Abstract
Background
The purpose of this study was to investigate the correlation between TRMT6 mRNA expression levels and clinicopathological features in primary HCC patients and to evaluate their prognostic value.
Material/Methods
The clinical information and the mRNA sequencing data of the patients with primary hepatocellular carcinoma (HCC) were extracted from The Cancer Genome Atlas (TCGA) Liver Cancer database. The correlation between the clinicopathological features and the expression of TRMT6 was analyzed by t test and chi-square test. The overall survival (OS) and recurrence-free survival (RFS) were estimated using the Kaplan-Meier method and Cox regression models. Gene set enrichment analysis (GSEA) was used to explore the potential mechanisms of TRMT6 dysregulation in primary HCC patients.
Results
Compared to normal tissues, TRMT6 was significantly upregulated in primary HCC tissues. Kaplan-Meier survival curves revealed that higher TRMT6 expression was associated with reduced RFS (p=0.0146) and OS (p=0.0224) in HCC patients. Moreover, multivariable Cox regression analysis indicated that TRMT6 upregulation independently predicted poor RFS (HR: 1.871, 95% CI: 1.204, 2.905, p=0.005) and OS (HR: 2.176, 95% CI: 1.234, 3.836, p=0.007). Gene Set Enrichment Analysis (GSEA) indicated that primary HCC samples in the TRMT6 high expression group were enriched for the G2M checkpoint, spermatogenesis, and MYC target genes.
Conclusions
TRMT6 was upregulated in HCC tissues, and higher TRMT6 expression levels was correlated with reduced OS and RFS in patients with primary HCC. TRMT6 might be a promising prognostic biomarker for poor clinical outcomes in primary HCC patients.
MeSH Keywords: Carcinoma, Hepatocellular; Prognosis; tRNA Methyltransferases
Background
Liver cancer is one of the most prevalent malignancies and the third leading cause of cancer-related mortality in China [1]. Hepatocellular carcinoma (HCC), the predominant pathological type of liver cancer, accounts for approximately 80% to 90% of cases [2]. With the advancement of medical technology, there are a variety of ways to help patients with HCC achieve long-term survival, including surgical resection, molecular-targeted therapy, liver transplantation, radiofrequency ablation and chemoembolization. However, despite these advances, clinical outcome for most patients with HCC remains poor [3], and the recurrence rate of extrahepatic and locoregional is high. Several studies have demonstrated that the 5-year recurrence rates are approximately 60~80% for patients with HCC [4,5], with both initiation and metastasis being the primary factors affecting the prognosis of patients with HCC. Therefore, early diagnosis and identification of prognostic biomarker are crucial for improving survival in HCC patients. Currently, alpha-fetoprotein (AFP) is the most widely used biomarker for early clinical diagnosis of liver cancer. However, with relatively low sensitivity and specificity in clinical practice, its clinical value is not satisfactory [6]. Research on the molecular mechanisms related to HCC development and progression has received extensive attention in recent years [7], and the genetic and epigenetic alterations have also received substantial attention in scientific research. Chemical modifications of RNA are very common in human diseases; over 100 types of these modifications have been identified, and methylation is the main modification [8].
RNA methylation is closely associated with multiple diseases, including tumors and metabolic disease [9,10]. N1-methyladenosine (M1A), a recently identified mRNA modification of N6-methyladenosine, affects mRNA localization, stability, translation, and splicing [11]. M1A is dependent on the TRMT6/TRMT61A methyltransferase complex [12].
Despite these findings, few studies have focused on the biological role and clinical significance of TRMT6. The purpose of this study was to perform bioinformatics analysis, and to examine mRNA expression levels of TRMT6 in HCC patients based on data obtained from The Cancer Genome Atlas (TCGA) database. We attempted to evaluate the prognostic value of TRMT6 by exploring the potential relationship between the expression of TRMT6 and the clinicopathological features of primary HCC patients.
