Summary
Background
Transmission of multidrug-resistant Candida auris infection has been reported in the USA. To better understand its emergence and transmission dynamics and to guide clinical and public health responses, we did a molecular epidemiological investigation of C auris cases in the USA.
Methods
In this molecular epidemiological survey, we used whole-genome sequencing to assess the genetic similarity between isolates collected from patients in ten US states (California, Connecticut, Florida, Illinois, Indiana, Maryland, Massachusetts, New Jersey, New York, and Oklahoma) and those identified in several other countries (Colombia, India, Japan, Pakistan, South Africa, South Korea, and Venezuela). We worked with state health departments, who provided us with isolates for sequencing. These isolates of C auris were collected during the normal course of clinical care (clinical cases) or as part of contact investigations or point prevalence surveys (screening cases). We integrated data from standardised case report forms and contact investigations, including travel history and epidemiological links (ie, patients that had shared a room or ward with a patient with C auris). Genetic diversity of C auris within a patient, a facility, and a state were evaluated by pairwise differences in single-nucleotide polymorphisms (SNPs).
Findings
From May 11, 2013, to Aug 31, 2017, isolates that corresponded to 133 cases (73 clinical cases and 60 screening cases) were collected. Of 73 clinical cases, 66 (90%) cases involved isolates related to south Asian isolates, five (7%) cases were related to South American isolates, one (1%) case to African isolates, and one (1%) case to east Asian isolates. Most (60 [82%]) clinical cases were identified in New York and New Jersey; these isolates, although related to south Asian isolates, were genetically distinct. Genomic data corroborated five (7%) clinical cases in which patients probably acquired C auris through health-care exposures abroad. Among clinical and screening cases, the genetic diversity of C auris isolates within a person was similar to that within a facility during an outbreak (median SNP difference three SNPs, range 0–12).
Interpretation
Isolates of C auris in the USA were genetically related to those from four global regions, suggesting that C auris was introduced into the USA several times. The five travel-related cases are examples of how introductions can occur. Genetic diversity among isolates from the same patients, health-care facilities, and states indicates that there is local and ongoing transmission.
Introduction
Candida auris is an emerging, pathogenic yeast. It causes invasive infection with high crude mortality,1,2 and is often multidrug resistant, with some isolates resistant to all three major classes of antifungals.3 Unlike other Candida species, C auris can spread readily in health-care settings, causing outbreaks.4,5 The abilities of C auris to colonise skin and other body sites and to persist in the health-care environment facilitate its transmission.6
C auris was first reported in 2009, after being identified in external ear discharge from a patient in Japan.7 Since then, invasive C auris infections and outbreaks have been reported in more than 30 countries on six continents.4,8 This rapid emergence has raised the question of whether C auris has spread between regions. Whole-genome sequencing (WGS) analyses of isolates from five countries identified four major C auris clades associated with geographical regions, suggesting independent emergence of C auris in four global regions (Africa, South America, east Asia, and south Asia).3 The clades were phylogenetically supported and were separated by tens of thousands of genomic differences, known as single nucleotide polymorphisms (SNPs). Within each clade, isolates were highly related, often differing by less than 100 SNPs.3
In 2016, the first seven patients with C auris were reported in the USA.9 The first known US patient with C auris was infected from 2013, and was identified through retrospective review. The patient was transferred to the USA from a health-care facility in the United Arab Emirates. The other six patients, all of whom had C auris isolates collected in 2015 or 2016, had no known health-care exposure abroad. By May, 2017, 122 patients with C auris from seven states had been reported.10
In June, 2016, the US Centers for Disease Control and Prevention (CDC) issued a clinical alert to US health-care facilities that recommended that suspected and confirmed C auris cases be reported to state and local health departments and the CDC.11 In response, epidemiological and molecular investigation of the reported C auris cases and their associated isolates was done to better understand the emergence and transmission dynamics of C auris in the USA.
Methods
Study design and participants
In this molecular epidemiological survey, we retrospectively identified cases by reviewing isolate collections and microbiology records from the CDC’s Mycotic Diseases Branch laboratory and reference laboratories to identify patients with C auris infections. We also prospectively identified cases that had been newly reported by state health departments. Isolates were taken from patients in ten states (California, Connecticut, Florida, Illinois, Indiana, Maryland, Massachusetts, New Jersey, New York, and Oklahoma) in the USA, to be confirmed as C auris infections or colonisations.
