Abstract
Surfaces within the neonatal intensive care unit (NICU), especially those handled frequently by hospital staff, provide sources of gut-colonizing bacteria for hospitalized infants, in addition to those acquired perinatally from maternal sources such as breastmilk. In comparison to bacteria, very little is known about potential sources of colonizing fungi in the NICU setting. Thus, the objective of this study was to characterize fungal communities (mycobiomes) of potential colonization sources for neonates hospitalized in a large university NICU. We hypothesized that the unit surfaces would contain different mycobiomes than those of human-associated (breastmilk) sources. We characterized mycobiomes of NICU surfaces of multiple individual patient care areas as well as those of breastmilk samples by sequencing the internal transcribed spacer region 2 (ITS2) of the fungal rDNA locus. We found that, across all samples, Candida and Saccharomyces species were the most prevalent taxa and had the greatest relative abundances. Breastmilk samples had significantly higher fungal alpha-diversities than NICU surface samples and fungal community compositions (beta diversities) differed significantly between the two sample types. Mycobiome compositions were predominantly driven by the relative abundances of three fungal taxa: Candida albicans, Candida parapsilosis, and Saccharomyces cerevisiae. In total, 21 individual fungal taxa showed significantly greater relative abundances in breastmilk as compared to NICU surfaces, with three being of particular interest to human health: Candida glabrata, Candida tropicalis, and Cryptococcus neoformans. Since no fungal DNA was detected when whole breastmilk was used as the DNA template, as opposed to breastmilk subjected to cell lysis during the DNA isolation procedure, our results indicate that DNA is from fungal cells and is not cell-free DNA. In summary, both NICU surfaces and human breastmilk harbor distinct fungal communities that could provide a source of fungi for the developing infant gut mycobiota. In particular, Candida and Saccharomyces species are abundant and prevalent for both of these potential sources that infants are exposed to.
Keywords: Mycobiome, Breastmilk, Newborn Intensive Care, Hospital Surfaces, Fungi
1. Introduction
Exposure to commensal gut bacteria during early life has been associated with the development and function of many biological systems including metabolism, immunity, and the brain (Arrieta et al., 2015; Cho et al., 2012; Cox et al., 2014; Diaz Heijtz et al., 2011). Extensive epidemiological surveys have improved our understanding of how factors such as birth mode and diet shape the bacterial microbiome in early life (Mueller et al., 2015a; Mueller et al., 2015b). In the U.S., it is estimated that ~10% of infants require hospitalization in a newborn intensive care unit (NICU) immediately after birth. This creates a significant population of infants at risk for potential health effects from abnormal maturation of gut microbial communities due to disruption in the normal pace of colonization (La Rosa et al., 2014), as well as exposure to atypical environmental sources. Indeed, recent studies have shown that as compared to healthy term-born infants, premature infants have delayed bacterial microbiome maturation (La Rosa et al., 2014) as well as compositional differences (increased Proteobacteria and decreased Bifidobacteria) in their gut bacterial communities (Costello et al., 2013).
Emerging research has shown that NICU surfaces and breastmilk harbor bacteria that provide sources of microbes for gut microbiome development in hospitalized infants. Gut bacterial microbiomes of these infants are similar to those of surfaces of the NICU (Brooks et al., 2014), and deep metagenomic sequencing approaches have confirmed that specific strain transmission occurs between NICU surfaces and the infant gut (Brooks et al., 2017). In addition, unpasteurized breastmilk provides a source of microbes for infant gut microbiome development, with bacterial diversity and compositional changes being driven by amount of breastmilk intake in a dose dependent manner (Pannaraj et al., 2017).
Very little data exist regarding other microbial kingdoms present among microbial sources that hospitalized infants are exposed to. In particular, we lack knowledge about fungi, the dominant eukaryotes in the human gut (Parfrey et al., 2014), which have been found to modulate inflammation, immunity, and metabolism in animal models (Everard et al., 2014; Iliev et al., 2012; Jiang et al., 2017). Importantly, fungi are prevalent colonizers of the gut during early infancy, and cause both superficial mucosal as well as invasive infections, particularly in hospitalized infants (Bendel, 2011). In this study, we sought to identify potential sources of fungi for early gut mycobiome development in hospitalized infants. In addition, we compared fungal communities of different sources to determine the extent to which they may provide different potential fungal colonizers.
2. Materials and Methods
2.1. Sample collection.
Environmental samples were collected from surfaces within 9 individual patient care areas of the University of Minnesota Masonic Children’s Hospital NICU (Minneapolis, MN). Samples from each surface were obtained in duplicate, on four different days and over a period of 4 weeks, using sterile swabs moistened with sterile, DNA- and RNA-free water (Invitrogen, Grand Island, NY). Swabs were vigorously rubbed over the following environmental sites: stethoscope diaphragm, incubator porthole latches (heretofore referred to as handles), computer mouse, and computer keyboard. Sites were chosen because they are frequently touched by hospital staff and, in the case of incubator handles, also by parents. Of note, hospital personnel wear clean gloves when handling incubator handles and stethoscopes; but no gloves are worn when touching computer surfaces. Inoculated swabs were placed into a sterile tube and stored at −20°C for no longer than 48 h, and subsequently subjected to DNA isolation (described below).
