Abstract
The 3′ poly(A) tail of mRNAs is fundamental to regulating eukaryotic gene expression. Shortening of the poly(A) tail, termed deadenylation, reduces transcript stability and inhibits translation. Nonetheless, the mechanism for poly(A) recognition by the conserved deadenylase complexes, Pan2–Pan3 and Ccr4–Not, is poorly understood. Here we provide a model for poly(A) RNA recognition by two DEDD deadenylase enzymes, Pan2 and the Ccr4–Not nuclease Caf1. Crystal structures of S. cerevisiae Pan2 in complex with RNA show that, surprisingly, Pan2 does not form canonical base-specific contacts. Instead, it recognizes the intrinsic stacked, helical conformation of poly(A) RNA. Using a fully reconstituted biochemical system, we show that disruption of this structure, for example by guanosine incorporation into poly(A), inhibits deadenylation by both Pan2 and Caf1. Together, these data establish a paradigm for specific recognition of the conformation of poly(A) RNA by proteins that regulate gene expression.
Almost every eukaryotic messenger RNA (mRNA) has a 3′ polyadenosine (poly(A)) tail, which is a major determinant of gene expression1. The poly(A) tail protects the transcript body from exonucleolytic degradation and is bound by the essential cytoplasmic poly(A) binding protein Pab1 (PABPC1)2,3, which in turn is important for efficient translation initiation4 and transcript stability5. Canonical cytoplasmic mRNA decay is initiated by controlled poly(A) tail shortening, or deadenylation, followed by decapping and further exonucleolytic decay6,7. This leads to the repression of gene expression by reducing transcript half-life and by inhibiting translation initiation through the release of Pab1 (PABPC1)8. Given the importance of the poly(A) tail in multiple steps of gene expression, it is unsurprising that deadenylation is highly regulated, playing important roles in multiple physiological processes including development, response to stress, and the circadian clock9.
In eukaryotes, deadenylation is primarily carried out by two conserved multi-protein complexes: Pan2–Pan3 and Ccr4–Not10. Pan2–Pan3 is a heterotrimeric complex consisting of two Pan3 subunits and one Pan2 subunit, the latter containing an exonuclease domain11–13. Ccr4–Not contains seven core protein subunits, including two exonuclease enzymes, Caf1 (Pop2, CNOT7, or CNOT8) and Ccr4 (CNOT6 or CNOT6L)10. The specification of transcripts for deadenylation can be mediated through adapter proteins that interact with both target RNA sequences and the enzyme complexes. Nonetheless, it is largely unknown how the individual nucleases, in particular Caf1 and Pan2 of the DEDD exonuclease family, recognize the poly(A) tail, why the deadenylases do not degrade the entire mRNA, and how recently identified non-A nucleotides in the poly(A) tail14,15 affect the activity of deadenylase complexes.
Here we investigate the mechanistic basis of deadenylation specificity. Crystal structures of the Pan2 exonuclease in complex with RNA, together with biochemical and biophysical studies, demonstrate that Pan2 specificity is determined by recognition of a specific conformation adopted by poly(A) RNA. Pan2 and the structurally-related DEDD family deadenylase Caf1 are inhibited by the disruption of this unique structure. This contrasts with canonical mechanisms of sequence-specific RNA recognition used, for example, by cytoplasmic and nuclear poly(A) binding proteins16,17, where the proteins form contacts with base-specific functional groups. Thus, our data are consistent with the stacked, helical structure of polyadenosines acting as a determinant of recognition in certain conserved regulators of gene expression.
Results
3′ guanosines inhibit the Pan2 exonuclease
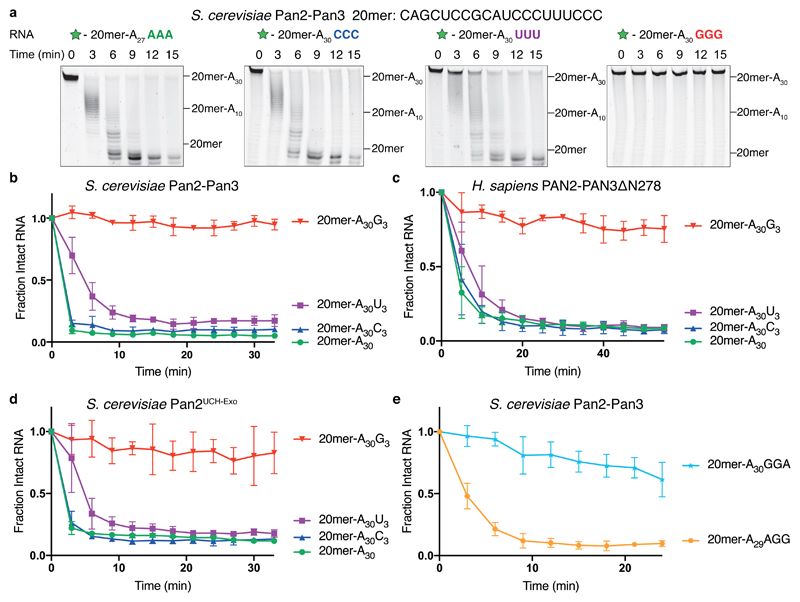
To determine the specificity of the Pan2–Pan3 deadenylation complex, we carried out in vitro assays (Supplementary Fig. 1a) with recombinant proteins on 5′ fluorescently-labeled RNA substrates, which consist of a 3′ UTR-like sequence of 20 non-A nucleotides (20mer) and a downstream 30-nucleotide poly(A) tail with different 3′ termini. Specifically, for each RNA substrate, the three terminal nucleotides (positions -2, -1, and +1, where cleavage occurs between -1 and +1) were replaced by other sequences. Reaction products were analyzed by denaturing polyacrylamide gel electrophoresis.
Relative to an RNA with only adenosines in the 3′ tail, we found that 3′ guanosines, but not cytosines or uracils, strongly inhibited Saccharomyces cerevisiae and Homo sapiens Pan2–Pan3 (Fig. 1a-c). Consistent with an ability to remove non-adenosine nucleotides, Pan2–Pan3 also degraded the upstream 3′ UTR-like sequence. To elucidate whether Pan3 is required for the observed nucleotide specificity, we performed assays with a construct of Pan2 including only the pseudoubiquitin C-terminal hydrolase and exonuclease domains (residues 461–1115, hereafter referred to as Pan2UCH-Exo, Supplementary Fig. 1b-c). The UCH domain was included in the construct because it had been proposed that a UCH loop folds into the active site to contact the RNA substrate12. Pan2UCH-Exo recapitulated the specificity of the full complex (Fig. 1d), suggesting that 3′ nucleotide specificity is intrinsic to the Pan2 exonuclease.
Figure 1. 3′ guanosines inhibit the Pan2 exonuclease.
a, Denaturing RNA gels showing deadenylation by recombinant S. cerevisiae Pan2–Pan3 on 5′ 6-FAM-labeled (green star) RNA substrates consisting of a 20mer non-poly(A) sequence (shown above) followed by a poly(A) tail of 30 adenosines. Where indicated, the substrate contains three additional non-A nucleotides at the 3′ end. These gels are representative of identical experiments performed 3 times. Uncropped gel images are shown in Supplementary Data Set 1. b-e, Analysis of deadenylation on poly(A) substrates with different 3′ nucleotides. Disappearance of the intact substrate was quantified by densitometry of the fluorescently labeled, full-length RNA. Data points were normalized to time = 0, and are connected by straight lines for clarity. Assays were carried out in triplicate (n = 3 independent experiments), the data points shown represent the mean, and error bars represent standard deviation. Assays are shown for full-length S. cerevisiae Pan2–Pan3 (b, e); H. sapiens PAN2–PAN3∆N278 (c); and S. cerevisiae Pan2UCH-Exo (residues 461-1115) (d).
We next investigated how different 3′ guanylation patterns affected Pan2 deadenylation activity. If the two nucleotides 5′ of the cleavage site were guanosines (-GGG, -GGA), there was a stronger inhibitory effect than if only one of these positions was a guanosine (-AGG, -AGA, -GAA) (Supplementary Fig. 1d-f). The identity of the nucleotide 3′ of the cleavage site was less important. For instance, 20mer-A30GGA was deadenylated more slowly than 20mer-A30GG despite both containing two consecutive guanosines near the 3′ end (Fig. 1e). Thus, Pan2 exhibited specificity against guanosines in a number- and position-dependent manner.