Cases were classified as clinical or screening on the basis of how they were identified. A clinical case was defined as isolation of a C auris sample from a patient that was collected during the normal course of clinical care, to diagnose or treat disease.12 These cases involved isolates from sterile sites, such as blood, and non-invasive sites, such as the respiratory tract or urine, even if no clinical infection was present. A screening case was defined as isolation of a C auris sample from a patient that was collected as part of a contact investigation or point prevalence survey in health-care facilities, to identify the burden of C auris colonisation. Patients were screened by swabbing the axilla and groin or, in some patients, nostrils and wounds.
As part of the public health response, all reported cases prompted epidemiological investigations, including collection of demographic and clinical information and travel history about the patient. State and local health departments used contact investigations to identify and screen patients who shared a room or ward with patients known to have C auris.
Because this work was part of an ongoing public health response, it was determined to be non-research public health practice by CDC officials who are responsible for human participant protection, and this study was therefore not subject to review by institutional review boards.
Procedures
C auris isolates and screening swabs were sent to the Mycotic Diseases Branch laboratory, and we cultured swabs for C auris in a Salt Sabouraud Dulcitol enrichment broth.6 Details of C auris culture and testing are described in the appendix. Isolates were confirmed to be C auris by matrix-assisted laser desorption ionisation (MALDI)–time of flight mass spectrometry with a MALDI Biotyper (Bruker Daltonik; Bremen, Germany) and the MicrobeNet MALDI database (CDC; Atlanta, GA, USA). Isolates from clinical cases were prioritised above screening cases for antifungal susceptibility testing. Details on these methods and the defined threshold for resistance to echinocandins, fluconazole, and amphotericin B are described in the appendix.
We whole-genome sequenced isolates that were confirmed to be C auris, and we quantified SNPs with the Northern Arizona SNP Pipeline;13 details on DNA extraction, sequencing, and SNP analysis are described in the appendix. All raw reads in this study have been submitted to the National Center for Biotechnology Information’s Sequence Read Archive (BioProject ID PRJNA493622).
We did phylogenetic analyses with selected sequences from isolates from the USA and those previously sequenced from other countries (Colombia, India, Japan, Pakistan, South Africa, South Korea, and Venezuela)3 to assess sequence similarity. For cases that had several associated isolates, the isolate with the earliest collection date was included in the analysis.
Only clinical cases were used to assess the genetic diversity of isolates within the USA, whereas both clinical and screening cases were used to assess genetic diversity within an individual, within a facility, and within each state, for assessment of transmission.
Statistical analysis
Maximum parsimony and neighbour-joining phylogenetic trees were constructed by use of MEGA14 version 7 and the ape15 version 5.0 R package. We did a principal component analysis (PCA) to identify clusters on the basis of SNP differences, by use of the adegenet16 version 2.1.0 R package (appendix).
We estimated genetic diversity by examining pairwise SNP differences. For any sample of isolates, n (n–1)/2 pairwise SNP differences were generated, where n was the number of isolates in the sample. For genetic diversity of C auris within a person, we selected patients in whom several isolates were recovered from specimens that were collected on the same day, and pairwise differences were generated and averaged to yield the mean SNP difference within a person. For genetic diversity of C auris within a facility, we selected and compared patients who had received health care and acquired C auris within a single facility. For genetic diversity of C auris in a state, pairwise differences were generated among isolates of all cases and between cases that had been identified as epidemiologically linked (ie, sharing a room or ward at the same time).
Role of the funding source
The funding arm of the CDC had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Between May 11, 2013, and Aug 31, 2017, the CDC received 385 C auris isolates from the ten US states that were reporting cases; all cases except one occurred after July, 2015 (appendix). Of these 385 isolates, 294 (76%) isolates were whole-genome sequenced; the remainder were not sequenced because of resource constraints. From April, 2017, priority for whole-genome sequencing was given to isolates only from incident cases because several isolates from the same patient were often received. These 294 sequenced isolates were taken from 133 patients, which included 73 clinical cases and 60 screening cases. Sex was reported for 128 (96%) patients, and age was reported for 124 (93%) patients (table). 67 (52%) patients with reported sex were male and 61 (48%) patients were female. The majority of patients (79 [64%]) with reported age were aged at least 65 years. The most common sources of specimens for C auris diagnosis were skin (55 [41%] patients), blood (37 [28%] patients), and urine (15 [11%] patients). Because screening cases were identified primarily by swabbing the axilla and groin, 54 (98%) of 55 skin specimens were from screening cases. The facility where the specimen was collected was reported for 118 (89%) patients. The majority of patients (79 [67%]) were diagnosed at an acute care facility, and 43 independent health-care facilities were represented in our analysis.
