Abstract
Background
Tick selenoproteins are involved in regulating oxidative and endoplasmic reticulum stress during prolonged tick feeding on mammalian hosts. How selenoproteins are activated upon tick-borne pathogen infection is yet to be defined.
Methods
To examine the functional role of selenoprotein K in Borrelia burgdorferi infection within the tick host Ixodes scapularis, RNA interference (RNAi)-based gene silencing was performed.
Results
Selenoprotein K is an endoplasmic reticulum (ER)-resident protein and a component of the ERAD complex involved in ER homeostasis. A qRT-PCR assay revealed the significant upregulation of selenogene K (selenoK) expression in B. burgdorferi-infected tick tissues. Silencing of the selenoK transcript significantly depleted B. burgdorferi copies within the infected tick tissues. Upon selenoK knockdown, another component of the ERAD complex, selenoprotein S (selenoS), was significantly upregulated, suggesting a compensatory mechanism to maintain ER homeostasis within the tick tissues. Knockdown of selenoK also upregulated ER stress-related unfolded protein response (UPR) pathway components, ATF6 and EIF2.
Conclusions
The exact mechanisms that contribute to depletion of B. burgdorferi upon selenoK knockdown is yet to be determined, but this study suggests that selenoK may play a vital role in the survival of B. burgdorferi within the tick host.
Electronic supplementary material
The online version of this article (10.1186/s13071-019-3548-y) contains supplementary material, which is available to authorized users.
Keywords: Tick, Ixodes scapularis, Borrelia burgdorferi, Lyme disease, Selenoproteins, ER stress
Background
In the USA, reported vector-borne diseases are predominantly tick-borne; for example, there are approximately 329,000 cases of Lyme disease annually in the USA. A recent CDC study based upon vector-borne disease cases reported to the National Notifiable Disease Surveillance System from 2004 to 2016 revealed 491,671 cases of tick-borne infectious diseases (76.5% of all vector-borne diseases) (http://ww.cdc.gov/mmwr) [1]. In the USA, Lyme disease-causing B. burgdorferi spirochetes are primarily harbored by the blacklegged tick I. scapularis [2].
Tick blood-feeding also generates toxic levels of reactive oxygen species (ROS) that could damage lipids, proteins and DNA, and promote mutation, cellular dysfunction and cell death. To successfully feed and survive, ticks must somehow prevent these detrimental effects and promote the beneficial aspects of ROS, which suggests that there are precise regulatory strategies for maintaining appropriate ROS levels both within the tick and possibly at the tick-host interface. Our previously published studies have shown an adaptive coevolutionary process that has enabled tick-borne pathogen survival by manipulating an antioxidant defense system associated with selenium including a full set of selenoproteins and other antioxidants [3–12]. Generation of ROS is among the first lines of host defense against invading microbes [13, 14].
Selenoproteins exhibit diverse biological functions such as detoxification of peroxides, regeneration of reduced thioredoxin and reduction of oxidized methionine residues by oxidation of the selenium (Se−) active site [15–17]. In the last decade, significant progress has been made in clarifying the functions and physiological roles of vertebrate selenoproteins; new selenoprotein families have been identified, and new functions have been assigned to previously characterized selenoproteins. Some of the newer specific functions of selenoproteins involve removal of hydrogen peroxide, repair of oxidatively damaged proteins, control of cytoskeleton/actin assembly, protein folding and mitigating ER stress as part of the endoplasmic reticulum associated degradation (ERAD) complex [17], among others. Tick selenoproteins have been shown to play an important role in mitigating oxidative stress [3–12], pathogen colonization [5, 7–9, 11, 18–20] and microbiota maintenance [9, 10, 12].
A study using a tick-pathogen model has shown that the initial stage of transmission of a pathogen by an arthropod vector is influenced by gene expression changes in both vectors and pathogens [21]. However, there is a lack of knowledge on how gene expression within the vector is altered by the presence of the vector-borne pathogen. Therefore, the present study was designed to test the hypothesis that B. burgdorferi infection activates upregulation of selenogene K expression in order to survive within the tick host, I. scapularis, before transmission to the mammalian host. To test this hypothesis, a qRT-PCR assay was used to determine the expression of selenoK in B. burgdorferi-infected I. scapularis tissues over the course of a feeding period. An RNA interference approach also was used to silence selenogene K expression in B. burgdorferi-infected ticks to examine pathogen survival within the tick vector.
Methods
Materials
All common laboratory supplies and chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA), Fisher Scientific (Grand Island, NY, USA) or Bio-Rad (Hercules, CA, USA) unless otherwise specified.