Using TAIL-seq, a recent study demonstrated that single guanosines were intermittently incorporated into poly(A) tails at low frequency (up to 5% of mRNAs for some classes of transcripts) in human cells14,15. To test the effect of a single interrupting guanosine on Pan2 activity, we compared deadenylation of two different RNAs that were differentially labeled (Supplementary Fig. 1g). While one RNA contained only adenosines in the 3′ tail (red), the other contained the same poly(A) tail interrupted by a single guanosine (blue). Strikingly, this two-color deadenylation assay showed that one guanosine was sufficient to inhibit Pan2. This suggests that, within a pool of RNAs, physiologically relevant single-guanosine incorporation into the poly(A) tail can inhibit Pan2 activity.
Guanosines inhibit both exonucleases of the Ccr4–Not complex
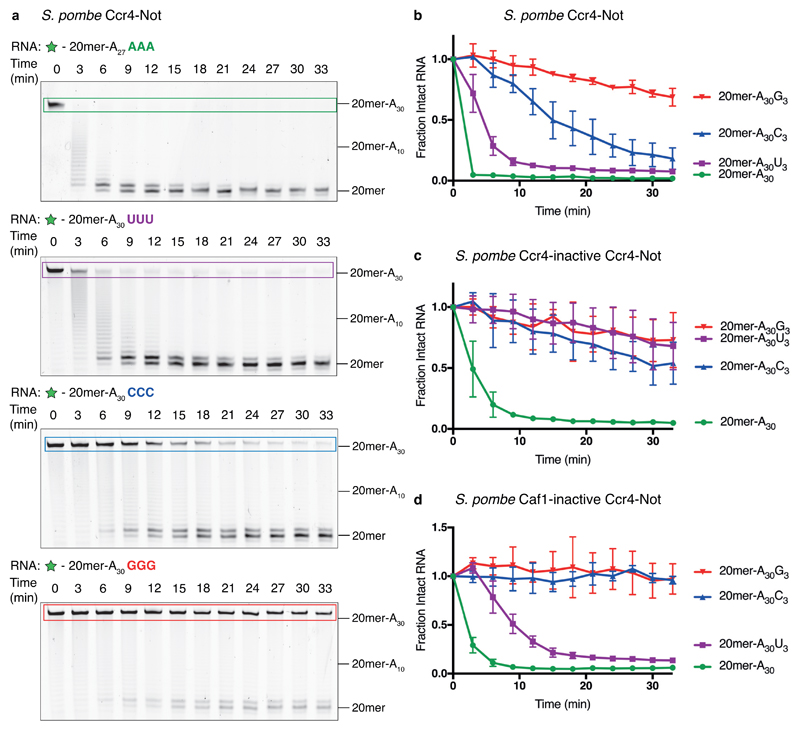
To determine the nucleotide specificity of the other major deadenylases, Caf1 and Ccr4 in the context of the full Ccr4–Not complex, we carried out deadenylation assays with recombinant Schizosaccharomyces pombe Ccr4–Not18. Wild-type Ccr4–Not was moderately inhibited by 3′ cytosines and strongly inhibited by 3′ guanosines (Fig. 2a-b). Thus, the wild-type Ccr4–Not complex has a different nucleotide specificity compared to Pan2–Pan3.
Figure 2. Ccr4–Not is inhibited by 3′ guanosines.
a, Denaturing RNA gels showing deadenylation by recombinant S. pombe Ccr4–Not on 5′ 6-FAM-labeled (green star) RNA substrates consisting of a 20mer non-poly(A) sequence (See Fig. 1a) followed by 30 adenosines. Where indicated, the substrate contains three additional non-A nucleotides at the 3′ end. These gels are representative of identical experiments performed 3 times. Uncropped gel images are shown in Supplementary Data Set 1. b-d, Analysis of deadenylation on poly(A) substrates with different 3′ nucleotides. Disappearance of the intact substrate was quantified by densitometry of the fluorescently labeled, full-length RNA. Data points were normalized to time = 0, and are connected by straight lines for clarity. Assays were carried out in triplicate (n = 3 independent experiments), the data points shown represent the mean, and error bars represent standard deviation. Assays are shown for wild-type S. pombe Ccr4–Not (b), Ccr4-inactive Ccr4–Not (c) and Caf1-inactive Ccr4–Not (d).
To determine the specificities of the individual nucleases, we carried out deadenylation assays using recombinantly expressed and purified Ccr4–Not complexes with catalytic point mutations in either Ccr4 or Caf1. The complex containing a catalytically inactive Ccr4 (only Caf1 active) showed stringent nucleotide specificity and was strongly inhibited by all substrates containing three terminal non-A nucleotides (Fig. 2c). The complex containing a catalytically inactive Caf1 (only Ccr4 active) was strongly inhibited by terminal guanosines and cytosines but was relatively unaffected by uracils (Fig. 2d). Hence, Ccr4 likely accounts for the ability of the wild-type complex to rapidly remove terminal uracils (Fig. 2b). Therefore, within the Ccr4–Not complex, the individual nucleases Caf1 and Ccr4 have distinct specificities. Strikingly, all tested deadenylases (Pan2, Caf1 and Ccr4) were strongly inhibited by 3′ guanosines.
Pan2 contacts the ribophosphate backbone of poly(A) RNA
Several crystal structures have previously been reported for Pan2 and Caf1, but none of these contained RNA in the active site. To elucidate the mechanistic basis of Pan2 nucleotide specificity, we crystallized a catalytically inactive mutant (E912A) of S. cerevisiae Pan2UCH-Exo (Supplementary Fig. 2a-c) which diffracted to 3.0 Å resolution (Table 1). We determined a structure of apo Pan2UCH-Exo, which was similar to a previously determined structure (PDB: 4Q8H)13 with a backbone RMSD of 0.51 Å (Supplementary Fig. 2d).
Table 1. Data collection and refinement statistics.
| Pan2UCH-Exo(6R9I) | Pan2UCH-Exo-A7(6R9J) | Pan2UCH-Exo-AAGGAA (6R9M) | Pan2UCH-Exo-AAGGA (6R9O) | Pan2UCH-Exo-AAUUAA (6R9P) | Pan2UCH-Exo-AACCAA (6R9Q) | |
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | I222 | I222 | I222 | I222 | I222 | I222 |
| Cell dimensions | ||||||
| a, b, c (Å) | 91.37, 117.07, 255.32 |
91.48, 116.41, 257.83 |
91.41, 118.10, 257.28 |
90.76, 117.59, 256.57 |
91.88, 117.09, 255.89 |
92.19, 117.32, 256.66 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 106.41-3.00 (3.08-3.00) | 71.95-3.33 (3.42-3.33) | 86.17-3.33 (3.42-3.33) | 85.50-3.32 (3.41-3.32) | 106.45-2.98 (3.06-2.98) | 72.49-3.08 (3.16-3.08) |
| Rmerge* | 0.093 (1.761) | 0.095 (1.791) | 0.097 (1.527) | 0.085 (1.512) | 0.065 (1.207) | 0.086 (1.733) |
| I/σI* | 12.4 (1.3) | 8.3 (1.0) | 7.3 (1.2) | 12.0 (1.0) | 13.5 (1.1) | 12.7 (1.1) |
| CC1/2* | 0.998 (0.774) | 0.990 (0.590) | 0.983 (0.649) | 0.998 (0.624) | 0.999 (0.726) | 0.917 (0.638) |
| Completeness (%)* | 100.0 (99.9) | 99.5 (98.6) | 100.0 (99.9) | 100.0 (99.8) | 100.0 (99.1) | 100.0 (100.0) |
| Redundancy* | 6.6 (6.6) | 4.6 (4.5) | 6.5 (6.0) | 6.5 (6.3) | 6.6 (6.2) | 6.6 (6.3) |
| Refinement | ||||||
| Resolution (Å) | 3.00 | 3.33 | 3.33 | 3.32 | 2.98 | 3.08 |
| Observed reflections | 184847 | 94264 | 135553 | 135414 | 188815 | 172231 |
| Unique reflections | 27902 | 20507 | 20875 | 20744 | 28662 | 26217 |
| Rwork / Rfree | 0.2434/0.2895 | 0.2472/0.2982 | 0.2369/0.2953 | 0.2545/0.3092 | 0.2610/0.3086 | 0.2605/0.2970 |
| No. atoms | ||||||
| Protein | 4542 | 4458 | 4586 | 4457 | 4548 | 4590 |
| Ligand | / | A7 RNA: 107 | AAGGAA RNA: 87 | AAGGA RNA: 87 | AAUUAA RNA: 81 | AACCAA RNA: 81 |
| B-factors | ||||||
| Protein | 113.69 | 148.9 | 150.6 | 156.5 | 131.1 | 126.6 |
| Ligand | / | A7 RNA:241.3 | AAGGAA RNA: 254.9 | AAGGA RNA:296.6 | AAUUAA RNA: 258.6 | AACCAA RNA: 275.8 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.003 | 0.001 | 0.002 | 0.002 | 0.004 | 0.002 |
| Bond angles (°) | 0.651 | 0.419 | 0.507 | 0.483 | 0.804 | 0.484 |
Values in parentheses are for highest-resolution shell.
