Abstract
The treatment of individual patients in cardiology practice increasingly relies on advanced imaging, genetic screening and devices. As the amount of imaging and other diagnostic data increases, paralleled by the greater capacity to personalize treatment, the difficulty of using the full array of measurements of a patient to determine an optimal treatment seems also to be paradoxically increasing. Computational models are progressively addressing this issue by providing a common framework for integrating multiple data sets from individual patients. These models, which are based on physiology and physics rather than on population statistics, enable computational simulations to reveal diagnostic information that would have otherwise remained concealed and to predict treatment outcomes for individual patients. The inherent need for patient-specific models in cardiology is clear and is driving the rapid development of tools and techniques for creating personalized methods to guide pharmaceutical therapy, deployment of devices and surgical interventions.
With precision medicine emerging as the future for cardiology, the diagnostic and therapeutic assessment of patients with cardiac disease increasingly relies on advanced imaging technology, genetic profiling, pharmaceuticals and medical devices1. However, at present, the choice of treatment still depends largely on the outcome of empirical clinical studies in which the effects of different therapeutic options are compared statistically between large groups of patients with similar cardiac pathologies. The increasing level of detail in the diagnostic data has uncovered more and more interindividual variability in pathophysiology2. The growing realization that patient groups are less uniform has led clinical researchers to stratify patients into smaller and more numerous subgroups3,4. In addition, the choice and possible gradation of therapeutic interventions have increased alongside the exponential advances in medical technology. Consequently, reaching the level of significance to identify better treatments for cardiac disease would be difficult in the conventional empirical setup of clinical trials5.
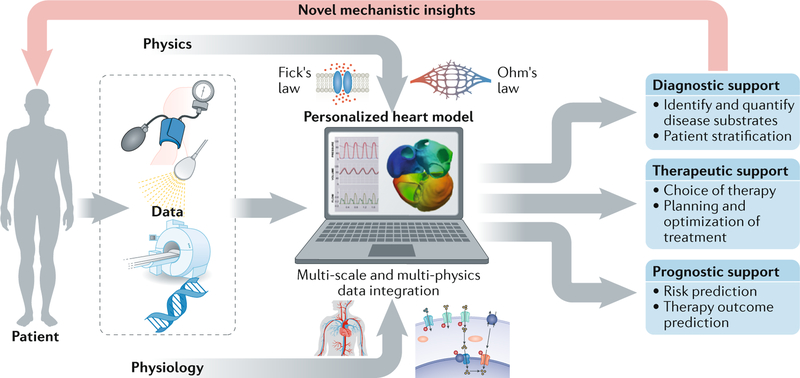
An alternative to the empirical route is to integrate the patient’s diagnostic data using physiological and physical principles important for cardiac function and to use analytical approaches to characterize most accurately the underlying disease and tailor the treatment for an individual’s pathology. Computational models of the heart are increasingly used to address this issue by providing a common framework to integrate multiple data sets from individual patients (FIG. 1). Often, these mechanistic models are complemented with population-based computational techniques, such as atlas-based estimations of global cardiac geometry or regional myocardial fibre orientations, and with principles of control theory, such as the rules of tissue adaptation that control global cardiac geometry through mechano-feedback6–9. These techniques are instrumental in providing reasonable parameter estimates when insufficient patient-specific clinical data are available to constrain the model and in ensuring that these parameters stay within physiological ranges. This integrative and mechanistic power enables computational simulations to reveal novel pathophysiological insights that would otherwise have remained concealed and eventually to predict the optimal treatment option for an individual patient. In contrast to the epidemiological approach, the integrative biophysical approach is primarily based on cause–consequence relationships. A patient-specific simulation is obtained by adjusting a well-chosen set of relevant parameters in the computational model, so that model simulation and clinical measurements agree. The capacity of the model to replicate validation data sets provides confidence that the model can be used to make reliable predictions and simulate the most likely status of a patient given the available measurements. The resulting ‘virtual patient’ can be used for further improvement of diagnosis and for in silico optimization and planning of a treatment.
Fig. 1. How can computational models improve current cardiology care?
A computational model of the human heart and circulation enables synergistic integration of multiple diagnostic data obtained with the use of different clinical modalities (such as echocardiography, MRI, electrocardiography, genetics and blood-pressure measurements) in one personalized heart simulation on the basis of widely accepted physical and physiological principles. The personalized integrative nature of such a virtual-patient simulation adds value to the existing clinical workflow by offering more quantitative and objective insight in the underlying disease substrates of a patient. In addition, the model provides a platform for virtual evaluation and optimization of a therapy.
As the field of cardiology is entering a new era of individualized precision care, patient-specific cardiac modelling might be an important technology to pave the way for personalized medicine10,11. The inherent need for patient-specific models in cardiology is clear. Patients routinely have distinct pathologies, which limits the efficacy of a ‘one-size-fits-all approach’. Patient-specific computational models provide a framework that addresses the challenges of representing the pathophysiology of the individual patient and provides a platform to test multiple therapies and determine the optimal therapy for a specific patient at a specific time. The need for patient-specific modelling is driving the rapid development of tools and techniques for creating personalized models to guide pharmaceutical therapy, deployment of devices and surgical interventions. This Review demonstrates the potential of cardiac models to represent a paradigm shift from population-based clinical decision-making to a true personalization of care that is based on an individual’s specific physiology and pathophysiology. We focus on multi-scale physiology-driven simulations of cardiac mechanics and electrophysiology. The parallel and important field of fluid simulations in the ventricles, coronary arteries and across the valves has been reviewed previously12,13. Clinical cardiac simulation studies are built on a substantial investment in imaging and modelling methods that are described in more technical reviews14,15. In addition, this Review summarizes how personalized cardiac models can increase cost savings and time efficiency of preclinical and clinical trials in cardiology and thereby shorten the path of novel diagnostic and therapeutic strategies to clinical application.
Modelling human cardiac cells
Using biophysical models to link protein function to emergent cellular physiology.
The first computational models of cardiac physiology were cellular models that provided a physiologically and physically constrained framework for quantitatively combining measurements and models of protein function into emergent cellular phenotypes. In the past 3 decades, the modelling of ventricular myocytes has moved from modelling generic mammalian cardiac myocytes by combining data from multiple species to species-specific modelling16,17. Cellular models that include K+, Na+ and Ca2+ channels (FIG. 2a), as well as physiological processes, such as pH regulation, β-adrenergic stimulation and Ca2+ homeostasis have been developed18,19. The functional properties of ion channels, buffers, pumps and transporters are represented in mathematical models that can be used in preclinical and human studies to link, for example, changes in channel function caused by mutations, drugs or physiological regulation to cellular and organ function. As an example, Fernandez-Chas and colleagues used detailed models of cellular membrane electro-physiology and Ca2+ handling in rabbit and human cardiac myocytes to predict the effect of doxorubicin and its metabolite doxorubicinol on the action potential and Ca2+ transient20.
Fig. 2. Modelling cardiac cells, channel mutations and drug response.
a | Schematic showing the level of detail in a cardiac cell model. The model is separated into the transverse (T)-tubule, subsarcolemma, cytosol, junctional sarcoplasmic reticulum (JSR) and network sarcoplasmic reticulum (NSR). The model shows the different ionic currents: Na+ current (INa), K+ currents (transient outward K+ current (Ito), rapid component of the delayed rectifier K+ current (IKr), slow component of the delayed rectifier K+ current (IKs) and inward rectifier K+ current (IK1)), Na+–K+ exchange current (INaK), Ca2+ currents from L-type Ca2+ channels (ICaL, ICaNa and ICaK), current from the membrane-bound Ca2+ pump (ICa), cytosol and subsarcolemmal Na+–Ca2+ exchanger currents (INaCa) and Ca2+ currents (ICa) from the ryanodine receptor and sarcoplasmic reticulum Ca2+ pump. The model includes intracellular Ca2+ fluxes within the sarcoplasmic reticulum, fluxes of Na+, K+ (IK) and Ca2+ between the subsarcolemmal space and the cytosol, and Ca2+ buffers (calsequestrin (CASQ), calmodulin (CALM), troponin C1 (TNNC1), Ca2+/calmodulin-dependent protein kinase (CaMK), anionic sarcoplasmic reticulum binding site for Ca2+ (BSR) and anionic sarcolemmal binding site for Ca2+ (BSL)). b | Schematic of the model of the structure of a wild-type Na+ channel in its three possible states: closed, open (O), and inactivated (fast (F) or slow (S)). The structure of the channel is changed with the addition with the ΔKPQ mutation. c | Effects of a channel inhibitor compound on the Na+ channel current. The current is calculated by: the channel conductance (gNa); proportion of channels in the open (Po), fast-inactivated or slow-inactivated states; voltage across the cell membrane (Vm); Nernst potential for Na+ (ENa); and the proportion of channels that are not inhibited by the compound (γ). The proportion of channels that are not inhibited by the compound is defined by a Hill equation with cooperativity (n) and the half-inhibition concentration (IC50).
Biophysical models of human cardiac cells are now available to simulate the electrophysiology and Ca2+ dynamics in the sinoatrial node and in atrial and ventricular myocytes, as well as tension generation in the ventricles21–31. These models have facilitated the development of studies with four different aims: modelling the effect of channel mutations on cellular and organ function, linking molecular drug effects to emergent cellular and organ function, simulating acute ischaemia, and translating molecular and preclinical physiological studies into clinical readouts.
Effects of protein mutations on cellular and organ function.
Since the identification of the first channelopathy-associated genes, the list of specific mutations associated with the risk of arrhythmia has grown substantially32,33. However, moving from a correlation between a mutation and the disease to a causative relationship as a precursor to identifying new therapeutic targets first requires finding a mechanistic explanation of the link between the mutation, the change in protein function and the increased risk of arrhythmia. In the case of channel mutations, the characterization of the mutated protein in an expression system often allows a specific mutation to be linked to the protein function34. However, this experimental characterization of the protein does not show whether a specific mutation causes a clinical phenotype. Consequently, linking a change in protein function to an increased risk of arrhythmia and to clinical electrocardiogram (ECG) morphologies remains challenging.