Bioinformatics analysis of selenoprotein K
The full-length gene sequence of I. scapularis selenoK (GenBank: XM_002403043.1) was obtained from the NCBI database. Corresponding protein sequences for I. scapularis selenoK (NCBI Accession number: XP_002403087.1) were aligned with selenoK protein sequences from other organisms such as Amblyomma maculatum, A. americanum, Gallus gallus, Rattus norvegicus, Mus musculus, Equus caballus, Homo sapiens, Macaca mulatta, Pan troglodytes and Pongo abelii by using Clustal Omega [22] for multiple sequence alignment. selenoK protein sequences aligned by Clustal Omega were refined by eye and presented by Jalview v.2.7 [23]. Additionally, the freely available online tool SECISsearch3 (http://seblastian.crg.es/) was used for the prediction of the SECIS (Selenocysteine Insertion Sequence) element in the tick selenoK nucleotide sequence, required for amino acid selenocysteine (Sec) insertion during translation [24]. Subcellular localization of tick selenoK was also predicted by using the freely available online algorithm DeepLoc-1.0 (http://www.cbs.dtu.dk/services/DeepLoc/index.php) which predicts the subcellular localization of eukaryotic proteins [25].
Ticks and tissue dissections
Ticks were purchased from the Oklahoma State University Tick Rearing Facility. Adult male and female I. scapularis were kept according to standard practices [26] and maintained in the laboratory as described in our previously published work [12]. Unfed female adult I. scapularis were infected with laboratory grown B. burgdorferi strain B31.5A19 using the capillary feeding method at Tulane National Primate Research Center [27]. Naturally B. burgdorferi-infected adult I. scapularis were collected from field (Kingston, Rhode Island) and maintained in the laboratory using standard procedures [28]. Rabbit was used as host for tick blood-feeding. The blood-fed adult female I. scapularis were dissected within 60 min of removal and collection from the rabbit. Tick tissues were dissected and washed in M-199 buffer as described previously [12]. Salivary glands and midguts from individual I. scapularis were stored in RNAlater (Life Technologies, Carlsbad, CA, USA) at − 80 °C until used.
Total RNA isolation, cDNA synthesis, dsRNA preparation and transcriptional expression
The methods to extract total RNA, cDNA synthesis, double-stranded RNA for selenoK, irrelevant gene LacZ, and qRT-PCR assays were performed as described previously [9, 12]. RNA was extracted from individually-dissected tick tissues (salivary gland, midgut), and cDNA was synthesized from each of them to check whether they were infected or not. Only tissues which were found infected were pursued for further work. The concentration of dsRNA used was 1000 ng/µl. One microliter of dsRNA was injected into the tick hemocoel. To investigate the role of selenoK in tick feeding success and pathogen survival, 45 unfed adult female capillary-fed (B. burgdorferi culture) ticks were injected with 1000 ng of selenoK-dsRNA into the hemocoel using a Hamilton syringe fitted with a 33-gauge needle [29]. As a control, a total of 45 unfed adult capillary-fed (B. burgdorferi culture) ticks were injected with 1000 ng of lacZ-dsRNA (an irrelevant control dsRNA). After the injection of dsRNA, ticks were kept at 37 °C overnight under high humidity to confirm tick survival. Ixodes scapularis ticks were then placed on rabbit ears [30]. Ixodes scapularis naturally infected with B. burgdorferi were collected from Rhode Island to determine the role of selenoK silencing on pathogen infection. A total of 90 female adult ticks were divided into two groups: one group of 45 female unfed ticks was injected with irrelevant dsRNA-lacZ; the second with selenoK dsRNA followed by tick infestation on rabbit ears as described earlier.
PCR-based detection of B. burgdorferi
Borrelia burgdorferi was detected in the tick tissues using the flaB gene in a PCR assay [31]. The flaB gene and other primers used in the experiments are provided in Additional file 1: Table S1. The PCR conditions were followed from a previous study with slight modification [31]: 1 cycle of 94 °C for 5 min; 50 cycles of 94 °C for 30 s, 50 °C for 30 s and 68 °C for 1 min; and 1 cycle of 72 °C for 8 min.
Quantification of B. burgdorferi in I. scapularis tissues
The principle of quantification of B. burgdorferi load was followed from our studies established to quantify the load of the spotted fever group rickettsia, Rickettsia parkeri, in tick tissues [9]. Borrelia burgdorferi load in I. scapularis tissues were estimated by quantifying the number of copies of B. burgdorferi flagellin gene, flaB [32] present per copy of housekeeping gene Rps4 [30]. A list of all primers used is provided in Additional file 1: Table S1. The gene flaB from B. burgdorferi and Rps4 from I. scapularis were amplified, purified, sequenced and verified prior to further assays. Their standard curves were determined by qRT-PCR based on serially-diluted PCR products. qRT-PCR conditions were as follows: 50 °C for 3 min, 95 °C for 10 min; followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s.
