-6xHis plasmid of synthesized oligonucleotides encoding for the HRASLS2 / PLAAT2 amino acid sequence using codons optimized for bacterial expression. This plasmid puts expression of PLAAT2 under the control of a T7 promotor and Lac operator. The nucleotide sequence encoding for HRASLS2 / PLAAT2 inserted into the pET-6xHis plasmid was as follows: ATGGCGCTGGCGCGTCCGCGTCCGCGTCTGGGTGACCTGATCGAAATCTCTCGTTTCGGT TACGCGCACTGGGCGATCTACGTTGGTGACGGTTACGTTGTTCACCTGGCGCCGGCGTCTGAAATCGCGGGTGCGGGTGCGGCGTCTGTTCTGTCTGCGCTGACCAACAAAGCGATCGTTAAAAAAGAACTGCTGTCTGTTGTTGCGGGTGGTGACAACTACCGTGTTAACAACAAACACGACGACCGTTACACCCCGCTGCCGTCTAACAAAATCGTTAAACGTGCGGAAGAACTGGTTGGTCAGGAACTGCCGTACTCTCTGACCTCTGACAACTGCGAACACTTCGTTAACCACCTGCGTTACGGTGTTTCTCGTTCTGACCAGGTTACCGGTGCGGTTACCACCGTTGGTGTTGCGGCGGGTCTGCTGGCGGCGGCGTCTCTGGTTGGTATCCTGCTGGCGCGTTCTAAACGTGAACGTCAGTAA.
Electro-competent EcN cells were transformed with pPLAAT2 by electroporation to give pPLAAT2-EcN. BL21Gold(DE3) and DH5α were transformed either with pQE-80L1 (QIAGEN) empty vector (to give pEcN, pBL21, and pDH5α), pQE-80L1-At (pNAPE-BL21, and pNAPE-DH5α) or pPLAAT2 (pPLAAT2-BL21 and pPLAAT2-DH5α).
Bacterial culture:
Bacteria were grown either in a nutrient Luria-Bertani (LB) broth supplemented with 0.1 mg/mL ampicillin or in M9 minimal medium supplemented with 0.1 mg/mL ampicillin, a carbon source (0.4% casamino acids or 0.4% glucose), 0.1 mM calcium chloride and 2.0 mM magnesium sulfate to produce optimal amount of NAPEs. Cultures were allowed to grow under aerobic conditions overnight at 37°C and 200 rpm (Benchmark INCU-Shaker™ 10L) then were diluted (1:50) in the same medium used for initial culture and continued to grow at 37°C and 200 rpm until they reached an optical density (OD600) of 0.5–0.6. OD600 was measured on Eppendorf BioSpectrometer® Basic (1 OD600 defined as 8×108 CFU/mL). Then isopropyl-β-D-thiogalactoside (IPTG, 1mM) was added to induce NAPE expression and the cultures were allowed to grow overnight under aerobic conditions at 28°C or 37°C at 200 rpm. After recording the optical density, cells were harvested by centrifugation at 5000 g and 4°C for 15 min and the pellet was resuspended in 1 mL of 5M NaCl.
Use of exogenous fatty acids for NAPE production:
Initial cultures were grown in M9 minimal medium supplemented with 0.1 mg/mL ampicillin, 0.4% casamino acids, 0.1 mM calcium chloride and 2.0 mM magnesium sulfate overnight under aerobic conditions at 37°C and 200 rpm. Free fatty acids were prepared as separate stock solutions in ethanol that were diluted by sterile M9 minimal medium (concentration of ethanol did not exceed 2% of total culture volume), in triplicates. After overnight growth, cultures were used as inoculum to test different concentrations (0.01, 0.025, 0.05, 0.1, 0.25, 0.5 and 1.0%) of Tween 80, stearic acid (C18:0) or oleic acid (C18:1n-9). Control medium contained no fatty acid. To determine which FFA would be preferentially used, we tested different concentrations (0–0.5%) of a mixture containing equal amounts of five FFAs: stearic acid (C18:0), oleic acid (C18:1n-9), linoleic acid (C18:2n-6), arachidonic acid (C20:4n-6) and docosahexadecanoic acid (C22:6n-3). Cultures continued to grow under aerobic conditions at 37°C and 200 rpm until they reached an optical density (A600) of 0.5–0.6. Then IPTG (1mM) was added and the cultures were allowed to grow overnight at 28°C at 200 rpm. After measurement of the optical density, cells were harvested by centrifugation at 5000 g for 15 min and the pellet was resuspended in 1 mL of 5M NaCl.