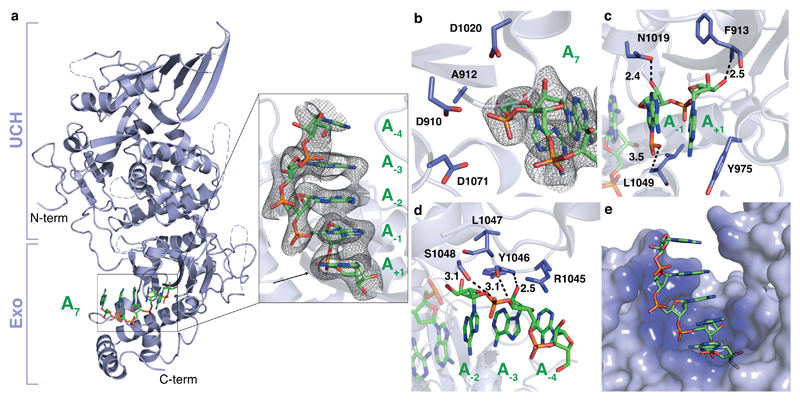
Next, we soaked A7 RNA into crystals of apo Pan2UCH-Exo, which diffracted to 3.3 Å resolution (Table 1). Electron density for five nucleotides is visible (Fig. 3a) and the RNA is bound in the active site of Pan2 without any substantial rearrangements of the protein (Supplementary Fig. 2e). The scissile phosphate bond of the RNA substrate is positioned to face the acidic amino acid side chains of the DEDD motif, but no catalytic metal ions19 are observed in the active site, consistent with the buffer conditions and the E912A mutation (Fig. 3b). Superposition with a CAF1 structure containing two divalent metal ions (PDB: 3G0Z)20 suggests that the conformation of RNA in the Pan2 structure is compatible with binding two catalytic metal ions in the Pan2 active site (Supplementary Fig. 3a).
Figure 3. Pan2 contacts the ribophosphate backbone of poly(A) RNA.
a, Crystal structure of Pan2UCH-Exo E912A (cartoon, blue) bound to A7 RNA (sticks, green). Unmodeled loops are represented by dashed lines. Inset: close-up view of the Pan2 exonuclease active site. The electron density represents a feature-enhanced map contoured to 1.3σ. Nucleotides are numbered relative to the scissile bond, which is indicated by an arrow. b, The electron density for the scissile phosphate bond and A7 substrate are shown in the Pan2 active site. The mesh represents electron density from a feature-enhanced map contoured to 1.8σ. c-d, Protein-RNA interactions between Pan2UCH-Exo E912A and oligo(A). Putative hydrogen bonds are indicated by dashed lines and interatomic distances are labeled in Ångstroms. e, Surface representation of Pan2UCH-Exo E912A (blue) bound to A7 (green). Pan2UCH-Exo E912A is colored by proximity to A7 from dark (<3 Å) to light (>7 Å) blue.
We observe several contacts between Pan2 and RNA. First, the terminal adenine is π-stacked against the aromatic ring of Y975 at the base of the active site (Fig. 3c). It would not be possible to accommodate an additional ribonucleotide 3′ to this adenosine, in agreement with the exo- (not endo-) nucleolytic activity of Pan2. Second, many of the RNA 2′ and 3′ hydroxyl groups, as well as phosphates, were in position to hydrogen bond with the Pan2 main chain (F913, Y1046, and L1049) or side chain atoms (N1019 and S1048, Fig. 3c-d). Strikingly, almost all contacts to the substrate RNA were mediated through the ribophosphate backbone (Fig. 3e, Supplementary Fig. 3b and Supplementary Movie 1).
The lack of base-specific interactions between Pan2UCH-Exo and adenines was surprising given that Pan2 displayed strong specificity against guanosines (Fig. 1). Upon closer examination, we found that oligo(A) formed a single-stranded A-form RNA helix in the Pan2 active site, where the adenine bases were π-stacked in an offset parallel structure and the RNA backbone geometry was consistent with a C3′-endo ribose pucker (Fig. 3a, e)21. Given that there are no canonical base-specific contacts, these results are consistent with Pan2 recognizing the conformation of poly(A) via backbone interactions, rather than directly recognizing the bases.
Since Caf1 is structurally homologous to the Pan2 exonuclease, it may also recognize the stacked, helical conformation of poly(A). We performed structural analysis on Caf1 to assess this. Docking of oligo(A) into the active site of a previously reported H. sapiens CAF1 structure by superposition20 shows no major clashes with the protein and is consistent with CAF1 recognizing the conformation of poly(A) using a similar mechanism to Pan2 (Supplementary Fig. 4a-b). However, poly(A) appears to be more buried within the CAF1 active site with amino acid residues in closer proximity to the bases (Supplementary Fig. 4a). These could potentially form base-specific contacts, providing an additional layer of specificity in CAF1 and resulting in its stricter preference for adenosine compared to Pan2 (Fig. 1, 2). Thus, recognition of the stacked, helical conformation of poly(A) as well as base-specific contacts likely contribute to CAF1 specificity for poly(A).
Guanosines disrupt the structure of poly(A) RNA in the Pan2 active site
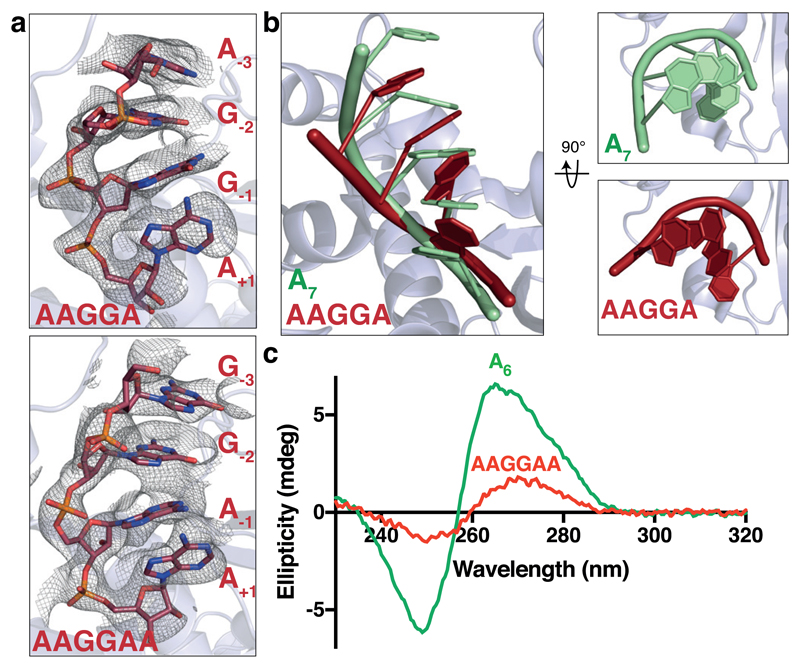
To understand why guanosines inhibit Pan2, we soaked AAGGA or AAGGAA RNAs into Pan2UCH-Exo crystals, which diffracted to 3.3 Å resolution (Table 1). We observed density for the four 3′ nucleotides of each of these RNAs (Supplementary Fig. 5 and Supplementary Movie 2). Binding of the guanosine-containing RNAs did not induce substantial rearrangements in Pan2 relative to the oligo(A)-bound Pan2 (Supplementary Fig. 2e). In contrast, both AAGGA and AAGGAA show disrupted parallel base-base π-stacking (Fig. 4a and Supplementary Fig. 5a-b). Moreover, the helical conformation of the ribophosphate backbone of guanosine-containing RNAs was distorted compared to that of oligo(A) (Fig. 4b). Finally, the terminal adenosines of G-containing RNAs appear to be more poorly stacked on Y975 at the base of the active site (Supplementary Fig. 5c-d).