Statistical analysis
All data were expressed as mean values ± standard error of the mean (SEM) unless otherwise indicated. Statistical significance between two experimental groups or their respective controls was determined by a t-test (P-value, < 0.05). P < 0.05 was considered statistically significant. Comparative differences among multiple experimental groups were determined by analysis of variance with P-values < 0.05 considered statistically significant (GraphPad Prism 6.05; GraphPad Software, La Jolla, CA, USA). Transcriptional expression levels were determined using Bio-Rad CFX MANAGER v.3.1, and the gene expression values obtained were considered statistically significant if a P-value of 0.05 was obtained when compared with the control.
Results and discussion
Bioinformatics analysis
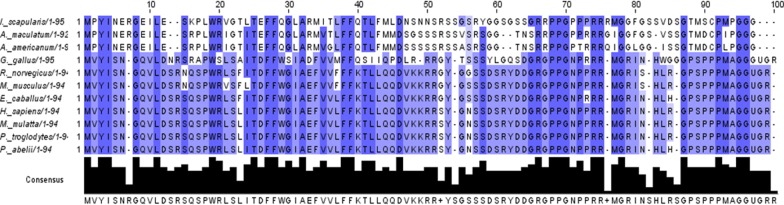
Selenoprotein K is a small (~ 10 kDa) and single spanning transmembrane protein localized on the ER membrane [33]. It contains the selenocysteine residue near the c-terminus in the cytosolic region, which remains in a highly disordered state [34]. Ixodes scapularis, Amblyomma maculatum, and A. americanum selenoK sequences (Fig. 1) demonstrate 77% amino acid identity with each other but share only 38–40% amino acid identity with Homo sapiens. Interestingly, many of the assembled transcripts in our unpublished RNA-Seq data are novel and not found in the I. scapularis annotation. This is not surprising, since the genome of I. scapularis has 50% coverage. Based on bioinformatics analysis, I. scapularis contains most of the selenogenes which have already been characterized in A. maculatum [5–7, 9, 10]. Interestingly, I. scapularis selenoK has 77% amino acid identity with that of A. maculatum suggesting conserved functions. In this work, the selenocysteine insertion element (SECIS) of tick selenoK was also predicted by algorithm SECISSearch3 [24]. The SECIS element is a hairpin loop structure required in a selenoprotein transcript for incorporation of selenocysteine (Sec) residue during translation. SECISSearch3 algorithm has been developed to detect SECIS elements in prospective selenogene sequences by using different parameters [24]. SECISSearch3 prediction is the combined output of three sources: infernal, used by the program cmsearch, SECIS model, used by the program Covels and the third source is the original program SECISSearch [35, 36]. Submission of I. scapularis selenoK sequence to the SECISSearch3 algorithm resulted into output data (Additional file 2: Figure S1). Generally, vertebrates have an infernal score of > 20 and a Covel score of > 15; in other species these scores are generally lower. The Covel score provides a quantitative measure of how better fit is the prediction to the SECIS model. An RNAfold algorithm is run to calculate the minimum free energy of SECIS structures among those suggested. Start to end positions of the SECIS element, and the strands on which it is located, is also given in the output data. The marker for SECIS element prediction quality is based on the experience of manual analysis of thousands of SECIS elements, and is graded as A, B or C in decreasing order of prediction quality. In our output data, infernal and Covel scores of the selenoK SECIS element were 19.62 and 21.35, respectively (Additional file 2: Figure S1). In our data, the grade provided by the algorithm to tick selenoK prediction was A, indicating the most trustworthy prediction possible under the algorithm parameters.
Fig. 1.
Multiple sequence alignment of selenoK. Tick selenoK sequences (as shown above, I. scapularis, A. maculatum and A. americanum) demonstrate 77% amino acid identity with each other but share only 38–40% amino acid identity with Homo sapiens
In the present work, subcellular localization of tick selenoK was predicted by DeepLoc-v.1.0 server which uses neural network algorithms based on the experimental localization of Uniprot proteins [25]. To include the protein sorting pathways into the algorithm, a hierarchical tree with multiple nodes was also designed. Each node of the tree represents a binary attempt to assign the protein right pathway from high to low in a hierarchical classification. As shown in Additional file 3: Figure S2, the probability score of tick selenoK is much higher at the ER/Golgi node than other organelles. Thus, our data suggest that the localization of tick selenoK is in the endoplasmic reticulum ER/Golgi membrane (Additional file 3: Figure S2). A previous study on Drosophila has also reported selenoK integration into both of these cell organelles [37]. Attention plot (Alpha) in Additional file 3: Figure S2 indicated that the specific interspersed region of the selenoprotein K (residue number ~ 20–40 from the N-terminal) contributes in binding to ER/Golgi membrane.