Simulations provide an increasingly important link between the characterization of a protein function in expression systems and clinical indices. Biophysical models of the wild-type channels can be adapted to represent the change that a specific mutation introduces in channel kinetics (FIG. 2b). The model of the mutated channel can be introduced into a whole-cell model to predict the effects of this mutation on emergent cellular function and to link the effects to the ECG. Four years after the identification of the first channelopathy-associated gene, models of the cardiac myocyte were used to link changes in Na+ channel function caused by a ΔKPQ mutation to cellular electrophysiology35. This work was primarily used to study patients with a long QT (LQT) interval associated with mutations in the Na+ channel, referred to as type 3 LQT syndrome (LQT3) but has since been extended to other LQT mutations with simulations to study LQT1 (REFS36,37), LQT2 (REFS38,39), LQT3 (REFS35,40–45) and LQT8 (REFS46–51) as well as the related inherited cardiomyopathies Brugada syndrome44,52,53 and short QT syndrome54,55. As an example, Moreno and colleagues used biophysical models to link a KCNQ1 missense mutation that had been associated with functional changes in channels expressed in HEK293 cells to the clinically observed shortened QT interval54.
Biophysical models capturing the complex cellular electrophysiology, combined with models representing measured changes in channel kinetics, can be used to demonstrate that a specific channel mutation is sufficient to explain the observed cellular and clinical phenotypes. This approach has been extended to provide mechanistic explanations for the clinical presentation of channel mutations in the context of altered heart rate or sympathetic stimulation36,56,57. In these cases, biophysical models explicitly represent the effects of altered heart rate or sympathetic stimulation and isolate these effects from the effects of the channel mutation, which allows the characterization of the interdependence of these effects. Additionally, models have been used to explain specific clinical manifestations of channelopathies caused by multiple channel mutations or genetic mosaicism58,59. The development of models that characterize the effect of a specific mutation at the scale of a channel, cell and organ has also provided a platform for predicting how drugs mitigate potential pathologies associated with a mutation60.
In the past 10 years, models of cardiac myocyte contraction have also been developed to simulate the biochemical reactions between the sarcomeric proteins61,62. These models are used to predict the effects of mutations on sarcomeric proteins and the emergent changes in tension development63,64.
Effects of drugs on cellular and organ function.
Models are now created to predict how drugs act on specific channels or on combinations of channels to affect cardiac cellular and tissue electrophysiology65. Drug effects on ion channels can be characterized in expression systems that provide a dose–response curve for a given compound on a given channel. The resulting dose–response curve can be used to scale the number of functioning channels (FIG. 2c). The effects of drugs on the channel can then be introduced into the whole-cell model to predict emergent therapeutic or toxic effects of drugs on cellular physiology. In this way, models provide a framework for combining preclinical data from binding assays, stem cell models, expression systems or animal studies within the context of known human physiology. The identification of novel therapies through computational models is still speculative and has yet to translate into novel therapies. The FDA Comprehensive In vitro Proarrhythmia Assay (CiPA) initiative is, however, currently leading the way for the translation of computer models into screening tools for cardiac toxicity52,66.
Building on the successful adoption in drug development and guidelines of physiology-based pharmacokinetic models that simulate drug distribution in the blood for a given dosing regimen52, biophysical cardiac model simulations are increasingly applied to predict the functional response of the heart to novel compounds. Simulations have demonstrated that they can be used to combine drug effects across multiple channels for arrhythmogenic risk prediction, account for biological variation in model predictions, test hypothesized adverse pathways, and predict how a compound and its metabolite influence preclinical and human electrophysiology20,67–70.These models are aiming to predict measures of drug efficacy or drug-related risk by using data that are available early in the drug-development process. As an example, in a 2014 study linking data from binding assays to results from a clinical study on QT interval, the models could correctly classify proarrhythmic drugs with a sensitivity and specificity of up to 79% and 100%, respectively71.
Effects of acute ischaemia on cellular and organ function.
Ischaemia has a prominent role in sudden cardiac death (SCD)72. The capacity to monitor the development of local acute ischaemia in patients is limited. Experimentally, transient ischaemia can be readily induced and monitored at the cellular and organ scales. In this context, modelling and simulation provide a framework for linking the effects of ischaemia on cellular function observed in animal studies with whole-organ function in human hearts73. Acute ischaemia is characterized by hyperkalaemia, hypoxia, and acidosis74. The effects of these changes on individual proteins have been recorded and characterized in biophysical protein models. In turn, these protein-scale models have been integrated into models of human ventricular myocytes to predict how changes in single proteins alter phenotypes at the cellular scale75–77. In human tissue models, ischaemia is recapitulated by a combination of increased extracellular K+ concentrations, introduction of an ATP-sensitive K+ current, and a decrease in the conductivity of the L-type Ca2+ and fast Na+ channels. These models have been used to investigate drug effects and the effect of non-transmural ischaemic regions on ECG morphology78,79. The work by Kazbanov and colleagues is noteworthy for providing a plausible mechanistic explanation for the decreasing dominant activation frequency during ischaemia from whole-heart epicardial activation patterns recorded during open-heart surgery80. The use of models to analyse complex activation sequences is likely to increase with the growing use of inverse-ECG methods and the large and complex data sets collected from whole-heart activation recordings.
Translating molecular and preclinical physiological studies into clinical readouts.
The fourth application of human cardiac cell models is linking the changes in protein or cellular physiology observed or hypothesized from animal models to cellular changes in human cells and predicting whole-organ human clinical phenotypes. This application has particular relevance for cardiac myocyte Ca2+ handling, given that in vivo measurements are not available and that few studies have characterized Ca2+ regulation in human myocytes. Models can be used to extrapolate changes observed in preclinical studies in Ca2+ dynamics and Ca2+ regulation into human contexts. Models have been successfully used to predict the effect of L-type Ca2+ channel relocation in patients with heart failure with increased arrhythmogenic risk and to explain the role of a decrease in the function of sarcoplasmic/endoplasmic reticulum Ca2+ ATPase on action potential alternans in patients with atrial fibrillation (AF) or the role of Ca2+ dysregulation in the increase in arrhythmia complexity and substrate81–83. Analogously to genetic and drug studies, the characterization of physiological and pathological regulation of channels can be done in expression systems and interpreted within the context of human physiology75. In the past 5 years, the advances and availability of pluripotent stem cell-derived cardiac myocytes have also led to the development of models tailored to recapitulate these cells. Paci and colleagues have developed models that describe atrial-like and ventricular-like stem cell phenotypes84. These models have been used to interpret toxicology screens and to facilitate the translation of findings from stem cells into mature cardiac myocytes and are likely to have a role in accelerating the analysis and design of stem-cell patches for regenerative medicine85–88.
Modelling the heart of patients
Model generation.
The advent of personalized and precision medicine in cardiology has generated a growing need to mechanistically link molecular treatments with functional outcomes at the cellular and organ scales. The use of biophysical models provides an efficient means to evaluate preclinical observations within the context of human physiology, thereby accelerating the translation of benchtop findings to the bedside. The development of patient-specific electrophysiological heart models became possible through the availability of cardiac images from patients, usually MRI or CT, and was substantially propelled by the movement towards personalized medicine. Clinical MRI scans with a contrast agent (late gadolinium enhancement) are used to visualize the structural remodelling in atria and ventricles and, therefore, have been used in computational modelling89,90. Atlases of ventricular geometry and shape have been assembled by averaging 3D cardiac image data sets from individuals and generating a mean 3D cardiac image or shape representative of cardiac anatomy91,92.
Given that patient-specific myocardial fibre orientations cannot currently be acquired in routine clinical practice, these data need to be estimated for inclusion in the individualized heart models. To date, two methodologies have been developed for estimating fibre orientations on the basis of the individual geometry of the ventricles. The first methodology is an atlas-based approach, in which the patient’s fibre orientation is estimated by morphing an atlas of heart anatomy generated from ex vivo MRI and diffusion tensor magnetic resonance imaging (DT-MRI) onto the patient’s heart anatomy7,93. The second approach uses a rule-based fibre orientation estimation derived from histological and DT-MRI data94. Utilization of patient-specific models for clinical applications emphasizing the use of noninvasive approaches, as described below, often necessitates the use of averaged human electrophysiology, for both normal and remodelled tissue, given that electrophysiological properties cannot be acquired noninvasively95–97. By contrast, some studies have aimed at the personalization of electrophysiological properties through invasive electro-anatomical mapping measurements98–100.
The success of modelling the ventricles as well as their pathologies and treatments has been mirrored in models of the atria. 3D models of human atria have been created to represent the atrial anatomy by using surface or volumetric models and have mainly been used to simulate AF101–111. Most models incorporate atrial geometries created from multimodality imaging102,103,110,111. Fibre structure has also been incorporated in these models, most often by using a rule-based approach; DT-MRI has also become available to estimate fibre orientations in the human atria112–114. Models of atrial electrophysiology can be generated by fitting clinical measurements from individual patients or using averaged human values115,116.