Breastmilk samples from mothers that delivered preterm at the University of Minnesota Medical Center were obtained from a biorepository of de-identified samples that had been collected as part of University of Minnesota Institutional Review Board protocol 1305M33583. The milk was collected into sterile microfuge tubes and then stored at −20°C for no longer than 48 h prior to being subjected to DNA isolation (described below).
2.2. DNA isolation.
DNA was isolated from swabs of NICU surfaces and from 1–2 ml of human milk using the PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA) following the manufacturer’s recommended protocol, with the following exceptions. Samples were incubated at 70°C for 10 min following addition of solution C1. To elute DNA from the spin columns, DNA- and RNA-free sterile water was used rather than solution C6. DNA samples were stored at −20°C until use for DNA amplicon generation by PCR.
2.3. qPCR.
To detect and quantify fungal DNA present in NICU surface and breastmilk samples, probe-based qPCR was employed as previously described (Heisel et al., 2015) using a LightCycler 480 (Roche, Indianapolis, IN). Briefly, fungal DNA was amplified from isolated sample DNA using universal fungal primers (UNI1 and UNI2) that target the fungal rDNA locus. Reactions were performed in duplicate and the results were averaged.
Breastmilk samples were additionally evaluated for the presence of cell-free fungal DNA by qPCR using whole milk (not subjected to cell lysis and DNA isolation procedures) as the template, along with the universal fungal primers as described above, using SYBR Green (Roche) for amplification detection and quantification.
2.4. Fungal amplicon generation and preparation for sequencing.
Fungal ITS2 was amplified from isolated sample DNA by conventional PCR using dual-barcoded ITS2-specific primers as previously described (Heisel et al., 2017). Briefly, 150 μl of reaction mix was prepared for each sample and equally divided into three 50-μl wells to which was added 1 μl of KAPA HiFi polymerase (KAPA Biosystems, Wilmington, MA) following the manufacturer’s recommendations. Following PCR, sample-specific amplicons were pooled into 150 μl of total solution and cleaned using 180 μl of SPRISelect (Beckman Coulter, Brea, CA) to select for amplicons of ~150 bp and larger. Cleaned amplicons were eluted in 35 μl of RNA- and DNA-free sterile water and amplicon concentrations were determined using the Qubit dsDNA HS kit and a Qubit2 reader (Invitrogen/Molecular Probes, Eugene, OR). All samples were pooled with equivalent amounts of DNA into a single solution for submission for sequencing.
2.5. ITS2 sequencing.
Cleaned and pooled amplicons were submitted to the University of Minnesota Genomics Center (Minneapolis, MN) for library preparation and sequencing. Library preparation was performed using the TruSeq Nano kit (Illumina, San Diego, CA). Sequencing was performed using the Illumina MiSeq platform using V2 2×250 bp paired-end chemistry. Sequences were deposited and hosted on the Minnesota Supercomputing Institute (University of Minnesota, Minneapolis, MN) system servers.
2.6. Sequence Analysis.
Sequences were demultiplexed and subjected to basic QC (adapter primers trimmed, joining of paired reads, removing low-quality reads [average sequence quality < 35]) using SHI7 (Al-Ghalith et al., 2018). Raw sequence reads were aligned to a fungal-specific ITS2 database using the BURST program (Al-Ghalith and Knights, 2017). The ITS2 alignment database was generated by BURST using the ITS RefSeq database hosted by the National Center for Biotechnology Information (NCBI, Washington, D.C.). Sequencing results were analyzed using R (https://www.r-project.org). Samples containing fewer than 100 sequence reads were discarded. Sequences from duplicate NICU surface samples were combined. This combinatorial approach was validated by the finding that beta diversity distances between replicate samples were less than those between non-replicate samples (p<0.001, Wilcoxon rank sum test), indicating that replicate samples had more similar mycobiomes than did non-replicates. In addition to the base R package, the vegan package (Oksanen) was used for access to more biologically targeted statistical analyses such as Permutational Multivariate Analysis of Variance Using Distance Matrices (PERMANOVA) and phylogenetic distance calculations such as the Bray-Curtis dissimilarity index.
2.7. Data Availability.
Sequence data for all samples in this project is available in the NCBI Sequence Read Archive (SRA) database under BioProject ID PRJNA505509.