It has been suggested that all of the characterized selenoproteins can reduce ROS levels in the cell. However, at the physiological level, their specific roles are yet to be discovered in arthropod vectors like ticks. selenoK lacks the defined redox motif C-X-X-Sec (X = any amino acid) found in other known selenoproteins. Thus, at this time, it cannot be firmly stated that selenoK has an antioxidant role in vivo. It could be speculated that selenoK is part of the protein complex, which demonstrates its antioxidant property (Additional file 3: Figure S2). The exact cellular localization and function of tick selenoprotein in hematophagy and pathogen infection has yet to be determined.
Temporal expression during ingestion of the blood meal
Our recently published work showed the pathogen-induced expression of several tick selenoproteins [7, 9]. Several studies implicated the involvement of selenoK in mitigating endoplasmic reticulum (ER) stress generated upon microbial infection and elevated oxidative stress [38–40]. These and bioinformatics results prompted us to determine the the functional role of selenoK in B. burgdorferi infection of the tick vector. The B. burgdorferi infection level of capillary-fed and naturally-infected I. scapularis was determined using flaB (flagellin) gene PCR. The B. burgdorferi infection level in capillary-fed ticks and field-collected ticks ranged between 10–20%. Borrelia burgdorferi multiplies in midgut tissues before trafficking to the salivary glands. Therefore, the time-dependent expression of ER-resident selenoK, selenoS and UPR pathway genes ATF6 and EIF2 were examined in B. burgdorferi-infected midgut tissues. Transcriptional expression of selenoK significantly increased from 2- to 20-fold in tick tissues from day 2 to day 8 of blood-feeding (2 days, t = 4.627, P = 0.0098; 4 days, t = 10.94, P = 0.0004; 6 days, t = 24.8, P < 0.0001; 8 days, t = 3.525, P = 0.0243) (Fig. 2) supporting its critical role in B. burgdorferi colonization of tick tissues. Other upregulated ER-stress related genes during blood-feeding are selenoS (8 days, t = 3.462, P = 0.0258), ATF6 (2 days, t = 3.404, P = 0.0272; 6 days, t = 7.505, P = 0.0017; 8 days, t = 3.142, P = 0.0348), EIF2 (4 days, t = 2.812, P = 0.0482). selenoK is an ER-resident selenoprotein and one of the constituents of the ERAD complex which has a role in mitigating ER stress [41, 42]. Previous studies have also revealed that ER stress increases during pathogen infection and elevated oxidative stress [38–40]. According to one estimate, 70–80% of oxidative stress in the cell is due to mitochondria and mitochondrial ROS, and pathogen infection can disturb the ER homeostasis by disrupting protein folding and ER calcium ion balance which may result in elevated ER stress levels [43]. A previous study has already shown a physical and biochemical interaction between the ER and mitochondria [43, 44]. Since selenoK is an ER-resident selenoprotein and is one of the constituents of the ERAD complex which has a role in mitigating ER stress [41, 42], the significant upregulation of selenoK provided the basis of suggesting a functional role of selenoK in B. burgdorferi colonization within the tick. Other ER-stress related genes considered in this study during blood-feeding were SelS, ATF6 and EIF2. Unfolded protein response (UPR) sensor related genes ATF6 and EIF2 also demonstrated upregulation during blood-feeding.
Fig. 2.
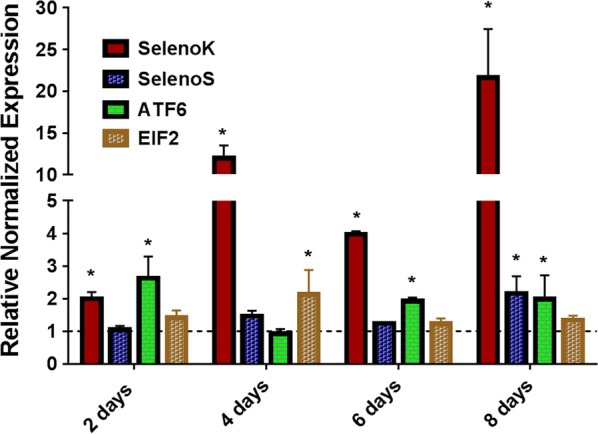
Temporal expression of ER-stress related genes in Borrelia burgdorferi-infected ticks. ATF6 and EIF2 are unfolded protein response related sensor genes while selenoK and selenoS are components of the ERAD complex involved in ER stress mitigation through the ERAD pathway. Gene expression of the mentioned genes was normalized with that of clean tick midgut gene expression (indicated as 1 on the y-axis). Statistically significant gene expression values (P < 0.05) are indicated with * (asterisk). Rps4 was used as a housekeeping gene. Abbreviations: ATF6, activating transcription factor 6; EIF2, eukaryotic initiation factor 2 (eIF2)
SelenoK knockdown and B. burgdorferi infection
Injection of selenoK-dsRNA into I. scapularis female ticks resulted in an 80–90% reduction in corresponding transcript levels in the salivary gland (SG) and midgut (MG) tissues (Fig. 3a). One of the limitations of this study was that the B. burgdorferi infection rate was 10–20%, hence it was necessary to check every tick for infection and to quantify the infection level. In capillary-fed ticks, the infection level varied from tick to tick, whereas naturally-infected ticks had a more uniform level of infection. Interestingly, selenoK silencing significantly upregulated the expression of selenoS, suggesting a compensatory mechanism in this tick species, also shown in a recent study from our laboratory [9].
