Abstract
Exome sequencing in the context of developmental disorders is a useful technique, but variants found need to be interpreted in the context of detailed phenotypic information. Whole gene deletions and loss-of-function-mutations in the HNRNPU gene have been associated with intellectual disability and seizures in some patients. However, a unifying syndromic phenotype has not been previously elucidated.
Here, we report a total of seven patients (six patients identified through the Wellcome Trust Deciphering Developmental Disorders study, with one additional patient), who have heterozygous de novo mutations in HNRNPU. These were found via trio-based exome sequencing. All but one of the mutations is predicted to cause loss-of-function. These patients have dysmorphic features in common, including prominent eyebrows, long palpebral fissures, overhanging columella and thin upper lip. All patients have developmental delay and intellectual disability (ID), ranging from moderate to severe. Seizures are common from early childhood. These initially occur in the context of febrile episodes.
This series demonstrates common phenotypic features, including emerging dysmorphism, associated with heterozygous HNRNPU mutations. This allows us to define a novel neurodevelopmental syndrome, with a likely mechanism of haploinsufficiency.
Keywords: HNRNPU, seizures, behaviour, aggressive outbursts, trio exome sequencing, intellectual disability
INTRODUCTION
Exome sequencing has proven to be an effective tool in the diagnosis of rare developmental disorders [Wright et al.,2015]. Increasingly, this method is utilised in clinical practice. However, the use of genomic data comes with its own challenges. The volume of variants that can be generated necessitates sophisticated filtering systems to identify those likely to be pathogenic. This means there is an increasing emphasis on detailed and accurate phenotyping to enable correct interpretation of sequencing results.
The importance of this approach can be seen with exome sequencing studies of patients with epilepsy [Carvill et al., 2013; de Kovel et al., 2016; Epi4K Consortium, 2013]. This condition displays marked genetic heterogeneity. Given the rarity of many epilepsy-associated conditions, it can be difficult to determine the pathogenicity of variants in genes found using an exome-wide testing method. Therefore, identifying a more detailed phenotype associated with a particular gene allows for better interpretation of sequencing results and more confidence in predictions of pathogenicity.
Overlapping chromosomal microdeletions can be useful in the initial identification of phenotypes associated with haploinsufficiency of a particular gene. Patients with 1q44 deletions have previously been described with a phenotype of developmental delay (particularly speech), microcephaly, hypogenesis/agenesis of the corpus callosum and seizures. Analysis of the smallest region of overlap has identified the HNRNPU (Heterogeneous Nuclear Ribonucleoprotein U) gene as a candidate for the epilepsy and intellectual disability (ID) phenotype associated with this microdeletion [Caliebe et al., 2010; Thierry et al., 2012].
HNRNPU loss-of-function mutations have been associated with epilepsy and developmental delay in individual patients as part of larger cohorts [Carvill et al., 2013; de Kovel et al., 2016; Epi4K Consortium, 2013; Hamdan et al., 2014; Need et al., 2012]. Recently, two series confirming the association of HNRNPU mutations with epilepsy and ID have been published [Bramswig et al., 2017., Depienne et al., 2017]. However, the phenotypic spectrum still remains to be delineated. In particular, a common dysmorphic phenotype has not yet been fully defined.
Here, we report seven patients with de novo mutations in HNRNPU. Six of these patients were identified through the Wellcome Trust Deciphering Developmental Disorders (DDD) study [Wright et al., 2015] with an additional patient identified via MatchMaker exchange [Philippakis et al., 2015]. We demonstrate common phenotypic features, including facial dysmorphism, ID, seizures, and behavioural problems. This previously-undescribed pattern of features allows us to define a novel neurodevelopmental syndrome.
MATERIALS AND METHODS
Patients 1 to 6 were identified through the DDD study. Recruitment to this study was via UK regional Clinical Genetics centres after routine referral. The affected person and their parents underwent trio-based exome sequencing and analysis as previously published [Wright et al,. 2015]. Array-based comparative genomic hybridization (aCGH) was also used to analyse copy number variation. Patient 7 was recruited via a local Clinical Genetics centre in Melbourne, Australia, and had an uninformative Single Nucleotide Polymorphism (SNP) array. For this patient, whole exome sequencing and data processing were performed by Genomics Platform at the Broad Institute of Harvard and MIT (Broad Institute, Cambridge, MA, USA). Whole exome sequencing was performed using Illumina exome capture (38 Mb target) and the data was processed through a pipeline based on Picard. SNPs and insertions/deletions (indels) were jointly called across all samples using Genome Analysis Toolkit (GATK) HaplotypeCaller package version 3.4.
RESULTS
Patient reports
Patient 1 (DECIPHER ID: 258995)
Patient 1 is a female, born to non-consanguineous parents. The family history was unremarkable. The pregnancy was complicated by gestational diabetes. She was born at 38 weeks gestation by emergency caesarean section. Her birth-weight was on the75th centile. She walked independently after the age of two years. Her first words were at the age of five years.