Figure 4. Guanosines disrupt the conformation of oligo(A).
a, Crystal structure of Pan2UCH-Exo E912A (blue) bound to AAGGA (top) or AAGGAA (bottom) RNA (red). Close-up views of the RNA bound in the active site are shown with feature-enhanced maps contoured to 1.8σ. b, Superposition of A7 (green) and AAGGA (red) RNAs. The bases in G-containing RNAs are positioned at an angle relative to each other, disrupting the stacking geometry. c, Circular dichroism spectra of A6 (green) and AAGGAA (red) RNAs. These spectra are representative of identical experiments performed 2 times.
The amino acid corresponding to Y975 is a conserved aromatic residue in all Pan2 orthologs. We mutated this amino acid to an alanine and found that the mutant Pan2 had a much lower overall activity and a reduced ability to discriminate against guanosines (Supplementary Fig. 6). Base stacking on Y975 is therefore likely important for the activity and specificity of Pan2. Together, these data suggest that guanosines disrupt the stacked, helical conformation of poly(A) in the Pan2 active site.
The intrinsic structure of poly(A) in solution is disrupted by guanosines
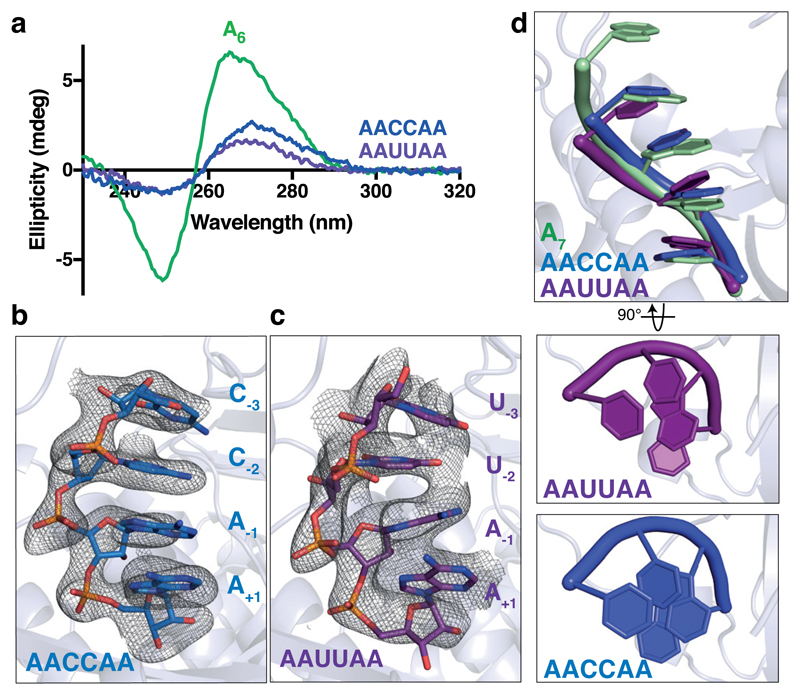
To examine the effect of guanosines on the conformation of poly(A) RNA in solution in the absence of protein binding, we used circular dichroism spectroscopy (CD). In CD studies of nucleic acids, the signal provided by chiral atoms is sensitive to the higher order geometry adopted by the polynucleotide22. The conformation of poly(A) gives rise to a characteristic spectrum with a maximum at ~264 nm and a minimum at ~249 nm (Fig. 4c), suggestive of a higher order, π-stacked structure23,24. This is consistent with previous experimental observations by CD25,26.
Introducing two guanosines into an oligo(A) RNA disrupted the CD signature and thus the helical conformation (Fig. 4c and Supplementary Fig. 7a). Structural changes occurred both 5′ and 3′ of the guanosines, as the reduction in signal was greater when the two guanosines were introduced in the middle of an oligo(A) RNA compared to introduction at the 5′ or 3′ end (Supplementary Fig. 7b). Therefore, oligo(A) RNA forms a π-stacked helical structure in solution in the absence of protein, and this intrinsic structure is disrupted by guanosines.
Cytosines and uracils can adopt a stacked, helical structure in the Pan2 active site
To understand the effect of cytosines and uracils on the solution conformation of poly(A), we repeated CD studies on oligo(A) RNAs containing interrupting cytosines or uracils. The introduction of Cs or Us into an oligo(A) also disrupts the stacked, helical signature characteristic of poly(A) to a similar degree as guanosines (Fig. 5a).
Figure 5. Cytosines and uracils stack to form a helical conformation in the Pan2 active site.
a, Circular dichroism spectra of A6 (green), AAUUAA (purple), and AACCAA (blue) RNAs. These spectra are representative of identical experiments performed 2 times. b-c, Crystal structure of Pan2UCH-Exo E912A (light blue) bound to AACCAA (dark blue, b) or AAUUAA (purple, c) RNA. The electron density of RNA bound in the active site is visualized with feature-enhanced maps contoured to 1.8σ. d, Superposition of A7 (green), AAUUAA (purple), and AACCAA (blue) RNAs. The bases in C- and U-containing RNAs are relatively well-stacked compared to G-containing RNAs (Fig. 4b) and are thus permissive to form the single-stranded RNA helix required for Pan2 activity.
Next, we soaked AACCAA and AAUUAA RNAs into the active site of Pan2UCH-Exo crystals. These diffracted to 3.1 and 3.0 Å resolution respectively (Table 1). Similar to the guanosine-containing RNAs, we observed four 3′ nucleotides and RNA soaking does not cause substantial changes in protein conformation. In contrast to AAGGA and AAGGAA, however, the bases are reasonably well-stacked (Fig. 5b-c), and the conformation of the ribophosphate backbone is more similar to that of the oligo(A) substrate (Fig. 5d). Therefore, the structures suggest that Pan2 can act on C- and U-containing RNAs because the cytidines and uridines can stack to adopt the single-stranded helical conformation in the Pan2 active site.
Base stacking is required for Pan2 and Caf1 deadenylation activity
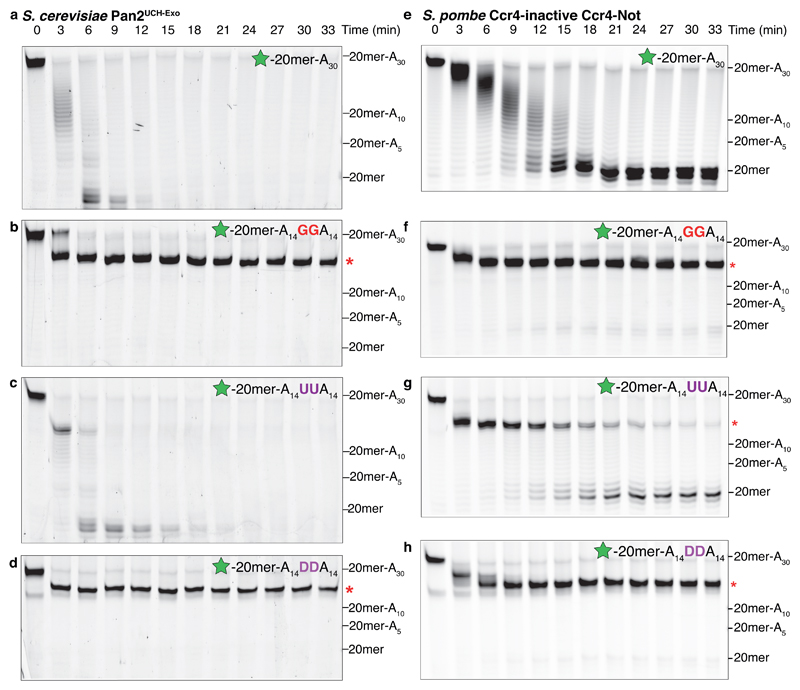
The disrupted stacking observed in guanosine-containing RNAs, both in the crystal structure and in CD experiments, suggests that guanosines may exert a long-range inhibitory effect on RNA backbone conformation and thus Pan2 deadenylation. To determine the range of guanosine influence on the poly(A) tail, we performed in vitro deadenylation assays with RNAs containing two guanosines in the middle of an A30 poly(A) tail. Relative to an RNA with only adenosines in the 3′ tail, Pan2UCH-Exo and Pan2–Pan3 were strongly inhibited when the guanosines were in the -4 and -5 positions (Fig. 6a-b, Supplementary Fig. 8a-c). Since the guanosines were distal to the scissile phosphate bond, Pan2 inhibition likely occurs due to an altered local RNA conformation.
Figure 6. Nucleotide base stacking is required for Pan2 and Caf1 deadenylase activity.