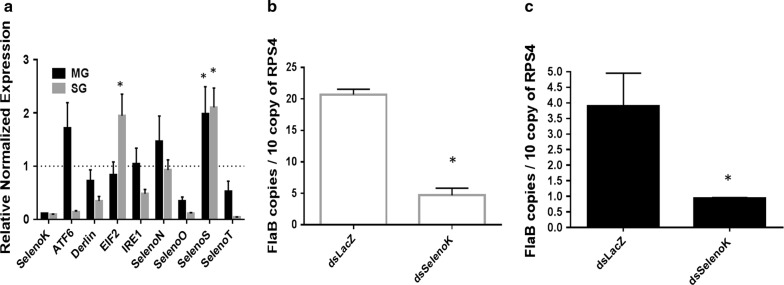
Fig. 3.
Effect of selenoK knockdown on Borrelia burgdorferi infection (capillary-fed induced infection). a Compensatory expression of ER stress genes when selenoK is knocked down in B. burgdorferi-infected tick tissues. selenoS, ATF6 and EIF2 demonstrate compensatory expression. selenoS and selenoK localized on ER membrane and are components of the ERAD, selenoS demonstrates compensation for selenoK in both tissues (SG, MG). b, c Impact of selenoK knockdown on B. burgdorferi infection in SG (b) and MG (c) tissues. Statistically significant gene expression values (P < 0.05) are indicated with * (asterisk). Rps4 was used as a housekeeping gene. Abbreviations: ATF6, activating transcription factor 6; EIF2, eukaryotic initiation factor 2; IRE1, inositol-requiring enzyme; Rps4, ribosomal protein S4; SG, salivary gland; MG, midgut
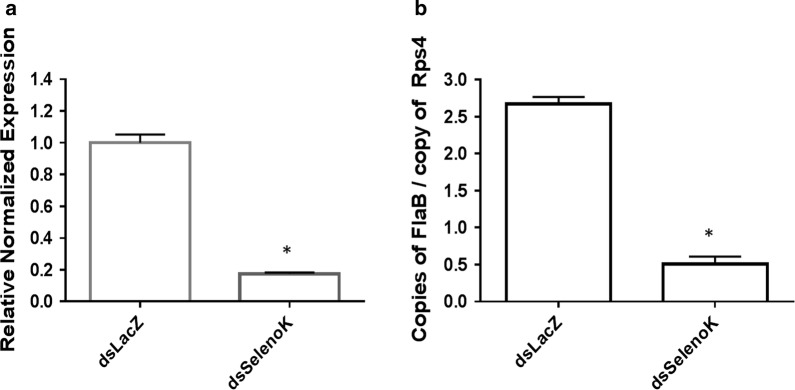
It has been reported that two cellular functions exist for selenoK, including ER stress mitigation by participation in the ER-associated protein degradation (ERAD) pathway, and regulation of Ca2+ flux from the ER [34]. Its speculative role in ERAD is based on the evidence that its transcripts were upregulated during ER stress [33], and later an ER stress response element (ERSE) was identified in the promoter of human selenoK [33]. Studies have demonstrated that selenoK is bound within the ERAD complex along with selenoS, p97 (VCP; valosin-containing protein) and the Derlins [33, 45]. In the present study, upregulation of ATF6 and EIF2 indicated an elevation in ER stress (Fig. 3a, for EIF2, t = 3.099, P = 0.0363), and upregulation of selenoS (~ 2 folds) in both SG and MG (Fig. 3a; for MG tissue, t = 3.143, P = 0.0347; for SG tissue, t = 3.055, P = 0.0379) indicate a partial compensatory mechanism (Fig. 3a). Borrelia burgdorferi load was significantly altered in selenoK knockdown salivary gland and midgut tissues (Fig. 3b, c; F(3,8) = 158.7, P < 0.0001). One-way ANOVA using multiple comparison was used with Tukeyʼs test. SelenoS also is one of the components of the ERAD complex required to mitigate ER stress. Previous studies have shown that selenoK knockout mice appeared healthy, fertile and without any ER stress [33], probably because of redundancy or compensation [34]. Silencing of selenoK showed significant reduction in B. burgdorferi copies in both SG and MG tissues (Fig. 3b, c; for SG, t = 11.32, P = 0.0003; for MG, t = 4.508, P = 0.0108). The uneven infection level of B. burgdorferi in capillary-fed ticks prompted us to use naturally infected ticks to confirm these results (Fig. 4a, b). Upon selenoK knockdown in ticks, naturally infected I. scapularis ticks also demonstrated reduction in B. burgdorferi load (Fig. 4a, selenoK knockdown, t = 15.68, P < 0.0001; Fig. 4b, bacterial load reduction, t = 15.82, P < 0.0001).