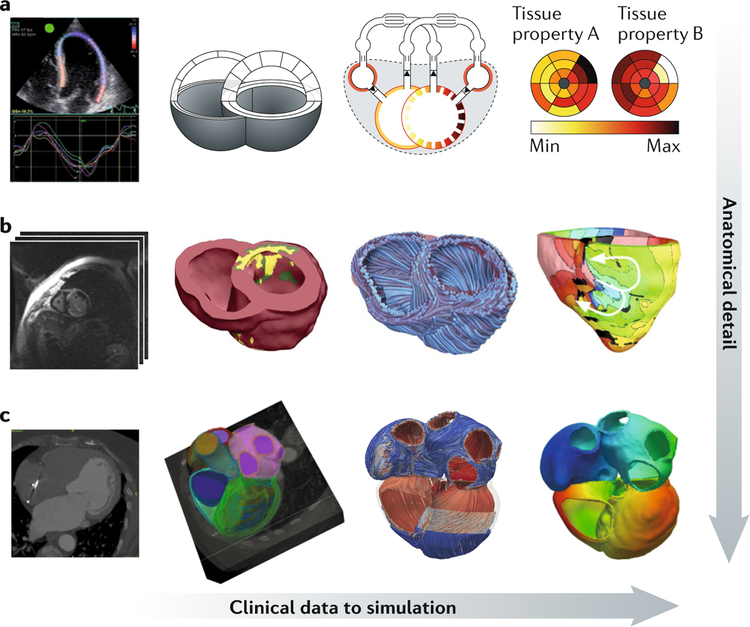
FIGURE 3, which presents cardiac model generation from clinical images or imaging information, illustrates the variety of imaging modalities that can be used as model inputs and highlights the different degrees of model complexity that can be reached depending on the problems addressed by the simulations.
Fig. 3. Linking clinical data to computer models of the heart with increasing levels of anatomical detail.
a | Left to right: echocardiography strain measurements are used to constrain a simplified anatomical model, linked to a closed-loop cardiovascular system representation that can generate model predictions of tissue properties. b | Left to right: multimodality MRI data (cine, late gadolinium enhancement, anatomical) can be used to create anatomical models of the ventricles, highlighting structural remodelling (shown in the middle image with labels for viable tissue (pink), border zone (green) and infarct (yellow)); fibre fields can be added to the model and simulations of arrhythmia performed. c | Left to right: cardiac CT data segmented to label ventricular and atrial myocardium; fibre fields are generated across the four cardiac chambers, and simulations of atrial and ventricular physiology are performed. Max, maximum; Min, minimum. Part b is adapted from REF.95, Springer Nature Limited.
Ventricular arrhythmias.
The vision to use computational ventricular electrophysiological models in the clinic stems from a number of studies using a single model of the human ventricle to understand the mechanisms by which arrhythmias arise and are sustained in the human heart. The aim of these studies included understanding the mechanisms underlying ventricular arrhythmias, such as the characteristics of ventricular fibrillation under a variety of conditions including heart failure, the effect of afterdepolarizations on propagation and the maintenance of torsades de pointes78,80,117–121. Furthermore, the model has been successfully used to reveal the origin of specific characteristics of the ECG such as the notched T waves, the effects of ionic current changes, and the propagation pattern such as left bundle branch block, as well as to detect instabilities in the QT interval, a predictor of the onset of ventricular tachycardia in patients with acute myocardial infarction122–125. Combined with an image-based torso representation, the human heart model has been used to evaluate the effect of heart position on ECG morphology in patients with heart failure and the signatures of left ventricular (LV) hypertrophy in the ECG126,127. Although these simulation studies do not involve patient populations, their insight has been pivotal in advancing human cardiac electrophysiology modelling and the field of cardiac arrhythmogenesis.
Applying patient-specific heart or ventricular modelling to populations of patients has become the new frontier of computational cardiology. Arevalo and colleagues conducted a retrospective study of 41 patients after myocardial infarction and with left ventricular ejection fraction (LVEF) < 35% to determine the patient’s propensity to develop infarct-related ventricular arrhythmias and, therefore, to assess the risk of SCD95. This noninvasive approach to SCD risk stratification was termed the virtual-heart arrhythmia risk predictor (VARP). The comparison of the predictive capabilities of VARP with those of other clinical risk assessment metrics, including LVEF — the current clinical stratifier for SCD — revealed that only VARP outcome was significantly associated with arrhythmic risk in this cohort. Of note, the noninvasive nature of VARP is an additional advantage over clinical testing that entails risks of vascular access, sedation and induction of ventricular arrhythmias requiring defibrillation in already-weakened patients with cardiomyopathy. VARP has also been successfully applied in the past 2 years for the analysis of data from patients after myocardial infarction and with LVEF >35%96.
Patient-specific electrophysiology models have also been employed to predict noninvasively the target for catheter ablation, a procedure based on the delivery of heat or cold to destroy the ability of cardiac tissue to generate and conduct electrical signals locally. Zhu and colleagues demonstrated that heart models could be used to localize noninvasively accessory pathways in patients with Wolff–Parkinson–White syndrome128. In a retrospective analysis of 13 patients, Ashikaga and colleagues demonstrated that the prediction of ablation targets for infarct-related ventricular tachycardia could also result in lesions that are much smaller than those executed in the clinic129. Finally, a biophysically detailed heart–torso modelling approach has been used to predict the optimal location of the implantable cardioverter–defibrillator leads in a patient with congenital heart disease130. The study demonstrated that defibrillation shock energy could be reduced if computational simulations were used as part of the device implantation planning.
Atrial arrhythmias.
The growing burden of AF and the difficulties associated with the personalized treatment of the disease has led the field of cardiac computational modelling to focus on understanding how functional and structural remodelling result in the turbulent propagation associated with AF116,131. Atrial computational models have made major contributions in understanding how intrinsic atrial structural and electrophysiological heterogeneities predispose to atrial arrhythmias132–135. Examples of such contributions include investigating the role of pulmonary vein triggers in initiating paroxysmal AF, determining the mechanisms of lone paroxysmal AF that arise from inherited ion channel dysfunction, establishing the mechanisms for atrial alternans, elucidating the role of the ganglionic plexi in initiating AF and revealing the contribution of the electrical uncoupling between the endocardial and epicardial layers83,136–140. In the past couple of years, the understanding of the role of fibrosis in maintaining persistent AF has led to a number of new computational studies aimed at determining how the remodelled atrial structure alters AF dynamics. These studies include the assessment of the different representations of fibrosis and the spatial resolution of clinical measurement needed to detect re-entrant drivers that sustain AF141–143.
Predictive approaches that combine clinical imaging and computational modelling and can be applied for the treatment of AF have also been developed. The first attempt at a clinical application of human 3D atrial modelling was in the context of pacing for AF termination144,145. In the past decade, catheter-based AF ablation has become one of the next big frontiers in the use of 3D computational modelling to guide patient therapy. For example, human atrial models have been used to suggest how to optimize AF ablation, explore strategies to minimize the size of ablation lesions and study the effect of gaps in ablation lines103,146. Atrial models that incorporate the ganglionic plexi and the resulting AF have also been developed and have already been used successfully in a patient cohort to guide ablation for paroxysmal AF147. Although pulmonary vein isolation is currently the recommended treatment for patients with persistent AF and fibrosis, no reliable ablation option is available. Therefore, simulation approaches can make a great difference for this specific application by providing guidance for ablation in addition to the standard pulmonary vein isolation. A number of human atrial models have been developed for the sole purpose of guiding ablation in the fibrotic substrate, primarily through the detection of regions of high-dominant frequency or locations of the re-entrant drivers perpetuating AF110,111,142,147,148. Models of atrial fibrosis have already shown early preliminary success in providing guidance in substrate ablation; these results have raised expectations that atrial models could be used to predict optimal AF ablation strategies in a patient-specific manner149,150.
Of note, atrial and ventricular models for clinical applications that aim at noninvasive assessment of risk of arrhythmias or noninvasive computational prediction of ablation targets cannot include patient-specific electrophysiology, given that acquisition of these parameters is currently invasive. Using averaged human electrophysiology associated with the given atrial or ventricular disease in the models will entail a certain level of model uncertainty. Further model uncertainty could result from the low resolution of clinical scans. In the past 7 years, studies have started to assess the level of uncertainty in the models and to take steps to out-line the potential clinical applicability of the different modelling approaches151–158.
Heart failure therapy.
Cardiac resynchronization therapy (CRT), a clinically important device therapy for patients with dyssynchronous heart failure, has been the subject of many computational modelling studies. These studies addressed the absence of a response to CRT, which is often a multifactorial problem related to the large interindividual variability in underlying disease patterns and therapy delivery159. Computer models covering this complexity in an integrative way and at different scales have been frequently used to investigate ways to optimize patient selection and therapy delivery160. Simulations of the cardiovascular system have been used to provide representative models for specific patient groups, giving insight into disease-specific mechanisms. By contrast, personalized 3D models have aimed at capturing an individual’s anatomy and pathophysiology to provide insight into disease mechanisms and therapy outcomes for individual patients. In regard to patient selection for CRT, computational studies have demonstrated that several electromechanical aspects of the patient’s baseline physiology and pathophysiology can determine the response to CRT. Most consistently, the presence of a marked right-to-left ventricular-activation delay has been shown to be an absolute pre-requisite for response to CRT, which is well-represented by the criteria of QRS widening and left bundle branch block morphology of the QRS complex in current clinical guidelines for CRT161–166. Other modelling studies have found that a prolonged intrinsic atrioventricular conduction time increases the likelihood of acute haemodynamic improvement with CRT because biventricular pacing at a shorter atrioventricular delay increases LV filling time and thereby preload167. The presence of a myocardial infarct modulates, but does not necessarily preclude, an improvement in ventricular function after CRT164,168,169. Simulations also revealed that the size and location of the infarct are important baseline characteristics, with the observation that the acute haemodynamic response to CRT decreases with increasing infarct size and with a more lateral location168,169. Another electromechanical modelling study showed that CRT response is decreased when the length-dependence of tension generation — the innate myocardial-tissue property responsible for the Frank–Starling mechanism at the organ level — is compromised170,171.