Denaturing RNA gels showing deadenylation by (a-d) S. cerevisiae Pan2UCH-Exo or (e-h) S. pombe Ccr4-inactive Ccr4–Not on 5′ 6-FAM-labeled (green star) RNAs consisting of a 20mer non-poly(A) sequence (see Fig. 1a) followed by the indicated tail sequence. RNAs either had no additional nucleotides (a, e), two guanosines (b, f), two uracils (c, g), or two dihydrouracils (abbreviated D, panels d, h) in the middle of the poly(A) tail. Red asterisks indicate the point of inhibition. Both Pan2 and Caf1 were strongly inhibited by guanosines and dihydrouracils interrupting a poly(A) tail. These gels are representative of identical experiments performed 2 times. Uncropped gel images are shown in Supplementary Data Set 1.
To further test the effect of disrupted base stacking on Pan2 activity, we designed an RNA substrate containing two dihydrouracils (DHU) within the poly(A) tail. DHU differs from uracil only in that it contains a C-C single bond between C5 and C6 instead of a C=C double bond (Supplementary Fig. 8d). Therefore, it does not adopt a planar conformation and disrupts stacking in oligonucleotides27. While the introduction of uracils had a mild inhibitory effect on Pan2 activity and caused a pause in deadenylation, the introduction of DHUs strongly inhibited Pan2 activity (Fig. 6c-d and Supplementary Fig. 8e-f). Pan2 was also strongly inhibited when the DHUs were distal to the active site (Supplementary Fig. 8g), suggesting long-range disruption of the RNA conformation. Thus, Pan2 activity requires the RNA substrate to adopt a locally stacked, helical conformation, and guanosines or DHUs strongly inhibit Pan2 activity by disrupting this conformation.
To test if Caf1 also recognizes the conformation of the RNA substrate and is thereby inhibited by guanosines (Fig. 2c), we repeated the deadenylation assays with a Ccr4-inactive mutant of Ccr4–Not (only Caf1 active). Similar to Pan2, Caf1 was strongly inhibited by guanosines or dihydrouracils interrupting a poly(A) tail (Fig. 6e-h), suggesting that base stacking and the unique conformation of poly(A) are also required for Caf1 activity. Caf1 showed more stringent specificity than Pan2 as interrupting uracils are more inhibitory (Fig. 6g). Thus, consistent with our structural superposition, the stacked, helical conformation of poly(A) as well as base-specific contacts likely contribute to the nucleotide specificity of Caf1.
Discussion
The poly(A) tail is found on almost every eukaryotic mRNA and controls gene expression at a post-transcriptional level. Despite being one of the most ubiquitous RNA sequences in the eukaryotic cell, it was not known how polyadenosines are specified by highly conserved deadenylase enzymes. Here we demonstrate that the major deadenylase complexes exhibit nucleotide specificity. Furthermore, we investigate the mechanistic basis of poly(A) recognition by the DEDD-family deadenylases Pan2 and Caf1. We show that the intrinsic conformation of poly(A) RNA contributes towards its recognition by these deadenylases.
Specificities of deadenylases prevent degradation of 3′ UTRs
The conformation of poly(A) RNA contributes to the specificity of both Caf1 and Pan2, but Caf1 (Fig. 2c) is more stringent against non-A nucleotides than Pan2 (Fig. 1d). Consistent with this, Pan2 can degrade the upstream 20mer RNA in vitro, whereas Ccr4–Not stops at the end of the poly(A) tail. The higher stringency of Caf1 can be explained by the proximity of amino acid side chains to the modeled oligo(A) substrate, which may additionally form base-specific interactions with adenines (Supplementary Fig. 4b). Ccr4 also has higher specificity for adenosines compared to Pan2 (Fig. 2d), which is likely mediated by base-specific contacts in the Ccr4 active site that select against pyrimidines and guanosines28.
The different stringencies of Pan2, Caf1, and Ccr4 may be evolutionarily linked to their in vivo roles. In a biphasic model of deadenylation29, Pan2–Pan3 removes the distal portion of the poly(A) tail, whereas Ccr4–Not removes adenosines proximal to the 3′ UTR. Caf1 and Ccr4 are hence more likely to encounter non-A nucleotides and may require more stringent base-specific contacts for recognizing adenosines to ensure they only act on the poly(A) tail. In contrast, in this model, Pan2 would most likely only encounter poly(A). Thus, it may rely on less stringent mechanisms, such as RNA conformation alone, to recognize the distal poly(A) tail. We speculate that the nucleotide specificity of deadenylases is related to their physiological roles, and the higher stringency of Ccr4–Not nucleases prevents degradation of the 3′ UTR. Inhibition of deadenylase complexes by guanosines may prevent spurious degradation of 3′ UTRs, allowing conventional mRNA decay to proceed.
Guanosines inhibit deadenylation
It was recently demonstrated by TAIL-seq that non-adenosine nucleotides can be incorporated into the poly(A) tails of some transcripts in human cells14,15. The addition of 3′ uracils to short poly(A) tails by TUT4 and TUT730 may stimulate decay through recruitment of the Lsm1-7–Pat1 complex to enhance decapping31,32 or through preferential degradation by Dis3L233,34. We show that uridylation has only a mild effect on both Pan2–Pan3 and Ccr4–Not, suggesting that 3′ uridylation alone may not strongly influence deadenylation.
A recent report identified TENT4A and TENT4B as non-canonical poly(A) polymerases in human cells responsible for the regulated addition of guanosines into poly(A) tails15. Transcripts with more frequently guanylated poly(A) tails showed increased stability14. We found that 3′ guanosines can strongly inhibit S. cerevisiae and H. sapiens Pan2–Pan3 and S. pombe Ccr4–Not in a fully reconstituted in vitro system (Fig. 1b-c, Fig. 2b). Guanylation of poly(A) tails is relatively infrequent in vivo and guanosines are usually incorporated as individual nucleotides14,15. Nevertheless, our in vitro two-color deadenylation assay (Supplementary Fig. 1g) shows that a single guanosine may be sufficient to inhibit Pan2 activity in the cell where polyadenylated transcripts are in vast excess over transcripts with modified poly(A) tails. This is consistent with observations obtained with CNOT6L and CNOT7 isolated by immunoprecipitation15. Our data therefore suggest that the stability of guanylated transcripts is increased because 3′ guanosines inhibit both of the major deadenylases, thereby slowing this rate-limiting step of mRNA decay7, resulting in longer transcript half-lives and thus a likely increase in gene expression.
The intrinsic conformation of poly(A) can be recognized by regulatory proteins
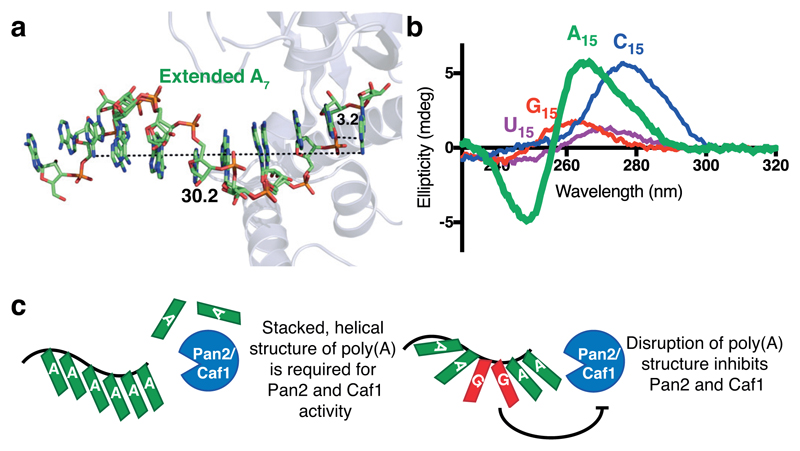
The structure of oligo(A) and poly(A) RNA has been investigated by numerous methods25,26,35–40. From these studies and a crystal structure of A3 RNA36, a single-stranded helical structure for poly(A) with nine nucleotides per turn was previously proposed. When we computationally extended the five adenosines visible in our crystal structure, they formed a helix whose parameters are consistent with this previously proposed model (Fig. 7a). Intriguingly, the tendency of poly(A) to form the characteristic single-stranded RNA helix was unique in CD studies, as all other polyribonucleotides displayed either no significant structure (poly(U), poly(G)) or a different conformation (poly(C)) (Fig. 7b)24,41. Since no other polyribonucleotide exhibits the same conformational properties in solution, our results suggest that the unique structure adopted by poly(A) tails is fundamental to its role in biological systems.
Figure 7. Model for the recognition of the intrinsic poly(A) structure by Pan2 and Caf1 deadenylases.
a, Extended oligo(A) helix bound to Pan2, modeled by duplicating and superposing the observed A5, showing the helical conformation adopted by oligo(A) in the active site. Distances are shown in Ångstroms. b, Circular dichroism spectra of A15 (green), U15 (purple), C15 (blue), and G15 (red) RNAs. These spectra are representative of identical experiments performed 2 times. c, Proposed model for the recognition of poly(A) RNA by the Caf1 and Pan2 DEDD family deadenylases.