Fig. 4.
Effect of selenoK knockdown on Borrelia burgdorferi infection (naturally-infected ticks) a Knockdown level of selenoK in infected tick tissue. b Impact of selenoK knockdown on Borrelia load in ticks. Statistically significant gene expression values (P < 0.05) are indicated with * (asterisk). Rps4 was used as a housekeeping gene
Conclusions
This proof-of-concept study suggests a role for the ER stress machinery in B. burgdorferi colonization and survival inside ticks. Our data provide a deeper insight into the possible role of selenoK in the pathogen colonization of tick vectors. The nymphal ticks play an epidemiologically important role in the infection of humans with B. burgdorferi and a detailed mechanistic study to investigate the role of ER-resident selenoproteins in pathogen infection and transmission is still needed.
Additional files
Additional file 1: Table S1. Gene-specific PCR and qRT-PCR primers used in this study.
Additional file 2: Figure S1. SECIS prediction for Ixodes scapularis selenoK (XM_002403043.1) by SECISsearch3 algorithm. SECISsearch3 predicts the potential SECIS (selenocysteine insertion sequences) element for eukaryotes required for translation of selenoprotein from its mRNA.
Additional file 3: Figure S2. Prediction of subcellular localization of tick selenoprotein K (XP_002403087.1) by DeepLoc-1.0 algorithm which predicts its localization in the ER/Golgi membrane.
Acknowledgements
We thank Latoyia Downs for her technical assistance in pathogen quantification and critical comments.
Abbreviations
- ERAD
endoplasmic reticulum associated degradation
- ATF6
activating transcription factor 6
- EIF2
eukaryotic initiation factor 2
- IRE1
inositol-requiring enzyme
- Rps4
ribosomal protein S4
- SG
salivary gland
- MG
midgut
- qRT-PCR
quantitative real-time polymerase chain reaction
- ROS
reactive oxygen species
Authors' contributions
Conceived and designed the experiments: DK and SK. Performed the experiments: DK and SK. Analyzed the data: DK and SK. Contributed reagents/materials/analysis tools: DK, ME, TNM and SK. Wrote the paper: DK and SK. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Institutes of General Medical Sciences #P20 and GM103476, USDA NIFA award #2017-67017-26171 & 2016-09395, and US Department of State award # 2017-67016-26864. The funders had no role in study design, data collection, analysis, decision to publish or manuscript preparation.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. Raw data are available from the corresponding author upon request.
Ethics approval and consent to participate
All animal experiments were conducted in strict accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, USA. The protocol for tick blood-feeding on sheep was approved by the Institutional Animal Care and Use Committee of Southern Mississippi University (protocol #18022801and #15101501.1).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Deepak Kumar, Email: Deepak.Kumar@usm.edu.
Monica Embers, Email: members@tulane.edu.
Thomas N. Mather, Email: tmather@uri.edu
Shahid Karim, Email: Shahid.Karim@usm.edu.