Studies combining model simulations with experimental and clinical measurements of pacing-induced changes in cardiac electromechanics were the first to show quantitatively the importance of right ventricular myocardial function for CRT response, which was explained by the increase in right ventricular workload during pacing through interventricular interaction172,173. In the past 5 years, simulations of CRT in virtual patients have been used to devise a myocardial strain-based diagnostic index that quantifies a combination of electrical and mechanical substrates amenable to CRT164. Interestingly, the same study illustrated the added clinical value of a simulation-based design of the diagnostic index by showing that it accurately identified patients with a more favourable outcome after CRT, including those with intermediate electrocardiographic criteria for whom CRT response was less certain according to the current guideline criteria only.
In silico studies on CRT have also contributed to the optimization of therapy delivery. Many computational studies investigated the effect of LV-pacing location on acute haemodynamic response to CRT174–178. Despite the use of different modelling approaches, these studies consistently reported that the lateral LV free wall was the optimal LV pacing location in noninfarcted failing hearts. In the presence of a myocardial infarction, simulation studies have demonstrated that pacing remotely from the infarct zone and multisite pacing using a quadripolar lead were more efficient than conventional CRT168,169,179. Furthermore, the beneficial effect of endocardial LV pacing over epicardial LV pacing was attributed to its relatively faster and, therefore, more synchronous ventricular activation180. In addition to the number and location of pacing sites, the timing of lead stimulation, configurable through atrioventricular and ventriculo-ventricular pacing delay settings, has also been the subject of in silico optimization studies167,175,176,181–183. Optimal atrioventricular and ventriculo-ventricular delay settings were shown to be highly patient-specific and even to change over time for the same patient176. The application of virtual-patient simulations for prediction of the response to CRT is less mature. Several personalized modelling studies have shown the feasibility of patient-specific prediction of acute haemodynamic CRT response in small numbers of patients175–177,179,184–187. These successful proof-of-principle studies have demonstrated the translational potential of this emergent technology and brought it an important step closer to being adopted as an actual support tool for clinical decision-making in the field of pacing therapy.
The future of computational models
The advances in cardiac research reviewed above demonstrate the enormous progress made to date in using computational approaches to address both the mechanisms of cardiac dysfunction and the issues related to the clinical application of therapies for cardiac disease. Computational models of the heart now cover most of the biophysical complexity of the individual patient’s cardiac pathology. As a result, models linking cellular electrophysiology, myocardial tissue mechanics, and system haemodynamics have become promising platforms for virtual-patient simulations and for in silico evaluation of novel diagnostic and therapeutic strategies188. These biophysically detailed models of the heart are having an increasingly important role in the road to personalized medicine in cardiology clinics189,190. In this time of growing regulatory and administrative burdens that restrict the ability to develop new drugs, medical devices and diagnostic technologies, simulations of virtual patients and therapy provide the means for the pharmaceutical and biomedical device industries to reduce the developmental costs of innovation and the time to market189. Models allow fast evaluation of medical device settings, modes of therapy delivery and patient-selection criteria through relatively low-cost simulation and are thereby poised to complement, augment and even fully replace conventional preclinical and clinical device development and evaluation steps.
Although simulations have made inroads in the clinic, a number of barriers to their widespread adoption remain. Simulation applications in cardiology are currently targeted at a narrow range of heart diseases for which the effect of treatment is well-characterized, imaging and invasive measurements are routinely made and treatment can be evaluated with a short-term measure of success. To broaden the application beyond these conditions and extend the use of simulations in cardiology will require specific challenges spanning model creation, speed and physiological detail of simulations and the communication of model predications to cardiologists to be addressed. Currently, the customization of cardiac models to represent a specific patient or make a representative model of a specific pathology focuses on how parameters can be inferred directly from clinical data. Although this matter is important to ensure that models are informed by the clinical data available, the desired data will not be available in many potential clinical applications or simply cannot be measured in a clinical context or with sufficient accuracy. Whereas preclinical measurements can be used to inform on unknown or patient model parameters, the uncertainty introduced by these parameters is poorly characterized191. Improved methods for translating preclinical measurements into patient-specific models would substantially improve our ability to create models of a broader range of complex pathologies and robustly and reliably use this information to inform clinical decisions. In turn, this framework for linking animal research to the clinic would shorten development times and improve experimental design of pre-clinical studies. Another approach to this problem of model parameterization is to tailor model complexity, for example the number of model parameters, to the clinical application192. This approach has been particularly well-developed in cardiac cell models, so that it is large enough to describe the cardiac pathology of interest and small enough to estimate the model parameters reliably from the scarce clinical data193–195.
Personalizing models to patient-specific data requires large numbers of simulations to be performed. The speeds of current simulations limit the ability to create models reliably on clinical timelines and to perform simulation studies on > 40–50 patients185. This limitation makes the advancement of algorithms and approaches for high-speed simulations of critical importance to enable cardiac modelling to become a routine clinical tool.
Although new developments in algorithms and implementation have improved simulation speed, new modelling tools and techniques still need to be developed to incorporate fine-grained cardiac structural and functional data in the models and enable models to run in a period of time appropriate for clinical applications196,197. Research efforts need to be invested in dramatically improving the scalability of heart models and simulation standards, in modularizing and interfacing multiple model levels, as well as in preserving and curating models and data in easy-to-access repositories198–202.
All the applications of simulations in cardiology described in this Review focused on predicting short-term responses to treatment such as prolongation in action potential, termination of arrhythmia or pacing-induced improvement of cardiac pump function. However, the primary interest of cardiologists is not in the patient’s immediate response after a procedure, but how the heart of the patient will respond in the long term. This limitation of the models poses a physiological and simulation challenge. Simulations of cardiac tissue remodelling and growth are still in their early stages because they are driven by rule-based phenomenological models and require further development of theory and implementation8,9,203. At the same time, the physiology and drivers of remodelling and growth are often characterized by pathway topology and not quantitative dynamics, which limits their utility in quantitative predictive cardiac modelling. The ability to predict how the heart will remodel in response to disease progression or to a treatment is an essential element in making long-term predictions related to clinical outcome and intervention success and will substantially increase the applications of simulations in cardiology.
Finally, communicating model results to cardiologists and caregivers in a simple format and under the right conditions will be critical for the adoption of model predictions in clinical decision-making. The development of electrophysiological and electromechanical models of the heart currently requires a great amount of expertise in several different fields including numerical analysis, computer science, cardiac electrophysiology, mechanics and image processing. Translating the models to the clinic has necessitated the development of interfaces that allow model utilization by clinical non-experts. Even though displaying results of simulations in an office environment presents simple requirements for interface development — a cardiologist should be able to manipulate and examine the results of simulations — the utilization of model predictions in an environment in which procedures are performed, such as in the surgical theatre or catheter laboratory, poses a particular challenge204,205. The generation of model predictions for procedure guidance requires the integration of results from different clinical devices, such as electroanatomical mapping systems (including CARTO (Biosense Webster) and EnSite NavX (St. Jude Medical/Abbott)), x-ray fluoroscopy or MRI206,207. This application will require working with the technical support staff of the devices to integrate the results of the model with imaging data sets and to dynamically guide the clinical procedure.
Conclusions
Computer models in cardiology have an enormous potential to become the new quantitative approach for detecting and treating cardiac disease given the capacity of these models to provide low-cost, low-risk, rapid and integrative analysis of a patient’s physiology and pathology with predictions that are free from subjective assessments. The discipline of computational cardiology will continue to grow and develop, driving new clinical developments and personalization of cardiac care.
Key points.
Computational models of the heart have an important and growing role in cardiology, enabling patients to be diagnosed and treated on the basis of their specific pathophysiology.
Simulations provide the link between the effects of genetic mutations, physiological regulations or drugs on protein function and emergent cellular and tissue function or clinical phenotypes.
Models representing an individual patient or a specific pathology are now used to identify the mechanisms underpinning a disease, improve patient selection and predict clinical outcomes.
Predictive modelling also contributes to the development of new diagnostics and devices and to the tailoring of therapies for individual patients.
Translational barriers remain regarding model personalization, speed and detail of the simulations and how to communicate model predictions to cardiologists within a clinical environment.
Acknowledgements
S.A.N. acknowledges support from the UK Engineering and Physical Sciences Research Council (EP/M012492/1, NS/ A000049/1 and EP/P01268X/1), the British Heart Foundation (PG/15/91/31812 and PG/13/37/30280) and King’s Health Partners London National Institute for Health Research (NIHR) Biomedical Research Centre. J.L. acknowledges support from the Dr. Dekker Program of the Dutch Heart Foundation (grant 2015T082) and the Netherlands Organisation for Scientific Research (NWO-ZonMw, VIDI grant 016.176.340). N.A.T. acknowledges support from Leducq Foundation and from the NIH (grants DP1-HL123271 and R01HL116280). M. Strocchi (King’s College London, UK) provided assistance with creating the four-chamber heart images in figure 3.
S.A.N. has received support from Abbott, Boston Scientific, Edwards Lifesciences, Pfizer, Roche and Siemens. J.L. has received support from Medtronic.