3. Results
3.1. Surfaces within the NICU and human breastmilk contain fungal DNA.
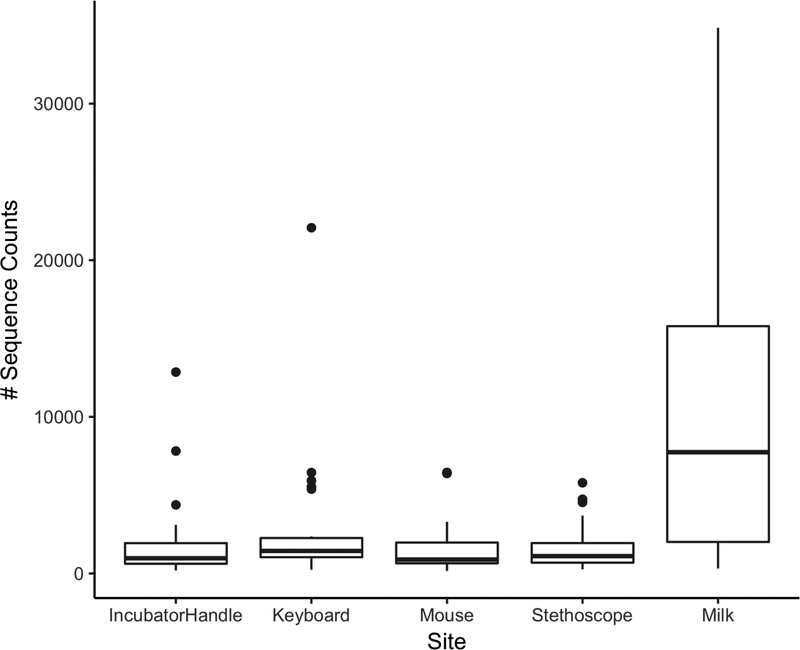
We hypothesized that fungal cell numbers are very low on hospital surfaces that receive regular cleaning with antimicrobial solutions. Thus, we used sensitive fungal-specific PCR-based approaches to detect fungi and generate DNA amplicons for sequencing to characterize fungal communities of NICU surfaces and breastmilk. We found that NICU surfaces and breastmilk samples contain fungal DNA by both qPCR and sequencing. (Table 1). As expected, fungal DNA amounts as determined by qPCR were low (Cq values of all samples were below the limit of quantification (Heisel et al., 2015)). Although samples did not give signals that allowed quantification, signals were nonetheless present for most samples and were greater than those of the negative (no template) controls (n=20), most of which failed to produce a signal after 40 qPCR cycles. In addition, the majority of the NICU surface, and all of the breastmilk, samples produced fungal sequence reads using a different set of primers targeting fungal ITS2 (Table 1). Breastmilk samples generated substantially greater numbers of sequencing reads (mean of 11,500 sequences/sample) than did any of the NICU surfaces (mean range of 1600–3000 sequences/sample) (Table 1, Figure 1), suggesting that breastmilk contained higher fungal loads than did the NICU surfaces.
Table 1:
Fungal DNA detection on NICU surfaces and in breastmilk.
| Sample type (number) | Sample number positive by qPCR1 (%) | Mean sequence reads/sample (range, number of samples sequenced)2 |
|---|---|---|
| Computer mouse (30) | 22 (73) | 1738 (range 166–6447, n=26) |
| Computer keyboard (30) | 21 (70) | 2986 (range 238–22075, n=22) |
| Incubator handles (31) | 21 (68) | 1938 (range 196–12858, n=28) |
| Stethoscope diaphragm (32) | 20 (61) | 1609 (range 268–5791, n=25) |
| Breastmilk (6) | 6 (100) | 11538 (range 320–34839, n=6) |
| “No template” control (20) | 2 (10) | 50 (range 1–143, n=4) |
| Breastmilk - no lysis control (12) | 0 (0) | NA |
Cq values indicated fungal DNA amounts above the limit of detection
NICU surfaces and breastmilk sequencing information is included only for samples that passed quality control as described in Materials and Methods
Figure 1.
Breastmilk samples yield significantly more fungal sequencing reads than NICU surfaces. Box-whisker plot of the number of sequence reads from each sample type. Mean sequence reads for each sample type are noted in Table 1.
3.2. Fungal DNA detected in breastmilk is due to the presence of fungal cells.
To determine if the fungal DNA in breastmilk represents DNA from fungal cells versus free fungal DNA, whole breastmilk samples were analyzed by qPCR. We found that none of the whole breastmilk samples generated a qPCR signal (Table 1). In contrast, qPCR signals were obtained for all of these breastmilk samples, including replicates, which had been subjected to cell lysis as part of the DNA isolation process (mean Cq 35.0), as well as from whole breastmilk not subjected to cell lysis and spiked with 300 ng of isolated fungal DNA (data not shown). In addition, negative controls using water instead of sample solutions did not generate a signal, indicating that PCR and lysis reagents were not contaminated with fungal DNA. These results indicate that the fungal DNA detected in breastmilk by qPCR was contained within fungal cells and was not cell-free DNA.