Table:
Main characteristics of the 133 clinical and screening cases in the USA
| New York (n=59) |
New Jersey (n=54) |
Illinois (n=10) |
Massachusetts (n=4) |
California (n=1) |
Connecticut (n=1) |
Florida (n=1) |
Indiana (n=1) |
Maryland (n=1) |
Oklahoma (n=1) |
Total (n=133) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex* | |||||||||||
| Male | 27 (47%) | 29 (56%) | 4 (44%) | 2 (50%) | 1 (100%) | 0 | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 67 (52%) |
| Female | 30 (53%) | 23 (44%) | 5 (56%) | 2 (50%) | 0 | 1 (100%) | 0 | 0 | 0 | 0 | 61 (48%) |
| Age† | |||||||||||
| <21 years | 1 (2%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (1%) |
| ≥21 years to ≤64 years | 21 (36%) | 13 (26%) | 7 (78%) | 0 | 0 | 1 (100%) | 0 | 1 (100%) | 1 (100%) | 0 | 44 (35%) |
| ≥65 years | 36 (62%) | 37 (74%) | 2 (22%) | 1 (100%) | 1 (100%) | 0 | 1 (100%) | 0 | 0 | 1 (100%) | 79 (64%) |
| Case status | |||||||||||
| Clinical | 44 (75%) | 17 (31%) | 3 (30%) | 3 (75%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 73 (55%) |
| Screening | 15 (25%) | 37 (69%) | 7 (70%) | 1 (25%) | 0 | 0 | 0 | 0 | 0 | 0 | 60 (45%) |
| Specimen source | |||||||||||
| Skin | 14 (24%) | 34 (63%) | 6 (60%) | 1 (25%) | 0 | 0 | 0 | 0 | 0 | 0 | 55 (41%) |
| Blood | 25 (42%) | 10 (19%) | 1 (10%) | 0 | 0 | 0 | 0 | 0 | 1 (100%) | 0 | 37 (28%) |
| Urine | 4 (7%) | 5 (9%) | 2 (20%) | 1 (25%) | 1 (100%) | 0 | 1 | 0 | 0 | 1 (100%) | 15 (11%) |
| Other‡ | 16 (27%) | 5 (9%) | 1 (10%) | 2 (50%) | 0 | 1 (100%) | 0 | 1 (100%) | 0 | 0 | 26 (20%) |
| Facility type§ | |||||||||||
| Acute care | 44 (83%) | 23 (47%) | 2 (33%) | 4 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 1 (100%) | 79 (67%) |
| Long-term care | 9 (17%) | 26 (53%) | 4 (67%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 39 (33%) |
Data are n (%) in each state, where the specimen was collected.
Because of missing data, sex was only reported for 128 (96%) of the 133 cases; data were missing from New York (two cases), New Jersey (two cases), and Illinois (one case).
Because of missing data, age was only reported for 124 (93%) of the 133 cases; data were missing for New York (one case), New Jersey (four cases), Illinois (one case), and Massachussetts (three cases).
Most commonly included nostrils, sputum, and wound.
Because of missing data, the facility was only reported for 118 (89%) of the 133 cases; data were missing from New York (six cases), New Jersey (five cases), and Illinois (four cases).
Isolates from clinical cases were prioritised above screening cases for antifungal susceptibility testing. Of 99 isolates tested, 88 (89%) isolates were resistant to fluconazole, 33 (33%) isolates were resistant to amphotericin B, and six (6%) isolates were resistant to echinocandins. 39 (39%) isolates were resistant to two antifungal classes.
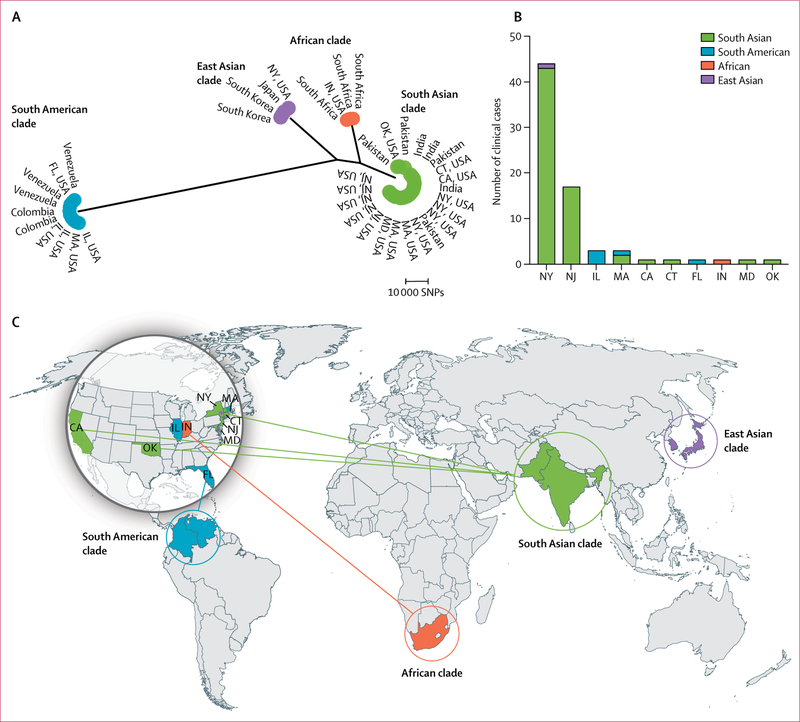
Phylogenetic analysis revealed that all isolates from the USA were from one of the four major clades (figure 1). Of the 73 clinical cases, 66 (90%) patients had isolates with sequences that clustered to the south Asian clade. The isolates with sequences that clustered to the south Asian clade included 43 of 44 New York cases, all 17 New Jersey cases, two of three Massachusetts cases, and individual cases from California, Connecticut, Maryland, and Oklahoma. Five (7%) patients had isolates with sequences that clustered to the South American clade, which included all three cases from Illinois and individual cases from Florida and Massachussetts. The isolate from a patient in Indiana clustered to the African clade, and one New York isolate clustered to the east Asian clade.