Footnotes
Competing interests
N.A.T. declares no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Cardiology thanks B. Rodriguez and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Antman EM & Loscalzo J Precision medicine in cardiology. Nat. Rev. Cardiol 13, 591–602 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Loscalzo J, Kohane I & Barabasi AL Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol. Syst. Biol 3, 124 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyon A et al. Distinct ECG phenotypes identified in hypertrophic cardiomyopathy using machine learning associate with arrhythmic risk markers. Front. Physiol 9, 213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horiuchi Y et al. Identifying novel phenotypes of acute heart failure using cluster analysis of clinical variables. Int. J. Cardiol 262, 57–63 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Stanley K Design of randomized controlled trials. Circulation 115, 1164–1169 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Gilbert K et al. Atlas-based computational analysis of heart shape and function in congenital heart disease. J. Cardiovasc. Transl Res 11, 123–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vadakkumpadan F, Arevalo H, Ceritoglu C, Miller M & Trayanova N Image-based estimation of ventricular fiber orientations for personalized modeling of cardiac electrophysiology. IEEE Trans. Med. Imag 31, 1051–1060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witzenburg CM & Holmes JWA Comparison of phenomenologic growth laws for myocardial hypertrophy. J. Elast 129, 257–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arts T, Lumens J, Kroon W & Delhaas T Control of whole heart geometry by intramyocardial mechano-feedback: a model study. PLoS Comput. Biol 8, e1002369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niederer SA & Smith NP Using physiologically based models for clinical translation: predictive modelling, data interpretation or something in-between? J. Physiol 594, 6849–6863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trayanova NA, Boyle PM & Nikolov PP Personalized imaging and modeling strategies for arrhythmia prevention and therapy. Curr. Opin. Biomed. Eng 5, 21–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris PD et al. Computational fluid dynamics modelling in cardiovascular medicine. Heart 102, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor CA & Figueroa C Patient-specific modeling of cardiovascular mechanics. Annu. Rev. Biomed. Eng 11, 109–134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamata P et al. Images as drivers of progress in cardiac computational modelling. Prog. Biophys. Mol. Biol 115, 198–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crozier A et al. Image-based personalization of cardiac anatomy for coupled eSlectromechanical modeling. Annu. Rev. Biomed. Eng 44, 58–70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo C & Rudy Y A model of the ventricular cardiac action potential. Depolarization, repolarization, and their interaction. Circ. Res 68, 1501–1526 (1991). [DOI] [PubMed] [Google Scholar]
- 17.Fink M et al. Cardiac cell modelling: observations from the heart of the cardiac physiome project. Prog. Biophys. Mol. Biol 104, 2–21 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Heijman J, Volders PG, Westra RL & Rudy Y Local control of β-adrenergic stimulation: effects on ventricular myocyte electrophysiology and Ca2+-transient. J. Mol. Cell. Cardiol 50, 863–871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lascano EC et al. Role of CaMKII in post acidosis arrhythmias: a simulation study using a human myocyte model. J. Mol. Cell. Cardiol 60, 172–183 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Chas M, Curtis MJ & Niederer SA Mechanism of doxorubicin cardiotoxicity evaluated by integrating multiple molecular effects into a biophysical model. Br. J. Pharmacol 175, 763–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri A, Fantini M, Wilders R & Severi S Computational analysis of the human sinus node action potential: model development and effects of mutations. J. Physiol 595, 2365–2396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Tusscher KHWJ & Panfilov AV Alternans and spiral breakup in a human ventricular tissue model. Am. J. Physiol. Heart Circ. Physiol 291, H1088–H1100 (2006). [DOI] [PubMed] [Google Scholar]
- 23.O’Hara T, Virág L, Varró A & Rudy Y Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput. Biol 7, e1002061 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grandi E, Pasqualini FS & Bers DM A novel computational model of the human ventricular action potential and Ca transient. J. Mol. Cell. Cardiol 48, 112–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priebe L & Beuckelmann DJ Simulation study of cellular electric properties in heart failure. Circ. Res 82, 1206–1223 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Grandi E et al. Human atrial action potential and Ca2+ model: sinus rhythm and chronic atrial fibrillation. Circ. Res 109, 1055–1066 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koivumäki JT, Korhonen T & Tavi P Impact of sarcoplasmic reticulum calcium release on calcium dynamics and action potential morphology in human atrial myocytes: a computational study. PLoS Comput. Biol 7, e1001067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maleckar MM, Greenstein JL, Trayanova NA & Giles WR Mathematical simulations of ligand-gated and cell-type specific effects on the action potential of human atrium. Prog. Biophys. Mol. Biol 98, 161–170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nygren A et al. Mathematical model of an adult human atrial cell: the role of K+ currents in repolarization. Circ. Res 82, 63–81 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Courtemanche M, Ramirez RJ & Nattel S Ionic mechanisms underlying human atrial action potential properties: insights from a mathematical model. Am. J. Physiol 275, H301–H321 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Land S et al. A model of cardiac contraction based on novel measurements of tension development in human cardiomyocytes. J. Mol. Cell. Cardiol 106, 68–83 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Wang Q et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell 80, 805–811 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Wilde AAM & Behr ER Genetic testing for inherited cardiac disease. Nat. Rev. Cardiol 10, 571–583 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Adsit GS, Vaidyanathan R, Galler CM, Kyle JW & Makielski JC Channelopathies from mutations in the cardiac sodium channel protein complex. J. Mol. Cell. Cardiol 61, 34–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clancy CE & Rudy Y Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature 400, 566–569 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Saucerman JJ, Healy SN, Belik ME, Puglisi JL & McCulloch AD Proarrhythmic consequences of a KCNQ1 AKAP-binding domain mutation: computational models of whole cells and heterogeneous tissue. Circ. Res 95, 1216–1224 (2004). [DOI] [PubMed] [Google Scholar]
- 37.O’Hara T & Rudy Y Arrhythmia formation in subclinical (“silent”) long QT syndrome requires multiple insults: quantitative mechanistic study using the KCNQ1 mutation Q357R as example. Heart Rhythm 9, 275–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ficker E et al. Novel characteristics of a misprocessed mutant HERG channel linked to hereditary long QT syndrome. Am. J. Physiol. Heart Circ. Physiol 279, H1748–H1756 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Choe CU et al. C-Terminal HERG (LQT2) mutations disrupt IKr channel regulation through 14–3-3ε. Hum. Mol. Genet 15, 2888–2902 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Clancy CE, Tateyama M, Liu H, Wehrens XHT & Kass RS Non-equilibrium gating in cardiac Na+ channels: an original mechanism of arrhythmia. Circulation 107, 2233–2237 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Flaim SN, Giles WR & McCulloch AD Arrhythmogenic consequences of Na+ channel mutations in the transmurally heterogeneous mammalian left ventricle: analysis of the I1768V SCN5A mutation. Heart Rhythm 4, 768–778 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Wehrens XHT, Abriel H, Cabo C, Benhorin J & Kass RS Arrhythmogenic mechanism of an LQT-3 mutation of the human heart Na+ channel α-subunit: a comptutational analysis. Circulation 102, 584–590 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Bankston JR et al. A novel LQT-3 mutation disrupts an inactivation gate complex with distinct rate-dependent phenotypic consequences. Channels 1, 273–280 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Vecchietti S et al. In silico assessment of Y1795C and Y1795H SCN5A mutations: implication for inherited arrhythmogenic syndromes. Am. J. Physiol. Heart Circ. Physiol 292, H56–H65 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Ahrens-Nicklas RC, Clancy CE & Christini DJ Re-evaluating the efficacy of beta-adrenergic agonists and antagonists in long QT-3 syndrome through computational modelling. Cardiovasc. Res 82, 439–447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel WH et al. Proarrhythmic defects in Timothy syndrome require calmodulin kinase II. Circulation 118, 2225–2234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Splawski I et al. Ca V 1. 2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism 119, 19–31 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Boczek NJ et al. Novel Timothy syndrome mutation leading to increase in CACNA1C window current. Heart Rhythm 12, 211–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung RJ et al. Beta-adrenergic modulation of arrhythmogenesis and identification of targeted sites of antiarrhythmic therapy in Timothy (LQT8) syndrome: a theoretical study. Am. J. Physiol. Heart Circ. Physiol 298, H33–44 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Zhu ZI & Clancy CE L-Type Ca2+ channel mutations and T-wave alternans: a model study. Am. J. Physiol. Heart Circ. Physiol 293, H3480–H3489 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Faber GM, Silva J, Livshitz L & Rudy Y Kinetic properties of the cardiac L-type Ca2+ channel and its role in myocyte electrophysiology: a theoretical investigation. Biophys. J 92, 1522–1543 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fermini B et al. A new perspective in the field of cardiac safety testing through the comprehensive in vitro proarrhythmia assay paradigm. J. Biomol. Screen 21, 1–11 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Verkerk AO et al. Role of sequence variations in the human ether-a-go-go-related gene (HERG, KCNH2) in the Brugada syndrome. Cardiovasc. Res 68, 441–453 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Moreno C et al. A new KCNQ1 mutation at the S5 segment that impairs its association with KCNE1 is responsible for short QT syndrome. Cardiovasc. Res 107, 613–623 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Hancox JC, Whittaker DG, Du C, Stuart AG & Zhang H Emerging therapeutic targets in the short QT syndrome. Expert Opin. Ther. Targets 22, 439–451 