References
- 1.Rosenberg R, Lindsey NP, Fischer M, Gregory CJ, Hinckley AF, Mead PS, et al. Vital Signs: trends in reported vectorborne disease case-United States and territories, 2004–2016. NMWR. 2018;67:496–501. doi: 10.15585/mmwr.mm6717e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro ED, Wormser GP. Lyme Disease in 2018: what is new (and what is not) JAMA. 2018;320:635–636. doi: 10.1001/jama.2018.10974. [DOI] [PubMed] [Google Scholar]
- 3.Karim S, Singh P, Ribeiro JMC. A deep insight into the sialotranscriptome of the Gulf Coast tick, Amblyomma maculatum. PLoS ONE. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karim S, Ribeiro JM. An insight into the sialome of the lone star tick, Amblyomma americanum, with a glimpse on its time dependent gene expression. PLoS ONE. 2015;10:e0131292. doi: 10.1371/journal.pone.0131292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamson SW, Browning RE, Budachetri K, Ribeiro JM, Karim S. Knockdown of selenocysteine-specific elongation factor in Amblyomma maculatum alters the pathogen burden of Rickettsia parkeri with epigenetic control by the Sin3 histone deacetylase corepressor complex. PLoS ONE. 2013;8:e82012. doi: 10.1371/journal.pone.0082012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamson SW, Browning RE, Singh P, Nobles S, Villarreal A, Karim S. Transcriptional activation of antioxidants may compensate for selenoprotein deficiencies in Amblyomma maculatum (Acari: Ixodidae) injected with selK-or selM-dsRNA. Insect Mol Biol. 2014;23:497–510. doi: 10.1111/imb.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budachetri K, Crispell G, Karim S. Amblyomma maculatum SECIS binding protein 2 and putative selenoprotein P are indispensable for pathogen replication and tick fecundity. Insect Biochem Mol Biol. 2017;88:37–47. doi: 10.1016/j.ibmb.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budachetri K, Kumar D, Karim S. Catalase is a determinant of the colonization and transovarial transmission of Rickettsia parkeri in the Gulf Coast tick Amblyomma maculatum. Insect Mol Biol. 2017;26:414–419. doi: 10.1111/imb.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budachetri K, Kumar D, Crispell G, Beck C, Dasch G, Karim S. The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of Rickettsia parkeri in the gulf coast tick vector. Microbiome. 2018;6:141. doi: 10.1186/s40168-018-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budachetri K, Karim S. An insight into the functional role of thioredoxin reductase, a selenoprotein, in maintaining normal native microbiota in the Gulf Coast tick (Amblyomma maculatum) Insect Mol Biol. 2015;24:570–581. doi: 10.1111/imb.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crispell G, Budachetri K, Karim S. Rickettsia parkeri colonization in Amblyomma maculatum: the role of superoxide dismutases. Parasit Vectors. 2016;9:291. doi: 10.1186/s13071-016-1579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar D, Budachetri K, Meyers VC, Karim S. Assessment of tick antioxidant responses to exogenous oxidative stressors and insight into the role of catalase in the reproductive fitness of the Gulf-Coast tick (Amblyomma maculatum) Insect Mol Biol. 2016;25:283–294. doi: 10.1111/imb.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman JS. The immune responses of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 14.Ha E-M, Oh C-T, Ryu J-H, Bae Y-S, Kang S-W, et al. An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell. 2005;8:125–132. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Gromer S, Eubel JK, Lee BL, Jacob J. Human selenoproteins at a glance. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves MA, Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell Mol Life Sci. 2009;66:2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narasimhan S, Sukumaran B, Bozdogan U, Thomas V, Lian X, DePonte K, et al. A tick antioxidant facilitates the Lyme disease agent’s successful migration from mammalian host to the arthropod. Cell Host Microbe. 2007;2:7–18. doi: 10.1016/j.chom.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kocan KM, Zivkovic Z, Blouin EF, Naranjo V, Almazan C, Mitra R, de la Fuente J. Silencing of genes involved in Anaplasma marginale-tick interactions affects the pathogen developmental cycle in Dermacentor variabilis. BMC Dev Biol. 2009;9:42. doi: 10.1186/1471-213X-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De la Fuente J, Blouin EF, Manazno-Roman R, Naranjo V, Almazan C, Perez de la Lastra JM, et al. Functional genomic studies of tick cells in response to infection with the cattle pathogen. Anaplasma marginale. Genomics. 2007;90:7120722. doi: 10.1016/j.ygeno.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Hovius JW, van Dam AP, Fikrig E. Tick-host-pathogen interactions in Lyme borreliosis. Trends Parasitol. 2007;23:434–438. doi: 10.1016/j.pt.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Sievers F, Higgins D. Multiple sequence alignment using Clustal Omega [version 1; not peer reviewed] F1000Research. 2015;4(ISCB Comm J):511. [Google Scholar]
- 23.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariotti M, Lobanov AV, Guigo R, Gladyshev VN. SECISearch3 and Seblastian: new tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res. 2013;41:e149. doi: 10.1093/nar/gkt550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armenteros JJ, Sonderby CK, Sonderby SK, Nielsen H, Winther O. DeepLoc: prediction of protein subcellular local- ization using deep learning. Bioinformatics. 2017;33:3387–3395. doi: 10.1093/bioinformatics/btx431. [DOI] [PubMed] [Google Scholar]