Measurement of NAPEs by LC/MS/MS:
Extraction and methylamine hydrolysis of NAPEs were performed as previously described28. The bacterial pellet from a 20 ml culture or 1 ml fecal homogenate were mixed with 6 mL pre-chilled Folch solution (chloroform/methanol, 2:1 containing 0.01% BHT), followed by addition of 1 nmol C17:0 NAPE (10μL of 0.1mg/ml C17:0‐NAPE in chloroform) and 3 mL of water. The mixture was vortexed for 5 min, incubated in ice-bath for 30 min, and centrifuged at 4,000 g for 5 min. The resulting lower (chloroform) layer was carefully collected into a new tube. The aqueous phase was re-extracted with 4 mL of chloroform. The mixture was vortexed for 2 min and centrifuged at 4,000 g for 5 min. The organic phases were combined and dried under nitrogen. The extracted lipids were reconstituted in 1 mL chloroform and loaded onto a Sep-Pak silica cartridge pre-equilibrated with 4mL methanol then 8 mL chloroform. Lines were washed by 8 mL methanol: chloroform (1:9) then NAPEs were eluted by 8 mL of Folch solution (chloroform/methanol, 2:1). The lipids were dried under nitrogen and saved at −20°C before methylamine hydrolysis. Dried lipid extracts were dissolved in 200 μL of methylamine reagent (methylamine solution (40 wt. %)/ methanol/1-butanol (4/4/1)) and mixed well. The reaction was incubated at 53°C for 1 h and the resulting solution was used for MS analysis.
NAPE concentration was quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after methylamine hydrolysis28. Samples were analyzed on a ThermoFinnigan Quantum electrospray ionization triple quadrapole mass spectrometer operating in negative ion mode, equipped with ThermoPal Surveyor autosampler. Volume of injection was 10μl. The mobile phase consisted of solvent A (1mM triethylammonium acetate in water), and solvent B (1mM triethylammonium acetate in acetonitrile). The lipids were chromatographed on a Kinetex C18 column (50 mm × 2.10 mm, 2.6 um, 100 Å; Phenomenex, Torrance, CA) with a constant flow rate of 250 uL/min. After 0.5 min hold at 1% solvent B, the solvent was gradient ramped to 99% solvent B over 4.5 minutes, held at 99% solvent B for 1.5 min and return to 1% solvent B over 0.5 min and held for 2 min before the next injection. The electrospray needle was maintained at −3300 V. The ion-transfer tube was operated at −35 V and 270 °C. The tube lens voltage was set to −180 V.
For multiple reaction monitoring (MRM) mode, mass transitions with the product ion of m/z 79.1 at the specified parent ion (Table 1) at a collision energy of 50 eV were monitored. The chromatographic results were processed in Xcaliber software (ThermoFinnigan) using 13-point Gaussian smoothing. Linear regression and correlation analysis was performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, San Diego, CA).
Table (1):
Multiple reaction monitoring (MRM) parameters
| Species | Precursor ion | Product ion |
|---|---|---|
| C12:0 GP-NAE | 396.2 | 79.1 |
| C14:1 GP-NAE | 422.3 | 79.1 |
| C14:0 GP-NAE | 424.3 | 79.1 |
| C15:1 GP-NAE | 436.3 | 79.1 |
| C15:0 GP-NAE | 438.3 | 79.1 |
| C16:1 GP-NAE | 450.3 | 79.1 |
| C16:0 GP-NAE | 452.3 | 79.1 |
| C17:0 GP-NAE | 466.3 | 79.1 |
| C17cy GP-NAE | 464.3 | 79.1 |
| C18:3 GP-NAE | 474.3 | 79.1 |
| C18:2 GP-NAE | 476.3 | 79.1 |
| C18:1 GP-NAE | 478.3 | 79.1 |
| C18:0 GP-NAE | 480.3 | 79.1 |
| C19cy GP-NAE | 492.3 | 79.1 |
| C20:4 GP-NAE | 500.3 | 79.1 |
| C24:6 GP-NAE | 524.3 | 79.1 |
Animal study:
Four to six week-old male C57BL/6J mice were individually housed in the Vanderbilt University animal facility in a 12-hour light/12-hour dark cycle. They were initially fed either a chow diet (LabDiet 5001) or a refined High-Fat Diet (Research Diets D12492, with 60% kcal% fat, 5.24 kcal/g) and acclimated for 10 days. On experimental day 0, mice started receiving their assigned treatment. Depending on the study, the treatment groups received standard drinking water either with 0.125% gelatin (vehicle), with 0.125% gelatin and 5×109 CFU/ml EcN (EcN), with 0.125% gelatin and 5×109 CFU/ml pNAPE-EcN, or with 0.125% gelatin and 5×109 CFU/ml pPLAAT2-EcN. Drinking water with various treatments were changed 3 times per week and treatment was provided for a total of 2 weeks (through experimental day 14). After ending bacterial treatment, mice remained on the same diet during the follow-up period. Feces were collected before, during and after bacterial treatment.