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Clancy CE & Rudy Y Na+ channel mutation that causes both Brugada and long-QT syndrome phenotypes: a simulation study of mechanism. Circulation 105, 1208–1213 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu J, Kato K, Delisle BP & Horie M A molecular mechanism for adrenergic-induced long QT syndrome. J. Am. Coll. Cardiol 63, 819–827 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Hu D et al. Dual variation in SCN5A and CACNB2b underlies the development of cardiac conduction disease without Brugada syndrome. Pacing Clin. Electrophysiol 33, 274–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Priest JR et al. Early somatic mosaicism is a rare cause of long-QT syndrome. Proc. Natl Acad. Sci. USA 113, 11555–11560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreno JD et al. Ranolazine for congenital and acquired late iNa-linked arrhythmias: In silico pharmacological screening. Circ. Res 113, e50–e61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campbell SG, Lionetti FV, Campbell KS & McCulloch AD Coupling of adjacent tropomyosins enhances cross-bridge-mediated cooperative activation in a Markov model of the cardiac thin filament. Biophys. J 98, 2254–2264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Land S & Niederer SA A spatially detailed model of isometric contraction based on competitive binding of troponin I explains cooperative interactions between tropomyosin and crossbridges. PLoS Comput. Biol 11, e1004376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sewanan LR, Moore JR, Lehman W & Campbell SG Predicting effects of tropomyosin mutations on cardiac muscle contraction through myofilament modeling. Front. Physiol 7, 473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dewan S, McCabe KJ, Regnier M, McCulloch AD & Lindert S Molecular effects of cTnC DCM mutations on calcium sensitivity and myofilament activation-an integrated multiscale modeling study. J. Phys. Chem. B 120, 8264–8275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z et al. Improving the in silico assessment of proarrhythmia risk by combining hERG (Human Ether-à-go-go-related gene) channel–drug binding kinetics and multichannel pharmacology. Circ. Arrhythm. Electrophysiol 10, e004628 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Sager PT, Gintant G, Turner JR, Pettit S & Stockbridge N Rechanneling the cardiac proarrhythmia safety paradigm: a meeting report from the Cardiac Safety Research Consortium. Am. Heart J 167, 292–300 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Johnstone RH et al. Uncertainty and variability in models of the cardiac action potential: Can we build trustworthy models? J. Mol. Cell. Cardiol 96, 49–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sarkar AX, Christini DJ & Sobie EA Exploiting mathematical models to illuminate electrophysiological variability between individuals. J. Physiol 590, 2555–2567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mirams GR et al. Simulation of multiple ion channel block provides improved early prediction of compounds’ clinical torsadogenic risk. Cardiovasc. Res 91, 53–61 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Oliveira BL & Niederer S A biophysical systems approach to identifying the pathways of acute and chronic doxorubicin mitochondrial cardiotoxicity. PLoS Comput. Biol 12, e1005214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mirams GR et al. Prediction of thorough QT study results using action potential simulations based on ion channel screens. J. Pharmacol. Toxicol. Methods 70, 246–254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zipes DP & Wellens HJJ Sudden cardiac death. Circulation 98, 2334–2351 (1998). [DOI] [PubMed] [Google Scholar]
- 73.Ferrero JM, Trenor B & Romero L Multiscale computational analysis of the bioelectric consequences of myocardial ischaemia and infarction. Europace 16, 405–415 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Morena H et al. Comparison of the effects of regional ischemia, hypoxia, hyperkalemia, and acidosis on intracellular and extracellular potentials and metabolism in the isolated porcine heart. Circ. Res 46, 634–646 (1980). [DOI] [PubMed] [Google Scholar]
- 75.Jones DK, Peters CH, Tolhurst SA, Claydon TW & Ruben PC Extracellular proton modulation of the cardiac voltage-gated sodium channel, NaV1.5. Biophys. J 101, 2147–2156 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Du Chun YUN et al. Acidosis impairs the protective role of hERG K+ channels against premature stimulation. J. Cardiovasc. Electrophysiol 21, 1160–1169 (2010). [DOI] [PubMed] [Google Scholar]
- 77.Dutta S, Mincholé A, Quinn TA & Rodriguez B Electrophysiological properties of computational human ventricular cell action potential models under acute ischemic conditions. Prog. Biophys. Mol. Biol 129, 40–52 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Dutta S et al. Early afterdepolarizations promote transmural reentry in ischemic human ventricles with reduced repolarization reserve. Prog. Biophys. Mol. Biol 120, 236–248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Potse M, Coronel R, Falcao S, LeBlanc AR & Vinet A The effect of lesion size and tissue remodeling on ST deviation in partial-thickness ischemia. Heart Rhythm 4, 200–206 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Kazbanov IV et al. Effect of global cardiac ischemia on human ventricular fibrillation: insights from a multi-scale mechanistic model of the human heart. PLoS Comput. Biol 10, e1003891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sanchez-Alonso JL et al. Microdomain-specific modulation of L-type calcium channels leads to triggered ventricular arrhythmia in heart failure. Circ. Res 119, 944–945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Narayan SM, Bayer JD, Lalani G & Trayanova NA Action potential dynamics explain arrhythmic vulnerability in human heart failure: a clinical and modeling study implicating abnormal calcium handling. J. Am. Coll. Cardiol 52, 1782–1792 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang KC & Trayanova NA Mechanisms of arrhythmogenesis related to calcium-driven alternans in a model of human atrial fibrillation. Sci. Rep 6, 36395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paci M, Hyttinen J, Aalto-Setala K & Severi S Computational models of ventricular- and atrial-like human induced pluripotent stem cell derived cardiomyocytes. Ann. Biomed. Eng 41, 2334–2348 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Koivumaki JT et al. Structural immaturity of human iPSC-derived cardiomyocytes: in silico investigation of effects on function and disease modeling. Front. Physiol 9, 80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lei CL et al. Tailoring mathematical models to stem-cell derived cardiomyocyte lines can improve predictions of drug-induced changes to their electrophysiology. Front. Physiol 8, 986 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paci M, Hyttinen J, Rodriguez B & Severi S Human induced pluripotent stem cell-derived versus adult cardiomyocytes: an in silico electrophysiological study on effects of ionic current block. Br. J. Pharmacol 172, 5147–5160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harding SE Large stem cell-derived cardiomyocyte grafts: cellular ventricular assist devices? Mol. Ther 22, 1240–1242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ukwatta E et al. Myocardial infarct segmentation from magnetic resonance images for personalized modeling of cardiac electrophysiology. IEEE Trans. Med. Imag 35, 1408–1419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ukwatta E et al. Image-based reconstruction of three-dimensional myocardial infarct geometry for patient-specific modeling of cardiac electrophysiology. Med. Phys 42, 4579–4590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suinesiaputra A, McCulloch AD, Nash MP, Pontre B & Young AA Cardiac image modelling: breadth and depth in heart disease. Med. Image Anal 33, 38–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X et al. Information maximizing component analysis of left ventricular remodeling due to myocardial infarction. J. Transl Med 13, 343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ringenberg J et al. Corrigendum to “effects of fibrosis morphology on reentrant ventricular tachycardia inducibility and simulation fidelity in patient-derived models”. Clin. Med. Insights Cardiol 8, 51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bayer JD, Blake RC, Plank G & Trayanova NA A novel rule-based algorithm for assigning myocardial fiber orientation to computational heart models. Ann. Biomed. Eng 40, 2243–2254 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arevalo HJ et al. Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models. Nat. Commun 7, 11437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deng D, Arevalo HJ, Prakosa A, Callans DJ & Trayanova NA A feasibility study of arrhythmia risk prediction in patients with myocardial infarction and preserved ejection fraction. Europace 18, iv60–iv66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sanchez C et al. Sensitivity analysis of ventricular activation and electrocardiogram in tailored models of heart-failure patients. Med. Biol. Eng. Comput 56, 491–504 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Ranjan R et al. Personalized MRI-based modeling predicts ventricular tachycardia vulnerability in patients receiving primary prevention ICDs [abstract 16247]. Circulation 134, A16247–A16247 (2016). [Google Scholar]
- 99.Relan J et al. Coupled personalization of cardiac electrophysiology models for prediction of ischaemic ventricular tachycardia. Interface Focus 1, 396–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Corrado C et al. Personalized models of human atrial electrophysiology derived from endocardial electrograms. IEEE Trans. Biomed. Eng 64, 735–742 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Vigmond EJ, Ruckdeschel R & Trayanova N Reentry in a morphologically realistic atrial model. J. Cardiovasc. Electrophysiol 12, 1046–1054 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Virag N et al. Study of atrial arrhythmias in a computer model based on magnetic resonance images of human atria. Chaos 12, 754–763 (2002). [DOI] [PubMed] [Google Scholar]
- 103.Dang L et al. Evaluation of ablation patterns using a biophysical model of atrial fibrillation. Ann. Biomed. Eng 33, 465–474 (2005). [DOI] [PubMed] [Google Scholar]
- 104.Vigmond EJ et al. The effect of vagally induced dispersion of action potential duration on atrial arrhythmogenesis. Heart Rhythm 1, 334–344 (2004). [DOI] [PubMed] [Google Scholar]
- 105.Freudenberg J, Schiemann T, Tiede U & Hohne KH Simulation of cardiac excitation patterns in a three-dimensional anatomical heart atlas. Comput. Biol. Med 30, 191–205 (2000). [DOI] [PubMed] [Google Scholar]
- 106.Harrild D & Henriquez C A computer model of normal conduction in the human atria. Circ. Res 87, 25–36 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Seemann G et al. Heterogeneous three-dimensional anatomical and electrophysiological model of human atria. Phil. Trans. A Math. Phys. Eng. Sci 364, 1465–1481 (2006). [DOI] [PubMed] [Google Scholar]