Together, our data demonstrate that the Pan2 exonuclease has nucleotide specificity but does not directly recognize its RNA substrate through canonical sequence-specific interactions. Instead, the Pan2 active site recognizes the intrinsic solution conformation of poly(A) RNA. We hypothesize that cytosines and uracils disrupt the solution conformation of poly(A), but are permissive to form a stacked, A-form helix in the Pan2 active site. This would be accompanied with an entropic cost associated with the ordering of cytosines and uracils into the oligo(A)-like conformation. On the other hand, guanosines or dihydrouracils strongly inhibit Pan2 and Caf1 as they are unable to adopt the stacked, helical structure of poly(A) RNA. Caf1 likely recognizes the stacked, helical conformation of poly(A) RNA, and interacts with adenosines via base-specific contacts (Supplementary Fig. 4b), giving rise to greater nucleotide specificity (Fig. 2c). We propose a model whereby Caf1 and Pan2 exploit the tendency of poly(A) to form a stacked, helical structure, enabling the enzymes to distinguish between poly(A) and non-poly(A) tails (Fig. 7c). This mechanism of recognition is in contrast with a structure of oligo(A)-bound RRMs 1 and 2 of poly(A) binding protein16, where poly(A) is recognized by base-specific contacts and not by its unique conformation.
To our knowledge, the ability of Pan2 to read the specific conformation of poly(A) is a novel mode of protein-RNA recognition. This binding mechanism is reminiscent of indirect readout of specific DNA sequences, for example by the Trp repressor-DNA complex42. The unique physicochemical properties of polyadenosines, resulting in a distinct structure, may have evolutionary implications in the conservation of poly(A) at the 3′ end of mRNAs. Our results establish a novel paradigm for recognition of the conformation of the ubiquitous poly(A) sequence by proteins. Reading of the stacked, helical structure of poly(A) RNA may be important not only in the regulation of mRNA stability, but also in other biological contexts such as quality control in translation43,44 or polyadenylation45,46.
Methods
Protein Purification
S. cerevisiae Pan2–Pan3 was purified after co-transformation and co-overexpression of the subunits in S. cerevisiae as described in a previous study11. In brief, the complex containing a C-terminal Strep-II tag was purified using StrepTactin affinity chromatography, followed by cation exchange chromatography and size exclusion chromatography.
S. cerevisiae His8-Pan2UCH-Exo (residues 461–1115), H. sapiens PAN2, and H. sapiens PAN3∆N278-SII (residues 279–887) were synthesized with codon optimization and cloned into pACEBac1 (Epoch Life Science Inc.). His8-Pan2UCH-Exo E912A, His8-Pan2UCH-Exo Y975A, and His8-Pan2UCH-Exo E912A Y975A were generated using the QuickChange™ Site-directed Mutagenesis System (Stratagene, La Jolla, USA). For H. sapiens PAN2–PAN3∆N278-SII, constructs were amplified and assembled by Gibson assembly into a modified pBIG1 as described in the biGBac protocol47. pACEBac1 or pBIG1 constructs were transformed into DH10EMBacY E. coli. Bacmid DNA was purified and used to transfect Sf9 cells as described previously18. The virus from the first passage was harvested 72 h after transfection, supplemented with FBS to 50%, and stored at 4°C. The virus was further amplified by infecting Sf9 cells (2 × 106 cells/ml) at a 1:50 v/v ratio. The virus from the second passage was harvested by centrifugation at 1000 rpm for 5 min, and the supernatant was stored at 4°C. Large-scale infections were carried out in 500 ml cultures of Sf9 cells (2 × 106 cells/ml) at a 1:100 v/v ratio using virus from the second passage. Cultures were harvested after 48 h or 72 h (as determined by optimization studies) by centrifugation at 4000 rpm for 20 min and cell pellets were flash frozen.
To purify H. sapiens PAN2–PAN3∆N278, 1 liter cell pellets were thawed and lysed by sonication at 4°C in lysis buffer (100 mM HEPES pH 8.0, 300 mM NaCl, 2 mM DTT, 2 µg/ml DNase I, 2 µg/ml RNase A, supplemented by cOmplete protease inhibitor tablets (Roche) and 0.5 mM PMSF). The lysate was clarified by centrifugation at 235,400 × g for 30 min, and was incubated with 4 ml 50% Strep-Tactin sepharose resin (IBA) for 2 hours on a rotating platform at 4°C. The resin was washed with a high salt buffer (50 mM HEPES pH 8.0, 1 M NaCl, 2 mM DTT) and protein was eluted with an elution buffer (50 mM HEPES pH 8.0, 200 mM NaCl, 2 mM DTT, 20 mM desthiobiotin). The eluate was diluted eight-fold with buffer A (25 mM HEPES pH 8.0, 2 mM DTT), loaded onto a pre-equilibrated 5 ml HiTrap Q column (GE), washed with buffer A containing 125 mM NaCl, and eluted with a step to 400 mM NaCl. The eluate was pooled and loaded onto an S200 26/60 gel filtration column (GE) pre-equilibrated in size exclusion buffer (20 mM HEPES pH 8.0, 200 mM NaCl, 1 mM TCEP). The eluate was pooled, concentrated using a 100 kDa MWCO Vivaspin 20 centrifugal concentrator (GE), aliquoted, and flash frozen until further use.
To purify S. cerevisiae His8-Pan2UCH-Exo, His8-Pan2UCH-Exo E912A, and His8-Pan2UCH-Exo Y975A, 2 liter cell pellets were thawed, lysed by sonication, and clarified as above. Cell lysate was incubated with 6 ml 50% Ni-NTA Agarose resin (Qiagen) for 2 hours on a rotating platform at 4°C. The resin was washed with a low imidazole buffer (50 mM HEPES pH 8.0, 200 mM NaCl, 30 mM imidazole, 2 mM DTT) and eluted with a high imidazole buffer (50 mM HEPES pH 8.0, 200 mM NaCl, 500 mM imidazole, 2 mM DTT). The eluate was diluted eight-fold with buffer A (25 mM HEPES pH 8.0, 2 mM DTT), loaded onto a pre-equilibrated 5 ml HiTrap Q column (GE), washed with buffer A containing 30 mM NaCl, and eluted with a 30-1000 mM NaCl gradient over 30 column volumes. For His8-Pan2UCH-Exo and His8-Pan2UCH-Exo Y975A, the His8 tag was removed by overnight cleavage at 4°C with PreScission protease. The cleaved protein was purified by anion exchange chromatography as above. Fractions were pooled and loaded onto an S200 26/60 gel filtration column (GE) pre-equilibrated in size exclusion buffer. The eluate was pooled, concentrated using a 30 kDa MWCO Vivaspin 20 centrifugal concentrator (GE), aliquoted, and flash frozen until further use.
S. pombe Ccr4–Not and associated catalytic mutants were overexpressed by the MultiBac protocol48 and purified as previously described18.
Unless otherwise indicated, all column purification steps were carried out on an ÄKTA Pure (GE). At each stage, the purity and composition of purified fractions were analyzed by the separation of fractions on a NuPage 4-12% Bis-Tris SDS polyacrylamide gel (Invitrogen). The gels were visualized by Coomassie staining with InstantBlue (Expedeon).
Deadenylation Assays
Deadenylation assays were carried out as described in a previous protocol49 in 50 µl in 20 mM PIPES pH 6.8, 10 mM KCl, 30 mM NaCl, 2 mM Mg(OAc)2, 0.1 mM TCEP at 22°C. The enzymes were prepared at 10× concentrations (500 nM S. cerevisiae Pan2–Pan3, Ccr4–Not, Ccr4-inactive Ccr4–Not, and Caf1-inactive Ccr4–Not; 1 µM S. cerevisiae Pan2UCH-Exo; 5 µM S. cerevisiae Pan2UCH-Exo Y975A and H. sapiens PAN2–PAN3∆N278) in 20 mM HEPES pH 7.5, 300 mM NaCl, 2 mM Mg(OAc)2, 0.5 mM TCEP and added to the deadenylation reaction.
20mer-A30 (20mer: CAGCUCCGCAUCCCUUUCCC) with varied 3′ ends and intervening nucleotides were synthesized with a 5′ 6-FAM fluorophore label (Integrated DNA Technologies or, for 20mer-A14DDA14, Dharmacon).