3.3. Mycobiomes of NICU surfaces and breastmilk are different.
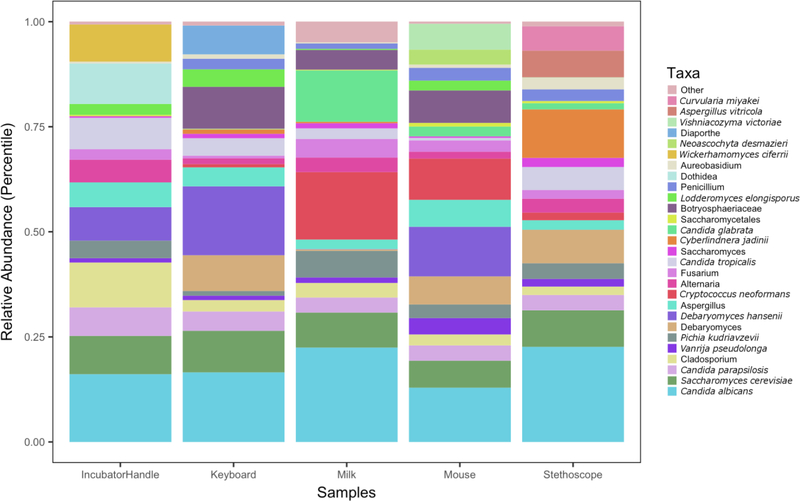
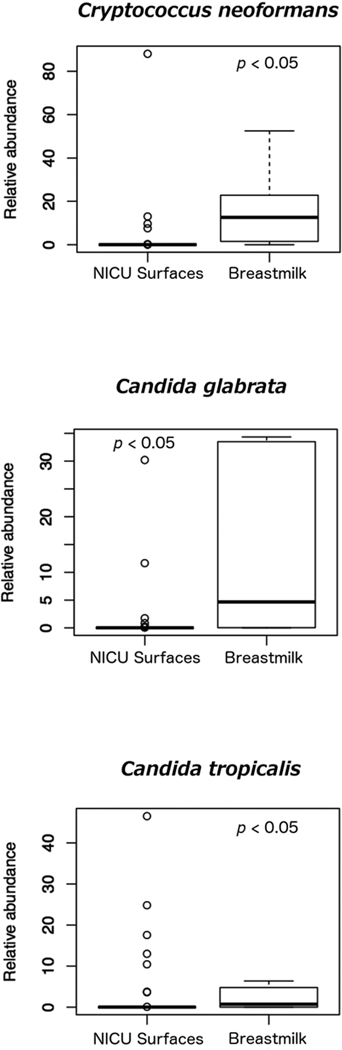
We characterized fungal communities of NICU surfaces and breastmilk samples by using amplicon-based sequencing of the fungal ITS2. Overall, mycobiomes in both sample types were diverse (Figure 2). Taxa that were both prevalent and abundant in breastmilk were Candida albicans, Candida parapsilosis, Cryptococcus neoformans, Saccharomyces cerevisiae, and Candida glabrata (Tables 2 and 3). For NICU surfaces, the most prevalent and abundant taxa also included Candida albicans, Candida parapsilosis, and Saccharomyces cerevisiae. In contrast to breastmilk, NICU surface mycobiomes did not exhibit a substantial presence of Cryptococcus neoformans, but did contain Vanrija pseudolonga (homotypic synonym Cryptococcus pseudolongus (Liu et al., 2015)). Overall, the relative abundances of 21 taxa differed significantly between NICU surfaces and breastmilk (Figure 3 and Supplemental Figure 6, p < 0.05). Taxa of particular interest due to their association with human disease that differed between the two groups were Candida glabrata, Candida tropicalis, and Cryptococcus neoformans (Figure 3). All three taxa had greater relative abundances in breastmilk than on NICU surfaces.
Figure 2.
Relative abundances of fungal taxa on NICU surfaces and in breastmilk. Mean fungal sequence reads are displayed as percentages of the total number of sequence reads for each sample type. The relative abundance taxonomy plots for individual samples from each site are shown in Supplementary Figures 1–5.