On assessment at the age of 15 years, her head circumference was on the 75th centile, weight between the 98th and 99.6th centile and height between the 9th and 25th centiles.
She had special educational needs and moderate ID. She developed epilepsy at the age of approximately one year. Her seizures were frequent, primarily absences, and could be triggered by fevers. Dysmorphic features included a short philtrum, thin upper lip, deep-set eyes and long eyelashes. Her gait was wide-based and she was noted to have hand-flapping movements. Magnetic resonance imaging (MRI) of the brain and echocardiogram were normal.
Patient 2 (DECIPHER ID: 260453)
Patient 2 is a male, born to non-consanguineous parents. He was noted to have two maternal uncles with mild learning difficulties and a maternal cousin with autism. During pregnancy, cardiac abnormalities were found on the 20 week antenatal ultrasound. He was born at 37 weeks gestation. Birth-weight was between the 2nd and 9th centiles. He was found to have transposition of the great vessels, tricuspid atresia and a ventricular septal defect. He had pulmonary artery banding on the first day of life and has subsequently undergone multiple cardiac operations. He walked independently, and spoke his first words after the age of two years.
He had special educational needs, and has moderate ID. He was noted to be emotionally labile and to anger easily. He did not have any seizures or hand-flapping movements. He also developed a brain abscess at the age of eight years and had recurrent ear infections. He has been diagnosed with Tourette syndrome.
On assessment at the age of 14 years, his head circumference was between the 2nd to 9th centiles, weight between 9th and 25th centiles and height between the 2nd to 9th centiles. His facial features were thought to resemble his family. He had a left single transverse palmar crease.
He is now attending an adult education college. He has learning difficulties, particularly regarding communication and social skills. He uses a wheelchair and scooter for mobility. He cannot walk or propel his wheelchair for long distances.
MRI brain as a child demonstrated small periventricular areas with high T2 signal, thought to be non-specific, but possibly due to previous ischaemic events. He was additionally found to have maternal uniparental disomy (UPD) of chromosome nine (previously published by King et al., 2014). It is not certain whether this is significantly contributing to his phenotype.
Patient 3 (DECIPHER ID: 265865)
Patient 3 is a female, born to non-consanguineous parents. She had a paternal cousin with development delay and seizures. The pregnancy was complicated by a maternal renal infection. She was born at term, with a birth-weight between the 9th and 25th centiles.
She walked independently after the age of four years and her first words were at the age of 18 months. She was able to speak a few words by the age of three years. She was also noted to be hypotonic as an infant.
She was thought to have autistic traits. She had special educational needs and severe ID. She developed epilepsy at the age of one year, initially febrile seizures, which have now resolved. On assessment, at the age of 12 years, her height was on the 9th centile and weight on the 96th centile. Her head circumference was previously on the 25th centile at the age of three-and-a-half years. She had dysmorphic features including synophrys with thick eyebrows and a short nose. She had hand flapping when excited. Her echocardiogram was normal.
Patient 4 (DECIPHER ID: 268390)
Patient 4 is a female, born to non-consanguineous parents. The family history was unremarkable. The pregnancy was uncomplicated. She was born at 38 weeks gestation, with a birth-weight on the 2nd centile.
She crawled at two years nine months of age, and walked independently at the age of approximately five years. Her first words were at around four-and-a-half years of age, and she had limited communication with Makaton sign language prior to this.
Drooling has been a long-standing problem. Initially this was treated with hyoscine patches, but she later required bilateral trans-tympanic neuronectomy. This gave a good result initially, but drooling has started to worsen again. She has also previously had conductive hearing loss with vent insertion. She had a single seizure at the age of approximately five years. She had special educational needs. She displayed good comprehension, but her words remained unclear and speech was limited to short sentences.
On assessment at the age of 6-and-a-half years, her head circumference was on the 0.4th centile, weight between the 25th and 50th centiles, and height between the 0.4th and 2nd centiles.
She had dysmorphic features including: low-set, posteriorly rotated ears, a mild conductive hearing impairment, strabismus, anteverted nares, a prominent metopic ridge, epicanthic folds, smooth philtrum, thin vermillion border and two-three toe cutaneous syndactyly bilaterally (Fig. 3). She is hypermobile, particularly affecting the upper limbs. She had hand-flapping behaviours, and a generally pleasant demeanour. She was also myopic. MRI brain scan was normal at two years of age.
Figure 3.