Material and Methods
Bioinformatics analysis based on the TCGA database
The TCGA database is a collaboration between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) that has been made publicly available, containing multidimensional maps of key genomic changes in 33 types of cancer. This genomic information helps the cancer research community to improve the prevention, diagnosis, and treatment of cancer [13]. Clinical information and mRNA sequencing data in the TCGA Liver Cancer (TCGA-LIHC) database and primary HCC patients were obtained from the Genomic Data Commons Portal (https://portal.gdc.cancer.gov/). These patient data are publicly available, and the samples are de-identified before publication. In the TCGA-LIHC cohort, mRNA expression profiles and clinical information for 424 patients were downloaded. After excluding patients with missing data, we collected 410 tissue samples from 361 patients with TRMT6 mRNA gene expression levels, including 312 single HCC tissues samples and 49 paired tissues samples (HCC tissues and normal tissues from the same patient) and clinicopathological information for 361 patients (244 males and 117 females) diagnosed with primary HCC.
Gene set enrichment analysis (GSEA)
GSEA is a computational approach to identifying significantly enriched or depleted groups of genes [14,15]. To gain insight into the molecular mechanism related to the TRMT6 gene, the GSEA tool was used to analyze the association between TRMT6 expression and HCC expression profiles in the TCGA-LIHC cohort. In our study, the genomic expression profiles of 361 HCC patients were classified into high (n=181) and low (n=180) expression groups based on the median value of TRMT6 mRNA expression. The h.all.v6.2.symbols.gmt data set was obtained from the GSEA website, and the number of random permutations was set at 1000 runs.
Statistical analysis
Statistical analyses were performed using R (v.3.5.1) software. Quantitative data are reported as the means ± standard deviation (SD), and categorical data are reported as counts (percentage). The cut-off value for TRMT6 mRNA expression levels was determined based on its median value. Differences in TRMT6 mRNA expression levels between HCC and normal liver tissues were assessed by using unpaired and paired t tests. Unpaired 2-sample t test and chi-square analysis were used to evaluate the association between the clinicopathological features and the TRMT6 mRNA expression levels. The differences in OS and RFS were estimated by using the Kaplan-Meier method based on log-rank tests. The effects of the clinical characteristics and the mRNA expression levels of TRMT6 on the prognosis of HCC patients were calculated by univariable and multivariable Cox regression models. Potential covariates from basic characteristics were adjusted using multivariable analysis. In addition, we performed sensitivity analysis by repeating the 2 main analyses using TRMT6 expression as a continuous number or including covariates with p values less than 0.1. P value <0.05 was considered statistically significant.
Results
mRNA expression levels of TRMT6 and clinical characteristics of HCC patients based on the TCGA database
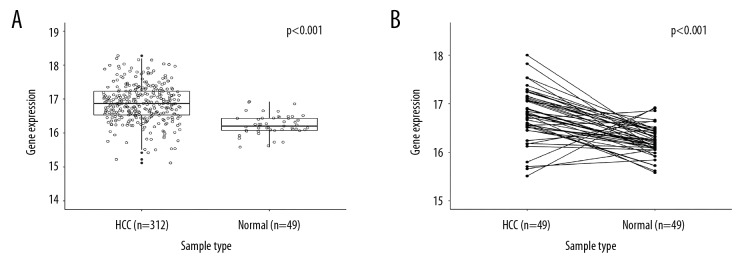
As shown in Figure 1A, the expression levels of TRMT6 were significantly higher in the 312 single HCC tissues than in the 49 normal tissues (unpaired 2-sample t test, P<0.001), and results were consistent with the 49 matched tissue samples from the HCC patients (paired t test, P<0.001) (Figure 1B). We next investigated the relationship between the TRMT6 mRNA expression and the clinical characteristics of patients with HCC. Race, fibrosis, and histological grade were significantly different between high and low expression groups in patients with HCC (p<0.001, p=0.016, and p<0.001, respectively); however, gene group was not significantly associated with age, gender, BMI, Child-Pugh grade, or clinical stage (Table 1).
Figure 1.