200 nM RNA was used in each reaction, except in assays with H. sapiens PAN2–PAN3∆N278 and S. cerevisiae Pan2UCH-Exo Y975A, where 500 nM RNA was used. Reactions were stopped at the indicated time points by mixing 4 µl reaction with 4 µl 2× denaturing loading dye (86% formamide, 0.01% w/v bromophenol blue, 10 mM EDTA pH 7.4). RNA markers of known tail length were run concurrently to assess the tail lengths of products from the assay. To count individual bases, the 20mer-A30 substrate was digested by alkaline hydrolysis: 400 nM RNA was incubated in a buffer containing 50 mM NaHCO3 pH 9.0, and 5 mM EDTA at 95°C for 30 min. The digested RNA was mixed with the 20mer substrate as a marker. Samples were loaded onto 20% Tris-borate-EDTA (TBE) polyacrylamide (19:1 acrylamide: bis-acrylamide) gels containing 7 M urea and separated by application of 350 V for 1 hour in 1× TBE running buffer. Gels were visualized with a Typhoon FLA-7000 (473 nm excitation, 520 nm fluorescence) and the photomultiplier tube was adjusted to prevent saturation of the detector.
To perform quantitative analysis, deadenylation reactions were carried out in triplicate and visualized as above. The bands corresponding to intact species were boxed (Supplementary Fig. 1a) and their intensities integrated in FIJI50. Because the molarity of the fluorophore is equal to the RNA concentration, the integrated fluorescence is proportional to RNA concentration. The intensities were divided by the signal at time = 0 to represent the fraction of intact RNA at each time point. These were plotted against time and straight lines were used to connect the points for clarity in Graphpad Prism. Error bars were plotted to represent standard deviation.
For two-color deadenylation assays, 20mer-A30 was synthesized with a 5′ Alexa Fluor-647 fluorescent label and 20mer-A14GA15 was synthesized with a 5′ 6-FAM label (Integrated DNA Technologies). Reactions with 50 nM S. cerevisiae Pan2UCH-Exo contained each RNA at 100 nM. Quenched aliquots were analyzed by denaturing polyacrylamide gel electrophoresis as above, and gels were scanned at the appropriate wavelengths (473 nm excitation, 520 nm fluorescence for 6-FAM; 650 nm excitation, 670 nm fluorescence for Alexa Fluor-647). The resulting images were subjected to identical contrast adjustments, overlaid, and falsely colored for clarity.
Crystallization of Protein-RNA Complexes
Purified Pan2UCH-Exo E912A (final concentration 42.5 µM) was used in crystallization experiments performed in 96-well sitting drops (100 nl protein + 100 nl reservoir). Hits were obtained and the best diffracting crystallization condition was further optimized by incubation of the purified protein (final concentration 37.3 µM) in 96-well sitting drops (200 nl protein + 200 nl reservoir) containing a titration of ammonium phosphate (final concentration 0.10 to 0.32 M). Diffraction quality crystals were obtained in a condition containing 0.14 to 0.32 M ammonium phosphate.
RNAs (A7, AAGGA, AAGGAA, AACCAA, AAUUAA) were synthesized by Integrated DNA Technologies and resuspended in TE buffer (5 mM Tris pH 8.0, 0.5 mM EDTA pH 7.4). Crystals were soaked in RNA-containing buffer (25 mM HEPES pH 8.0, 0.28 M ammonium phosphate, 5 mM A7, AAGGA, and AAGGAA or 2 mM AACCAA and AAUUAA) for different time periods for optimization. Crystals were soaked in A7 for 16 hours, in AAGGA and AAUUAA for 5 hours, and in AAGGAA for 2 hours. AACCAA was added to the crystallization droplet and incubated for 5 hours. The soaked crystals were cryoprotected in RNA-containing buffer supplemented with 25% ethylene glycol before plunging into liquid nitrogen.
X-ray Data Collection, Model Building, and Refinement
Diffraction data for apo Pan2UCH-Exo and Pan2UCH-Exo-A7 were recorded at 100 K at Diamond Light Source beamline I03 with a Pilatus3 6M detector (Dectris) at 0.9763 Å. Diffraction data from Pan2UCH-Exo-AAGGA, Pan2UCH-Exo-AAGGAA, Pan2UCH-Exo-AACCAA, and Pan2UCH-Exo-AAUUAA crystals were recorded at 100 K at Diamond Light Source beamline I04-1 with a Pilatus 6M-F (25Hz) detector (Dectris) at 0.9159 Å. Data collection statistics are provided in Table 1. Diffraction data were indexed and integrated using DIALS within the Xia2 pipeline51. All datasets were scaled and merged with AIMLESS as part of CCP4i252. Resolution cut-off was determined by CC1/2 > 0.5 and I/σI > 1.053. The apo structure was solved by molecular replacement using Phaser with Neurospora crassa apo Pan2UCH-Exo (PDB: 4CZW12) as a search model and by further manual model building. Subsequent structures were refined against the apo structure. RNA in the active site was visualized first by mFo-DFc maps, built using RCrane 54, and further positioned in Coot55 with the help of feature-enhanced maps56. The structure of the protein-RNA complex was iteratively refined using PHENIX.refine and maps were calculated in PHENIX57. Final refinement statistics are presented in Table 1. The Ramachandran statistics for apo Pan2UCH-Exo are 92.4% favored, 0.0% outliers; for Pan2UCH-Exo-A7 are 94.7% favored, 0.0% outliers; for Pan2UCH-Exo-AAGGA are 96.0% favored, 0.0% outliers; for Pan2UCH-Exo-AAGGAA are 95.2% favored, 0.0% outliers; for Pan2UCH-Exo-AACCAA are 94.8% favored, 0.0% outliers; for Pan2UCH-Exo-AAUUAA are 91.4% favored, 0.0% outliers. Structural figures were rendered in PyMOL58.
Structural Analysis
Superposition analysis for calculation of backbone RMSD was carried out between the apo Pan2UCH-Exo structure and a previously solved structure of apo Pan2UCH-Exo (PDB: 4Q8H13) with PyMOL58. Structure-based alignments were carried out using SSM and LSQ superposition implemented in Coot55. Multiple sequence alignments were carried out using the T-Coffee suite59 and visualized using JalView60. Map coefficients were generated in CCP4 using the FFT tool61. Protein-ligand interactions were visualized using LigPlot+62. Maps and protein structures were visualized in PyMOL58.
Circular Dichroism Spectroscopy
RNAs (A5, AAGGA, A6, AAGGAA, AAUUAA, AACCAA, A8G2, A6G2A2, A4G2A4, A2G2A6, G2A8, A15, C15, U15, G15) were synthesized by Integrated DNA Technologies and redissolved in DEPC-treated water. The RNAs were added to a circular dichroism buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 1 mM Mg(OAc)2, 1 mM TCEP) at final concentrations of 5.0 µM (Fig. 7b), 9.0 µM (Fig. 4c and 5a, and Supplementary Fig. 7a) or 25.0 µM (Supplementary Fig. 7b) in 200 µl. Circular dichroism spectra were obtained in a J-815 (Jasco) between 340-200 nm at 0.5 nm intervals. The spectra were obtained at 50 nm/min with standard (100 mdeg) sensitivity. For each RNA, nine individual spectra were obtained and averaged. A baseline of buffer alone was taken and subtracted from the obtained data, which was then plotted in Graphpad Prism.
Data Availability
The structures generated during the current study have been deposited in the wwPDB under accession codes 6R9I (apo), 6R9J (A7-bound), 6R9M (AAGGAA-bound), 6R9O (AAGGA-bound), 6R9P (AAUUAA-bound), and 6R9Q (AACCAA-bound). Source data for figures 1b-e, 2b-d, 4c, 5a, 7b, supplementary figures 1e-f, 2a, 6e-g, and 7a-b are available in tabular form with the paper online. Source data for figures 1a, 2a, and 6a-h are available in Supplementary Data Set 1 with the paper online. All annotated gels are available on Mendeley (http://dx.doi.org/10.17632/zkfsh9nftk.1). All other data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Chris Russo and Michael Webster for assistance and advice; Minmin Yu and Danguole Ciziene for help with crystallography; Mathias Girbig for biGBac constructs; Chris Johnson and Stephen McLaughlin for help with biophysics; and Martin Egli, Ditlev Brodersen, Chris Oubridge, Kiyoshi Nagai, and Manu Hegde for advice or comments on the manuscript. This work was supported by a Herchel Smith PhD Studentship from the University of Cambridge (to T.T.L.T.); the European Union’s Horizon 2020 research and innovation programme (ERC grant 725685) (to L.A.P.); and Medical Research Council grant MC_U105192715 (L.A.P.). We acknowledge Diamond Light Source for beamtime (proposal MX15916), and the staff of beamlines I24, I03, I04, and I04-1 for assistance with crystal testing and data collection.