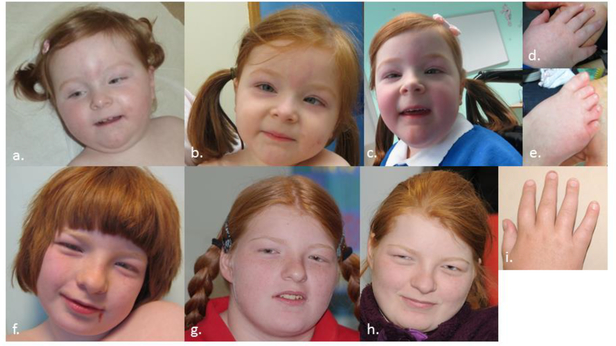
Patients 4 & 5. Progression of features with age. Patient 4 (a-e) shown age 2yr (a.), 5yr (b.), and 6yr (c.). Patient 5 (f-i) shown age 11yr (f.), 15yr (g.), and 18yr (h.) Note common facial dysmorphism including prominent eyebrows, long palpebral fissures, overhanging columella and thin upper lip. Both have tapering fingers (d. & i.). Patient 4 has short & broad toes with 2–3 syndactyly bilaterally (e.). [Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552–4833.]
Patient 5 (DECIPHER ID: 277603)
Patient 5 is a female, born to non-consanguineous parents. The family history was unremarkable. The pregnancy was uncomplicated. She was born at term, with a birth-weight between the 50th and 75th centiles. She walked independently at around two years of age. Her first words were after two years.
Her behaviour was aggressive at times, with violent episodes. She also had a diagnosis of autism. She had frequent ear infections and constipation. She had special educational needs and severe ID.
She developed frequent absence seizures before the age of one year. She also had generalised seizures, usually triggered by fevers, requiring anti-epileptic medications (Lamotrigine and Topiramate). On assessment at the age of 21 years, her head circumference was between the 75th and 91st centiles, weight greater than the 99.6th centile, and height between the 0.4th and 2nd centiles.
Dysmorphic features included prominent eyebrows, elongated palpebral fissures, epicanthic folds, hypertelorism, brachydactyly, and tapering fingers. She had hand flapping and wringing movements. She continues to have occasional absence seizures and is housed in a specialist centre for autism.
Patient 6 (DECIPHER ID: 263453)
Patient 6 is a female, born to non-consanguineous parents. The family history was unremarkable. The pregnancy was uncomplicated. She was born at 42 weeks gestation, with a birth-weight on the 9th centile. Her parents had concerns about her development from four months of age, as she had hypotonia and delayed milestones.
She did not walk independently until the age of three years. Her speech was delayed, particularly expressive. At age 11 years, she still had no formal words, but could communicate through signing and using a picture board on a computer pad. She has special educational needs, and attends a school for children with severe ID. She has a sociable, loving personality. She has repetitive mannerisms, such as hand-flapping movements, particularly when excited.
She developed epilepsy with generalised tonic-clonic seizures at the age of eight months. These were well-controlled on sodium valproate. Her seizures resolved and she was weaned off treatment aged six years.
On assessment at the age of 11 years, her head circumference was on the 75th centile, weight on the 75th centile, and height on the 2nd centile. Dysmorphic features included thick hair and eyebrows, prognathism, broad thumbs and great toes, and truncal obesity. She walked with a wide-based, unsteady gait, with hypermobility, especially at her ankles, requiring supportive footwear. MRI brain at the age of one year demonstrated delayed myelination, but no other abnormalities.
Patient 7
Patient 7 is a female, born to non-consanguineous parents. The family history was unremarkable. The mother had ovarian stimulation prior to pregnancy. This patient is one of a dizygous twin pair; the other twin is unaffected. The pregnancy was uncomplicated. This patient was born at 38 weeks gestation with a birth-weight on the 0.4th centile. She was admitted to the Special Care Baby Unit due to complications related to low birth-weight. She sat independently at 18 months of age, and walked independently at 6-and-a-half years. Her first words were at three years of age.
On assessment at the age of eight-and-a-half years, her head circumference was on the 2nd to 9th centile, weight on the 2nd centile, and height below the 0.4th centile. She did not have any dysmorphic features. She had moderate ID. She developed generalised seizures at the age of 18-months, which were initially triggered by fevers. She was treated with Levetiracetam. Her seizures resolved and she was weaned off treatment aged five years.
MRI brain scans at the ages of three and seven years showed non-progressive T2 and FLAIR hyperintense lesions involving the deep white matter bilaterally, with sparing of the occipital lobes and basal ganglia.
Sequencing results
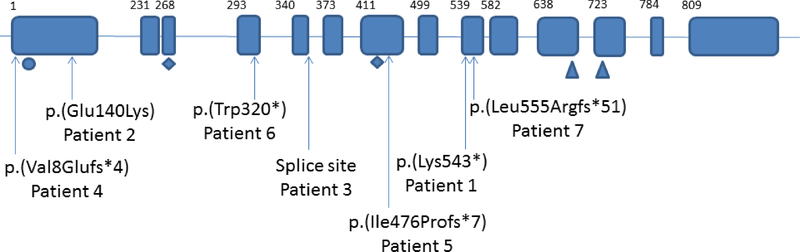
Each patient was found to have a heterozygous de novo mutation in HNRNPU (Table I; Fig. 1). Patient 2 was also previously noted to have complete maternal UPD of chromosome nine [King et al., 2014].
