Abstract
The performance of current microfluidic methods for exosome detection is constrained by boundary conditions and by fundamental limits to microscale mass transfer and to interfacial exosome binding. Here, we show that a microfluidic chip designed with self-assembled 3D herringbone nanopatterns can detect low levels of tumour-associated exosomes in plasma (10 exosomes μL−1, or approximately 200 vesicles per 20-μL spiked sample) that would otherwise be undetectable by standard microfluidic systems for biosensing. The nanopatterns promote microscale mass transfer, increase surface area and probe density to enhance the efficiency and speed of exosome binding, and permit drainage of the boundary fluid to reduce near-surface hydrodynamic resistance, thus promoting particle–surface interactions for exosome binding. We used the device for the detection, in 2-μL plasma samples from 20 ovarian cancer patients and from 10 age-matched controls, of exosome subpopulations expressing CD24, EpCAM, and FRalpha proteins, and suggest exosomal FRalpha as a potential biomarker for the early detection and progression monitoring of ovarian cancer. The nanolithography-free nanopatterned device should facilitate the use of liquid biopsies for cancer diagnosis.
Rapid progress in early-stage diagnostics and precision therapy calls for new tools for the ultrasensitive detection of disease-specific biological particles in bodily fluids, such as circulating tumor cells (CTCs) and extracellular vesicles (EVs)1–4. Exosomes, a subtype of EVs secreted by mammalian cells with a typical size range of 30–150 nm, have been implicated in many biological processes, including immune response, tumorigenesis and metastasis5, 6. Thus, circulating exosomes are emerging as a new paradigm of “liquid biopsy” for non-invasive cancer diagnosis4, 7. However, tumor-associated exosomes in biofluids can be very rare during early stages of the disease development. Thus, it is imperative to develop new biosensing capabilities for ultrasensitive and specific analysis of disease-associated exosome subtypes in the background of normal cell-derived vesicles. Current “gold standard” methods for exosome capture and characterization, such as ultracentrifugation (UC) and Western blot (WB), suffer from poor isolation efficiency, low sensitivity, time-consuming procedures and large sample consumption. Microfluidics has been recently exploited to leverage exosome analysis; but the progress in improving sensitivity, speed or multiplicity has been rather limited3, 8.
Biosensing largely relies on targets interacting with surface-immobilized probes for affinity capture. In such interfacial processes, mass transfer of targets to the surface and the equilibrium and kinetics of binding reactions are the fundamental factors that govern sensing performance9. Microfluidics and nano-engineering approaches have been extensively explored to address these limitations. Many micromixing methods, including herringbone mixer, have been developed to promote microscale mass transfer10, 11. Two-dimensional (2D) nano-engineering12 offers an effective means to enhance interfacial binding of biomolecules13, 14, exosomes8, and cells15. In addition to the limits of mass transfer and binding reaction, it was recently reported that flow stagnation and hydrodynamic resistance due to the non-slip condition at the liquid-solid interface restrict the convection of slow-diffusing particles to the surface, which significantly reduces binding efficiency16–18. To address these boundary effects, a microfluidic chip integrated with 3D nanoporous microposts was developed by micropatterning carbon-nanotube (CNT) posts inside microchannels. Compared to solid microposts, the nanoporous CNT microposts permit the drainage of liquid through the nanopores, greatly reducing the near-surface flow stagnation and hydrodynamic resistance to enhance surface binding of particles of 40 nm to ~15 μm16–18. These studies present a creative strategy for microfluidic integration of 3D nanostructures to overcome the boundary effects; however, fabrication of 3D CNT patterns is highly sophisticated and time consuming.
As discussed above, previous approaches sought to individually or partially overcome the fundamental limits in mass transfer, surface reaction, and boundary effects, which presents a major conceptual constraint in leveraging the biosensing performance. Here we report an integrated, comprehensive strategy that addresses these three limits simultaneously in one device. Our approach, termed multiscale integration by designed self-assembly (MINDS), exploits microfluidically engineered colloidal self-assembly (CSA)19 to achieve simple, large-scale integration of 3D nanostructured functional microelements. We used the MINDS to combine micro-patterning and 3D nanostructuring of a widely used functional microelement, herringbone mixer for flow manipulation and molecular recognition. We showed that this 3D nanostructured herringbone (nano-HB) addresses the aforementioned limits in one device (Fig. 1a) as it: 1) effectively promotes microscale mass transfer of bioparticles20, 21; 2) increases surface area and probe density to enhance binding efficiency and speed; and 3) permits drainage of the boundary layer of fluid through the pores of a nano-HB, which reduces near-surface hydrodynamic resistance16, 17 and enriches particles near the surface to enhance surface binding of particles. As a result, the nano-HB chip afforded an extremely low limit of detection of 10 exosomes μL−1 (200 exosomes per assay) for spiked-in standards and enabled quantitative detection of low-level exosome subpopulations in blood plasma that are otherwise indiscernible to the conventional flat-channel microfluidic chips. We demonstrated quantitative detection of circulating exosomal CD24, epithelial cell adhesion molecule (EpCAM), and folate receptor alpha (FRα) markers to detect ovarian cancer using only 2 μL of plasma, indicating the potential applications of our technology to liquid biopsy-based cancer diagnosis.
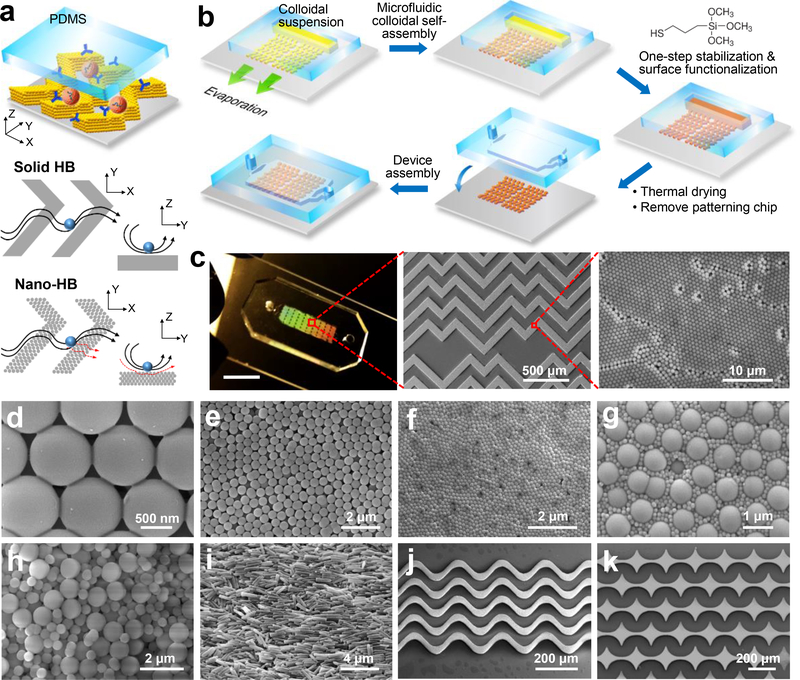
Fig. 1. Multiscale Integration by Designed Self-assembly (MINDS).
(a) Conceptual illustration of the MINDS strategy that improves biosensing by 3D nanostructuring of microfluidic elements, e.g., herringbone (HB) mixer. Conventional solid HB mixer creates microvortices to promote mass transfer of targets. A particle will experience hydrodynamic resistance near a solid surface that reduces direct surface contact. In a 3D nanoporous HB (nano-HB) chip, fluid near the surface can be drained through the porous structure (red dashed lines) to increase the probability of particle-surface collisions. (b) The workflow for fabricating a 3D nano-HB chip by MINDS. (c) A nano-HB chip fabricated with 960-nm silica colloids. The digital photo exhibits Bragg diffraction of light (scale bar, 5 mm). The SEM images show a high-quality, crack-free HB array patterned on a glass substrate with a crystalline nanoporous structure. (d) Silica necks were formed between the contacting particles by treatment with 5% 3-MPS. (e, f) SEM images of mono-assembled nano-HBs with silica colloids of 520 and 170 nm. (g) A well-ordered binary lattice composed of 170 and 960 nm silica colloids at equal mass ratio. (h) A randomly organized nano-HB co-assembled with 520 and 960 nm silica colloids at a 1:1 mass ratio. (i) An anisotropic microstructure assembled from silica nanorods with an averaged diameter of 238 ± 32 nm and length of 1.34 ± 0.26 μm. (j, k) Fabrication of crack-free, 3D nanostructured sinusoidal ribbon and concave diamond arrays using designed microfluidic channels to engineer evaporative self-assembly of 960-nm colloids.
Results
Designable 3D Nanostructuring of Functional Microelements by MINDS.
Fabrication of a MINDS-chip is illustrated in Fig. 1b and detailed in the Methods section. A nano-HB mixer was constructed by using a microchannel network to guide evaporation-driven CSA. A simple in situ treatment with (3-mercaptopropyl) trimethoxysilane (3-MPS) was developed to strengthen the assembled micropatterns and prime the surface for antibody conjugation in one step. After thermal drying and removing the patterning chip, the nano-herringbone pattern was aligned and sealed with an assay chip with ~50-μm high channels to construct a complete device. Fig. 1c displays a nano-HB chip fabricated by MINDS using 960-nm silica colloids, which exhibited Bragg scattering of light by the embedded colloidal nanostructures. Scanning electron microscopy (SEM) imaging confirmed high-fidelity replication of the channel geometry by the engineered CSA and the 3D nanoporous crystalline structures (Fig. 1c). The optimized 3-MPS treatment prevents the formation of cracks during thermal drying and minimizes the mechanical damages caused by removing the patterning chip, while preserving the nanoporosity. This is attributed to the formation of silica necks gluing the contacting nanoparticles (NPs) by hydrolysis and condensation reactions of 5% 3-MPS (Fig. 1d). High concentrations of 3-MPS (e.g., 10%) can be used to completely fill the pores in nano-HBs (Supplementary Fig. S1), which allows us to fabricate solid-HB chips used in the following studies. Meanwhile, 3-MPS functionalizes the surfaces with sulfhydryl groups for antibody conjugation via maleimide reaction. Thus, this one-step treatment method greatly simplifies device fabrication.