Footnotes
Author Contributions: T.T.L.T., J.A.W.S., and L.A.P. conceived the study. T.T.L.T. and J.A.W.S. purified proteins and prepared samples for crystallography. T.T.L.T., J.A.W.S., and C.H.H. collected crystallography data, and T.T.L.T. built atomic models and interpreted the structure with help from C.H.H. T.T.L.T. carried out all other experiments. T.T.L.T. and L.A.P. wrote the manuscript. All authors discussed and commented on the final manuscript.
Competing Interests Statement: The authors declare no competing interests.
References
- 1.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 3.Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 4.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 5.Brook M, Gray NK. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem Soc Trans. 2012;40:856–864. doi: 10.1042/BST20120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 7.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes & development. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 8.Webster MW, et al. mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol Cell. 2018;70:1089–1100. doi: 10.1016/j.molcel.2018.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 10.Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta. 2013;1829:561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Wolf J, et al. Structural basis for Pan3 binding to Pan2 and its function in mRNA recruitment and deadenylation. EMBO J. 2014;33:1514–1526. doi: 10.15252/embj.201488373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonas S, et al. An asymmetric PAN3 dimer recruits a single PAN2 exonuclease to mediate mRNA deadenylation and decay. Nat Struct Mol Biol. 2014;21:599–608. doi: 10.1038/nsmb.2837. [DOI] [PubMed] [Google Scholar]
- 13.Schafer IB, Rode M, Bonneau F, Schussler S, Conti E. The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase. Nat Struct Mol Biol. 2014;21:591–598. doi: 10.1038/nsmb.2834. [DOI] [PubMed] [Google Scholar]
- 14.Chang H, Lim J, Ha M, Kim VN. TAIL-seq: genome-wide determination of poly(A) tail length and 3' end modifications. Mol Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Lim J, et al. Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science. 2018;361:701–704. doi: 10.1126/science.aam5794. [DOI] [PubMed] [Google Scholar]
- 16.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 17.Aibara S, Gordon JM, Riesterer AS, McLaughlin SH, Stewart M. Structural basis for the dimerization of Nab2 generated by RNA binding provides insight into its contribution to both poly(A) tail length determination and transcript compaction in Saccharomyces cerevisiae. Nucleic Acids Res. 2017;45:1529–1538. doi: 10.1093/nar/gkw1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stowell JA, et al. Reconstitution of Targeted Deadenylation by the Ccr4-Not Complex and the YTH Domain Protein Mmi1. Cell Rep. 2016;17:1978–1989. doi: 10.1016/j.celrep.2016.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen KR, Jonstrup AT, Van LB, Brodersen DE. The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site. RNA. 2009;15:850–861. doi: 10.1261/rna.1489409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D-Biological Crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaeger JA, SantaLucia J, Jr, Tinoco J., Jr Determination of RNA structure and thermodynamics. Annu Rev Biochem. 1993;62:255–287. doi: 10.1146/annurev.bi.62.070193.001351. [DOI] [PubMed] [Google Scholar]
- 23.Bloomfield VA, Crothers DM, Tinoco I. Nucleic acids : structures, properties, and functions. University Science Books; 2000. [Google Scholar]
- 24.Saenger W. Principles of nucleic acid structure. Springer-Verlag; 1984. [Google Scholar]
- 25.Brahms J, Michelson AM, Van Holde KE. Adenylate oligomers in single- and double-strand conformation. Journal of molecular biology. 1966;15:467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- 26.Hashizume H, Imahori K. Circular dichroism and conformation of natural and synthetic polynucleotides. J Biochem. 1967;61:738–749. doi: 10.1093/oxfordjournals.jbchem.a128608. [DOI] [PubMed] [Google Scholar]
- 27.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res. 1996;24:1073–1079. doi: 10.1093/nar/24.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010;29:2566–2576. doi: 10.1038/emboj.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita A, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 30.Lim J, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159:1365–1376. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song MG, Kiledjian M. 3' Terminal oligo U-tract-mediated stimulation of decapping. Rna. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rissland OS, Norbury CJ. Decapping is preceded by 3' uridylation in a novel pathway of bulk mRNA turnover. Nature structural & molecular biology. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malecki M, et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. The EMBO journal. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faehnle CR, Walleshauser J, Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514:252–256. doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewey TG, Turner DH. Laser temperature-jump study of stacking in adenylic acid polymers. Biochemistry. 1979;18:5757–5762. doi: 10.1021/bi00593a002. [DOI] [PubMed] [Google Scholar]
- 36.Saenger W, Riecke J, Suck D. A structural model for the polyadenylic acid single helix. J Mol Biol. 1975;93:529–534. doi: 10.1016/0022-2836(75)90244-2. [DOI] [PubMed] [Google Scholar]
- 37.Smith BL, Gallie DR, Le H, Hansma PK. Visualization of poly(A)-binding protein complex formation with poly(A) RNA using atomic force microscopy. J Struct Biol. 1997;119:109–117. doi: 10.1006/jsbi.1997.3864. [DOI] [PubMed] [Google Scholar]
- 38.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys Rev Lett. 2007;98 doi: 10.1103/PhysRevLett.98.158103. 158103. [DOI] [PubMed] [Google Scholar]
- 39.Isaksson J, Acharya S, Barman J, Cheruku P, Chattopadhyaya J. Single-stranded adenine-rich DNA and RNA retain structural characteristics of their respective double-stranded conformations and show directional differences in stacking pattern. Biochemistry. 2004;43:15996–16010. doi: 10.1021/bi048221v. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Kolomeisky A, Meller A. Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel. Phys Rev Lett. 2010;104 doi: 10.1103/PhysRevLett.104.158101. 158101. [DOI] [PubMed] [Google Scholar]
- 41.Arnott S, Chandrasekaran R, Leslie AG. Structure of the single-stranded polyribonucleotide polycytidylic acid. J Mol Biol. 1976;106:735–748. doi: 10.1016/0022-2836(76)90262-x. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, et al. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 43.Juszkiewicz S, Hegde RS. Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol Cell. 2017;65:743–750 e744. doi: 10.1016/j.molcel.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arthur L, et al. Translational control by lysine-encoding A-rich sequences. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 46.Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weissmann F, et al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci U S A. 2016;113:E2564–2569. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sari D, et al. The MultiBac Baculovirus/Insect Cell Expression Vector System for Producing Complex Protein Biologics. Adv Exp Med Biol. 2016;896:199–215. doi: 10.1007/978-3-319-27216-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webster MW, Stowell JAW, Tang TTL, Passmore LA. Analysis of mRNA deadenylation by multi-protein complexes. Methods. 2017 doi: 10.1016/j.ymeth.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winter G. xia2: an expert system for macromolecular crystallography data reduction. Journal of Applied Crystallography. 2010;43:186–190. doi: 10.1107/S0021889809045701. [DOI] [Google Scholar]
- 52.Potterton L, et al. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr D Struct Biol. 2018;74:68–84. doi: 10.1107/S2059798317016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keating KS, Pyle AM. RCrane: semi-automated RNA model building. Acta Crystallogr D Biol Crystallogr. 2012;68:985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afonine PV, et al. FEM: feature-enhanced map. Acta Crystallogr D Biol Crystallogr. 2015;71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015 [Google Scholar]
- 59.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 60.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 61.Read RJ, Schierbeek AJ. A Phased Translation Function. J Appl Crystallogr. 1988;21:490–495. doi: 10.1107/S002188988800562x. [DOI] [Google Scholar]
- 62.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures generated during the current study have been deposited in the wwPDB under accession codes 6R9I (apo), 6R9J (A7-bound), 6R9M (AAGGAA-bound), 6R9O (AAGGA-bound), 6R9P (AAUUAA-bound), and 6R9Q (AACCAA-bound). Source data for figures 1b-e, 2b-d, 4c, 5a, 7b, supplementary figures 1e-f, 2a, 6e-g, and 7a-b are available in tabular form with the paper online. Source data for figures 1a, 2a, and 6a-h are available in Supplementary Data Set 1 with the paper online. All annotated gels are available on Mendeley (http://dx.doi.org/10.17632/zkfsh9nftk.1). All other data that support the findings of this study are available from the corresponding author upon reasonable request.