Table I.
HNRNPU mutations found in patient series, with predicted protein change shown.
| Patient number | HNRNPU mutation | Predicted protein change |
|---|---|---|
| 1 | c.1626_1627insA | p.(Lys543*) |
| 2 | c.418G>A | p.(Glu140Lys) |
| 3 | c.1117+1G>A | Splice donor site alteration |
| 4 | c.23del | p.(Val8Glufs*4) |
| 5 | c.1424_1425insTC | p.(Ile476Profs*7) |
| 6 | c.960G>A | p.(Trp320*) |
| 7 | c.1664del | p.(Leu555Argfs*51) |
Figure 1.
HNRNPU gene (transcript ENST00000283179.9, Human Genome Build GRCh37). Boxes indicate exons, lines introns. First amino acid of each exon shown above. Mutations from patients in this series shown below gene. Functional domains shown below gene: circle shows the SAP domain (putative DNA binding site), area between diamonds is B30.2/SPRY protein-protein interaction domain, triangles indicate RNA-binding RGG box. [Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552–4833.]
DISCUSSION
Gene function
HNRNPU is an RNA-binding protein, expressed in brain (particularly the cerebellum), heart, kidney and liver [Thierry et al., 2012]. It is largely localised to the nucleus, functioning as a mediator of alternative splicing and in transcriptional regulation [Geuens et al., 2016]. Reduced expression of Hnrnpu results in embryonic lethality in mice [Roshon et al., 2005]. Hnrnpu cardiac-specific knockout results in a rapidly progressive dilated cardiomyopathy, with widespread dysregulation of splicing [Ye et al., 2015].
The role of HNRNPU in transcriptional regulation can be mediated through an interaction with long noncoding RNA (lncRNA), which are increasingly recognised as playing a role in gene expression [Lin et al,. 2017]. For example, HNRNPU plays a crucial role in X-inactivation, by enabling chromosomal localisation of the lncRNA Xist, as well as being required for Xist-mediated function [Hasegawa et al., 2010].
Hnrnpu has also been shown, in mice, to give rise to circular RNA (ciRNA), with expression almost entirely in neurons. ciRNA have an emerging role in a number of areas including regulation of microRNA function and alternative splicing [Reddy et al., 2017]. HNRNPU is part of a larger family of hnRNP (Heterogeneous Nuclear Ribonucleoproteins). Interestingly, other hnRNP have been implicated in neurological disease and cancer [Geuens et al., 2016], emphasising the wide-ranging functions of these proteins.
HNRNPU-related syndrome
The patient series presented here clarifies and delineates the phenotypic features associated with HNRNPU mutations (Table II, Table III). Emerging dysmorphism includes a common facial appearance, with prominent eyebrows (4/7) elongated palpebral fissures (4/7), a prominent nasal bridge (3/7), overhanging columella (4/7) and a thin upper lip (6/7) (Fig. 2). There are facial changes with age, particularly with regard to the prominent nasal bridge (Fig. 3). The oldest patient in our series is now 21 years of age (Patient 5), and these changes are especially apparent in her. Of note, there are no significant associated complications antenatally, or during the neonatal period.
Table II.
Features of patients with de novo mutations in HNRNPU. Includes patients in case series presented as well as previously reported cases. Of note, patient in Need et al., 2012 not included as patient has additional variant and complex phenotype not accounted for by HNRNPU mutation.