- 108.Reumann M, Bohnert J, Seemann G, Osswald B & Dossel O Preventive ablation strategies in a biophysical model of atrial fibrillation based on realistic anatomical data. IEEE Trans. Biomed. Eng 55, 399–406 (2008). [DOI] [PubMed] [Google Scholar]
- 109.Aslanidi OV et al. 3D virtual human atria: a computational platform for studying clinical atrial fibrillation. Prog. Biophys. Mol. Biol 107, 156–168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McDowell KS et al. Mechanistic inquiry into the role of tissue remodeling in fibrotic lesions in human atrial fibrillation. Biophys. J 104, 2764–2773 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McDowell KS et al. Methodology for patient-specific modeling of atrial fibrosis as a substrate for atrial fibrillation. J. Electrocardiol 45, 640–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dossel O, Krueger MW, Weber FM, Wilhelms M & Seemann G Computational modeling of the human atrial anatomy and electrophysiology. Med. Biol. Eng. Comput 50, 773–799 (2012). [DOI] [PubMed] [Google Scholar]
- 113.Fastl TE et al. Personalized computational modeling of left atrial geometry and transmural myofiber architecture. Med. Image Anal 47, 180–190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pashakhanloo F et al. Myofiber architecture of the human atria as revealed by submillimeter diffusion tensor imaging. Circ. Arrhythm. Electrophysiol 9, e004133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Corrado C et al. A work flow to build and validate patient specific left atrium electrophysiology models from catheter measurements. Med. Image Anal 47, 153–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trayanova NA Mathematical approaches to understanding and imaging atrial fibrillation: significance for mechanisms and management. Circ. Res 114, 1516–1531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ten Tusscher KH, Hren R & Panfilov AV Organization of ventricular fibrillation in the human heart. Circ. Res 100, e87–101 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Keldermann RH et al. Effect of heterogeneous APD restitution on VF organization in a model of the human ventricles. Am. J. Physiol. Heart Circ. Physiol 294, H764–H774 (2008). [DOI] [PubMed] [Google Scholar]
- 119.Bayer JD, Lalani GG, Vigmond EJ, Narayan SM & Trayanova NA Mechanisms linking electrical alternans and clinical ventricular arrhythmia in human heart failure. Heart Rhythm 13, 1922–1931 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Van Nieuwenhuyse E, Seemann G, Panfilov AV & Vandersickel N Effects of early afterdepolarizations on excitation patterns in an accurate model of the human ventricles. PLoS ONE 12, e0188867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vandersickel N, de Boer TP, Vos MA & Panfilov AV Perpetuation of torsade de pointes in heterogeneous hearts: competing foci or re-entry? J. Physiol 594, 6865–6878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sadrieh A et al. Multiscale cardiac modelling reveals the origins of notched T waves in long QT syndrome type 2. Nat. Commun 5, 5069 (2014). [DOI] [PubMed] [Google Scholar]
- 123.Potse M et al. Similarities and differences between electrocardiogram signs of left bundle-branch block and left-ventricular uncoupling. Europace 14, v33–v39 (2012). Suppl. 5. [DOI] [PubMed] [Google Scholar]
- 124.Keller DU, Weiss DL, Dossel O & Seemann G Influence of I(Ks) heterogeneities on the genesis of the T-wave: a computational evaluation. IEEE Trans. Biomed. Eng 59, 311–322 (2012). [DOI] [PubMed] [Google Scholar]
- 125.Chen X, Hu Y, Fetics BJ, Berger RD & Trayanova NA Unstable QT interval dynamics precedes ventricular tachycardia onset in patients with acute myocardial infarction: a novel approach to detect instability in QT interval dynamics from clinical ECG. Circ. Arrhythm. Electrophysiol 4, 858–866 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nguyen UC et al. An in-silico analysis of the effect of heart position and orientation on the ECG morphology and vectorcardiogram parameters in patients with heart failure and intraventricular conduction defects. J. Electrocardiol 48, 617–625 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Bacharova L et al. The effect of reduced intercellular coupling on electrocardiographic signs of left ventricular hypertrophy. J. Electrocardiol 44, 571–576 (2011). [DOI] [PubMed] [Google Scholar]
- 128.Zhu X, Wei D & Okazaki O Computer simulation of clinical electrophysiological study. Pacing Clin. Electrophysiol 35, 718–729 (2012). [DOI] [PubMed] [Google Scholar]
- 129.Ashikaga H et al. Feasibility of image-based simulation to estimate ablation target in human ventricular arrhythmia. Heart Rhythm 10, 1109–1116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rantner LJ, Vadakkumpadan F, Spevak PJ, Crosson JE & Trayanova NA Placement of implantable cardioverter-defibrillators in paediatric and congenital heart defect patients: a pipeline for model generation and simulation prediction of optimal configurations. J. Physiol 591, 4321–4334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heijman J, Erfanian Abdoust P, Voigt N, Nattel S & Dobrev D Computational models of atrial cellular electrophysiology and calcium handling, and their role in atrial fibrillation. J. Physiol 594, 537–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Krummen DE et al. Mechanisms of human atrial fibrillation initiation: clinical and computational studies of repolarization restitution and activation latency. Circ. Arrhythm. Electrophysiol 5, 1149–1159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao J, Trew ML, Legrice IJ, Smaill BH & Pullan AJ A tissue-specific model of reentry in the right atrial appendage. J. Cardiovasc. Electrophysiol 20, 675–684 (2009). [DOI] [PubMed] [Google Scholar]
- 134.Aslanidi OV, Boyett MR, Dobrzynski H, Li J & Zhang H Mechanisms of transition from normal to reentrant electrical activity in a model of rabbit atrial tissue: interaction of tissue heterogeneity and anisotropy. Biophys. J 96, 798–817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu TJ et al. Role of pectinate muscle bundles in the generation and maintenance of intra-atrial reentry: potential implications for the mechanism of conversion between atrial fibrillation and atrial flutter. Circ. Res 83, 448–462 (1998). [DOI] [PubMed] [Google Scholar]
- 136.Gong Y et al. Mechanism underlying initiation of paroxysmal atrial flutter/atrial fibrillation by ectopic foci: a simulation study. Circulation 115, 2094–2102 (2007). [DOI] [PubMed] [Google Scholar]
- 137.Cherry EM, Ehrlich JR, Nattel S & Fenton FH Pulmonary vein reentry — properties and size matter: insights from a computational analysis. Heart Rhythm 4, 1553–1562 (2007). [DOI] [PubMed] [Google Scholar]
- 138.Chang KC, Bayer JD & Trayanova NA Disrupted calcium release as a mechanism for atrial alternans associated with human atrial fibrillation. PLoS Comput. Biol 10, e1004011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hwang M et al. Ganglionated plexi stimulation induces pulmonary vein triggers and promotes atrial arrhythmogenecity: In silico modeling study. PLoS ONE 12, e0172931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gharaviri A et al. How disruption of endo-epicardial electrical connections enhances endo-epicardial conduction during atrial fibrillation. Europace 19, 308–318 (2017). [DOI] [PubMed] [Google Scholar]
- 141.Roney CH et al. Modelling methodology of atrial fibrosis affects rotor dynamics and electrograms. Europace 18, iv146–iv155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vigmond E, Pashaei A, Amraoui S, Cochet H & Hassaguerre M Percolation as a mechanism to explain atrial fractionated electrograms and reentry in a fibrosis model based on imaging data. Heart Rhythm 13, 1536–1543 (2016). [DOI] [PubMed] [Google Scholar]
- 143.Roney CH et al. Spatial resolution requirements for accurate identification of drivers of atrial fibrillation. Circ. Arrhythm. Electrophysiol 10, e004899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Uldry L, Virag N, Jacquemet V, Vesin JM & Kappenberger L Optimizing local capture of atrial fibrillation by rapid pacing: study of the influence of tissue dynamics. Ann. Biomed. Eng 38, 3664–3673 (2010). [DOI] [PubMed] [Google Scholar]
- 145.Uldry L, Virag N, Lindemans F, Vesin JM & Kappenberger L Atrial septal pacing for the termination of atrial fibrillation: study in a biophysical model of human atria. Europace 14, 112–120 (2012). [DOI] [PubMed] [Google Scholar]
- 146.Ruchat P et al. A biophysical model of atrial fibrillation to define the appropriate ablation pattern in modified maze. Eur. J. Cardiothorac. Surg 31, 65–69 (2007). [DOI] [PubMed] [Google Scholar]
- 147.Li C et al. The spatiotemporal stability of dominant frequency sites in in-silico modeling of 3-dimensional left atrial mapping of atrial fibrillation. PLoS ONE 11, e0160017 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zahid S et al. Patient-derived models link re-entrant driver localization in atrial fibrillation to fibrosis spatial pattern. Cardiovasc. Res 110, 443–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zahid S et al. Feasibility of using patient-specific models and the “minimum cut” algorithm to predict optimal ablation targets for left atrial flutter. Heart Rhythm 13, 1687–1698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McDowell KS et al. Virtual electrophysiological study of atrial fibrillation in fibrotic remodeling. PLoS ONE 10, e0117110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dhamala J et al. Quantifying the uncertainty in model parameters using Gaussian process-based Markov chain Monte Carlo in cardiac electrophysiology. Med. Image Anal 48, 43–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Konukoglu E et al. Efficient probabilistic model personalization integrating uncertainty on data and parameters: Application to eikonal-diffusion models in cardiac electrophysiology. Prog. Biophys. Mol. Biol 107, 134–146 (2011). [DOI] [PubMed] [Google Scholar]
- 153.Wallman M, Smith NP & Rodriguez B Computational methods to reduce uncertainty in the estimation of cardiac conduction properties from electroanatomical recordings. Med. Image Anal 18, 228–240 (2014). [DOI] [PubMed] [Google Scholar]
- 154.Pathmanathan P, Shotwell MS, Gavaghan DJ, Cordeiro JM & Gray RA Uncertainty quantification of fast sodium current steady-state inactivation for multi-scale models of cardiac electrophysiology. Prog. Biophys, Mol. Biol 117, 4–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shotwell MS & Gray RA Estimability analysis and optimal design in dynamic multi-scale models of cardiac electrophysiology. J. Agr. Biol. Environ. Stat 21, 261–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dhamala J et al. Spatially adaptive multi-scale optimization for local parameter estimation in cardiac electrophysiology. IEEE Trans. Med. Imag 36, 1966–1978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Johnston BM, Coveney S, Chang ETY, Johnston PR & Clayton RH Quantifying the effect of uncertainty in input parameters in a simplified bidomain model of partial thickness ischaemia. Med. Biol. Eng. Comput 56, 761–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chang ET, Strong M & Clayton RH Bayesian Sensitivity Analysis of a cardiac cell model using a gaussian process emulator. PLoS ONE 10, e0130252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mullens W et al. Insights from a cardiac resynchronization optimization clinic as part of a heart failure disease management program. J. Am. Coll. Cardiol 53, 765–773 (2009). [DOI] [PubMed] [Google Scholar]