The MINDS strategy allows us to conveniently engineer the morphology of 3D nano-HBs. We have demonstrated the nano-HBs fabricated with a variety of nanomaterials, including mono-assemblies of silica colloids of 960, 520 and 170 nm (Fig. 1d-f), binary nanoparticle combinations (Fig. 1g, h), and silica nanorods (Fig. 3i and Supplementary Fig. S2). Co-assembly of multi-component mixtures was tuned to create complex morphologies via controlling assembly variables, including particle size ratio and volume fraction22, 23. Co-assembly of 170 and 960 nm NPs of an equal volume fraction results in a unique morphology of hexagonally packed large particles surrounded by small particles (Fig. 1g). In contrast, a 960/520 nm nano-HB was found to possess much higher structural disorder and porosity (Fig. 1h). In addition to HB patterns, we have demonstrated high-fidelity patterning of complex microelements, such as a sinusoidal channel array previously reported for immunocapture of CTCs24 (Fig. 1j) and an array of concave diamonds (Fig. 1k). These results suggest that utilizing the rich collection of nanomaterials and the flexibility of microfluidic design, the MINDS strategy affords broad applicability for microscale integration of designable 3D nanostructures to develop new multiscale integrated biosensing systems.
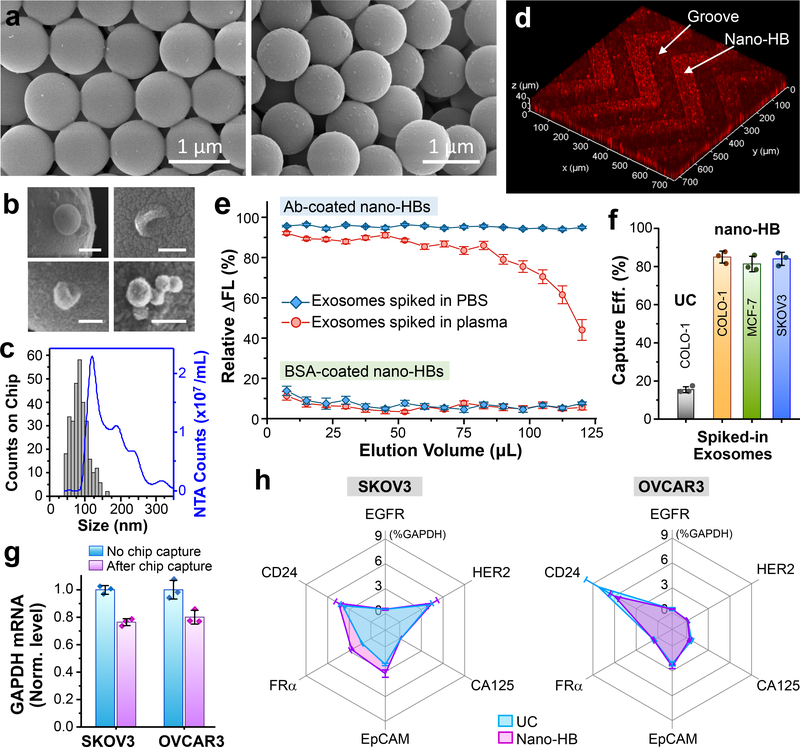
Fig. 3. 3D engineered Nano-HB chip affords efficient immunocapture of exosomes.
(a) SEM images showing minimal non-specific absorption and immunocapture of COLO-1 exosomes (105 μL−1) on a non-modified (left) or a mAb-coated device (right), respectively. (b) Typical sphere- or cup-shaped morphologies and the clusters of COLO-1 exosomes captured on nano-HBs. Scale bars: 100 nm. (c) Representative size distribution of nano-HB-captured COLO-1 exosomes (n > 300) measured by SEM, compared to that of NTA analysis of UC-isolated vesicles. (d) 3D confocal fluorescence microscopy showing exosomes captured inside the nano-HBs. DiO-stained COLO-1 cell-derived exosomes were spiked in human plasma (105 μL−1). (e) Immunological and non-specific capture of COLO-1 exosomes as a function of injected sample volume assessed using an mAb or BSA-coated nano-HB chip, respectively. DiO dye-stained exosomes were spiked in PBS or 10× diluted human plasma (106 μL−1) and injected continuously through the chips at 0.5 μL/min. 2 μL eluent was collected at the outlet every 15 min and measured by a micro-volume plate reader to determine the reduction in fluorescence signal with respect to that of the original sample. (f) Comparison of standard UC isolation and the nano-HB capture of fluorescently stained exosomes of various cancer cell lines spiked in healthy plasma (106 μL−1). Signal decrease caused by exosome immunocapture was measured as in (e) and subtracted by that of non-specific adsorption to calculate the percentage of the corrected signal reduction, i.e., the specific capture efficiency. (g) Capture efficiency of nano-HB chip measured by the GAPDH mRNA level. EVs isolated and concentrated from SKOV3 and OVCAR3 cell culture media by UC were spiked in PBS (106 μL−1) and 100 μL of the solution was run on the chip. Captured exosomes were eluted out for droplet digital PCR (ddPCR) quantification. The GAPDH mRNA levels in chip-captured exosomes were normalized by that in the same amount of EVs measured without chip capture. (h) Profiles of six mRNA markers measured in the chip-captured SKOV3 and OVCAR3 exosomes in comparison with the vesicles isolated and concentrated by UC. Exosome capture and elution on the nano-HB chip was performed as in (g) and the levels of individual mRNAs measured by ddPCR were normalized against GAPDH. Anti-CD81 mAb was used for exosome capture in all cases. Error bars, one S.D. (n = 3).
Fluidic and Surface Characterization of Nano-HB Chip for Bioparticle Sensing.
We conducted computational fluid dynamics (CFD) simulation to study the flow behavior in the nano-HB chip assembled from 960-nm colloids (see SI for the simulation details). Microfluidic HB structure has been widely used to promote microscale mass transfer by disrupting the laminar flow in microchannels10, 11, which is especially important for efficient capture of cells with low diffusivity20, 21. We adopted a staggered herringbone geometry of a 225 μm groove pitch and a 125 μm groove width to maximize particle-surface interactions, in accordance to the previous studies25, 26. A mathematical model of the porous domain was defined in order to resolve the nanoscale structure, while capturing the microscale flow pattern, with reasonable computational effort for this proof-of-concept study. Compared to the flat-channel chip, solid and nano-HB chips enhanced mixing of two co-flowing aqueous streams with almost the same performance. (Fig. 2a). Consistently, close examination of the flow behavior (Fig. 2b, c) visualizes the formation of microvortices by the HBs and the similar patterns of the transverse and streamwise flows in the two devices. In the nano-HB chip, the pressure-driven fluid can enter the nanoporous domain and exhibits the maximum flow rate in the corners and the thin surface layer, which was calculated to be less than 0.5% of that of the bulk flow (Fig. 2b and c insets). This explains that the 3D nanoporous structure does not significantly change the flow mixing behavior induced by the HBs (Fig. 2a). It is important to note that such small fluid drainage through the nanoporous domain can disrupt the stagnant boundary layer formed on a solid-HB due to the non-slip condition (Fig. 2b and c insets and Supplementary Fig. S3). This effect has been demonstrated to increase particle-surface collisions and thus greatly augment affinity capture of nano/micro-sized particles17, 18.
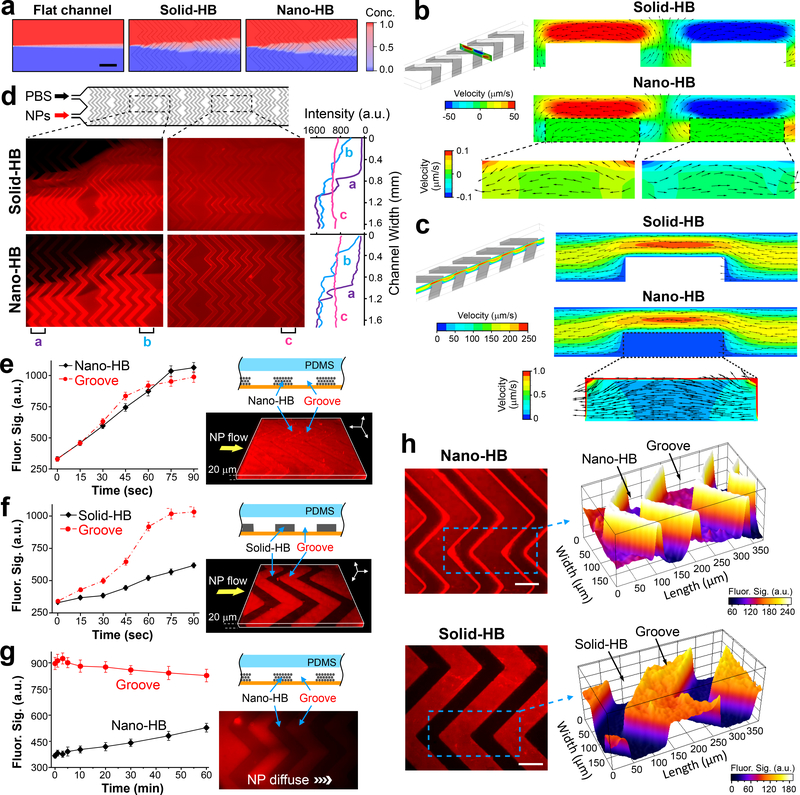
Fig. 2. Fluidic characterization of nano-HB chips.
(a) Simulation of mixing two streams of 50-nm nanoparticles (NPs) and water co-flowing at 1 μL/min in a flat-channel, solid-HB, or nano-HB device, respectively. Zoom-in view over the first two HB units was shown. Scale bar, 400 μm. (b, c) Simulation results showing the transverse flow profiles across the channel width (b) and the streamwise flow profiles along the channel length (c), respectively. Insets highlight the flow patterns inside the nanoporous domains. Color contours indicate the velocity magnitude, while vectors represent the flow direction. The flow rate was 0.5 μL/min. (d) Fluorescence images (left) and transverse intensity plots (right) of mixing two flows of 46-nm fluorescent NPs and PBS injected at 0.25 μL/min in parallel. The intensity plots were measured along the channel width at three positions on the images as indicated. (e, f) Time-lapse plots of fluorescence intensity measured in the HB structures and the open grooves when a solution of 46 nm fluorescent NPs was pumped through a nano- or solid-HB chip at 0.5 μL/min, respectively. 3D confocal fluorescence microscopy images (left) were acquired when the channels were filled with the NP solution. (g) Time-lapse monitoring of diffusion of 46-nm NPs from the grooves into nano-HB structures. Flow pumping was stopped when the NP solution entered the observation region. The fluorescence image (left) was acquired at t = 60 min. (h) Representative 2D confocal microscopic images of the nano-HB (top) and solid-HB structures (bottom), respectively, with the NP solution flowing through the channels. The images were acquired at approximately 15 μm below the HB surface. Partial surface plots of the images (right) were displayed for the areas indicated by the dashed rectangles. Scale bar, 100 μm. In (e-h), nano-HB chips were assembled with 960 nm silica colloids. a.u., arbitrary unit. Error bars, one standard deviation (S.D., n = 3).