Figure 1. Distribution of Candida auris clades within the USA.
(A) Maximum parsimony phylogenetic tree of isolates from Colombia, India, Japan, Pakistan, South Korea, South Africa, Venezuela, and clinical cases in the USA (up to the first five clinical cases from each US state). (B) The frequency of clinical cases in the USA by clade. (C) The geographical distribution of clades; solid lines indicate introductions that are associated with patients known to have received health care abroad. CA=California. CT=Connecticut. FL=Florida. IL=Illinois. IN=Indiana. MA=Massachusetts. MD=Maryland. NY=New York. NJ=New Jersey. OK=Oklahoma. SNPs=single-nucleotide polymorphisms.
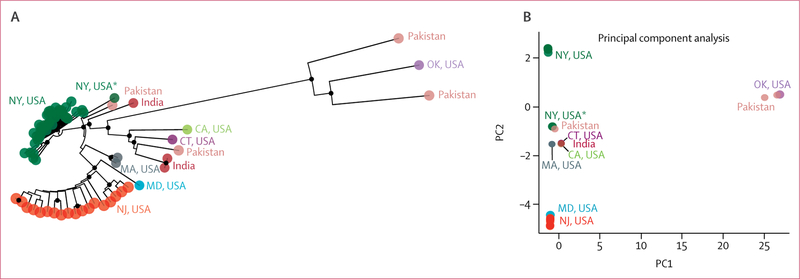
Because most isolates in the USA had sequences that clustered to the south Asian clade, we further examined this clade and identified genetically distinct strains in different states (figure 2). Isolates from 42 (98%) of the 43 clinical cases in New York that clustered to the south Asian clade—all 42 of which were collected from April 11, 2016, to May 10, 2017—clustered together by PCA, with SNP differences ranging from one to 26 SNPs (appendix). The remaining New York isolate was from a 2013 case that involved a patient with candidaemia who had a health-care exposure in the United Arab Emirates. This isolate was 32–45 SNPs different from the other New York cases and was distinct by PCA.
Figure 2. Identification of genetically distinct Candida auris strains in different US states.
(A) Neighbour-joining phylogenetic tree of the South Asian clade, to examine isolates from clinical cases from seven US states, India, and Pakistan. (B) The principal component analysis of genetic differences between these isolates. Circles at nodes indicate separations with a bootstrap value of at least 90%. CA=California. CT=Connecticut. FL=Florida. IL=Illinois. IN=Indiana. MA=Massachusetts. MD=Maryland. NY=New York. NJ=New Jersey. OK=Oklahoma. PC1=principal component 1. PC2=principal component 2. *The 2013 case in NY.
Although the geographic distance between New York and New Jersey facilities that reported cases of C auris was as close as 40 km, isolates from patients in New Jersey were genetically distinct from isolates from those in New York (figure 2; appendix). Comparison of isolates between the two states yielded a median SNP difference of 62 (range 41–88). No known transfers of patients were reported for those involved in these cases between New Jersey and New York facilities. The whole-genome sequence of the isolate from the patient in Maryland clustered with isolates from New Jersey cases. An epidemiological investigation revealed that the patient in Maryland, before diagnosis with a C auris infection, had an overlapping hospital stay with a patient with a C auris infection in a health-care facility in New Jersey, and isolates from both patients were ten SNPs different.