| Patient ID (DECIPHER ID) | 1 (258995) | 2 (260453) | 3 (265865) | 4 (268390) | 5 (277603) | 6 (263453) | 7 | Total (of features reported) |
|---|---|---|---|---|---|---|---|---|
| Genotype | c.1626_1627insA | c.418G>A | c.1117+1G>A | c.23del | c.1424_1425insTC | c.960G>A | c.1664del | - |
| Protein change (predicted) | p.(Lys543*) | p.(Glu140Lys) | splice donor variant | p.(Val8Glufs*4) | p.(Ile476Profs*7) | p.(Trp320*) | p.(Leu555Argfs*51) | - |
| Inheritance | De novo | De novo | De novo | De novo | De novo | De novo | De novo | - |
| Additional genetic finding | n/a | Complete UPD (mat) chr 9 | n/a | n/a | n/a | n/a | n/a | - |
| Age last assessed | 15yr | 14yr | 12yr | 6yr | 21yr | 11yr | 8.5yr | - |
| Sex | F | M | F | F | F | F | F | - |
| Pregnancy | NAD | Congenital cardiac disease 20 week scan | Maternal renal infection 16 weeks gestation | NAD | NAD | NAD | Ovarian stimulation, dizygotic twins | - |
| Birth | Emergency caesarean section | NAD | NAD | NAD | NAD | NAD | NAD | - |
| Neonatal unit | n/a | 18 days | n/a | n/a | n/a | n/a | Yes | - |
| Gestation (weeks) | 38 | 37 | 40 | 38 | 40 | 42 | 38 | - |
| Feeding Difficulties | No | Yes | No | No | No | No | No | - |
|
Additional
cranio-facial features |
Synophrys, deepset eyes, long eyelashes, slightly coarse | Pointed chin | Low anterior hairline, synophrys, strabismus, short upturned nose, coarse | Epicanthic folds, low set posteriorly rotated ears, upturned nose, smooth philtrum | Hypertelorism | Thick hair, prognathism | n/a | - |
| Other features | Puffy dry skin over hands and feet, wide based gait | Transposition of the great vessels, tricuspid atresia, VSD. Left single palmar crease | Spinal lordosis | Bilateral 2–3 toe cutaneous syndactyly, hypermobility. | Brachydactyly, tapering fingers | Broad thumbs and great toes, truncal obesity | n/a | - |
| Prominent eyebrows | No | Yes | Yes | No | Yes | Yes | No | 4/7 (57%) |
| Elongated PF | No | Yes | No | Yes | Yes | Yes | No | 4/7 (57%) |
| Prominent nasal bridge | No | Yes | No | No | Yes | Yes | No | 3/7 (43%) |
| Overhanging columella | No | Yes | No | Yes | Yes | Yes | No | 4/7 (57%) |
| Thin upper lip | Yes | Yes | No | Yes | Yes | Yes | Yes | 6/7 (86%) |
| Patient ID (DECIPHER ID) | 1 (258995) | 2 (260453) | 3 (265865) | 4 (268390) | 5 (277603) | 6 (263453) | 7 | Total (of features reported) |
| ID | Moderate | Moderate | Moderate | Severe | Severe | Severe | Moderate | 7/7 (100%) |
| Seizures | Yes | None | Yes | Single seizure only | Yes | Yes | Yes | 5/7 (71%) |
| Age onset of seizures | 5yr | n/a | 1yrr | 5yr | <1yr | 8 months | 18 months | n/a |
| Febrile seizures | Yes | No | Yes | No | Yes | No | Yes | 4/7 (57%) |
| EE | No | No | No | No | No | No | No | 0/7 (0%) |
| Delayed development | Yes | Yes | Yes | Yes | Yes | Yes | Yes | 7/7 (100%) |
| Sat independently | 9 months | 10 months | 18 months | 2–2.5yr | 15 months | 15 months | 18 months | n/a |
| Walked independently | 2–2.5yr | 2–2.5yr | 4–5yr | 5yr | 22 months | 39 months | 6.5yr | n/a |
| First words | Over 5yr | 2–2.5yr | 18 months | 5yr | 2–2.5yr | not achieved aged 11 yr | 3yr | n/a |
| Behaviour | n/a | Angers easily, Tourette syndrome | Autism traits | Very sociable | Aggression with violent episodes | Very sociable | n/a | n/a |
| Hand flapping | Yes | No | Yes | Yes | Yes | Yes | No | 5/7 (71%) |
| Special educational needs | Yes | Yes | Yes | Yes | Yes | Yes | Mainstream school with additional support | 6/7 (86%) |
| Cranial MRI abnormal* | No | Yes* | No | No | No | n/a | Yes* | 2/7 (29%) |
Abbreviations: EE – epileptic encephalopathy. ID – intellectual disability. n/a- not applicable. NAD- no abnormalities detected. n/r- not reported. PF – palpebral fissures. UPD- uniparental disomy. VSD – ventricular septal defect.
MRI abnormalities: Patient 2 - Small periventricular areas with high T2 signal; Patient 7 - Non-progressive T2 and FLAIR hyperintensities in white matter bilaterally.
Table III.
Comparison of features in individuals with HNRNPU mutations compared with those reported in the literature. Note the patient previously reported by Hamdan et al., 2014 has been described in more detail by Bramswig et al., 2017
| Clinical features | This study cohort | Depienne et al.,2017 | Bramswig et al.,2017 | de Kovel et al., 2016 | Epi 4K-Consortium., 2013 | Carvill et al., 2013 | Need et al., 2012 | Total (of reported features) |
|---|---|---|---|---|---|---|---|---|
| Distinct facial appearance | 6/7 | n/r | 6/6 | 1/1 | 0/1 | n/r | n/r | 13/15 (87%) |
| ID | 7/7 | 7/7 | 6/6 | n/r | 1/1 | 1/1 | 1/1 | 23/23 (100%) |
| Seizures | 5/7 | 6/7 | 6/6 | 1/1 | 1/1 | 1/1 | 1/1 | 2½4 (88%) |
| Seizure onset prior to five years age | 6/6 | 6/6 | 5/5 | 1/1 | 1/1 | 1/1 | n/r | 20/20 (100%) |
| Febrile seizures | 4/7 | 5/6 | 2/5 | 1/1 | 1/1 | 0/1 | n/r | 13/21 (62%) |
| Delayed development | 7/7 | 7/7 | 5/5 | 1/1 | 1/1 | 1/1 | n/r | 22/22 (100%) |
| Speech impairment | 6/7 | 7/7 | 6/6 | 1/1 | 1/1 | n/r | n/r | 2½2 (95%) |
| Hand flapping | 5/7 | 1/7 | 1/6 | n/r | n/r | n/r | n/r | 7/20 (35%) |
| Abnormal MRI-brain | 2/7 | 3/5 | 4/4 | 1/1 | 1/1 | n/r | n/r | 11/18 (61%) |
Abbreviations: ID – intellectual disability. n/a- not applicable. n/r- not reported. PF – palpebral fissures.