TRMT6 was upregulated in primary HCC patients. (A) Comparison of TRMT6 gene expression with 361 cases of HCC tissues and 49 adjacent normal liver tissues. (B) Using the paired t test to assay the difference between 49 cases of primary HCC tissues and the adjacent normal tissues.
Table 1.
The clinical characteristics of primary HCC patients in high expression of TRMT6 group and low expression group.
| Characteristics | TRMT6 expression | t/χ2 | P-value | ||
|---|---|---|---|---|---|
| Low (<16.96) | High (≥16.96) | ||||
| N=180 | N=181 | ||||
| Age (mean ± SD) | 60.13±13.07 | 59.06±13.48 | 0.766 | 0.444 | |
| Gender | Female | 62 (34.44%) | 55 (30.39%) | 0.678 | 0.41 |
| Male | 118 (65.56%) | 126 (69.61%) | |||
| Race | Asian | 58 (32.22%) | 98 (54.14%) | – | 0.000 |
| White | 104 (57.78%) | 72 (39.78%) | |||
| Others | 18 (10.00%) | 11 (6.08%) | |||
| BMI | 27.00±6.75 | 25.33±9.91 | 1.788 | 0.075 | |
| Child-Pugh grade | A | 106 (58.89%) | 106 (58.56%) | – | 0.362 |
| B | 8 (4.44%) | 13 (7.18%) | |||
| C | 1 (0.56%) | 0 (0.00%) | |||
| Unknown | 64 (36.11%) | 63 (34.25%) | |||
| Clinical stage | I+II | 126 (70.00%) | 123 (67.96%) | 0.079 | 0.779 |
| III+IV | 43 (23.89%) | 45 (24.86%) | |||
| Unknown | 11 (6.11%) | 13 (7.18%) | |||
| Fibrosis (Ishak score) | 0 | 49 (27.22%) | 23 (12.71%) | 5.787 | 0.016 |
| 1–6 | 70 (38.89%) | 68 (37.57%) | |||
| Unknown | 61 (33.89%) | 91 (49.72%) | |||
| Histologic grade | G1+G2 | 131 (72.78%) | 93 (51.38%) | 18.557 | 0.000 |
| G3+G4 | 46 (25.56%) | 86 (47.51%) | |||
| Unknown | 3 (1.67%) | 2 (1.10%) | |||
High TRMT6 expression was associated with reduced RFS in patients with primary HCC
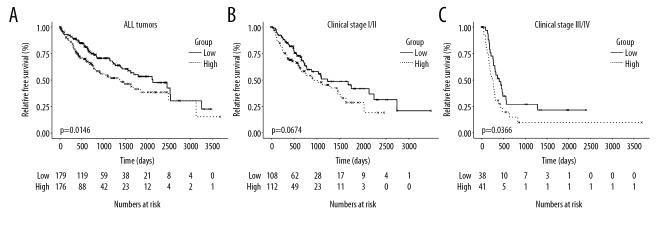
To investigate the association between recurrence-free survival time in HCC patients in the TRMT6 high and low expression groups, we created Kaplan-Meier plots of the RFS of primary HCC patients in both groups. As shown in Figure 2, high expression of TRMT6 was associated with poor RFS in primary HCC patients (p=0.0146, Figure 2A). The results from univariable Cox regression analysis showed that higher clinical stage and higher TRMT6 expression were associated with worse RFS. These results are consistent with those from subsequent multivariable analysis (Tables 2 and 3). In addition, we analyzed the RFS in different subgroups according to different clinical stage. High TRMT6 expression was not significantly associated with poor RFS in early stages (stage I/II) (P=0.0674, Figure 2B). Conversely, high TRMT6 expression was correlated with reduced RFS in advanced stages (stage III/IV) (P=0.0366, Figure 2C). Upregulated TRMT6 expression is likely an independent prognostic factor for poor RFS in patients with primary HCC, especially for patients with advanced disease stages.
Figure 2.
The RFS of all patients (A), early clinical stage patients (B), and clinical advanced stage patients (C) high expression of TRMT6 group and low expression of TRMT6 group.
Table 2.