The effects of permeability was studied by simulation because it is a critical property of a porous medium that measures the ability to transmit fluids18, 27, 28. With a 100-fold increase in permeability from 4.7 ×10−15 to 4.7 ×10−13 m2, no significant change occurs to the bulk flow in a nano-HB channel, while significantly more flow travels through the nanoporous domains (Supplementary Fig. S4a). Trajectories of 50 nm NPs in the flow were tracked across a unit of five HB elements. At the low permeability, ~7.6% of the particles can enter the nano-HBs, primarily on the corners; at a 100-fold permeability, this percentage increases to ~16% with more particles traveling through the nanostructures (Supplementary Fig. S4b). This increase in the particle-nanostructure interactions again supports the important role of fluid drainage by the nanoporous structure in promoting the boundary mass transport.
We experimentally assessed the enhancement of particle-surface interactions on 3D nano-HBs. The chips used here contain 25 μm tall nano-HBs assembled from 960-nm particles, which possess the pore sizes of >144 nm (15% of the particle size) estimated from the perfect face-cubic centered (FCC) structure. Fig. 2d shows that the solid- and nano-HB chips exhibited similar performance of creating the transverse flow across the channels to mix two streams of 46 nm fluorescent NPs and PBS. This observation agrees well with the simulation result that the porous structure does not significantly affect the ability of 960 nm HBs to create microvortex mixing (Fig. 2a). It is noted that the numerical simulation underestimates the flow penetration into the nano-HBs because the actual self-assembled colloidal crystals are more porous than the perfect FCC structure (Fig. 1c-f). To assess the flow penetration in the nano-HB, we injected the fluorescent NPs through a chip at a constant flow rate of 0.5 μL/min and monitored the fluorescence intensity inside the nanostructure and the open groove regions near the center of the microchannel. As shown in Fig. 2e and Supplementary Fig. S5a, the signals in the grooves and the nanostructures increased gradually and approached the steady levels within ~60 and ~90 seconds, respectively. On the contrary, only slight increase in fluorescence intensity was observed in the solid-HBs, attributed to the solution flowing above the microstructures (Fig. 2f, Supplementary Fig. S5b). We also used confocal fluorescence microscopy to verify the flow penetration. Representative 3D images constructed from scanning the HB patterns (25-μm tall) for a depth of 20 μm from the substrate (Fig. 2e & f) confirm the penetration of NPs into the surface layers of nano-HBs. For comparison, we carried out a diffusion experiment on the nano-HB chip in which the fluorescent NP solution was stopped once it entered the observation window and the diffusion of NPs from the groove into the nanostructures was monitored for 60 min (Fig. 2g). The fluorescence signal in the grooves decreased slowly, indicating that the NPs were diffusing downstream. The fluorescence level in the adjacent nanostructure increased at a rate much lower than that driven by the constant flow (Fig. 2e). These findings verify hydrodynamic flow as the primary mechanism driving the transport of NPs into nano-HB.
Moreover, it was seen that as opposed to solid-HBs, nano-HBs concentrate NPs in the stream on their surface and in the region beneath (Fig. 2h and Supplementary Fig. S5), in line with the simulated flow pattern in nano-HB (Fig. 2b, c). This is attributed to that the penetrating flow increases the flux of particles into the nano-HBs which retain NPs like a sieve. Particle enrichment at the binding interface will improve capture efficiency as it enhances surface interactions and favorably shifts the reaction equilibrium. We also verified that the analyte particles and the particulate objects in human plasma will not be permanently trapped inside nano-HBs to block the flow penetration and cause non-specific signals (Supplementary Fig. S6). This could be attributed to the unique porous structure of nano-HB in which the “through pores” defined by the packed colloids are all interconnected to form a 3D “nanochannel” network without dead ends. Overall, our results reflect the advantages of our 3D nano-engineering approach—not only does it increase binding surface area, but also overcomes the boundary effects for bioparticle sensing. Slight flow penetration was also observed in the CNT microposts with ~80 nm pores, which greatly enhances capture of virus-sized particles, bacteria and cells16, 17.
In addition to mass transfer and boundary effects, surface properties, such as surface probe density, are important variables that govern the performance of biosensing9, 12. To this end, we assessed the functionalization of nano-HBs made of 960 nm silica colloids with anti-CD81 monoclonal antibody (mAb). Using 3D confocal fluorescence microscopy, we verified that the internal surface of 3D nano-HBs was conjugated with CD81 mAb (Supplementary Fig. S7a-c). Compared with a flat glass substrate, the nano-HB chip was found to increase the amount of immobilized antibodies by 3.25 folds (Supplementary Fig. S7d-e). Such enhancement in probe density and accessible surface areas will promote the thermodynamics and kinetics of surface binding, and thus improve the sensitivity and speed of target capture and detection. Collectively, our fluidic and surface characterization studies suggest that the nano-HB chip enabled by the MINDS strategy provides a designable multi-scale engineered system to improve biosensing by overcoming the fundamental limits in mass transfer, surface reaction, and boundary effects in one device. As a proof-of-concept, we evaluated the device as a diagnostic exosome analysis system for liquid biopsy-based detection of cancer.
Evaluation of Exosome Immunocapture on Nano-HB Chip.
We first assessed the nano-HB chip for exosome immunocapture using the well-characterized EV standards isolated from a colon cancer COLO-1 cell line8. As opposed to the BSA-coated chip, an anti-CD81 monoclonal antibody (mAb) modified chip was shown to capture much more exosomes (Fig. 3a) from a 20 μL spiked-in sample (105 μL−1 in PBS), indicating good immunocapture specificity. The captured exosomes display the typical spherical or cup shapes and form the clusters (Fig. 3b), consistent with the previous findings29, 30. We estimated the sizes of vesicles captured on nano-HB chips (n > 300) from ten SEM images, which displayed a notably smaller size distribution of 40 to 160 nm than that of UC-isolated EVs as measured by nanoparticle tracking analysis (NTA) (Fig. 3c). The size range of EVs observed with the nano-HB immunocapture agree with that of the standard magnetic bead immunocapture31. To investigate exosome capture in complex matrix, we performed nano-HB capture of DiO dye-stained COLO-1 EVs spiked in a 10-fold diluted healthy human plasma. With confocal fluorescence microscopy, COLO-1 exosomes were seen to be captured mostly within the top layers of several microns deep with a decreasing density gradient inward (Fig. 3d and Supplementary Fig. S8). Such spatial distribution of captured exosomes is in line with the simulated flow patterns in nano-HB (Fig. 2b, c), confirming flow penetration into the 3D nanopore structures. Given that unlabeled plasma exosomes were also captured, our observation indicates that the binding events on the walls do not block the nano-HBs, presumably due to its large number of pores and the enhanced transport of nanovesicles driven by the penetrating flow.
We evaluated the capture capacity and efficiency of nano-HB chips by monitoring the amount of exosomes depleted from the sample solution flowing through the nano-HB chip32. To this end, DiO dye-stained COLO-1 EVs were spiked in either PBS or 10-fold diluted patient plasma at 106 μL−1 and injected continuously into the BSA- or CD81 mAb-coated chips at 0.5 μL/min. 2 μL of eluent was sampled at the outlet every 15 min and measured with a micro-volume plate reader to determine the decrease of fluorescence intensity (ΔFL) caused by exosome depletion relative to the signal level of the sample before capture (Fig. 3e). A very small ΔFL was observed on the BSA-coated chips (5.5 ± 0.98% of original sample for PBS and 5.3 ± 1.1% for plasma media), indicating minimal matrix effects on the nano-HB chip. Exosome capture on the Ab-modified chips constantly resulted in a high ΔFL (95 ± 2%) for spiked PBS over 250 min injection, i.e., an accumulated sample volume of 125 μL. The plasma was estimated to contain >1010 EVs per mL by UC and NTA. These unlabeled vesicles and other particulate species in plasma compete for the flow access into nano-HBs and/or the surface binding sites. Despite the vast background, a consistent ΔFL of 86% to 92% was observed for capturing exosomes spiked in diluted plasma over up to 82 μL volume loaded, which remained at 75% at 100 μL. Thus, the current nano-HB chip affords sufficient capacity for sensitive and quantitative analysis of up to 10 μL undiluted plasma. If needed, the capture capacity can be readily expanded by scaling up the chip size or the number of channels.
Based in the measurements in Fig 3e, the specific capture efficiency of our nano-HB chip can be estimated by subtracting the non-specific ΔFL measured by the BSA-coated chip from that obtained for exosome immunocapture to calculate the percentage of the corrected ΔFL. Using this approach, we determined the specific capture efficiency of the nano-HB chip for fluorescently stained EVs of various cancer cell lines spiked in healthy plasma: COLO-1, 85.0 ± 3.2%; MCF-7, 81.3 ± 4.1%; and SKOV3, 84.1 ± 3.5% (Fig. 3f). For comparison, standard UC isolation only recovered 15.5 ± 1.7% of fluorescent COLO-1 EVs spiked in healthy plasma in line with the previously reported values33, 34.
The capture performance of nano-HB chip was further evaluated for downstream analysis of mRNA contents of exosomes using the commercial droplet digital PCR (ddPCR) technique. Here we were focused on two ovarian cancer (OvCa) cell lines, SKOV3 and OVCAR3, which are commonly used in vitro cell culture models representative of high-grade serous OvCa, the most aggressive subtype that accounts for nearly two-thirds of all OvCa-related deaths35. Designed for non-invasive cancer diagnosis using small sample volumes (<100 μL, Fig. 3e), our microfluidic device has limited preparative sample processing capacity for bulk mRNA analysis; thus we used EVs isolated and concentrated by UC for on-chip immunocapture to yield sufficient exosomes for off-chip RNA extraction, cDNA synthesis, and ddPCR, following the established protocols4, 36 (see the Methods for more discussion). To assess the capture efficiency, 100 μL UC-concentrated SKOV3 and OVCAR3 EVs (106 μL−1) were run on the chips and the captured exosomes were eluted out for ddPCR quantification of GAPDH mRNAs (Fig. 3g). The GAPDH mRNA levels in chip-captured exosomes were measured to be 76.5 ± 2.6% for SKOV3 and 80.1 ± 5.0% for OVCAR3 relative to the initial samples. The capture efficiency for SKOV3 exosomes is slightly lower than that measured by the fluorescence detection method in Fig. 3f, which may be attributed to the RNA loss and degradation during multi-step exosome elution and RNA extraction. To assess the nano-HB chip for exosomal mRNA profiling in comparison to UC isolation, we detected six tumor-associated markers that have been identified in OvCa-derived exosomes4, human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), FRα, CA-125, EpCAM, and CD24. The combined assay of on-chip purification and ddPCR analysis reported an mRNA pattern consistent to that of the UC-purified SKOV3 and OVCAR3 EVs, respectively (Fig. 3h and Supplementary Fig. S9). These results further demonstrated the validity of the nano-HB chip for exosome immunocapture. High capture efficiency of the nano-HB chip compares favorably with the recently reported methods32, 33, 37, which permits sensitive exosome detection.