Within the south Asian clade, isolates from patients in California, Connecticut, Massachussetts, and Oklahoma were genetically distinct from the New York and New Jersey clusters by PCA (figure 2). Two of three patients in Massachussetts had isolates with sequences that clustered to the South Asian clade. The isolate from the third case, which was identified in a recipient of a lung transplant, belonged to the South American clade and was highly related to the isolates from patients in Illnois (with two SNPs or fewer). Epidemiological investigation revealed that the patient in Massachussetts received the organ from an Illnois donor, providing evidence of a donor-derived infection.
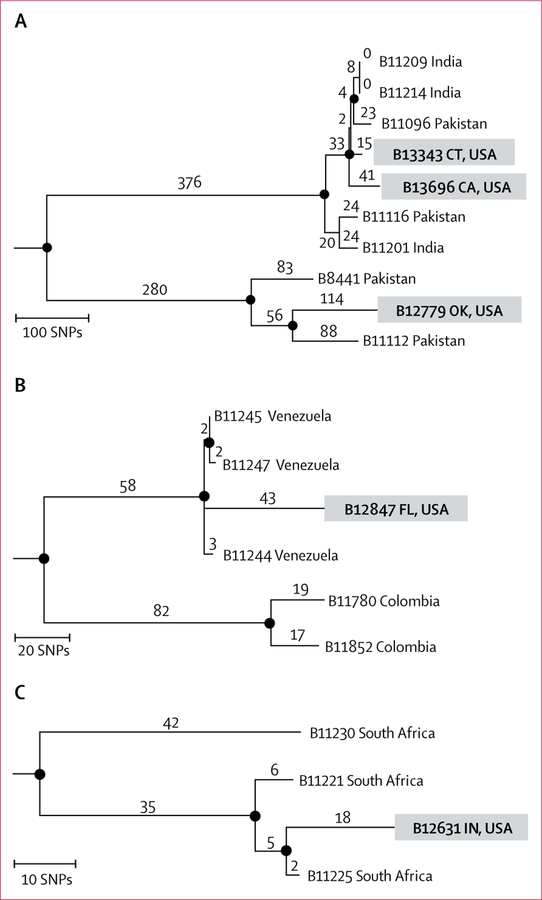
Of the 73 clinical cases, five (7%) patients had received health care outside the USA before C auris diagnosis. Genomic data corroborated results from epidemiological investigations of the cases in California, Connecticut, Florida, Indiana, and Oklahoma and suggested that C auris was acquired abroad in these cases. Notably, transmission was not identified from contact investigations of these cases.
The patient in California had received inpatient medical care in India 21 months before C auris diagnosis in the USA. The patient in Connecticut had been directly transferred from a hospital in the same city in India as the patient in California; C auris was identified within 1 month of arrival in the USA. Additionally, the patient in Oklahoma had been admitted to a hospital in Pakistan 3 months before diagnosis. The genomic sequences of the California, Connecticut, and Oklahoma isolates clustered with those of isolates from India and Pakistan, within the south Asian clade (figure 3). The sequence of the Oklahoma isolate clustered within a subclade that contained only Pakistani isolates. The patient in Florida had been admitted to a hospital in Venezuela immediately before their transfer to a hospital in Florida in 2015. A blood culture isolate in this patient was identified in Venezuela as Candida haemulonii, which is a common misidentification of C auris; however, the isolate was not available for confirmatory identification. C auris was subsequently isolated from the patient’s urine in 2017, and the genetic sequence of the isolate clustered with those of isolates from Venezuela. The patient in Indiana had been admitted to a hospital in South Africa immediately before transfer to a hospital in Indiana, and the genetic sequence of this patient’s isolate clustered with that of South African isolates.
Figure 3. Identification of five US travel-related Candida auris cases.
Phylogenetic trees of the South Asian (A), South American (B), and African (C) clades, to examine isolates from clinical cases in patients known to have received medical care abroad before diagnosis of C auris infection in the USA. Numbers above branches indicate SNPs. Circles at nodes indicate separations with a bootstrap value of at least 90%. CA=California. CT=Connecticut. FL=Florida. IN=Indiana. OK=Oklahoma. SNPs=single-nucleotide polymorphisms.