Univariable analysis of RFS and OS in patients with primary HCC.
| RFS | OS | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| TRMT6 expression (high vs. low) | 1.437 (1.051, 1.965) | 0.023 | 1.539 (1.086, 2.180) | 0.015 | |
| Age (continuous) | 0.995 (0.983, 1.008) | 0.456 | 1.012 (0.998, 1.025) | 0.099 | |
| Gender (Female vs. Male) | 1.016 (0.729, 1.416) | 0.925 | 1.204 (0.844, 1.716) | 0.306 | |
| Race | 1 | 1.161 (0.558, 2.413) | 0.69 | 0.585 (0.302, 1.134) | 0.112 |
| 2 | 1.281 (0.620, 2.647) | 0.504 | 0.786 (0.416, 1.487) | 0.459 | |
| 3 | Reference | Reference | |||
| BMI (continuous) | 0.978 (0.952, 1.005) | 0.113 | 1.002 (0.973, 1.031) | 0.908 | |
| Child-Pugh grade | A | 0.820 (0.114, 5.897) | 0.843 | 0.470 (0.065, 3.412) | 0.455 |
| B | 1.052 (0.132, 8.367) | 0.962 | 0.737 (0.091, 5.975) | 0.775 | |
| C | Reference | Reference | |||
| Tumor stage (I+II vs. III+IV) | 0.392 (0.280, 0.549) | <0.001 | 0.420 (0.290, 0.608) | <0.001 | |
| Fibrosis (0 vs. 1–6) | 0.812 (0.529, 1.245) | 0.339 | 1.263 (0.762, 2.091) | 0.365 | |
| Histologic grade (G1+G2 vs. G3+G4) | 0.862 (0.627, 1.185) | 0.36 | 0.958 (0.668, 1.375) | 0.817 | |
Table 3.
Multivariate analysis of RFS and OS in patients with primary HCC.
| RFS | OS | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| TRMT6 expression (high vs. low) | 2.226 (1.313, 3.774) | 0.003 | 3.051 (1.553, 5.996) | 0.001 | |
| Age (continuous) | 0.996 (0.977, 1.015) | 0.674 | 1.027 (1.001, 1.054) | 0.042 | |
| Gender (Female vs. Male) | 0.730 (0.423, 1.260) | 0.258 | 0.612 (0.309, 1.214) | 0.160 | |
| Race | 1 | 0.208 (0.057, 0.753) | 0.017 | 0.143 (0.047, 0.440) | <0.001 |
| 2 | 0.775 (0.229, 2.619) | 0.682 | 0.316 (0.111, 0.897) | 0.030 | |
| 3 | Reference | Reference | |||
| BMI (continuous) | 0.947 (0.900, 0.996) | 0.034 | 1.038 (1.005, 1.073) | 0.024 | |
| Child-Pugh grade | A | 1.421 (0.613, 3.294) | 0.413 | 0.853 (0.361, 2.016) | 0.718 |
| B | Not applicable | Not applicable | |||
| C | Reference | Reference | |||
| Tumor stage (I+II vs. III+IV) | 0.425 (0.252, 0.716) | 0.001 | 0.490 (0.252, 0.953) | 0.036 | |
| Fibrosis (0 vs. 1–6) | 0.620 (0.326, 1.177) | 0.144 | 1.414 (0.671, 2.980) | 0.362 | |
| Histologic grade (G1+G2 vs. G3+G4) | 0.765 (0.475, 1.232) | 0.27 | 0.657 (0.356, 1.216) | 0.181 | |
Upregulated expression of TRMT6 was associated with poor OS in primary HCC patients
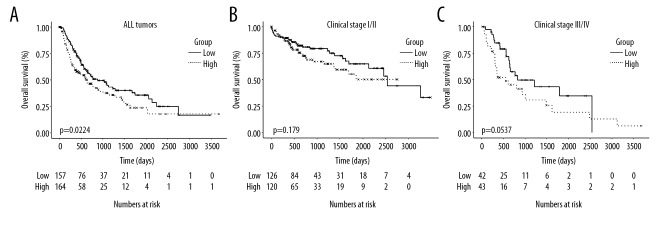
A log-rank test showed that primary HCC patients with high mRNA expression levels of TRMT6 exhibited significantly shorter OS than those with low expression (P=0.0224, Figure 3A). Both univariable and multivariable analyses showed that clinical stage and TRMT6 expression significantly influence OS (Tables 2 and 3). In addition, we analyzed OS in the different subgroups based on different clinical stages. High expression of TRMT6 was not significantly associated with poor OS in early stages (stage I/II) (P=0.179, Figure 3B). Conversely, a trend towards statistical significance was found in advanced stages (stage III and stage III/IV) (P=0.0537, Figure 3C), potentially due to the relatively small number of clinical cases in this study. Therefore, overexpression of TRMT6 remained a negative prognostic indicator in terms of OS in patients with primary HCC (multivariable analysis, hazard ratio=2.176, 95% confidence interval: 1.234–3.836, P=0.007).