Study approval:
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of Vanderbilt University.
Statistics:
Statistical analysis was performed using GraphPad Prism 7 for Windows (GraphPad Software, San Diego, CA). A P value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgement:
This work was supported in part by a grant from the National Institutes Health /National Center for Complementary and Integrative Health (R01 AT007830). E. coli Nissle 1917 was a gift from ArdeyPharm.
Abbreviations:
- NAPE
N-acyl-phosphatidylethanolamine
- NAE
N-acyl-ethanolamide
- LB
Luria-Bertani media
- M9g
M9 minimal medium+ glucose
- M9a
M9 minimal medium+ casaminoacids
- NAPE-PLD
NAPE-hydrolyzing phospholipase D
- C20:4NAE
N-arachidonyl-ethanolamide
- C16:0NAE
N-palmitoyl-ethanolamide
- C18:1NAE
N-oleoyl-ethanolamide
- C16:1NAPE
N-hexadecaenoyl-PE
- C16:0NAPE
N-hexadecanoyl-PE
- C17cycNAPE
N-methylenehexadecanoyl-PE
- C18:1NAPE
N-octadecaenoyl-PE
- C18:0NAPE
N-octadecanoyl-PE
- C20:4NAPE
N-arachidonyl-PE
- EcN
E. coli Nissle 1917
- pEcN
EcN transformed with empty expression plasmid
- pNAPE-EcN
EcN transformed with a plasmid for A. thaliana NAPE synthase
- PLAAT2
phospholipase A/acyltransferase 2
- pPLAAT2-EcN
EcN transformed with a plasmid for human PLAAT2/ HRASLS-2
- OD600
optical density at 600nm
- IPTG
isopropyl-β-D-thiogalactoside
- CFU
Colony Forming Unit
- LC/MS/MS
Liquid chromatography-tandem mass spectrometry
- MRM
multiple reaction monitoring
Footnotes
Conflicts of Interest: SSD, LG and ZC have a patent pending for the use of engineered bacteria expressing NAPE or NAE for treating obesity.
References
- [1].Gillum MP, Zhang D, Zhang XM, Erion DM, Jamison RA, Choi C, Dong J, Shanabrough M, Duenas HR, Frederick DW, Hsiao JJ, Horvath TL, Lo CM, Tso P, Cline GW, and Shulman GI (2008) N-acylphosphatidylethanolamine, a gut- derived circulating factor induced by fat ingestion, inhibits food intake, Cell 135, 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fu J, Astarita G, Gaetani S, Kim J, Cravatt BF, Mackie K, and Piomelli D (2007) Food intake regulates oleoylethanolamide formation and degradation in the proximal small intestine, J Biol Chem 282, 1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Piomelli D (2013) A fatty gut feeling, Trends Endocrinol Metab 24, 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen Z, Zhang Y, Guo L, Dosoky N, De Ferra L, Peters S, Niswender KD, and Davies SS (2017) Leptogenic effects of N-acyl-phosphatidylethanolamine (NAPE) requires activity of N-acylphosphatidylethanolamine-hydrolyzing phospholipase D (NAPE-PLD), J Lipid Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen Z, Guo L, Zhang Y, Walzem RL, Pendergast JS, Printz RL, Morris LC, Matafonova E, Stien X, Kang L, Coulon D, McGuinness OP, Niswender KD, and Davies SS (2014) Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity, J Clin Invest 124, 3391–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mardian EB, Bradley RM, and Duncan RE (2015) The HRASLS (PLA/AT) subfamily of enzymes, J Biomed Sci 22, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Uyama T, Ikematsu N, Inoue M, Shinohara N, Jin XH, Tsuboi K, Tonai T, Tokumura A, and Ueda N (2012) Generation of N-acylphosphatidylethanolamine by members of the phospholipase A/acyltransferase (PLA/AT) family, J Biol Chem 287, 31905–31919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Faure L, Coulon D, Laroche-Traineau J, Le Guedard M, Schmitter JM, Testet E, Lessire R, and Bessoule JJ (2009) Discovery and characterization of an Arabidopsis thaliana N-acylphosphatidylethanolamine synthase, J Biol Chem 284, 18734–18741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marr AG, and Ingraham JL (1962) Effect of Temperature on the Composition of Fatty Acids in Escherichia Coli, J Bacteriol 84, 1260–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Green PR, Merrill AH Jr., and Bell RM (1981) Membrane phospholipid synthesis in Escherichia coli. Purification, reconstitution, and characterization of sn-glycerol-3-phosphate acyltransferase, J Biol Chem 256, 11151–11159. [PubMed] [Google Scholar]