- 160.Auricchio A & Prinzen FW Enhancing response in the cardiac resynchronization therapy patient: the 3B perspective — bench, bits, and bedside. JACC Clin. Electrophysiol 3, 1203–1219 (2017). [DOI] [PubMed] [Google Scholar]
- 161.Auricchio A, Lumens J & Prinzen FW Does cardiac resynchronization therapy benefit patients with right bundle branch block: cardiac resynchronization therapy has a role in patients with right bundle branch block. Circ. Arrhythm. Electrophysiol 7, 532–542 (2014). [DOI] [PubMed] [Google Scholar]
- 162.Kerckhoffs RC et al. Cardiac resynchronization: insight from experimental and computational models. Prog. Biophys. Mol. Biol 97, 543–561 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Leenders GE et al. Septal deformation patterns delineate mechanical dyssynchrony and regional differences in contractility: analysis of patient data using a computer model. Circ. Heart Fail 5, 87–96 (2012). [DOI] [PubMed] [Google Scholar]
- 164.Lumens J et al. Differentiating electromechanical from non-electrical substrates of mechanical discoordination to identify responders to cardiac resynchronization therapy. Circ. Cardiovasc. Imag 8, e003744 (2015). [DOI] [PubMed] [Google Scholar]
- 165.Bishop M et al. Three-dimensional atrial wall thickness maps to inform catheter ablation procedures for atrial fibrillation. Europace 18, 376–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tracy CM et al. 2012 ACCF/AHA/HRS focused update of the 2008 guidelines for device-based therapy of cardiac rhythm abnormalities: a report of the American College of Cardiology Foundation/ American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. [corrected]. Circulation 126, 1784–1800 (2012). [DOI] [PubMed] [Google Scholar]
- 167.Jones S et al. Cardiac resynchronization therapy: mechanisms of action and scope for further improvement in cardiac function. Europace 19, 1178–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Huntjens PR et al. Influence of left ventricular lead position relative to scar location on response to cardiac resynchronization therapy: a model study. Europace 16 (Suppl. 4), iv62–iv68 (2014). [DOI] [PubMed] [Google Scholar]
- 169.Kerckhoffs RC, McCulloch AD, Omens JH & Mulligan LJ Effects of biventricular pacing and scar size in a computational model of the failing heart with left bundle branch block. Med. Image Anal 13, 362–369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.ter Keurs HE, Rijnsburger WH, van Heuningen R & Nagelsmit MJ Tension development and sarcomere length in rat cardiac trabeculae. Evidence of length-dependent activation. . Circ. Res 46, 703–714 (1980). [DOI] [PubMed] [Google Scholar]
- 171.Niederer SA et al. Length-dependent tension in the failing heart and the efficacy of cardiac resynchronization therapy. Cardiovasc. Res 89, 336–343 (2011). [DOI] [PubMed] [Google Scholar]
- 172.Lumens J et al. Comparative electromechanical and hemodynamic effects of left ventricular and biventricular pacing in dyssynchronous heart failure: electrical resynchronization versus left-right ventricular interaction. J. Am. Coll. Cardiol 62, 2395–2403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van Everdingen WM et al. Echocardiographic prediction of cardiac resynchronization therapy response requires analysis of both mechanical dyssynchrony and right ventricular function: a combined analysis of patient data and computer simulations. J. Am. Soc. Echocardiogr 30, 1012–1020 (2017). [DOI] [PubMed] [Google Scholar]
- 174.Constantino J, Hu Y & Trayanova NA A computational approach to understanding the cardiac electromechanical activation sequence in the normal and failing heart, with translation to the clinical practice of CRT. Prog. Biophys. Mol. Biol 110, 372–379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Crozier A et al. The relative role of patient physiology and device optimisation in cardiac resynchronisation therapy: a computational modelling study. J. Mol. Cell Cardiol 96, 93–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee AW et al. Biophysical modeling to determine the optimization of left ventricular pacing site and AV/VV delays in the acute and chronic phase of cardiac resynchronization therapy. J. Cardiovasc. Electrophysiol 28, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Okada JI et al. Multi-scale, tailor-made heart simulation can predict the effect of cardiac resynchronization therapy. J. Mol. Cell Cardiol 108, 17–23 (2017). [DOI] [PubMed] [Google Scholar]
- 178.Pluijmert M et al. New insights from a computational model on the relation between pacing site and CRT response. Europace 18, iv94–iv103 (2016). [DOI] [PubMed] [Google Scholar]
- 179.Niederer SA et al. Biophysical modeling to simulate the response to multisite left ventricular stimulation using a quadripolar pacing lead. Pacing Clin. Electrophysiol 35, 204–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hyde ER et al. Beneficial effect on cardiac resynchronization from left ventricular endocardial pacing is mediated by early access to high conduction velocity tissue: electrophysiological simulation study. Circ. Arrhythm. Electrophysiol 8, 1164–1172 (2015). [DOI] [PubMed] [Google Scholar]
- 181.Hu Y, Gurev V, Constantino J & Trayanova N Efficient preloading of the ventricles by a properly timed atrial contraction underlies stroke work improvement in the acute response to cardiac resynchronization therapy. Heart Rhythm 10, 1800–1806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Reumann M et al. Computer model for the optimization of AV and VV delay in cardiac resynchronization therapy. Med. Biol. Eng. Comput 45, 845–854 (2007). [DOI] [PubMed] [Google Scholar]
- 183.Hu Y, Gurev V, Constantino J & Trayanova N Optimizing cardiac resynchronization therapy to minimize ATP consumption heterogeneity throughout the left ventricle: a simulation analysis using a canine heart failure model. Heart Rhythm 11, 1063–1069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sermesant M et al. Patient-specific electromechanical models of the heart for the prediction of pacing acute effects in CRT: a preliminary clinical validation. Med. Image Anal 16, 201–215 (2012). [DOI] [PubMed] [Google Scholar]
- 185.Kayvanpour E et al. Towards personalized cardiology: multi-scale modeling of the failing heart. PLoS ONE 10, e0134869 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Krishnamurthy A et al. Patient-specific models of cardiac biomechanics. J. Comput. Phys 244, 4–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Huntjens PR et al. Electrical substrates driving response to cardiac resynchronization therapy: a combined clinical-computational evaluation. Circ. Arrhythm. Electrophysiol 11, e005647 (2018). [DOI] [PubMed] [Google Scholar]
- 188.Augustin CM et al. Anatomically accurate high resolution modeling of human whole heart electromechanics: a strongly scalable algebraic multigrid solver method for nonlinear deformation. J. Comput. Phys 305, 622–646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Viceconti M, Henney A & Morley-Fletcher E In silico clinical trials: how computer simulation will transform the biomedical industry. Int. J. Clin. Trials 3, 37–46 (2016). [Google Scholar]
- 190.Prinzen FW et al. Innovation in cardiovascular disease in Europe with focus on arrhythmias: current status, opportunities, roadblocks, and the role of multiple stakeholders. Europace 20, 733–738 (2017). [DOI] [PubMed] [Google Scholar]
- 191.Niederer SA, Fink M, Noble D & Smith NP A meta-analysis of cardiac electrophysiology computational models. Exp. Physiol 94, 486–495 (2009). [DOI] [PubMed] [Google Scholar]
- 192.Holmes JW & Lumens J Clinical applications of patient-specific models: the case for a simple approach. J. Cardiovasc. Transl Res 11, 71–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Richter Y, Lind PG & Maass P Modeling specific action potentials in the human atria based on a minimal single-cell model. PLoS ONE 13, e0190448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bueno-Orovio A, Cherry EM & Fenton FH Minimal model for human ventricular action potentials in tissue. J. Theor. Biol 253, 544–560 (2008). [DOI] [PubMed] [Google Scholar]
- 195.Corrado C & Niederer SA A two-variable model robust to pacemaker behaviour for the dynamics of the cardiac action potential. Math. Biosci 281, 46–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Neic A et al. Efficient computation of electrograms and ECGs in human whole heart simulations using a reaction-eikonal model. J. Comput. Phys 346, 191–211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gurev V et al. A high-resolution computational model of the deforming human heart. Biomech. Model. Mechanobiol 14, 829–849 (2015). [DOI] [PubMed] [Google Scholar]
- 198.Niederer S, Mitchell L, Smith N & Plank G Simulating human cardiac electrophysiology on clinical time-scales. Frontiers Physiol 2, 14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Niederer SA et al. Verification of cardiac tissue electrophysiology simulators using an N-version benchmark. Phil. Trans. A Math. Phys. Eng. Sci 369, 4331–4351 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Land S et al. Verification of cardiac mechanics software: benchmark problems and solutions for testing active and passive material behaviour. Proc. Math. Phys. Eng. Sci 471, 20150641 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Smith LP et al. SBML Level 3 package: hierarchical model composition, version 1 release 3. J. Integr. Bioinform 12, 603–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cuellar A et al. The CellML 1.1 specification. J. Integr. Bioinform 12, 259 (2015). [DOI] [PubMed] [Google Scholar]
- 203.Kerckhoffs RC, Omens JH & McCulloch AD Mechanical discoordination increases continuously after the onset of left bundle branch block despite constant electrical dyssynchrony in a computational model of cardiac electromechanics and growth. Europace 14 (Suppl. 5), v65–v72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nolden M et al. The medical imaging interaction toolkit: challenges and advances: 10 years of open-source development. Int. J. Comput. Assist. Radiol. Surg 8, 607–620 (2013). [DOI] [PubMed] [Google Scholar]
- 205.Ayachit U The Paraview Guide: A Parallel Visualization Application (Kitware Inc, 2015). [Google Scholar]
- 206.Rhode KS et al. A system for real-time XMR guided cardiovascular intervention. IEEE Trans. Med. Imag 24, 1428–1440 (2005). [DOI] [PubMed] [Google Scholar]
- 207.Razavi R et al. Cardiac catheterisation guided by MRI in children and adults with congenital heart disease. Lancet 362, 1877–1882 (2003). [DOI] [PubMed] [Google Scholar]