Figure 3.
The OS of all patients (A), early clinical stage patients (B), and advanced clinical stage patients (C) high expression of TRMT6 group and low expression of TRMT6 group.
Sensitivity analysis for RFS and OS based on univariable and multivariable Cox regression analyses
These results are summarized in Table 4. The findings from all 3 methods were similar and the observed trends were consistent. Only 2 multivariable models related to RFS using TRMT6 expression as a continuous variable approached significance (p=0.073 and p=0.064, respectively).
Table 4.
Gene sets enriched in the samples of primary HCC in the high expression of TRMT6 group.
| Outcome | TRMT6 expression (high vs. low) | TRMT6 expression (continuous variable) | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| RFS | Univariable | 1.437 (1.051, 1.965) | 0.023 | 1.482 (1.087, 2.022) | 0.013 |
| Multivariablea | 1.871 (1.204, 2.905) | 0.005 | 1.537 (0.960, 2.461) | 0.073 | |
| Multivariableb | 2.226 (1.313, 3.774) | 0.003 | 1.691 (0.970, 2.949) | 0.064 | |
| OS | Univariable | 1.539 (1.086, 2.180) | 0.015 | 1.862 (1.327, 2.614) | <0.001 |
| Multivariablec | 2.176 (1.234, 3.836) | 0.007 | 2.730 (1.531, 4.869) | <0.001 | |
| Multivariableb | 3.051 (1.553, 5.996) | 0.001 | 3.279 (1.762, 6.099) | <0.001 | |
Model was adjusted for tumor stage based on selection of variables with p<0.1 in univariable analysis;
Model was adjusted for all potential confounders presented in Table 1;
Model was adjusted for age and tumor stage based on selection of variables with p<0.1 in univariable analysis.
TRMT6-related signaling pathways identified using GSEA
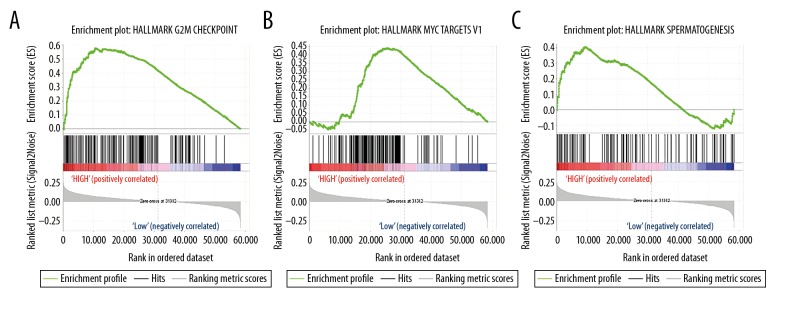
To elucidate the function of the TRMT6 gene in primary HCC patients, we performed GSEA analysis between TRMT6 gene expression and known oncogenic signaling pathways using TCGA data. Based on the false discovery rate (FDR) and the normalized enrichment score (NES), we selected 3 signaling pathways that were the most significantly enriched in the predefined hallmark gene (Table 5). As shown in Figure 4, GSEA analysis suggested that primary HCC samples in the TRMT6 high expression group were enriched in the G2M checkpoint (Figure 4A), spermatogenesis (Figure 4B), and MYC targets (Figure 4C). As expected, the TRMT6 gene may contribute to poor prognosis through these biological processes in HCC patients.