- [11].Zhang YM, and Rock CO (2008) Thematic review series: Glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis, J Lipid Res 49, 1867–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pech-Canul A, Nogales J, Miranda-Molina A, Alvarez L, Geiger O, Soto MJ, and Lopez-Lara IM (2011) FadD is required for utilization of endogenous fatty acids released from membrane lipids, J Bacteriol 193, 6295–6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Iqbal J, and Hussain MM (2009) Intestinal lipid absorption, American Journal of Physiology - Endocrinology and Metabolism 296, E1183–E1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].van den Berg B (2005) The FadL family: unusual transporters for unusual substrates, Curr Opin Struct Biol 15, 401–407. [DOI] [PubMed] [Google Scholar]
- [15].Black PN, and DiRusso CC (2003) Transmembrane movement of exogenous long-chain fatty acids: proteins, enzymes, and vectorial esterification, Microbiol Mol Biol Rev 67, 454–472, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Black PN (1990) Characterization of FadL-specific fatty acid binding in Escherichia coli, Biochim Biophys Acta 1046, 97–105. [DOI] [PubMed] [Google Scholar]
- [17].Kameda K, and Imai Y (1985) Isolation and characterization of the multiple charge isoforms of acyl-CoA synthetase from Escherichia coli, Biochim Biophys Acta 832, 343–350. [DOI] [PubMed] [Google Scholar]
- [18].Kameda K, and Nunn WD (1981) Purification and characterization of acyl coenzyme A synthetase from Escherichia coli, J Biol Chem 256, 5702–5707. [PubMed] [Google Scholar]
- [19].Boos W, Schaedel P, and Wallenfels K (1967) [Investigation on induction of lac-enzyme. 1. Induction process and permeation], Eur J Biochem 1, 382–394. [DOI] [PubMed] [Google Scholar]
- [20].Egel R (1988) The ‘lac’ operon: an irrelevant paradox?, Trends Genet 4, 31. [DOI] [PubMed] [Google Scholar]
- [21].Fu J, Oveisi F, Gaetani S, Lin E, and Piomelli D (2005) Oleoylethanolamide, an endogenous PPAR-alpha agonist, lowers body weight and hyperlipidemia in obese rats, Neuropharmacology 48, 1147–1153. [DOI] [PubMed] [Google Scholar]
- [22].Diep TA, Madsen AN, Holst B, Kristiansen MM, Wellner N, Hansen SH, and Hansen HS (2011) Dietary fat decreases intestinal levels of the anorectic lipids through a fat sensor, FASEB J 25, 765–774. [DOI] [PubMed] [Google Scholar]
- [23].DeCoffe D, Quin C, Gill SK, Tasnim N, Brown K, Godovannyi A, Dai C, Abulizi N, Chan YK, Ghosh S, and Gibson DL (2016) Dietary Lipid Type, Rather Than Total Number of Calories, Alters Outcomes of Enteric Infection in Mice, The Journal of infectious diseases 213, 1846–1856. [DOI] [PubMed] [Google Scholar]
- [24].Terrazzino S, Berto F, Dalle Carbonare M, Fabris M, Guiotto A, Bernardini D, and Leon A (2004) Stearoylethanolamide exerts anorexic effects in mice via down-regulation of liver stearoyl-coenzyme A desaturase-1 mRNA expression, FASEB J 18, 1580–1582. [DOI] [PubMed] [Google Scholar]
- [25].Paladini A, Fusco M, Cenacchi T, Schievano C, Piroli A, and Varrassi G (2016) Palmitoylethanolamide, a Special Food for Medical Purposes, in the Treatment of Chronic Pain: A Pooled Data Meta-analysis, Pain Physician 19, 11–24. [PubMed] [Google Scholar]
- [26].Sayd A, Anton M, Alen F, Caso JR, Pavon J, Leza JC, Rodriguez de Fonseca F, Garcia-Bueno B, and Orio L (2014) Systemic administration of oleoylethanolamide protects from neuroinflammation and anhedonia induced by LPS in rats, Int J Neuropsychopharmacol 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dalle Carbonare M, Del Giudice E, Stecca A, Colavito D, Fabris M, D’Arrigo A, Bernardini D, Dam M, and Leon A (2008) A Saturated N-Acylethanolamine Other than N-Palmitoyl Ethanolamine with Anti-inflammatory Properties: a Neglected Story…, Journal of Neuroendocrinology 20, 26–34. [DOI] [PubMed] [Google Scholar]
- [28].Guo L, Amarnath V, and Davies SS (2010) A liquid chromatography-tandem mass spectrometry method for measurement of N-modified phosphatidylethanolamines, Anal Biochem 405, 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.