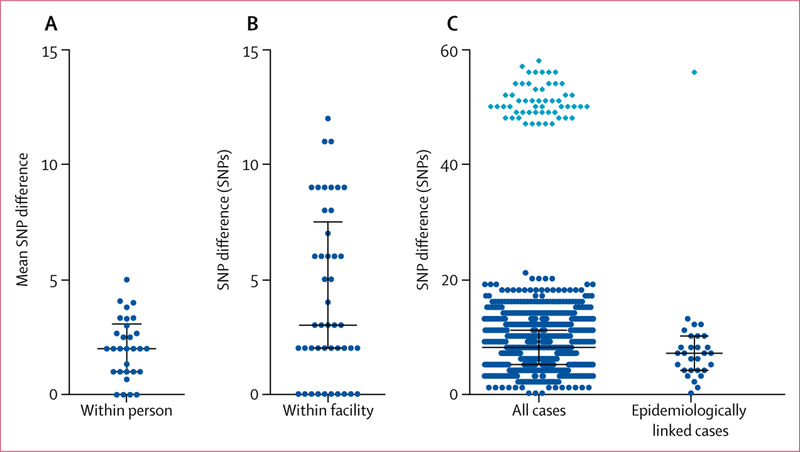
Of the 133 clinical and screening cases, 23 (17%) cases involved patients that had several samples collected on the same day. The mean C auris SNP difference within a patient ranged from zero to five SNPs (median two SNPs; figure 4). The largest difference, 12 SNPs, was between two isolates that had been taken from skin swabs of different body sites from a patient in New York.
Figure 4. Genetic diversity of Candida auris isolates taken from patients in the USA during transmission.
(A) The mean SNP difference among several isolates taken from the same person, in 23 patients in the USA. (B) The number of SNP differences for each pairwise comparison among ten patients at a facility in New Jersey with an outbreak of C auris. (C) The number of SNP differences for each pairwise comparison among all isolates from 42 patients in New York, excluding the 2013 case, and among the epidemiologically linked pairwise comparisons in New York. The light blue diamond indicates pairwise comparisons with a single screening case in New York in 2016. SNP=single-nucleotide polymorphism.
We examined isolates from ten screening cases in a facility in New Jersey with an ongoing outbreak of C auris (figure 4). As part of outbreak control measures, screening for C auris colonisation was done at admission and every 2 weeks during each hospital stay. The patients included in this analysis did not have evidence of C auris on admission but became colonised with C auris while at the facility. Analysis of the first isolate of each of the ten patients yielded a median SNP difference of three SNPs (range 0–12).
Because New York had the highest number of clinical cases in the USA, and because all but one of the isolates were related to the south Asian clade, we examined SNP differences among New York clinical and screening cases of the south Asian clade and between those that were epidemiologically linked (ie, patients who shared a room or a ward in a health-care facility at the same time). The 2013 case in New York was considered to be a separate introduction of C auris (figure 2) and thus was excluded. Isolates from the remaining 56 patients (42 clinical cases, 14 screening cases) yielded a median SNP difference of eight SNPs (range 0–58; figure 4). The isolate from one 2016 screening case was 47–58 SNPs different from the other New York isolates. Among the 29 pairs of epidemiologically linked cases, the median SNP difference was seven SNPs (0–56).
Discussion
We found that all C auris isolates in the patients assessed in the USA belonged to one of the four known clades (African, South American, east Asian, and south Asian), suggesting several introductions of C auris into the USA. This finding was further supported by the identification of five travel-related cases, in which genomic and epidemiological data corroborated patients acquiring C auris through health-care exposures outside the USA.
Genetic similarities among isolates from several cases within New York and New Jersey (figure 2; appendix) indicated ongoing transmission within each state. However, no transmission between these states was identified, which was unexpected, given the geographical proximity between the affected health-care facilities in the two states. These findings indicate that transmission is restricted to local networks of health-care facilities. The absence of transmission between the states was supported by an epidemiological investigation, which identified no patients being transferred between facilities in New York and New Jersey.
By contrast with New Jersey, where a single clade of genetically related isolates was identified, multiple introductions of C auris have occurred in New York. Although most New York cases had isolates related to the south Asian clade, the genetic sequence of one isolate, from a case in 2016, clustered with the east Asian clade (figure 2), and two isolates from the 2013 case (figure 2) and a 2016 screening case (figure 4) were genetically distinct, suggesting separate introductions. The remaining cases represented recent, albeit less immediate, transmission. Notably, genetic diversity among New York epide miologically linked cases (median seven SNPs) was similar to that for all New York cases (eight SNPs; figure 4), which was consistent with recent introduction and ongoing transmission.
Although phylogenetic analysis suggests multiple introductions of C auris into the USA, epidemiological associations with health care abroad were only identified for a few cases, for which no subsequent transmission was detected. Notably, patients involved in these cases were diagnosed with a bacterial multidrug-resistant organism before C auris diagnosis and were put on contact precautions. These precautions might have served a role in preventing subsequent transmission for these cases. Conversely, for the three states with the highest burden— New York, New Jersey, and Illinois— no index patients with links to health-care exposures abroad were uncovered. However, the absence of C auris in the USA before 2013, the genetic relatedness of isolates in Illinois, New Jersey, and New York to those from other global regions, and the high relatedness among isolates within each state are consistent with recent, unrecognised introductions from abroad followed by local transmission.