Table 5.
Gene sets enriched in the samples of primary HCC in the high expression of TRMT6 group.
| Gene set name | ES | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|
| HALLMARK_G2M_CHECKPOINT | 0.578 | 1.754 | 0.000 | 0.034 |
| HALLMARK_MYC_TARGETS_V1 | 0.441 | 1.879 | 0.000 | 0.031 |
| HALLMARK_SPERMATOGENESIS | 0.406 | 1.793 | 0.000 | 0.016 |
Figure 4.
Enrichment plots from GSEA. GSEA results showing G2M checkpoint (A), c-MYC target gene (B), and spermatogenesis (C).
Discussion
Liver cancer is one of the principal causes of cancer-related morbidity and mortality worldwide, resulting in approximately 782 000 newly diagnosed cases and 745 000 deaths in 2012 [16]. However, clinical outcomes for most HCC patients are poor. Therefore, identifying more efficient biomarkers for HCC diagnosis and prognosis is urgently needed.
Post-transcriptional regulation of gene expression at the RNA level, and dynamic and reversible RNA modifications are important ways to regulate gene expression [17]. Recently, the contribution of RNA methylation to HCC oncogenesis and prognosis have been reported by several studies [18,19]. M1A is a recently identified RNA modification that occurs on most different gene transcripts in eukaryotic cells at an estimated average transcript stoichiometry of 20% in humans, and it exerts a disruptive function on base pairing, leading to translational inhibition [20]. TRMT6, a member of the t-RNA methyltransferase 6 protein family, is localized to chromosomal 20p12.3. In eukaryotes, TRMT6 protein, together with the TRMT61A catalytic subunit, mediates methylation of adenosine residues at the N (1) position of subset of mRNAs [21,22]. Macari et al. demonstrated that depletion of TRM6/61 reduced proliferation and increased death in C6 glioma cells. In addition, TRM6/TRM61 mRNA expression was significantly upregulated in highly aggressive glioblastoma multiforme compared with grade II/III glioblastomas [23]. Yeon et al. reported that frameshift mutations in the TRMT6 gene might play a role in tumorigenesis through their inactivation in colorectal adenocarcinoma [24]. However, little is known about the clinical value or biological function of TRMT6 in HCC patients. The present study found that TRMT6 mRNA is clearly overexpressed in HCC tissues compared to normal tissues. Comparing the clinicopathological parameters with the TRMT6 mRNA expression levels in tissues of HCC patients, our study revealed statistically significant differences in HCC patients in race, fibrosis, and histological grade (P<0.001) between high and the low TRMT6 gene expression groups. Moreover, patients with high TRMT6 expression levels exhibited poor disease prognosis. These results demonstrate that TRMT6 gene expression plays a role in liver tumorigenesis and might predict poor clinical outcome in HCC patients. GSEA results suggested that TRMT6 plays a role in cell cycle progression, proliferation, apoptosis, and cellular transformation in primary HCC cells through the G2M checkpoint, spermatogenesis, and MYC signaling pathways. These signaling pathways, which play important roles in the carcinogenesis of multiple human tumors, are attracting increasing attention [25–27].
Sensitivity analysis determined the robustness of our assessment of considering TRMT6 expression as a true prognosis marker for HCC. In clinical practice, the method of dichotomizing continuous covariates is common in medical and epidemiological research for several reasons, both clinical and statistical. A simple risk classification into “high” versus “low” could be offered by binary variables to assist in making easy recommendations.
From the findings above, overexpression of TRMT6 in primary HCC tissues is correlated with poor OS and RFS in patients with primary HCC, suggesting TRMT6 as a potential prognostic biomarker of poor clinical outcomes in primary HCC. However, in vivo and in vitro detailed research and validation are still needed for a comprehensive understanding of the molecular mechanisms of the biological effects of TRMT6 in primary HCC.