Ultrasensitive Detection and Quantitative Protein Profiling of Exosomes.
The nano-HB chip combined with a sandwich exosome ELISA method8 was implemented and optimized for sensitive and specific detection of tumor-derived exosomes. CD81 provided high capture efficiency for exosomes derived from various cancer cell lines (Fig. 3e-g). Thus, our assay was designed to capture overall exosomes by anti-CD81 mAb and detect surface protein markers with specific mAbs, which is assisted by fluorogenic signal amplification to further enhance the sensitivity. With the COLO-1 exosome standards, we have systematically examined the critical conditions for the immunoassay and enzymatic signal amplification, including washing buffer, the concentration of reporter enzyme, and reaction time, to afford the maximal signal/noise ratio (Supplementary Fig. S10a-c). We also verified that there is no endogenous β-galactosidase activity from exosomes that would interfere with the enzymatic fluorogenic detection (Supplementary Fig. S10d), which could be due to that β-galactosidase is not present on the exosome membrane. The established protocol affords specific exosome detection on the Ab-modified chip, as verified by the low non-specific background obtained by using the PBS blank as the control sample and a control chip coated by BSA or normal human IgG (Supplementary Fig. S10e, f and Fig. S11a).
As the porous structure has significant effects on interfacial sensing, we evaluated the nano-HBs engineered with distinct morphologies, shown in Fig. 1d-i, for exosome detection (Fig. 4a). Substantial signal enhancement was observed shifting from the 170 nm HB with ~26 nm pores to the 960 nm HB with ~144 nm pores, owing to the enhanced flow penetration that augments exosome-surface interactions and the accessibility to the internal binding surface. The nanorod-HBs yielded a lower signal than that of 520 nm particles, which can be attributed to the lower porosity of well-aligned, tightly packed rods (Fig. 1i) compared to close-packed monodisperse spheres38. Random packing of 520 and 960 nm particles led to larger pores than the ordered mono-assemblies (Fig. 1h) and thereby a better detection sensitivity. Decreasing signal levels were obtained for the 520/170 nm and 960/170 nm structures, which agrees with the findings that increasing particle size ratio improves the order of packing, but reduce porosity and pore sizes22, 23. Compared to the close-packed array of monodispersed NPs, precise control of multi-component CSA is more difficult22 and highly porous, randomly-packed structures result in reduced mechanical strength. Thus, we selected the 960 nm HB for subsequent studies as it offers a combination of high sensitivity and ease of fabrication. In addition to the morphology, we have also optimized the geometry of nano-HBs. Using the MINDS strategy, the microscale geometry of nano-elements can be directly manipulated by designing the molding channels (Fig. 1j, k). Here we adopted a well-established HB design of 100 μm width and 225 μm pitch from the previous studies25, 26 and investigated the nano-HBs of 15–35 μm height embedded in a 50 μm tall channel. The 25 μm height was found to provide the best flow mixing efficiency and the highest signal/noise ratio for exosome detection (Supplementary Fig. S12).
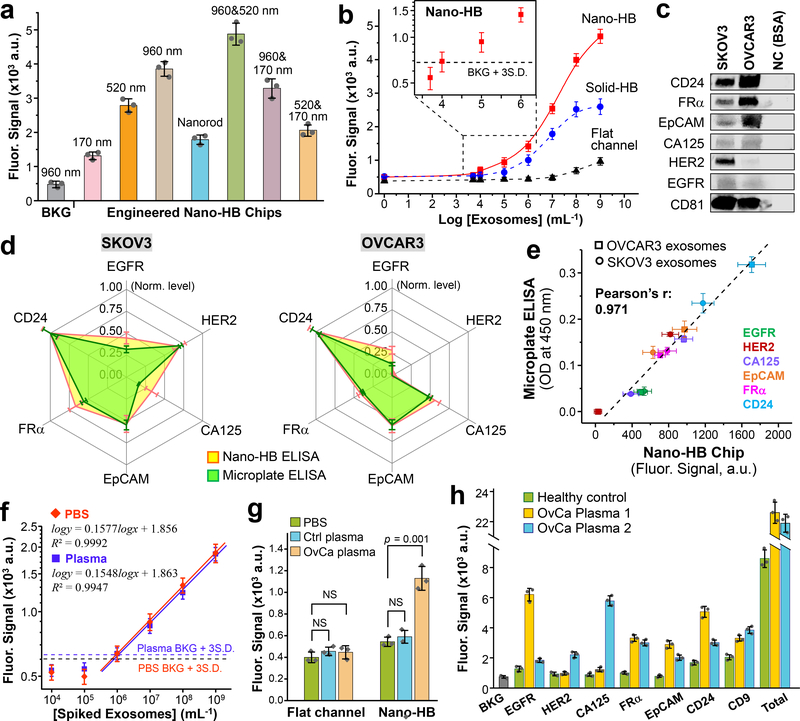
Fig. 4. Ultrasensitive detection of exosomes with the nano-HB chip.
(a) Engineering nano-HBs by the MINDS programs the sensitivity for detecting COLO-1 exosomes (105 μL−1). Statistic comparison between the 960 and 960/520 nm chips yielded p = 0.005. (b) Calibration curves for quantifying total exosomes by the flat-channel, solid-HB, and nano-HB chips. Serial 10× diluted COLO-1 exosome standards were used with a mixture of anti-CD9, CD63, and EpCAM mAbs for detection. (c) Western blot analysis of the protein markers in SKOV3 and OVCAR3 EVs with 10 μg BSA as negative control (NC). (d) Comparing the nano-HB chip and a standard microplate kit for ELISA detection of six proteins in SKOV3 and OVCAR3 exosomes. 20 μL, 105 μL−1 purified EVs were used for nano-HB assay, and 100 μL, 106 μL−1 EVs for microplate ELISA. All analyses were normalized against CD24 that was found most abundant. (e) The measurements of six targets in SKOV3 and OVCAR3 exosomes by the nano-HB chip and microplate-based ELISA correlated well. (f) Calibration curves for detecting the FRα+ subtype in UC-purified SKOV3 EVs spiked in PBS and a 10× diluted healthy plasma without detectable FRα. Total exosomes were captured with anti-CD81 mAb and the FRα+ subtype was detected with anti-FRα mAb. (g) High sensitivity of the nano-HB chip enables detection of FRα+ exosomes in an OvCa plasma sample, which is indiscernible to the conventional flat-channel chip. Two-tailed Student’s t-test was used at a significance level of p < 0.05. NS, not significant. (h) Protein profiling of exosomes directly in plasma from a control and two OvCa patients with nano-HB chips. Total exosomes were detected with a mix of CD9, CD63, and CD81 Abs. CD81 mAb was used for exosome capture in all cases. Error bars: one S.D. (n = 3).
With the optimized chip design and assay conditions, we calibrated and compared the nano-HB chips to the flat-channel and solid-HB devices for quantitative exosome detection (Fig. 4b and Fig. S13). A mixture of anti-CD9, CD63, and EpCAM mAbs was used to quantify the overall exosomes captured on chip (Supplementary Table S1). The flat-channel chip yielded a theoretical limit of detection (LOD) of 5.0 × 104 EVs μL−1, calculated from the mean blank signal plus three standard deviations (n = 3). The solid-HB chip afforded a much lower LOD of 3.2 × 102 μL−1, as a result of its substantially better mixing efficiency and larger surface area. 3D nano-HB chips further reduced the LOD to 10 μL−1 (i.e., 200 vesicles in a 20 μL sample, Fig. 4b inset), and expanded the dynamic range over four orders of magnitude. Considering the solid-HB also has nanostructured exterior surface (Supplementary Fig. S1), this direct comparison with the solid-HB assay supports that the enhancement in sensitivity arises from small boundary flow drainage through the nano-HBs that increases vesicle-surface interactions and the accessible binding surface area, while preserving the HB’s ability to enhance flow mixing in microchannels. Our data suggest that our nano-HB chip offers better exosome detection sensitivity than the existing methods3, 8, including a nano-plasmonic sensor that requires at least 3,000 exosomes per assay4.
The nano-HB chip-based assay was then adapted to detect exosomes derived by OvCa cell lines. We confirmed CD81 as the target for exosome immunocapture as it outperformed two commonly targeted exosomal markers, CD63 and CD9, when tested with the OVCAR3 exosomes (Supplementary Fig. S11a). CD81 was also found to be abundantly expressed in the OvCa patient-derived exosomes in our previous studies8, 39. We further verified the specificity of the anti-CD81 capture Ab for targeted immuno-detection of OvCa exosomes against free proteins (Supplementary Fig. S11b). The nano-HB assay was then assessed for quantitative profiling of OvCa-derived exosome subpopulations defined by six surface proteins, i.e., HER2, FRα, CA125, EpCAM, EGFR, and CD24. The presence of these biomarkers and high abundance of CD81 in UC-purified SKOV3 and OVCAR3 exosomes was verified by WB (Fig. 4c). Using the nano-HB assay and a commercial exosome ELISA kit, the levels of the individual exosomal markers were quantified and normalized against CD24 found to be most abundant. The exosomal protein patterns obtained by the on-chip fluorescence detection for each cell line showed high similarity with those by the microplate chromogenic ELISA (Fig. 4d) and agreed well with the previously reported results4. Regression analysis of all measured values for six markers in both SKOV3 and OVCAR3 exosomes resulted in a strong linear correlation between the two methods (Pearson’s r = 0.971), validating the nano-HB ELISA detection for quantitative exosome analysis (Fig. 4e).