These findings of multiple introductions are not unique to the USA: C auris strains from 14 hospitals in the UK also reportedly grouped to several clades.17,18 Given widespread global travel and the persistence of C auris in patients who have been colonised by this fungus and in health-care environments, we anticipate that other countries will have similar experiences. This series of events will probably lead to C auris losing a distinct phylogeographical structure as several clades get introduced into the same world regions.
One limitation of our analysis is that the numbers of SNPs that we describe are not directly comparable with other studies unless the same bioinformatics methods are used. The number of SNPs depend on many parameters, including which reference isolate is analysed, how SNPs are filtered, and what software are used. Thus, strict SNP cutoffs for determining transmission are difficult to establish. However, our results showed that SNP differences observed within a patient can be used as a point of reference to assess transmission. The observed median mean SNP difference within a patient was two SNPs (range 0–5) and the maximum SNP difference was 12 SNPs (figure 4), which was similar to the genetic diversity within a facility with an ongoing outbreak (median difference three SNPs, range 0–12). Based on these findings, SNP differences of fewer than 12 SNPs between patients are likely to represent recent transmission. Accordingly, two SNPs or fewer separated C auris isolates from the patient who had received a lung transplant in Massachussetts and the organ donor in Illinois, with documented evidence of transmission.19
Controlling transmission will require health-care facilities to remain alert in detecting C auris when it is first identified in a region. As concluded by other countries, prompt identification of index patients and timely implementation of infection control measures are essential for prevention of subsequent, widespread transmission.20 Recommended measures, including implementation of contact precautions, screening contacts, and thorough environmental cleaning and disinfection have been suggested.21 To address the possibility of C auris introduction through patients who receive health care abroad, the CDC has recommended that US facilities consider additional measures for patients who have had an overnight stay in a health-care facility outside the USA in the previous year in a country with documented C auris transmission.22 These measures include obtaining candida isolates from both sterile and non-sterile body sites during the normal course of clinical care and identifying the fungus, at a species level. Detailed recommendations and a list of countries reporting C auris cases can be found on the CDC website.
In summary, molecular epidemiology has provided key insights into the origination and transmission dynamics of C auris in US health-care facilities. These findings have already shaped current investigations and recommendations, including the revelation of travel-related cases. Ultimately, diligence in preparing for and identifying this new multidrug-resistant yeast will help contain its spread and transmission within the USA.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for studies published on or before July 19, 2018, with the search terms “Candida auris AND sequencing OR molecular epidemiology OR genomic epidemiology” without any language restrictions. This search identified 34 articles; 28 of these articles were primary research and, of these, five reported use of whole-genome sequencing (WGS) for typing and phylogenetic analysis. Sharma and colleagues used WGS to show that C auris isolates from four Indian hospitals were highly related and their findings were suggestive of clonal transmission. Similar results were reported for Pakistan, South Africa, Venezuela, and Colombia, where C auris isolates within each country were highly related. However, large genetic differences were observed between isolates in different global regions, indicating that C auris has substantial phylogeographical structure. A 2018 study reported that all C auris isolates from a large outbreak in London were genetically related to those from India and Pakistan and concluded the outbreak had an Asian origin. Finally, a report from a large teaching hospital in the USA described an environmental investigation after a C auris case was identified; isolates from the patient involved and from the health-care environment were also related to isolates from South Asia.
Added value of this study
To our knowledge, this report is the first national molecular epidemiological description of C auris infections in the USA. We used WGS on isolates collected from 133 patients with C auris from ten US states, and we integrated data from epidemiological investigations (ie, patients who had shared a room or ward with a patient with C auris) and their travel history. We showed that all C auris isolates taken from patients in the USA belonged to one of the four known clades (African, South American, East Asian, and South Asian), suggesting that C auris was introduced several times into the USA. We also identified five travel-related cases in which patients probably acquired C auris through health-care exposures outside the USA. To our knowledge, this is the first report to use both genomic and epidemiological data to examine a collection of isolates from multiple health-care facilities from several regions that also describes how C auris introductions into a country can occur. Further, by comparing the genetic diversity of C auris within a patient, within a health-care facility undergoing an outbreak, and within and between states, we show the transmission dynamics within the USA.
Implications of all the available evidence
In the setting of increasing international concern regarding the spread of antibiotic-resistant bacteria in health-care settings, C auris has emerged as a multidrug-resistant fungus that behaves much in the same way. We provide a description of how C auris has been introduced and is spreading within a country. As a result of this work, the US Centers for Disease Control and Prevention has issued additional recommendations for infection control in patients with recent health-care exposures abroad. Importantly, these recommendations can help other national public health programmes to respond to C auris.