Figure 2a.
Patients in presented series (age): a. Pt 1 (15yr) b. Pt 2 (14yr) c. Pt 4 (6yr) d. Pt 5 (11yr) e. Pt 6 (6yr) f. Pt 7 (10yr) Note dysmorphic features including prominent eyebrows, long palpebral fissures and thin upper lip.
There is significant developmental delay in all of the patients presented here. Speech appears to be disproportionately affected. All the patients in this series have moderate to severe ID. Most have special educational needs (6/7). Epilepsy is a frequent finding (5/7), and seizure onset is usually by the age of one year. Initial seizure presentation is in the form of febrile seizures progressing to afebrile seizures. There does not appear to be a predominance of a particular form of seizure, with patients presenting with a combination of absences, generalised and focal epilepsy. Developmental delay is apparent before the onset of seizures.
Two patients had a tendency to aggressive outbursts, with one demonstrating some violent behaviours. Interestingly, a patient with similar issues, requiring antipsychotic medication, has been reported [Epi4K Consortium, 2013]. In addition, 5/7 patients demonstrated hand-flapping behaviours. Therefore, behavioural abnormalities may also represent part of the HNRNPU-associated phenotype. Of note, diagnoses considered, prior to exome sequencing, in some of these patients included Smith-Magenis and Angelman syndromes, suggesting behavioural issues are a prominent presenting feature. Additionally, some of the patients in this cohort also had RAI1 testing as the behavioural phenotype was very suggestive.
One patient in our series (Patient 2) was also found to have maternal UPD of chromosome nine [King et al., 2014]. No other variants were identified. To the authors’ knowledge, there is no definite evidence of imprinting on this chromosome, and UPD 9 (mat) has been reported in a phenotypically normal individual [Bjorck et al., 1999]. Therefore, this is not thought likely to have a significant impact on phenotype. However, Patient 2 had a significant cardiac defect and appears to have a more severe phenotype than other patients in this series. It remains to be seen if the UPD9 (mat) is contributing to some of his clinical presentation.
All but one of the patients presented here have mutations predicted to result in loss-of-function. This suggests haploinsufficiency as a likely pathogenic mechanism. Indeed, studies of patients with 1q43q44 deletions have identified HNRNPU deletion as a possible cause of epilepsy and ID, as part of a wider syndrome including microcephaly and central nervous system anomalies [Caliebe et al., 2010; Thierry et al., 2012]. More recently, this has been confirmed in a series directly comparing the phenotypes of patients with 1q43q44 microdeletions to those with loss-of-function HNRNPU mutations [Depienne et al., 2017]. This study showed that changes in HNRNPU determine the epilepsy phenotype in 1q43q44 syndrome, and have a significant influence on the degree of ID.
A recent series of six individuals with HNRNPU mutations also demonstrates an ID and epilepsy phenotype [Bramswig et al., 2017]. Interestingly, all of these patients have severe ID, in contrast to our cohort, in which 4/7 patients have moderate learning difficulties. Speech impairment was prominent, in keeping with our findings. Two of these patients (individuals 3 and 4) have similar facial features to those seen in our cohort, further delineating the phenotypic spectrum associated with HNRNPU.
Three other patients (individuals 1, 2 and 6) in the series of Bramswig et al. [2017] have more severe craniofacial dysmorphism, including dental anomalies, which is not in keeping with our findings. Two of these patients (1 and 6) have missense mutations. Interestingly, the HNRNPU mutations found in these individuals are all within the large protein-protein interaction domain B30.2/SPRY, indicating a possible genotype-phenotype correlation. However, two patients in our cohort (Patients 3 and 6) also have mutations within this domain, but do not share the more severe phenotype. It is possible that the missense mutations seen in individuals 1 and 6 from Bramswig et al. [2017] result in an alternate pathogenic mechanism compared to the likely haploinsufficiency seen in our cohort. Therefore, the significance of these mutations, and the possible clustering in B30.2/SPRY, remains to be defined.