Conclusions
We observed that gene expression of TRMT6 is upregulated in primary HCC patients, and patients with high TRMT6 expression exhibit worse prognosis.
Footnotes
Source of support: Funding was provided by the Science and Technology Research Project of Kaifeng Municipal Science and Technology Bureau (1603077)
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 3.Tunissiolli NM, Castanhole-Nunes M, Biselli-Chicote PM, et al. Hepatocellular carcinoma: A comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev. 2017;18(4):863–72. doi: 10.22034/APJCP.2017.18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allaire M, Nault JC. Advances in management of hepatocellular carcinoma. Curr Opin Oncol. 2017;29(4):288–95. doi: 10.1097/CCO.0000000000000378. [DOI] [PubMed] [Google Scholar]
- 5.Dai WC, Cheung TT. Strategic overview on the best treatment option for intrahepatic hepatocellular carcinoma recurrence. Expert Rev Anticancer Ther. 2016;16(10):1063–72. doi: 10.1080/14737140.2016.1226136. [DOI] [PubMed] [Google Scholar]
- 6.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1(4):593–98. doi: 10.3892/mco.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanasekaran R, Bandoh S, Roberts R. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Res. 2016;5 doi: 10.12688/f1000research.6946.1. pii: F1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39(Database issue):D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N(6)-methyladenosine (m(6)A) in human cancers. Cancer Lett. 2017;408:112–20. doi: 10.1016/j.canlet.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S, Hartmann JD, Watzinger P, et al. A single N(1)-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci Rep. 2018;8(1):11904. doi: 10.1038/s41598-018-30383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441–46. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Xiong X, Zhang M, et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell. 2017;68(5):993–1005. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Kuehn H, Gould J, et al. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23(23):3251–53. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 15.Debrabant B. The null hypothesis of GSEA, and a novel statistical model for competitive gene set analysis. Bioinformatics. 2017;33(9):1271–77. doi: 10.1093/bioinformatics/btw803. [DOI] [PubMed] [Google Scholar]
- 16.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 17.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erstad DJ, Fuchs BC, Tanabe KK. Molecular signatures in hepatocellular carcinoma: A step toward rationally designed cancer therapy. Cancer. 2018;124(15):3084–104. doi: 10.1002/cncr.31257. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network. Ally A, Balasundaram M, Carlsen R, et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–41. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safra M, Sas-Chen A, Nir R, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551(7679):251–55. doi: 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Xiong X, Wang K, et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12(5):311–16. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 22.Ozanick S, Krecic A, Andersland J, et al. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11(8):1281–90. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macari F, El-Houfi Y, Boldina G, et al. TRM6/61 connects PKCalpha with translational control through tRNAi(Met) stabilization: Impact on tumorigenesis. Oncogene. 2016;35(14):1785–96. doi: 10.1038/onc.2015.244. [DOI] [PubMed] [Google Scholar]
- 24.Yeon SY, Jo YS, Choi EJ, et al. Frameshift mutations in repeat sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 genes in colon cancers. Pathol Oncol Res. 2018;24(3):617–22. doi: 10.1007/s12253-017-0287-2. [DOI] [PubMed] [Google Scholar]
- 25.Li B, Simon MC. Molecular pathways: Targeting MYC-induced metabolic reprogramming and oncogenic stress in cancer. Clin Cancer Res. 2013;19(21):5835–41. doi: 10.1158/1078-0432.CCR-12-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalsbeek D, Golsteyn RM. G2/M-phase checkpoint adaptation and micronuclei formation as mechanisms that contribute to genomic instability in human cells. Int J Mol Sci. 2017;18(11) doi: 10.3390/ijms18112344. pii: E2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babatunde KA, Najafi A, Salehipour P, et al. Cancer/Testis genes in relation to sperm biology and function. Iran J Basic Med Sci. 2017;20(9):967–74. doi: 10.22038/IJBMS.2017.9259. [DOI] [PMC free article] [PubMed] [Google Scholar]