Lastly, we optimized the nano-HB assay for analyzing clinical plasma samples. Specific detection of tumor-derived exosomes was assessed via probing the FRα+ subtype in the purified SKOV3 EVs spiked in both PBS and a healthy plasma with no detectable FRα+ exosomes (Fig. 4f). Analysis of spiked plasma demonstrated comparable analytical merits with that of spiked PBS, such as calibration sensitivity (i.e., the slope, 0.1548 vs. 0.1577) and LOD (1.0 × 103 vs. 0.9 × 103 total EVs μL−1). Such matched detection performance verifies that our optimized assay effectively reduces the matrix effects while maintaining high exosome binding efficiency. The LOD for the FRα+ subtype (103 total EVs μL−1) is in line with that for total exosomes (10 μL−1, Fig. 4b) because FRα+ exosomes were found to account for a small fraction of total SKOV3 exosomes based on the measured relative FRα mRNA level (2.7% of GAPDH). Such high sensitivity allowed us to detect a low level of FRα+ exosomes directly in 2 μL OvCa patient plasma, as verified by the measurements of a PBS blank and a control plasma (p < 0.001, Fig. 4g). On the contrary, the conventional flat-channel chip failed to detect such low-level FRα+ exosomes in the same sample under the same assay conditions. Profiling of circulating exosomes directly from healthy and OvCa plasma samples was also demonstrated (Fig. 4h), which identified different biomarker profiles between the OvCa cell lines (Fig. 4d) and the plasma and among the patients and the control. It is noted that some tumor-relevant exosomes, such as the FRα+ subtype, have quite low abundance despite a high level of total exosomes in the plasma. This can be attributed to small fractions of those subtypes in the overall exosomes derived by tumor cells, which are further reduced in patient biofluids by exosomes originated from non-cancer cells.
Accurate quantification of the fraction of a tumor-derived subtype in circulating exosomes is challenging. One of the challenges is that the molecular properties of tumor-associated exosomes are poorly understood so that the exosome number cannot be directly derived from quantifying their molecular contents40. Here we implemented a simple strategy to estimate the fraction of tumor-associated subtypes in circulating exosomes (Supplementary Fig. S14). Briefly, a series of exosome standards of known quantities are assayed to quantify the total exosomes with anti-CD81 capture mAb and a mix of anti-CD9 and CD63 mAbs for detection. The calibration plot establishes the correlation between exosome number and the measured level of CD9 and CD63. We assume that anti-CD81 captures all exosomes and that CD9 and CD63 combined can also detect all the captured exosomes and are evenly distributed on every vesicle. Based on this assumption, the subtype exosomes can be captured by specific mAbs and quantified by measuring their CD9 and CD63 level to calculate the exosome number from the calibration plot. Using this approach, we estimated the fraction of FRα+ exosomes to be 4.5 ± 0.8% for Patient #8 with a plasma exosome level of 6.4 × 1010 mL−1 (Supplementary Fig. S14). This value agrees closely with that estimated by the exosomal expression of FRα mRNA relative to GAPDH (2.7%, Fig. 3h) if we assume that FRα mRNAs are homogeneously distributed in the tumor-associated exosomes with the same stoichiometry as GAPDH mRNAs across the whole population. The accuracy of our method is limited by several key factors, such as heterogeneous distribution of CD9 and CD63, inaccurate numeration of total circulating exosomes by UC isolation and NTA, and lack of well-defined molecular properties of tumor exosomes. Nonetheless, our results suggest the level of clinically important exosomes, e.g., those from early-stage tumors, might be too low to be measured reliably with current capabilities, highlighting the necessity to develop ultrasensitive biosensing tools. Collectively, our studies suggest the superior sensitivity and clinical adaptability of our system, which could open new opportunities to advancing clinical applications of exosomes.
Clinical Analysis of Circulating Exosomes for Cancer Diagnosis.
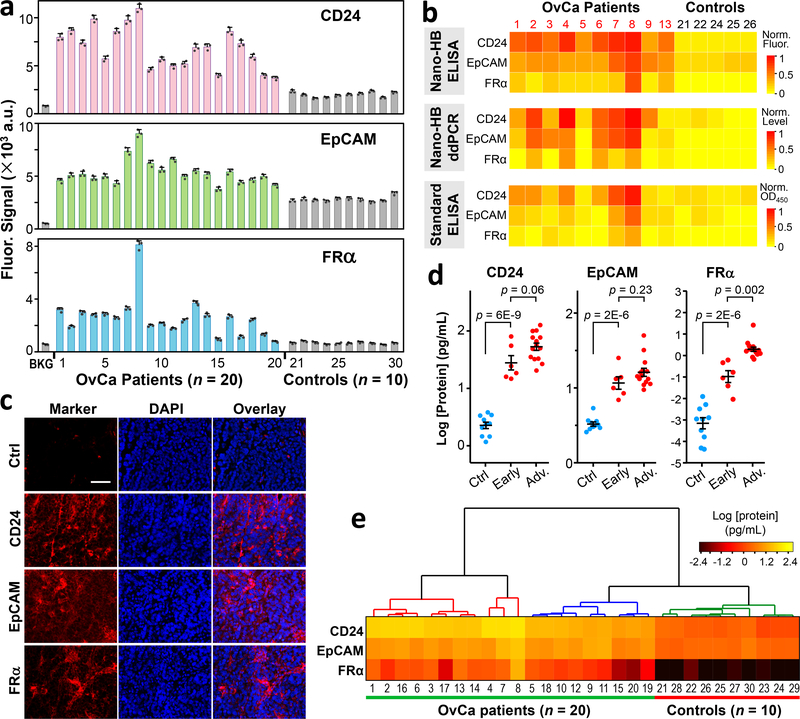
As a proof-of-concept, we adapted the nano-HB system to assess circulating exosomes as a non-invasive tool for diagnosis of OvCa. We examined plasma samples collected from OvCa patients (n = 20) and age-matched non-cancer controls (n = 10) (Supplementary Table S2). This sample size is sufficiently large to evaluate diagnostic accuracy with desired statistic errors (Table S3)4,29. Our exosomes profiling results show the universal presence of CD24 and EpCAM in exosomes from the OvCa cell lines and patient plasma, which is consistent with the previous studies4,39. FRα is a promising target for cancer diagnosis and treatment41–43. However, FRα levels were reported to be low in exosomes isolated from OvCa cell lines4 and has yet been studied in circulating exosomes for OvCa diagnosis. Interestingly, our method detected relatively abundant exosomal FRα in the OvCa cell lines and plasma samples (Fig. 4). Thus, FRα, CD24, and EpCAM were further investigated as the candidate biomarkers. Fig. 5a shows that CD24 and EpCAM were not only detected at high concentrations in OvCa exosomes, but also in the control samples. In contrast, FRα levels were much lower in OvCa exosomes but importantly almost undetectable in the control samples. We calibrated the nano-HB chip for protein immunoassay, which yielded a very low LOD for CD24 (100 fg/mL), EpCAM (10 fg/mL), and FRα (10 fg/mL), respectively (Supplementary Fig. S15). The calibration curves were used to convert the fluorescence signals to protein concentrations. Despite the large interpatient heterogeneity observed, the levels of three markers can be used to discriminate the OvCa patients from the controls (Supplementary Fig. S16, p = 1E-12 for CD24; p = 2E-10 for EpCAM and FRα, two-tailed Student’s t-test).
Fig. 5. Clinical profiling of circulating exosomes for diagnosis of ovarian cancer.
(a) Quantification of the exosomal levels of CD24, EpCAM, and FRα proteins directly from the plasma of OvCa patients (n = 20) and non-cancer controls (n = 10). Signals were subtracted by the corresponding background measursed with PBS to determine the protein levels. Error bars indicate S.D. (n = 3). (b) Comparison of the exosomal marker patterns measured with the nano-HB ELISA, the microplate ELISA, and the combined nano-HB capture and ddPCR of mRNAs. A subset of OvCa (n = 10) and control samples (n = 5) from (a) was assayed in triplicate and the mean values were normalized against CD24 in Patient #8 that showed the highest levels. For mRNA analysis, 100 μL plasma from each patient was diluted by 10 times and run through two 8-channel nano-HB chips to ensure fast and efficient exosome capture. (c) Typical images of immunofluorescence histological assays of the patient-matched tumor tissues (Patient #11). Scale bar, 50 μm. (d) Scatter dot plots of the plasma levels of three exosomal markers measured by the nano-HB assay for the patients of variable stages: control, early-stage (stage I/II), and advanced patients (stage III/IV). Error bars are the mean and one standard error of the mean (s.e.m.). Statistical comparison of three groups was performed by one-way ANOVA with post hoc Tukey’s test. Significance level was set at p < 0.05. (e) Heatmap constructed by non-supervised hierarchical clustering of the levels of exosomal CD24, EpCAM, and FRα recognizes the OvCa and control groups. Clustering analysis was performed with Ward linkage and Euclidean distance.
The diagnostic performance of the nano-HB assay was systematically cross-validated by several gold standard methods. EVs were isolated from 2–3 mL plasma samples (Table S2) following the established UC protocols31. A major size distribution of ~65–230 nm was determined for the purified EVs by NTA with a shoulder peak likely due to the aggregated vesicles. SEM demonstrates the characteristic round-cup morphology (Supplementary Fig. S17a). WB analysis confirmed high expression of exosomal CD81 in OvCa patients of various stages and detected a low level of FRα only in the stage-IV patient (Supplementary Fig. S17b). The EV particle number by NTA and the total EV protein by Bradford assay were compared between the OvCa patients and the controls and resulted in a p value of 0.13 and 0.001, respectively (two-tailed Student’s t-test, Supplementary Fig. S18). Thus, targeted analysis of tumor-associated exosomes by the nano-HB chip offers better diagnostic performance than these commonly applied approaches.
A subset of the plasma samples (10 OvCa patients and 5 controls) was analyzed by the combined assay of nano-HB capture and ddPCR and the microplate chromogenic ELISA kit to validate the nano-HB assay. The standard measurements of three exosomal markers at both mRNA and protein levels revealed similar expression patterns with the nano-HB assays, especially for the patients with relatively high marker levels (Fig. 5b). Statistical analysis yielded a reasonably strong correlation between the nano-HB data and those of the ddPCR (Pearson’s r = 0.806) and the microplate ELISA (Pearson’s r = 0.831). For both comparisons, a better linear correlation was observed for the high-concentration measurements and the reduced signal ratio at low concentrations, suggesting greater sensitivity of our method for clinical analysis of plasma exosomes (Supplementary Fig. S19). Moreover, compared to two standard methods, the nano-HB ELISA assay profoundly improved detection of the OvCa cases (Supplementary Fig. S20), while demanding only a fraction of sample volume (1/5) and assay time (~1/10). For instance, while 100 μL plasma from each patient was processed for exosomal mRNA analysis, FRα transcripts were not detected in 4 out of 10 OvCa patients (Supplementary Fig. S20 and S21). To test if circulating exosomes reflect the molecular profiles of OvCa tumor origin, we conducted immunofluorescence histological (IFH) studies of three patient-matched tumor tissues (Supplementary Table S2) and observed relatively high expression of three markers (Fig. 5c and Supplementary Fig. S22). The expression of three markers in these patients’ tissues was further verified by standard H&E staining and immunohistochemistry (IHC) analysis (Supplementary Fig. S23). The histological results suggest the correlation between the biomarker profiles of circulating exosomes and solid tumors, which supports potential applications of circulating exosomes as a non-invasive surrogate biopsy for tumors. Taken together, our validation studies corroborated the advantages of our technology for highly sensitive and specific analysis of tumor-derived circulating exosomes with minimal sample consumption, which are particularly compelling for the development of non-invasive tools for preclinical screening and early-stage diagnosis of cancer.