In addition, 4/6 individuals in the study of Bramswig et al. [2017] had cardiac abnormalities and ¾ renal abnormalities. Only one of our patients had cardiac abnormalities, and, as discussed, this may be associated with his UPD of chromosome 9. None of our patients had renal problems. Our findings therefore do not support the association of HNRNPU mutations with cardiac and/or renal abnormalities.
CONCLUSION
In summary, we present evidence that a neurodevelopmental syndrome with features including ID, seizures, behavioural abnormalities, and craniofacial dysmorphism, is associated with de novo loss of function mutations in HNRNPU. This delineation of the phenotype should aid identification of further clinically affected patients and allow for more accurate interpretation of results obtained through genomic testing.
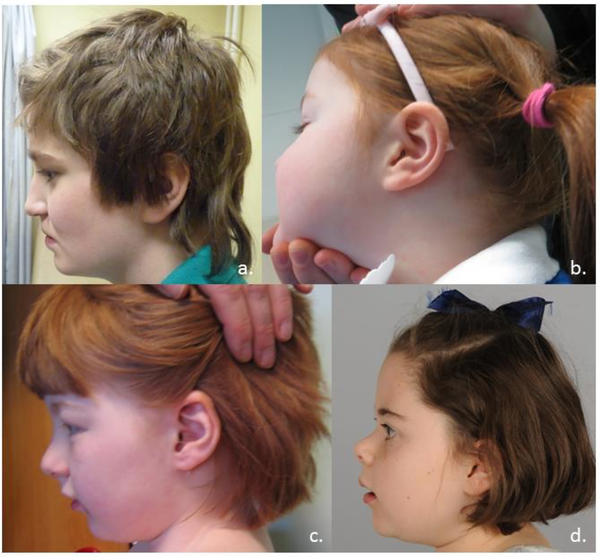
Figure 2b.
Patient profiles (age): a. Pt 2 (14yr) b. Pt 4 (6yr) c. Pt 5 (11yr) d. Pt 7 (10yr)
[Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552–4833.]
ACKNOWLEDGEMENTS
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009–003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
Sequencing and analysis of patient 7 was provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 to Daniel MacArthur and Heidi Rehm.
Footnotes
CONFLICT OF INTEREST
The authors do not have any conflict of interest to disclose.
CONSENT
Informed consent was obtained for all subjects for inclusion in this study.
REFERENCES
- Bjorck EJ, Anderlid BM, Blennow E. Maternal isodisomy of chromosome 9 with no impact on the phenotype in a woman with two isochromosomes: i(9p) and i(9q). Am J Med Genet 1999;87(1):49–52. [DOI] [PubMed] [Google Scholar]
- Bramswig NC, Ludecke H-J, Hamdan FF, Altmuller J, Beleggia F, Elcioglu NH, Freyer C, Gerkes EH, Demirkol YK, Knupp KG, Kuechler A, Li, Y, Lowenstein DH, Michaud JL, Park K, Stegmann APA, Veenstra-Knol HE, Wieland T, Wollnik B, Engels H, Strom TM, Kleefstra T, Wieczorek D. Heterozygous HNRNPU variants cause early onset epilepsy and severe intellectual disability. Hum Genet Published Online First: 9 April 2017.doi: 10.1007/s00439-017-1795-6 [DOI] [PubMed] [Google Scholar]
- Caliebe A, Kroes HY, van der Smagt JJ, Martin-Subero JI, Tönnies H, van’t Slot R, Nievelstein RA, Muhle H, Stephani U, Alfke K, Stefanova I, Hellenbroich Y, Gillessen-Kaesbach G, Hochstenbach R, Siebert R, Poot M. Four patients with speech delay, seizures and variable corpus callosum thickness sharing a 0.440 Mb deletion in region 1q44 containing the HNRPU gene. Eur J Med Genet 2010;53(4):179–185. [DOI] [PubMed] [Google Scholar]
- Carvill GL, Heavin SB, Yendle SC, Mc Mahon JM, O’Roak BJ, Cook J, Khan A, Dorschner MO, Weaver M, Calvert S, Malone S, Wallace G, Stanley T, Bye AME, Bleasel A, Howell KB, Kivity S, Mackay MT, Rodriguez-Casero V, Webster R, Korczyn A, Afawi Z, Zelnick N, Lerman-Sagie T, Lev D, Moller RS, Gill D, Andrade DM, Freeman JL, Sadleir LG, Shendure J, Berkovic SF, Scheffer IE, Mefford HC. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 2013;45(7):825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CGF, Brilstra EH, van Kempen MJA, van’t Slot R, Nijman IJ, Afawi Z, De Jonghe P, Djemie T, Guerrini R, Hardies K, Helbig I, Hendrickx R, Kanaan M, Kramer U, Lehesjoki A-EE, Lemke JR, Marini C, Mei D, Moller RS, Pendziwiat M, Stamberger H, Suls A, Weckhuysen S, EuroEPINOMICS RES Consortium, Koeleman BPC. Mol Genet Genomic Med 2016; 4(5): 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Nava C, Keren B, Heide S, Rastetter A, Passemard S, Chantot-Bastaraud S, Moutard M-L, Agrawal PB, VanNoy G, Stoler JM, Amor DJ, Billette de Villemeur T, Doummar D, Alby C, Cormier-Daire V, Garel C, Marzin P, Scheidecker S, de Saint-Martin A, Hirsch E, Korff C, Bottani A, Faivre L, Verloes A, Orzechowski C, Burglen L, Leheup B, Roume J, Andrieux J, Sheth F, Datar C, Parker MJ, Pasquier L, Odent S, Naudion S, Delrue