Table 2:
Top fungal taxa prevalence
| Taxa | Sample prevalence |
|---|---|
| Breastmilk | Total sample number = 6 |
| Candida albicans | 6 |
| Candida parapsilosis | 6 |
| Vanrija pseudolonga | 5 |
| Cryptococcus neoformans | 5 |
| Saccharomyces cerevisiae | 5 |
| Environment | Total sample number = 101 |
| Candida albicans | 93 |
| Order Saccharomycetales | 71 |
| Candida parapsilosis | 65 |
| Vanrija pseudolonga | 54 |
| Saccharomyces cerevisiae | 47 |
Table 3:
Top fungal taxa abundance
| Taxa | Mean relative abundance (percent) |
|---|---|
| Breastmilk | |
| Candida albicans | 28.5 |
| Cryptococcus neoformans | 17.0 |
| Candida glabrata | 12.9 |
| Saccharomyces cerevisiae | 8.8 |
| Candida krusei | 5.4 |
| Environment | |
| Candida albicans | 48.5 |
| Saccharomyces cerevisiae | 12.3 |
| Candida parapsilosis | 9.2 |
| Vanrija pseudolonga | 3.5 |
| Genus Cladosporium | 3.4 |
Figure 3.
Relative abundances of Cryptococcus neoformans, Candida glabrata, and Candida tropicalis are significantly higher for breastmilk as compared to NICU surfaces. Additional taxa that differed significantly between NICU surfaces and breastmilk are shown in Supplemental Figure 6.
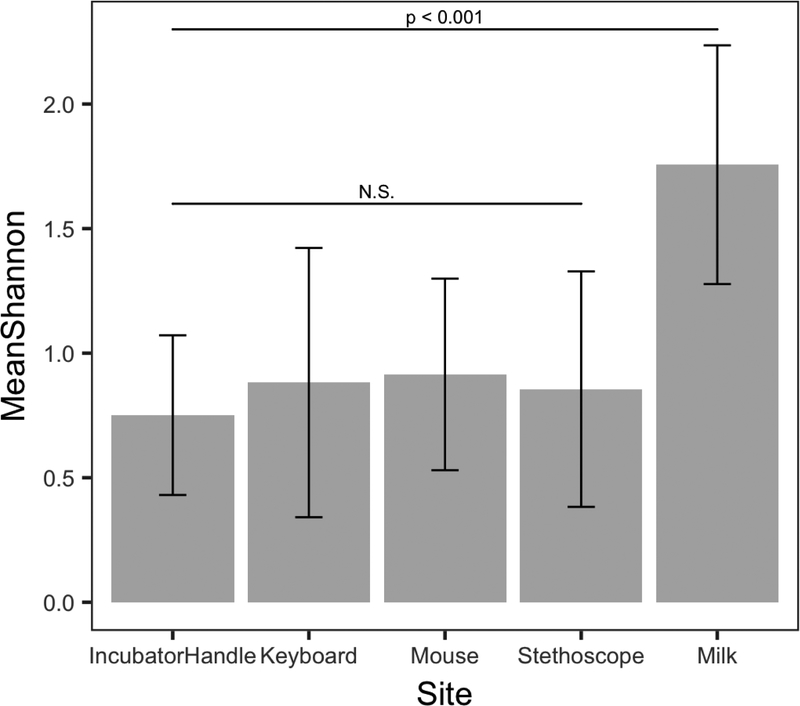
To assess the fungal diversity of individual sample types, we determined both the number of taxa/sample as well as calculated alpha diversity using the Shannon diversity index, which takes into account both the number of taxa as well as their relative abundances in a sample. Neither of these measures showed significant differences (p > 0.05) among NICU surface types (e.g. stethoscope versus computer keyboard) or among dates of collection. In contrast, fungal diversity (both Shannon indices (Figure 4) and number of taxa (data not shown)) was significantly different between NICU surfaces and breastmilk, with greater fungal diversity observed in breastmilk (all p-values < 0.001). This higher diversity is visible in the relative abundance plots of individual sample types (Supplemental Figures 1–5), where the NICU surfaces tend to be dominated by a few major taxa, whereas the milk samples show a more even distribution of taxa.
Figure 4.
Breastmilk has significantly higher fungal diversity than any NICU surface (p<0.001 for all comparisons). Mean Shannon diversity indices are shown and significant differences between sample types were determined using Student’s t-test. Fungal diversity of NICU surfaces did not significantly differ depending on the particular surface. N.S., not significantly different with p > 0.05 for all comparisons.