Statistic analyses of the nano-HB assay data in Fig. 5a were performed to quantitatively evaluate the diagnostic metrics of these exosomal markers. Analyses with receiver operating characteristic (ROC) curves revealed superior diagnostic accuracy for exosome profiling over NTA counting, with the area under the curve (AUC) for CD24, EpCAM, FRα, total protein, the three-marker set, and exosome counts determined to be 1.00, 1.00, 0.995, 1.00, 1.00, and 0.709, respectively (Supplementary Fig. S24). Probing exosomal CD24 and EpCAM has been documented for OvCa diagnosis with high accuracy4,39. While exosomal FRα showed slightly less diagnositic power than CD24 and EpCAM due to its lower expression, it was almost undetectable in non-cancer controls, indicating its good tumor specificity. We assessed the nano-HB assays of three exosomal markers for detecting the control, early-stage OvCa (stage I/II), and advanced (stage III/IV) groups using one-way ANOVA with post hoc Tukey’s test. As summarized in Supplementary Table S4, ANOVA detected overall significant difference among the three groups. Tukey’s multiple comparison tests between the groups revealed that exosomal FRα could significantly differentiate the early-stage OvCa from the advanced disease (p < 0.002), while CD24 (p = 0.06) and EpCAM (p = 0.23) could not (Fig. 5d and Supplementary Table S4). Recent studies revealed that the level of free FRα in serum and plasma was very low in normal and benign conditions and significantly elevated in malignant ovarian tumors with strong correlation with disease histotype, stage and grade42, 43. Our results, albeit in the limited sample size, should support the clinical relevance of FRα and suggest exosomal FRα as a potential marker for early OvCa detection that warrants further investigation. To assess the feasibility of our technology for multiplexed exosome profiling to identify disease fingerprints, we performed non-supervised hierarchical clustering analysis of the measured levels of three exosomal tumor markers (Fig. 5e). The resultant heatmap showed that the patients and controls were completely classified into distinct clusters by this three-marker combination, suggesting the potential of exosome profiling for OvCa diagnosis and stratification. Clinical promise of these biomarkers, however, needs rigorous validation with much larger cohorts.
Discussion
Microfluidic integration of nanostructures attracts enormous interests as it combines the advantageous micro- and nano-scale phenomena to immensely improve biosensing44, 45. However, device-scale microfluidic integration of 3D complex nanostructures has been a long-standing challenge. Standard nanofabrication techniques have been mainly used for small-scale 2D nano-patterning on planar surfaces. Existing 3D micro/nanofabrication techniques, such as proximity field nanopatterning46, multi-beam holographic lithography47, and femtosecond laser printing 48, require expensive, sophisticated facilities and labor-intensive, time-consuming fabrication procedures, which severely limits their scalability and practicality for routine applications. Nano-engineering of microfluidic sensing devices mostly involves surface modification of microfabricated elements with nanomaterials, yielding essentially 2D nanostructured sensing surfaces12, 32, 45, 49. Alternatively, CSA enables simple, cost-effective fabrication of 3D nanomaterials with complex morphologies and have received extensive applications to material synthesis and photonics44, 50. However, its bioanalytical applications remain largely underexplored. The MINDS strategy presents an enabling approach for developing new microfluidic nanosensing technologies. Compared to standard nanolithography, this bottom-up method affords several advantages: 1) simple and programmable 3D assembly of complex nanostructures; 2) inherent compatibility with standard soft lithography for designed assembly and integration of device-scale nanostructures; and 3) broad applicability for nanobiosensing because it combines flexible and scalable microfluidic engineering with a wide spectrum of available nanomaterials44.
Probing circulating exosomes has attracted rapidly increasing interests in clinical care, especially for non-invasive diagnosis and monitoring of patient response to treatment. Efficient isolation and specific measurement of disease-associated exosomes, often present in low concentrations in biofluids, remains challenging for conventional technologies. While microfluidics provides a uniquely suitable technology to surmount these challenges, existing microfluidic technologies have yielded limited improvement in exosome analysis3, 8. In contrast to these techniques, the MINDS approach provides a simple yet powerful multi-scale engineering strategy to simultaneously address the fundamental limits in mass transfer, reaction characteristics, and boundary effects. It was shown that 3D nano-engineering of a herringbone mixer by the MINDS approach preserves the HB’s performance in enhancing mass transfer, reduces the boundary flow resistance to promote surface-exosome interactions, and immensely increases the surface area for binding (Fig. 2). These distinct advantages result in an ultrasensitive system that permits rapid and specific immunosensing of tumor-related exosomes, e.g., FRα+ exosomes, which present at an undetectable concentration in biological samples for conventional methods. We showed that our technology can be adapted to exosome profiling in various cancer types, providing an enabling tool to facilitate the studies of biological functions and clinical relevance of exosomes.
Ovarian cancer is the fifth leading cause of cancer-related death among women in the US. It is most commonly diagnosed at a late clinical stage in 85% of patients, which has a 5-year survival rate for patients of ~20%51. Currently, there is no single method capable of detecting with high sensitivity and specificity, including CA125 which is the most commonly used ovarian tumor biomarker52. Tumor-derived exosomes in the bodily fluids, such as ascites and blood, hold the potential as diagnostic markers for non-invasive screening and early detection of cancer4,39. Relevant to future clinical applications, we have investigated alteration of CD24, EpCAM and FRα protein expression in OvCa-associated exosomes to demonstrate the feasibility for non-invasive biomarker profiling of cancer (Fig. 5). FRα is a glycosylphosphatidylinositol-anchored protein that mediates folate transport into cells. Owing primarily to its largely restricted expression profile in normal tissues and upregulated expression in a variety of solid tumors, including serous OvCa, FRα has been recently under intense investigation as a promising diagnostic and therapeutic target53. Despite its significant biological significance and clinical potential, presence of FRα in exosomes has not received appreciable attention yet. Here we showed that our method could detect low levels of FRα in circulating exosomes from early-stage OvCa patient plasma samples. The exosomal levels of FRα were significantly elevated in OvCa and could help detect early-stage malignances, which supports further investigations of clinical potential of exosomal FRα as a blood biomarker for sensitive and specific diagnosis of OvCa. Overall, our proof-of-concept clinical analyses should validate the ability of our technology for not only sensitive quantification of total circulating exosomes but also for exosome profiling to identify potential disease fingerprints for non-invasive OvCa diagnosis and stratification. In principle, our nano-HB chip is applicable to a broad spectrum of targets of biomedical significance, including circulating tumor cells20, 21, 26. Therefore, our method provides a useful tool to facilitate the development of new biosensing technologies and clinical biomarkers.
Methods
Reagents and Materials
Monodispersed silica colloids were ordered from Bangs Laboratories Inc. (3-Mercaptopropyl) trimethoxysilane (3-MPS), 4-Maleimidobutyric acid N-hydroxysuccinimide ester (GMBS), polyvinylpyrrolidone (PVP, MW~40,000), anhydrous ethanol, 1-pentanol (≥99%), and sodium citrate dihydrate (99%) were purchased from Sigma-Aldrich. Ammonia (28%) and tetra-ethyl orthosilicate (TEOS, 98%) were obtained from Fisher Scientific. The ELISA kits for EpCAM, CD24 and FRα were ordered from R&D Systems. Streptavidin conjugated β-Galactosidase (SβG), fluorescein-di-β-D-galactopyranoside (FDG), 3,3’-dioctadecyloxacarbocyanine perchlorate (DiO) lipophilic dye, Vybrant™ CM-DiO cell staining solution, and fluorescent nanoparticles (5% solid, Ex/Em 540/560 nm, actual size 46 nm) were purchased from Life Technologies. The detailed information of antibodies used in our studies was listed in Table S1 below. 1× phosphate-buffered saline solution (PBS) and SuperBlock buffer were from Mediatech, Inc and ThermoFisher Scientific, respectively. All other solutions were prepared with deionized water (18.2 MV-cm, Thermo Scientific). SβG and FDG were dissolved in PBS working solution (PBSW) at pH 7.4 which contain 0.5 mM DL-dithiothreitol (Sigma-Aldrich), 2 mM MgCl2 (Fluka Analytical), and 5% bovine serum albumin (BSA, Sigma-Aldrich).
Microfabrication of PDMS Chips
The MINDS approach uses a patterning chip for microfluidic colloidal self-assembly and an assay chip for exosome analysis, both made of PDMS (Figure 1b, main text). The PDMS chips were fabricated using standard photolithography, as detailed previously.8 All silicon molds were patterned using SU-8 photoresist (MicroChem) following the procedures recommended by the manufacturer. For the patterning chips, we used SU-8 2010 to pattern the herringbone channel arrays with the heights of 15 μm, 25 μm and 35 μm by controlling the spin-coating speed. For the assay chips, SU-8 2050 was employed to pattern the flow channels of 50 μm in height. The prepared silicon molds were pre-treated by trichloro(1H, 1H, 2H, 2H-perfluorooctyl) silane under vacuum for 4 h. 33 g PDMS mixture at a 10 (base material): 1 (curing agent) ratio was poured on the mold and cured in the oven at 70 °C for 4 h. PDMS slabs were peeled off from the molds and the access holes were punched.