Acknowledgments
We acknowledge the US Centers for Disease Control and Prevention Advanced Molecular Detection Initiative and Biotechnology Core Facility Branch for their continued support. The use of product names in this manuscript does not imply their endorsement by the US Department of Health and Human Services. The findings and conclusions in this Article are those of the authors and do not necessarily represent the views of the US Centers for Disease Control and Prevention.
Funding US Centers for Disease Control and Prevention
Footnotes
Declaration of interests
We declare no competing interests.
Contributor Information
Nancy A Chow, Mycotic Diseases Branch, Atlanta, GA, USA.
Lalitha Gade, Mycotic Diseases Branch, Atlanta, GA, USA.
Sharon V Tsay, Mycotic Diseases Branch, Epidemic Intelligence Service, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Kaitlin Forsberg, IHRC, Atlanta, GA, USA.
Jane A Greenko, New York State Department of Health, Albany, NY, USA.
Karen L Southwick, New York State Department of Health, Albany, NY, USA.
Patricia M Barrett, New Jersey Department of Health, Trenton, NJ, USA.
Janna L Kerins, Chicago Department of Public Health, Chicago, IL, USA.
Shawn R Lockhart, Mycotic Diseases Branch, Atlanta, GA, USA.
Tom M Chiller, Mycotic Diseases Branch, Atlanta, GA, USA.
Anastasia P Litvintseva, Mycotic Diseases Branch, Atlanta, GA, USA.
References
- 1.Morales-Lopez SE, Parra-Giraldo CM, Ceballos-Garzon A, et al. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 2017; 23: 162–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams E, Quinn M, Tsay S, et al. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis 2018; 24: 1816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lockhart SR, Etienne KA, Vallabhaneni S, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 2017; 64: 134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhary A, Sharma C, Meis JF. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 2017; 13: e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 2016; 5: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh RM, Bentz ML, Shams A, et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic healthcare surface. J Clin Microbiol 2017; 55: 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh K, Tamura T, Makimura K, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 2009; 53: 41–44. [DOI] [PubMed] [Google Scholar]
- 8.Saris K, Meis JF, Voss A. Candida auris. Curr Opin Infect Dis 2018; 31: 334–40. [DOI] [PubMed] [Google Scholar]
- 9.Vallabhaneni S, Kallen A, Tsay S, et al. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013–August 2016. MMWR Morb Mortal Wkly Rep 2016; 65: 1234–37. [DOI] [PubMed] [Google Scholar]
- 10.Tsay S, Welsh RM, Adams EH, et al. Notes from the field: ongoing transmission of Candida auris in health care facilities-United States, June 2016-May 2017. MMWR Morb Mortal Wkly Rep 2017; 66: 514–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. Global emergence of invasive infections caused by the multidrug-resistant yeast Candida auris June 24, 2016. https://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html (accessed Sept 28, 2018).
- 12.Centers for Disease Control and Prevention. Candida auris case definition https://wwwn.cdc.gov/nndss/conditions/candida-auris/case-definition/2018/ (accessed Sept 28, 2018).
- 13.Sahl JW, Lemmer D, Travis J, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom 2016; 2: e000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7·0 for bigger datasets. Mol Biol Evol 2016; 33: 1870–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popescu AA, Huber KT, Paradis E. ape 3·0: new tools for distance-based phylogenetics and evolutionary analysis in R. Bioinformatics 2012; 28: 1536–37. [DOI] [PubMed] [Google Scholar]
- 16.Jombart T, Ahmed I. adegenet 1·3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 2011; 27: 3070–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borman AM, Szekely A, Johnson EM. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 2017; 55: 563–67. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes J, Abdolrasouli A, Farrer RA, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 2018; 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azar MM, Turbett SE, Fishman JA, Pierce VM. Donor-derived transmission of Candida auris during lung transplantation. Clin Infect Dis 2017; 65: 1040–42. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhary A, Voss A, Meis JF. Multidrug-resistant Candida auris: “new kid on the block” in hospital-associated infections? J Hosp Infect 2016; 94: 209–12. [DOI] [PubMed] [Google Scholar]
- 21.Tsay S, Kallen A, Jackson BR, Chiller TM, Vallabhaneni S. Approach to the investigation and management of patients with Candida auris, an emerging multidrug-resistant yeast. Clin Infect Dis 2018; 66: 306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. Candida auris clinical update September, 2017. https://www.cdc.gov/fungal/candidaauris/c-auris-alert-09-17.html (accessed Sept 28, 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.