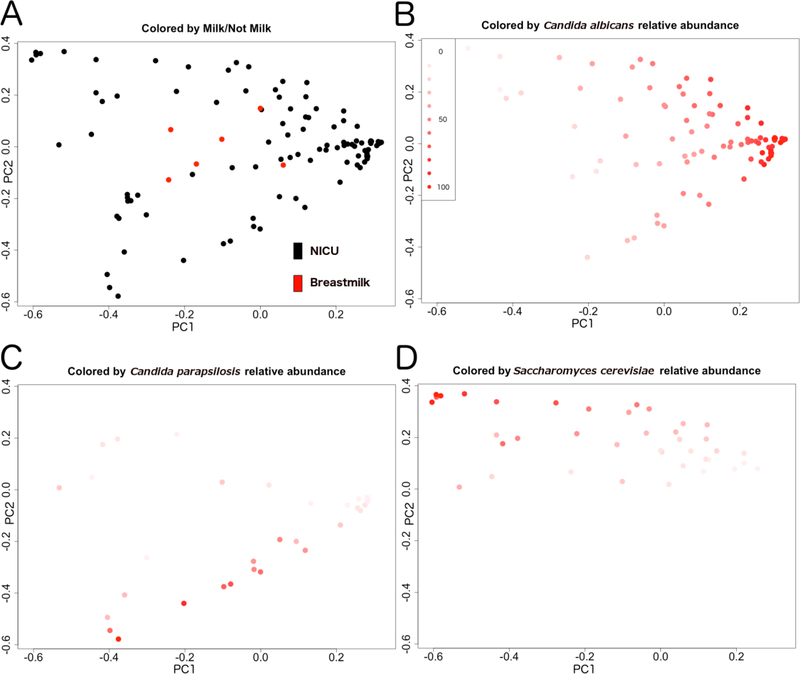
Beta diversity, or community compositional differences, between samples was assessed using the Bray-Curtis dissimilarity index. We found that fungal community structures of breastmilk differed significantly from those of NICU surfaces. A Principal Coordinates Analysis (PCoA) plot of between-sample Bray-Curtis distances of the species-level taxon relative abundances demonstrates these community differences visually, with breast milk fungal communities being clustered more centrally in the plot (red dots, Figure 5A). We identified the three most abundant taxa (Candida albicans, Candida parapsilosis, and Saccharomyces cerevisiae) across samples, and colored the plot points according to the relative abundances of these taxa in order to visualize covariation among them (Figure 5 B–D). The relative abundance of Candida albicans was strongly associated with the first principal axis of variation. Thus, Candida albicans appears to be the primary driver of the variation in the cohort. For example, the least diverse samples tended to be those with the highest Candida albicans abundance (negative correlation, Supplemental Figure 7).
Figure 5.
Beta-diversity comparisons of fungal communities. PCoA of Bray-Curtis distances for (A) breastmilk (red dots) and NICU surface (black dots) samples. (B-D) Each sample (dot) is colored based on relative abundances of Candida albicans (B), Candida parapsilosis (C), and Saccharomyces cerevisiae (D), with darkest red indicating highest relative abundance values and lightest red representing lowest relative abundance, as indicated in the common heatmap shown in panel B. The proportion of variance explained by each principal coordinate (all plots) is 30.6 for PC1 and 17.0 for PC2.
4. Discussion
To our knowledge, there has been only one previous study that analyzed fungal communities of NICU surfaces (Bokulich et al., 2013) and, in that study, DNA from most NICU surfaces failed to amplify with primers targeting ITS of the fungal rDNA region. However, the primers used by Bokulich et al (BITS and B58S3) also failed to produce amplicons in another study (Huseyin et al., 2017) in which several alternative ITS primer pairs were able to successfully amplify fungal DNA, including the primer pair utilized in our study (UNI1 and UNI2). For the small number of surfaces that did amplify in the study by Bokulich et al, four fungal taxa were identified and these were also found in the samples analyzed in our study (species Candida albicans and Saccharomyces cerevisiae, and genera Debaryomyces and Cryptococcus). A study of non-hospital (residential) surfaces also reported fungal communities on indoor surfaces that appeared to originate from both indoor and outdoor air sources (Adams et al., 2013). Similar to what is reported here, these indoor fungal communities did not significantly differ by surface type.
A majority of infants, including those born prematurely and hospitalized in NICUs, receive human breastmilk feedings during the first months of life. Breastmilk provides an important early source of microbes for the establishment of gut microbiota as well as nutrients and bioactive molecules that potentially modulate microbial community composition of the infant gut s (Ballard and Morrow, 2013; Mueller et al., 2015a). Only one prior study has reported the presence of fungi in breastmilk (Boix-Amoros et al., 2017). In this study of healthy lactating women, viable fungi were detected in breastmilk by culture, PCR, fluorescence-in-situ hybridization, and sequencing. The most abundant and prevalent fungal genera in breastmilk included Malassezia, Candida, and Saccharomyces. In the current study, we also identified Candida and Saccharomyces species as being abundant and prevalent. The finding that we did not identify Malassezia species in breastmilk may be related to the small sample size, differences in donor cohort (e.g. mothers delivering preterm versus at term), differences in collection methods (more or less skin microbes being present in the milk, for example), and/or differing fungal-specific primer sets used in our present study.
In our study, we found that Candida albicans, Candida parapsilosis, and Saccharomyces cerevisiae were the fungi most responsible for driving the structures of mycobiota on NICU surfaces and in breastmilk. Of these, Candida albicans and Candida parapsilosis are the predominant fungal colonizers of humans, as well as causes of superficial and invasive fungal infections. Relative abundances of Candida albicans and Candida parapsilosis are the primary drivers of mycobiome structures in the premature infant gut (Heisel et al., 2015). Consequently, results of our study provide evidence that Candida species that preferentially associate with infants are present in environmental mycobiota that neonates are initially exposed to, and their relative amounts are important determinants of overall mycobiome composition for NICU surfaces, breastmilk and the infant intestine.