Fabrication of Nano-Herringbone-Integrated Chips by MINDS
The process of MINDS based on microfluidically guided colloidal self-assembly is schematically illustrated in Figure 1b (main text). Briefly, a PDMS patterning chip containing the herringbone channel array was sealed to clean glass slides without surface treatment. After 10 min sonication, 10 μL 10% w/v aqueous suspensions of monodisperse silica colloids was injected into the large solution reservoir shared by all channels. For co-assembly of colloids with different sizes, the total particle concentration of the mixed suspension was kept to be 10% w/v. The colloidal solution spontaneously filled the microchannels and stopped at individual channel outlets, due to the surface tension. The solution reservoir was sealed with a small piece of PDMS such that solvent evaporation from the open reservoir induced colloidal self-assembly to fully pack the microchannels. Colloidal suspension in the reservoir was then replaced by 5% 3-MPS to strengthen the mechanical stability of the assembled colloidal structures. The packed device was then completely dried at 80 °C for 1 h on a hotplate. After carefully peeling off the patterning chip, the self-assembled herringbone structure was aligned and sealed with a UV-Ozone-treated PDMS assay layer under a stereoscope. The assay channel is 10 mm long, 2 mm wide, and 50 μm tall, containing ten units of five herringbones.
Synthesis of Colloidal Silica Nanorods
Monodisperse silica nanorods with controlled aspect ratio were synthesized following a published wet-chemical method54 with minor modifications. Briefly, a solution was prepared by completely dissolving 30 g PVP in 300 mL 1-pentanol. 30 ml anhydrous ethanol, 8.4 ml ultrapure water, 2 ml of 0.18 M aqueous solution of sodium citrate dihydrate, 6.75 ml of fresh ammonia and 3 ml of TEOS were sequentially added into the PVP solution with hand shaking. The well-mixed solution was left in the fumehood without stirring to let the reaction proceed at the room temperature for ~4 h. The reaction mixture was then transferred into 50-mL centrifuge tubes. Monodisperse silica nanorods were purified by sequential centrifugation and washing: centrifuge at 2000× g for 1 h and re-disperse the sediment in ethanol; repeat the procedure at 1500× g for 15 min with ethanol (3 times) and water (3 times); and finally four times at 700× g for 15 min and disperse the purified nanorods in ~40 mL water. Using transmission electron microscopy (TEM), the dimensions of nanorods were determined to have an average diameter of 238 ± 32 nm and length of 1.34 ± 0.26 μm (Figure S2).
SEM Characterization of Nano-HB Chips
SEM images were taken using the FEI Versa 3D Dual Beam SEM at the KU Microscopy and Analytical Imaging Lab. To visualize exosomes captured on the surfaces, the samples were treated with 2.5% glutaraldehyde in PBS for 30 min and then rinsed for 3×5 min to fix the exosomes. The samples were post-fixed for 15 min in 1% osmium tetroxide and rinsed 10 min with water. The samples were dehydrated in a graded series of ethanol (30%, 50%, 70%, 95% and 100%) for 2×10 min at each step. The dehydrated samples were coated with a gold thin film using a high resolution ion beam coater and imaged by SEM.
Confocal Imaging Characterization of Nano-HB Chips
Confocal images were taken with Olympus 3I spinning disk confocal epifluorescence TIRF inverted microscope, equipped with 405, 488, 561, and 642 nm solid state lasers. Objective used was 20× long working distance air lens. The laser intensity was 100% and exposure time 3000 ms. Image stacks were taken in a 1 μm interval along the z-axis, which ranged from the bottom of nano-HB structures to the top of flow channel. The obtained image stacks were fitted into 3D view photography using SlideBook version 5.5.
Numerical Simulation
Computational fluid dynamics (CFD) simulation was conducted using an ANSYS CFX R17.2 software to solve incompressible Newtonian fluid with Navier–Stokes equations through a finite volume approximation. Water properties and a diffusion rate of 5 × 10−11 m2/s were used to model the flow mixing of the nanoparticles suspended in water55. A solid-HB mixer with the identical dimensions of the experiments was modeled first, omitting the porous domain, as the ‘baseline’ design to characterize the mixing efficiency in nano-HBs. This model included a structured mesh utilizing swept prism elements, due to the slanted nature of the geometry. A mesh sensitivity analysis was conducted to ensure the results were independent of the discretization of the computational domain. A total of 4,078,302 elements were used to capture the mixing interface appropriately and to model the complex flow pattern. The porous HB nanostructure was modeled by defining a mathematical porous domain within the channel. Inside this porous domain a momentum loss model was used to represent the flow restriction mathematically, following Darcy’s law. For this simulation, a ‘superficial velocity’ formulation was used to represent the nanonoporous media with an isotropic, linear, loss coefficient applied (permeability). The linear losses represent viscous losses while quadratic losses represent inertial loss. A widely used permeability–porosity relation in the field of flow in porous media, Carman-Kozeny equation, was used to calculate the permeability for random close-packed monodisperse particles of 960 nm with a porosity of 0.35,56, 57 yielding a Permeability (linear loss coefficient) of 4.7 × 10−15 m2. Since the flow is laminar with viscous forces dominating the inertial losses were neglected. A small was defined in order to resolve the nanoscale structure, while capturing the microscale flow pattern, with reasonable computational effort for this proof-of-concept study.
Cells and Culture Conditions
The human ovarian cancer cell lines SKOV3 and OVCAR3 were cultured in RPMI-1640 media with 10% (v/v) vesicle-depleted FBS, recombinant insulin (7.5 μg/mL), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37 °C in a humidified atmosphere with 5% CO2. Vesicle-depleted FBS was prepared by centrifuging FBS for 18 h at 100,000× g and then passing the supernatant through a 0.22 μm filter. Cell lines were maintained between 40–80% confluency during which time the conditioned media was collected and replaced every 24 h. The collected media was immediately centrifuged at 2,000× g for 10 min to remove large cell debris and then stored at 4 °C until sufficient volume was pooled for exosome isolation.
Extracellular Vesicle (EV) Isolation
EVs were isolated by differential ultracentrifugation (UC) of approximately 300 mL of conditioned media for each cell line collected over a 1–2 week period. Pooled conditioned media was spun for 45 min at 10,000× g to pellet large vesicles. The supernatant was then spun at 100,000× g for 1 h to pellet EVs. The supernatant was carefully decanted to avoid disturbing the EV pellet until ~2 mL remained. The 2-mL samples from multiple tubes were pooled together into a 13.5 mL polypropylene Quick-Seal® centrifuge tube (Beckman Coulter) and then spun at 100,000× g for 1 h to pellet the washed EVs. The supernatant was quickly and carefully removed and the EV pellet was re-suspended in 100–200 μL of cold 0.22 μm-filtered PBS and stored in low-retention tubes at −80 °C. Isolated EVs were characterized with NTA using NanoSight LM10 to determine the size and concentrations. Protein concentration was measured using Bradford protein assay (Bio-Rad).
Exosome Immunoassay Using Nano-HB Chips
The nano-HB chip was surface functionalized with 3-MPS for antibody conjugation via maleimide reaction. Briefly, the chips were filled with 5% 3-MPS in anhydrous ethanol solution and reacted for 1 h at the room temperature. Excess silane was washed away with 70% ethanol for three times and 0.28 mg/mL GMBS was injected into the chip to modify the surface for 0.5 h. After washing with PBS, 0.1 mg/mL anti-CD81 antibody was flowed through the chip to coat the surface for 1 h at room temperature. The chips were blocked with 5% BSA for 2 h and stored at 4 °C prior to experiments.
The lyophilized standard EVs of colon cancer COLO-1 cell line was purchased from HansaBioMed, Ltd (Tallinn, Estonia) and reconstituted in water prior to use. EVs of ovarian cancer cell line SKOV3 and OVCAR3 were isolated from cell culture media by UC as described above. The stock solution was aliquoted and stored at −80 °C. We determined the concentration of the stock solution by NTA and freshly prepare the standards by spiking the stock solution into PBS at different concentrations.
To characterize the nano-HB chip for exosome sensing, UC-purified EVs of known concentrations were prepared by serial dilutions of the stock solution in 1 mL PBS or 100 μL 10-fold diluted plasma; 20 μL of the solution was pumped through the chip at a flow rate of 0.5 μL/min by a micro-syringe pump. The channel was then washed with PBS at 1 μL/min for 10 min, filled with a mixture of three biotinylated detection antibodies for CD63, CD81 and EpCAM (20 μg/mL each) for detection of exosomes (0.5 μL/min, 30 min), and then washed with 10 μL SuperBlock buffer for 10 min. SβG prepared in PBSW buffer (20 ng/mL) was introduced at 0.5 μL/min for 15 min as the reporter enzyme. Excess enzyme was washed away by another 10 μL SuperBlock buffer and then FDG in PBSW (500 μM) was introduced and reacted for 0.5 h in dark for fluorescence detection assisted by enzymatic signal amplification. Fluorescence images were taken using a Zeiss Axiovert A1 inverted fluorescence microscope equipped with a LED excitation light source (Thorlabs, Newton, NJ). The digital images were processed and analyzed using ImageJ (NIH, http://rsbweb.nih.gov/ij/) to measure the fluorescence intensity.
Exosome Analysis Using Microplate ELISA
The ExoTEST™ Ready-to-Use Kit were purchased from HansaBioMed, Ltd (Tallinn, Estonia). 100 μL, 106 μL−1 purified SKOV3 and OVCAR3 EVs were added into each well of a 96-well plate. The plate was sealed with parafilm and incubated at room temperature while shaking for 30 min, then transfered into a 4 °C fridge for overnight incubation (12 h). 200 μL of washing buffer was added into each well, mixed by shaking, and discard by pouring out. The washing step was repeated for 3 times. The same biotinylated detection antibody as in the chip analysis was diluted to 2 μg/mL and 100 μL was added into wells. The plate was sealed and incubated at room temperature while shaking for 20 min, then incubated for 2 h at 4 °C. The plate was washed as above for 3 times, followed by adding 100 μL of 1:5000 diluted HRP-streptavidin conjugate, shaking at room temperature for 15 min, and incubation at 4 °C for 1 h. Excess enzyme was washed away with the washing buffer for 3 times. Lastly, 100 μL of chromogenic substrate solution was added to each well and incubated for 10 min at room temperature in dark. The reactions were stopped by adding 100 μL of stop solution. Optical density was measured at 450 nm (OD450) on a CYTATION 5 imaging reader (BioTek) and subtracted by the background measured with PBS.