- 26.Patrick CD, Hair JA. Laboratory rearing procedures and equipment for multi-host ticks (Acarina: Ixodidae) J Med Entomol. 1975;12:389–390. doi: 10.1093/jmedent/12.3.389. [DOI] [PubMed] [Google Scholar]
- 27.Embers ME, Grasperge BJ, Jacobs MB, Philipp MT. Feeding of ticks on animals for transmission and xenodiagnoses in Lyme disease research. J Visual Exp. 2013;78:50617. doi: 10.3791/50617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mather TN, Mather ME. Intrinsic competence of three ixodid ticks (Acari) as vectors of the Lyme disease spirochete. J Med Entomol. 1991;27:646–650. doi: 10.1093/jmedent/27.4.646. [DOI] [PubMed] [Google Scholar]
- 29.Karim S, Ramakrishnan VG, Tucker JS, Essenberg RC, Sauer JR. Amblyomma americanum salivary glands: double-stranded RNA-mediated gene silencing of synaptobrevin homologue and inhibition of PGE2 stimulated protein secretion. Insect Biochem Mol Biol. 2002;34:407–413. doi: 10.1016/j.ibmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Karim S, Adamson SW. RNA interference in ticks: a functional genomics tool for the study of physiology. In: Jockusch EL, editor. Small RNAs: their diversity, roles, and practical uses. Storrs: Elsevier; 2012. [Google Scholar]
- 31.Stone BL, Russart NM, Gaultney RA, Floden AM, Vaughan JA, Brissette CA. The western progression of Lyme disease: infectious and nonclonal Borrelia burgdorferi sensu lato populations in Grand Forks County, North Dakota. Appl Environ Microbiol. 2014;81:48–58. doi: 10.1128/AEM.02422-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koci J, Simo L, Park Y. Validation of internal reference genes for real-time quantitative polymerase chain reaction studies in the tick, Ixodes scapularis (Acari: Ixodidae) J Med Entomol. 2013;50:79–84. doi: 10.1603/ME12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, et al. Selenoprotein K knockout mice exhibit deficient calcium flux in immune cells and impaired immune responses. J Immunol. 2011;186:2127–2137. doi: 10.4049/jimmunol.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitts MW, Hoffmann PR. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calcium. 2018;70:76–86. doi: 10.1016/j.ceca.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 36.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 37.Ben SB, Wang QY, Xia L, Xia JZ, Cui J, Wang J, et al. Selenoprotein dSelK in Drosophila elevates release of Ca2+ from endoplasmic reticulum by upregulating expression of inositol 1,4,5-tris-phosphate receptor. Biochemistry (Mosc). 2011;76:1030–1036. doi: 10.1134/S0006297911090070. [DOI] [PubMed] [Google Scholar]
- 38.Walker DH. Rickettsiae and rickettsial infections: the current state of knowledge. Clin Infect Dis. 2007;45:S39–S44. doi: 10.1086/518145. [DOI] [PubMed] [Google Scholar]
- 39.Pillich H, Loose M, Zimmer KP, Chakraborty T. Diverse roles of endoplasmic reticulum stress sensors in bacterial infection. Mol Cell Pediatr. 2016;3:9. doi: 10.1186/s40348-016-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shchedrina VA, Zhang Y, Labunskyy VM, Hatfield DL, Gladyshev VN. Structure function relations, physiological roles, and evolution of mammalian ER-resident selenoproteins. Antioxid Redox Signal. 2010;12:839–849. doi: 10.1089/ars.2009.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du S, Zhou J, Jia Y, Huang K. SelK is a novel ER stress-regulated protein andprotects HepG2 cells from ER stress agent-induced apoptosis. Arch Biochem Biophys. 2010;502:37–143. doi: 10.1016/j.abb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 42.Shchedrina VA, Everley RA, Zhang Y, Gygi SP, Hatfield DL, Gladyshev VN. Selenoprotein K binds multiprotein complexes and is involved in the regulation of endoplasmic reticulum homeostasis. J Biol Chem. 2011;286:42937–42948. doi: 10.1074/jbc.M111.310920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Csordas G, Hajnoczky G. SR/ER-mitochondrial local communication: calcium and ROS. Biochim Biophys Acta. 2009;1787:1352–1362. doi: 10.1016/j.bbabio.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierre N, Barbe C, Gilson H, Deldcque L, Raymacker JM, Francaux M. Activation of ER stress by hydrogen peroxide. Biochem Biophys Res Comm. 2014;450:459–463. doi: 10.1016/j.bbrc.2014.05.143. [DOI] [PubMed] [Google Scholar]
- 45.Lee JH, Park KJ, Jang JK, Jeon YH, Ko KY, Kwon JH, et al. Selenoprotein S-dependent selenoprotein K binding to p97 (VCP) protein is essential for endoplasmic reticulum-associated degradation. J Biol Chem. 2015;290:29941–29952. doi: 10.1074/jbc.M115.680215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Gene-specific PCR and qRT-PCR primers used in this study.
Additional file 2: Figure S1. SECIS prediction for Ixodes scapularis selenoK (XM_002403043.1) by SECISsearch3 algorithm. SECISsearch3 predicts the potential SECIS (selenocysteine insertion sequences) element for eukaryotes required for translation of selenoprotein from its mRNA.
Additional file 3: Figure S2. Prediction of subcellular localization of tick selenoprotein K (XP_002403087.1) by DeepLoc-1.0 algorithm which predicts its localization in the ER/Golgi membrane.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files. Raw data are available from the corresponding author upon request.