Despite having common taxa being primarily responsible for the overall structure of the community, we found that mycobiomes of NICU surfaces and breastmilk are different. On NICU surfaces, fungal compositions were homogenous, being largely dominated by a single taxon. In contrast, fungal communities of breastmilk were heterogeneous and had significantly greater relative abundances of many fungal taxa. The difference in community structures likely reflects distinct microbial growth opportunities offered by the particular environmental niches. For example, in the NICU, surfaces are cleaned regularly to inhibit growth and proliferation of microbes, whereas breastmilk is nutrient-rich and originates from a human source, which harbors its own microbial communities. Additionally, the surfaces selected for this study are all regularly touched by infant caregivers, thus, there could be transfer of skin fungi to abiotic surfaces.
Three human-associated fungi in particular, Cryptococcus neoformans, Candida glabrata, and Candida tropicalis, were observed to have significantly higher relative abundances in breastmilk as compared to NICU surfaces. We previously showed that Candida species, including C. albicans and C. parapsilosis, and Cryptococcus species were present in the oral and anal mycobiomes of term-born infants during the first month of life (Ward et al., 2017) and in the feces of premature infants (Heisel et al., 2015). Thus, there appears to be overlap between fungi present within infant mycobiomes along the alimentary tract and those that are enriched in breastmilk. In addition, we found that S. cerevisiae was abundant and prevalent in breastmilk, which may reflect dietary ingestion (bakers and/or brewers yeast) by the mother, with transmission from the maternal gut to the milk via the entero-mammary pathway, a mechanism proposed for breastmilk bacterial microbiome establishment (Rodriguez, 2014). Additional potential sources of breastmilk fungi include the skin of the areola/nipple and the infant oral cavity. Further studies are needed to determine if breastmilk truly harbors its own mycobiota and if specific fungal strains associated with breastmilk/breastfeeding are transmitted to the infant gut, as has been described for bacterial strains (Ferretti et al., 2018).
One limitation of our study is that highly sensitive DNA-based determinations of microbial presence do not provide information about organism viability. Thus, it is possible that our determinations of fungal taxa presence on NICU surfaces and in breastmilk represent non-viable fungal cells, viable fungal cells, or a combination of both. For breastmilk, we think that our results indicate the presence of at least some amount of viable fungal cells, based on the study of Boix-Amoros, et al (2017), which showed that viable fungi were recovered from 1 ml of breastmilk for 41% (17 out of 41) of the samples. We were not able to recover fungi from the breastmilk samples in our study, but this was likely due to the fact that we only had a very small quantity (100 ul) of breastmilk available for culturing. Another limitation of our study is that no sampling of the hands of hospital staff or family members was performed, so we were unable to evaluate the extent to which skin fungi are transferred to NICU surfaces.
In summary, NICU surfaces and human milk contain fungal DNA and, in particular, evidence of fungal species that have previously been shown to be abundant and prevalent in the infant gut. These potential reservoirs of fungi may provide an early source for mycobiome establishment in hospitalized newborn infants. Further studies are needed to determine if mother’s milk, as compared to other types of feeding, affects gut mycobiome composition and if mycobiomes established during NICU hospitalization are associated with relevant effects on health for infants and for the longer term.
Supplementary Material
Supplemental Figure 1. Relative abundance plot of fungal taxa identified from sequencing results of incubator handle samples.
Supplemental Figure 2. Relative abundance plot of fungal taxa identified from sequencing results of computer keyboard samples.
Supplemental Figure 3. Relative abundance plot of fungal taxa identified from sequencing results of computer mouse samples.
Supplemental Figure 4. Relative abundance plot of fungal taxa identified from sequencing results of stethoscope samples.
Supplemental Figure 5. Relative abundance plot of fungal taxa identified from individual breastmilk samples.
Supplemental Figure 6. Box-whisker plots showing fungal taxa (other than those shown in Figure 3) that had significantly different relative abundances on NICU surfaces as compared to breastmilk. Most taxa exhibited a higher abundance in breastmilk as compared to NICU surfaces. The one taxon found to have higher relative abundance in NICU surface samples was Cladosporium velox.
Supplemental Figure 7. Plot comparing Shannon’s diversity index to relative abundance of Candida albicans for each sample. Line drawn is the line of best fit, as determined using the linear modeling function in R. The correlation value (−0.35) is indicated on the figure.
Highlights.
Breastmilk and the NICU surfaces harbor mycobiomes that could provide early sources of colonizing fungi for hospitalized infants.