M-A, Le Caignec C, Vincent M, Isidor B, Renaldo F, Stewart F, Toutain A, Koehler U, Hackl B, von Stulpnagel C, Kluger G, Moller RS, Pal D, Jonson T, Soller M, Verbeek NE, van Haelst MM, de Kovel C, Koeleman B, Monroe G, van Haaften G, DDD study, Attie-Bitach T, Boutaud L, Heron D, Mignot C. Genetic and phenotypic dissection of 1q43q44 microdeletion syndrome and neurodevelopmental phenotypes associated with mutations in ZBTB18 and HNRNPU. Hum Genet 2017;136:463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi4K-Consortium & Epilepsy Phenome/Genome Project. De novo mutations in epileptic encephalopathies. Nature 2013;501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet 2016;135:851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Srour M, Capo-Chichi J-M, Daoud H, Nassif C, Patry L, Massicotte C, Ambalavanan A, Spiegelman D, Diallo O, Henrion E, Dionne-Laporte A, Fougerat A, Pshezhetsky AV, Venkateswaran S, Rouleau GA, Michaud JL. De novo mutations in moderate or severe intellectual disability. PLOS Genet 2014;10(10):e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 2010;19:469–476. [DOI] [PubMed] [Google Scholar]
- King DA, Fitzgerald TW, Miller R, Canham N, Clayton-Smith J, Johnson D, Mansour S, Stewart F, Vasudevan P, Hurles ME, DDD Study. A novel method for detecting uniparental disomy from trio genotype identifies a significant excess in children with developmental disorders. Genome Res 2014;24:673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Zhao X-Y, Li S, Yang G, Lin JD. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol Metab 2017;6:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, Meisler MH, Goldstein DB. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet 2012;49(6):353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, Dumitriu S, Dyke SOM, den Dunnen JT, Firth HV, Gibbs RA, Girdea M, Gonzalez M, Haendel MA, Hamosh A, Holm IA, Huang L, Hurles ME, Hutton B, Krier JB, Misyura A, Mungall CJ, Paschall J, Paten B, Robinson PN, Schiettecatte F, Sobreira NL, Swaminathan GJ, Taschner PE, Terry SF, Washington NL, Züchner S, Boycott KM, Rehm HL. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum Mutat 2015;36:915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS, O’Brien D, Pisat N, Weichselbaum CT, Sakers K, Lisci M, Dalal JS, Dougherty JD. A comprehensive analysis of cell type-specific nuclear RNA from neurons and glia of the brain. Biol Psychiatry 2017;81:252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshon MJ, Ruley HE. Hypomorphic mutation in hnRNP U results in post-implantation lethality. Transgenic Res 2005;14(2):179–192. [DOI] [PubMed] [Google Scholar]
- Thierry G, Bénéteau C, Pichon O, Flori E, Isidor B, Popelard F, Delrue MA, Duboscq-Bidot L, Thuresson AC, van Bon BW, Cailley D, Rooryck C, Paubel A, Metay C, Dusser A, Pasquier L, Béri M, Bonnet C, Jaillard S, Dubourg C, Tou B, Quéré MP, Soussi-Zander C, Toutain A, Lacombe D, Arveiler B, de Vries BB, Jonveaux P, David A, Le Caignec C. Molecular characterization of 1q44 microdeletion in 11 patients reveals three candidate genes for intellectual disability and seizures. Am J Med Genet A 2012;158A(7):1633–1630. [DOI] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, van Kogelenberg M, King DA, Ambridge K, Barrett DM, Bayzetinova T, Bevan AP, Bragin E, Chatzimichali EA, Gribble S, Jones P, Krishnappa N, Mason LE, Miller R, Morley KI, Parthiban V, Prigmore E, Rajan D, Sifrim A, Swaminathan GJ, Tivey AR, Middleton A, Parker M, Carter NP, Barrett JC, Hurles ME, FitzPatrick DR, Firth HV; DDD study. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015;385(9975):1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Beetz N, O’Keeffe S, Tapia JC, Macpherson L, Chen WV, Bassel-Duby R, Olson EN, Maniatis T. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc Natl Acad Sci USA 2015;9:112(23):E3020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]