Compositions of human-associated mycobiomes (breastmilk) are significantly different from those of abiotic (hospital) surfaces.
5. Acknowledgements
This study was supported in part by NIH award R21AI139730 (CAG) and a University of Minnesota Academic Health Center Faculty Research Development award (CAG).
Abbreviations:
- NICU
(Neonatal Intensive Care Unit)
- ITS
(Internal Transcribed Spacer Region)
- PERMANOVA
(Permutational Multivariate Analysis of Variance Using Distance Matrices)
- PCoA
(Principal Coordinates Analysis)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams RI, et al. , 2013. The diversity and distribution of fungi on residential surfaces. PLoS One. 8, e78866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghalith G, Knights D, BURST: an ultrafast optimal aligner for mapping large NGS data to large genome databases. 2017. [Google Scholar]
- Al-Ghalith GA, et al. , 2018. SHI7 Is a Self-Learning Pipeline for Multipurpose Short-Read DNA Quality Control. mSystems. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta MC, et al. , 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 7, 307ra152. [DOI] [PubMed] [Google Scholar]
- Ballard O, Morrow AL, 2013. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. 60, 49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendel CM, 2011. Infectious Diseases of the Fetus and Newborn Infant: Section IV Chapter 33: Candidiasis. Elsevier Saunders. [Google Scholar]
- Boix-Amoros A, et al. , 2017. Multiple approaches detect the presence of fungi in human breastmilk samples from healthy mothers. Sci Rep. 7, 13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, et al. , 2013. Surface microbes in the neonatal intensive care unit: changes with routine cleaning and over time. J Clin Microbiol. 51, 2617–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B, et al. , 2014. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B, et al. , 2017. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat Commun. 8, 1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, et al. , 2012. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 488, 621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello M, et al. , 2013. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Research. 41, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, et al. , 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 158, 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, et al. , 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl Acad Sci USA. 108, 3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, et al. , 2014. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. mBio. 5, e01011–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, et al. , 2018. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 24, 133–145 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel T, et al. , 2017. High-fat diet changes fungal microbiomes and inter-kingdom relationships in the murine gut. mSphere. 2, e00351–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisel T, et al. , 2015. Complementary amplicon-based genomic approaches for the study of fungal communities in humans. PLoS one. 10, e0116705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseyin CE, et al. , 2017. The Fungal Frontier: A comparative analysis of methods used in the study of the human gut mycobiome. Front Microbiol. 8, 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, et al. , 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 336, 1314–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang TT, et al. , 2017. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. 22, 809–816 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa PS, et al. , 2014. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci USA. 111, 12522–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, et al. , 2015. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol. 81, 85–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P, 1970. Cryobiology: the freezing of biological systems. Science. 168, 939–49. [DOI] [PubMed] [Google Scholar]
- Mazur P, 1977. The role of intracellular freezing in the death of cells cooled at supraoptimal rates. Cryobiology. 14, 251–72. [DOI] [PubMed] [Google Scholar]
- Mueller NT, et al. , 2015a. The infant microbiome development: mom matters. Trends Mol Med. 21, 109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller NT, et al. , 2015b. Prenatal exposure to antibiotics, cesarean section and risk of childhood obesity. Int J Obes (Lond). 39, 665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, 2018. vegan: Community Ecology Package. https://cran.r-project.org/web/packages/vegan/index.html [Google Scholar]
- Pannaraj PS, et al. , 2017. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 171, 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, et al. , 2014. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front Microbiol. 5, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, 2014. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr. 5, 779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TL, et al. , 2018. Development of the human mycobiome over the first month of life and across body sites. mSystems. 3, e00140–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Relative abundance plot of fungal taxa identified from sequencing results of incubator handle samples.
Supplemental Figure 2. Relative abundance plot of fungal taxa identified from sequencing results of computer keyboard samples.
Supplemental Figure 3. Relative abundance plot of fungal taxa identified from sequencing results of computer mouse samples.
Supplemental Figure 4. Relative abundance plot of fungal taxa identified from sequencing results of stethoscope samples.
Supplemental Figure 5. Relative abundance plot of fungal taxa identified from individual breastmilk samples.
Supplemental Figure 6. Box-whisker plots showing fungal taxa (other than those shown in Figure 3) that had significantly different relative abundances on NICU surfaces as compared to breastmilk. Most taxa exhibited a higher abundance in breastmilk as compared to NICU surfaces. The one taxon found to have higher relative abundance in NICU surface samples was Cladosporium velox.
Supplemental Figure 7. Plot comparing Shannon’s diversity index to relative abundance of Candida albicans for each sample. Line drawn is the line of best fit, as determined using the linear modeling function in R. The correlation value (−0.35) is indicated on the figure.
Data Availability Statement
Sequence data for all samples in this project is available in the NCBI Sequence Read Archive (SRA) database under BioProject ID PRJNA505509.