Quantitative Calibration of Standard Proteins
On-chip protein quantitative assay was carried out for CD24, EpCAM and FRα, respectively, using a DuoSet ELISA kit (R&D Systems, Table S1), which consist of capture antibody, standard protein and detection antibody. The capture antibody (0.1 mg/mL) was immobilized on the nano-HB chip via 3-MPS and GMBS modification as described above. After blocking with 5% BSA for 1 h, 20 μL of standard proteins in PBS was flowed through the nano-HB chip within 2 h. The non-captured proteins were washed away using PBST washing buffer (0.05% Tween-20, 1% BSA in PBS). 10 μL of biotinylated detection antibody (20 μg/mL) was introduced to label the captured proteins. After washing with PBST, SβG and FDG in PBSW solution was added subsequently to trigger the enzymatic fluorescence amplification reaction. The fluorescent signal was measured as described above and plotted as a function of protein concentrations.
Western Blot Analysis of EVs
Western blotting was performed using 4–12% precast polyacrylamide slab mini-gels (Tris-glycine pH 8.3) with Blot Module (Bio-Rad), following the standard protocol. 30–50 μg purified EVs were pretreated with RIPA lysis buffer with protease inhibitors on ice for at least 3 h and heated at 72 °C for 10 min after adding equal volume of 2× loading buffer. The electrophoresis was carried out at 125 V for 2 h, and then gels were electro-transferred to the cellulose membranes (0.2 μm) at 25 V for 2.5 h. The NC membrane was first blocked with Odyssey® Blocking Buffer (PBS), then incubated overnight at 4 °C in primary antibody (1:1000). The membranes were washed 3 times for 10 min each (1× PBS, 0.5% Tween 20, pH 7.4) and then immerged into goat anti-mouse IRDYE@ 800CW secondary antibody (1:10,000) for 60 min at room temperature. After that, the washing step was repeated three times. Imaging was performed using an Odyssey Fc Imaging System (LI-COR Biosciences).
mRNA Analysis of OvCa Cell-Derived EVs
NTA counting reported a typical EV concentration of 107-108 mL−1 in SKOV3 and OVCAR3 cell culture media, which is ~102 to 104 fold lower than in human plasma. Because of the low concentration of EVs, hundreds of milliliters of the culture media were processed by UC to obtain sufficient amount of vesicles for droplet digital PCR (ddPCR) analysis of six mRNA markers. For bulk analysis of UC-purified OvCa EVs, total RNA was extracted from 40–60 μg EVs by RNAqueous™-Micro Total RNA Isolation Kit (Ambion) and then was reverse transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), per the manufacturer’s protocol, respectively. PCR mix was prepared by mixing 1 μL of the obtained cDNA and the commercial pre-designed primer sets for each mRNA targets (PrimeTime® qPCR Assays, Integrated DNA Technologies) with QX200™ ddPCR™ EvaGreen® Supermix (Bio-Rad), following the manufacturer’s recommendations. ddPCR was performed using the QX200™ Droplet Digital™ PCR System (Bio-Rad). The PCR protocol was: 5 min at 95 °C; 30 s at 95 °C, 1 min at 55 °C for 40 cycles; 5 min at 4 °C, 5 min at 90 °C. Background signal was determined for each assay and subtracted in data analysis to eliminate non-specific amplification. Measurement of each mRNA target was repeated at least three times with different batches of purified EVs. The concentrations of target mRNAs were normalized against GAPDH.
The nano-HB chip is designed for diagnostic exosome analysis using non-invasive sample volumes, which can run 20 −100 μL sample with a reasonable processing time (40–200 min). Chip isolation of exosomes from such small volume of cell culture medium was found not to yield sufficient amount of exosomes to detect the low-frequency mRNA targets by ddPCR. Based on the bulk mRNA analysis of UC-isolated EVs, it was estimated that at least 10 mL culture media is needed for the nano-HB chip isolation and subsequent ddPCR analysis of the mRNA targets, which is beyond the sample processing capacity of the chip. Therefore, we used the UC-concentrated SKOV3 and OVCAR3 EVs to characterize the nano-HB capture for mRNA profiling of exosomes. To this end, 100 μL of the concentrated EVs (106 μL−1) was run on the chip modified with anti-CD81 capture antibody with protein G as the linker1. Captured exosomes were eluted out by flowing 20 μL Pierce™ IgG elution buffer (ThermoFisher) through the chip, which will dissociate capture antibodies from Protein G. We lysed the eluted exosomes, extracted total RNA, and performed ddPCR analysis of mRNA targets, as described above. Profiles of six mRNA markers measured in the chip-purified exosomes were compared with that of bulk analysis of UC-purified EVs.
Clinical Exosome ELISA Analysis Using Nano-HB Chips and the Microplate Kit
De-identified plasma samples from ovarian cancer patients and cancer-free individuals were provided from the KU Cancer Center’s Biospecimen Repository Core Facility (BRCF) along with accompanying clinical information (Supplementary Table S2). Blood specimens, i.e., plasma samples, were obtained from individuals enrolled under the repository’s Institutional Review Board (IRB)–approved protocol (HSC #5929) and following U.S. Common Rule. Once the patient provides written, informed consent in accordance with the BRCF’s IRB protocol, blood was collected by BRCF staff and processed for long-term storage at −80˚C. We estimated the required sample size for evaluating diagnostic accuracy via comparing the area under a ROC curve (AUC) with a null hypothesis value of 0.5. For conventional characterization of the samples, EVs were purified by UC and then characterized by NTA sizing, Bradford assay, and Western blot, following the protocols that we established.31 For microfluidic analysis, 2-μL plasma was used without any pre-treatment except 10 times dilution with PBS to permit convenient injection by a 50-μL syringe and to prevent channel clogging. Exosome assay and data acquisition followed the same processes as for exosome standards. We used CD81 mAb for capture and specific mAbs (20 μg/mL) for immuno-detection of CD24, EpCAM and FRα.
For analysis of plasma exosomes with the standard method, the ExoTEST™ Ready-to-Use Kit from HansaBioMed, Ltd (Tallinn, Estonia) were used. Ten OvCa patients and five controls listed in Table S2 were selected for standard microplate ELISA. 10 μL plasma samples were 10-fold diluted to 100 μL which were added into each well of a 96-well plate. The assays were carried out following the protocol described above.
Clinical Exosome mRNA Analysis
For mRNA analysis of exosomes in patient plasma, we have expanded the nano-HB chip to an 8-channel device to enhance the capacity and throughput for exosome capture. 100 μL plasma from each patient was diluted by 10 times and run through two 8-channel chips in 2 h to capture sufficient exosomes for subsequent ddPCR assays. Elution of the captured exosomes by 20 μL Pierce™ IgG elution buffer, exosome lysis, total RNA extraction, cDNA synthesis, and ddPCR assays were performed as described above.
Histological Analysis of Patient-Matched Tissues
The immunofluorescence histology analysis of patient-matched tissues was performed in the Histology Laboratory at the KU Medical Center following the standard protocol.31 Briefly, the fresh frozen ovarian cancer tissues were embedded in OCT compound in cryomolds and cut into 4 μm thick cryostat sections. The slides were fixed in ice cold acetone for 5 min and washed with PBST for 2 min after 30 min air dry. 1% BSA was used to block the tissue sections for 30 min. The same primary detection antibodies (1:100 dilution) as exosomes immunoassay were added and incubated at 4 °C overnight. After washing with PBST, 0.4 μM DAPI in PBS and secondary detection antibody goat anti-mouse IgG (1:100 dilution, Cy5 labeled) were incubated sequentially with the tissue sections. After staining, the tissue sections were mounted by coverslips and imaged using an upright epi-fluorescence microscope (Nikon Eclipse 80i) equipped with 20× or 40× objective, and three color filter sets (blue, green, red). Images were collected and merged in three-color using Metamorph and analyzed using ImageJ. Immunohistochemical (IHC) staining was performed according to the following procedure. Four micron paraffin sections were mounted on Fisherbrand Superfrost slides and baked for 60 min at 60 °C then deparaffinized. Epitope retrieval was performed using a Biocare Decloaking Chamber (pressure cooker) under pressure for 5 min, using pH 6.0 Citrate buffer followed by a 10 min cool down period. Endogenous peroxidase was blocked with 3% H2O2 for 10 min followed by incubation with a specific primary antibody for 30 min: 1:100 dilution of monoclonal EpCAM (MOC-31) (Abcam); 1:50 dilution of monoclonal CD24 (SN3) (ThermoFisher Scientific); ready-to-use monoclonal FOLR1 (26B3.F2) (Biocare Medical); and ready-to-use monoclonal p53 (DO-7) (Dako Agilent). This was followed by Envision+ anti-mouse secondary (Dako) for 30 min and DAB+ chromogen (Dako) for 5 min. IHC staining was performed using the IntelliPATH FLX Automated Stainer at room temperature. A light hematoxylin counterstain was performed, following which the slides were dehydrated, cleared, and mounted using permanent mounting media.
Statistical Analysis
We estimated the required sample size for ROC analysis of diagnostic accuracy, by comparing the area under a ROC curve (AUC) with a null hypothesis value of 0.5. Statistical calculation was performed at various type I error α and type II error β levels with an expected AUC of 0.8 and a sample allocation ratio of 0.5 (control/patient). The computed sample sizes were listed in Table S3. A total sample size of 30 (i.e., 10 non-cancer controls and 20 ovarian cancer patients) is sufficient for statistical evaluation of diagnostic accuracy.
To mitigate analytical variations, detection signals for biomarkers were corrected by the background measured in parallel. Each measurement was repeated at least three times. Mean, standard deviation, and standard error of mean were calculated with standard formulas. To compare the patient and control groups, two-tailed Student’s t-test was performed with the significance level of p < 0.05. Overall statistical significance of difference in the mean levels of exosomal CD24, EpCAM and FRα among the control, early-stage (stage I/II) and advanced (stage III/IV) groups was tested using one-way ANOVA. The significance of the difference between two groups was evaluated using the post hoc Tukey’s test (Table S4). An outlier in the FRα data was detected by the Grubbs’ test and excluded from the post hoc test. Clustering and ROC curve analyses were performed with the protein levels of exosomal CD24, EpCAM and FRα in Figure 5e (main text) and the NTA data in Figure S16, using the R package. For the combined three-marker set, the arithmetic average of the levels of three markers was used as an independent variable for calculation. Non-supervised hierarchical clustering analysis was performed with Ward linkage and Euclidean distance to generate a heatmap with the dendrogram. 95% confidence level was used for all statistical analysis.
Supplementary Material
Acknowledgements
We thank the microfabrication core facility at the KU COBRE Center for Molecular Analysis of Disease Pathways for device fabrication and KU Cancer Center’s Biospecimen Repository Core Facility funded in part by the National Cancer Institute Cancer Center Support Grant (P30 CA168524), Tara Meyer (KUMC) for the technical support for immunohistochemistry, Dr. Rashna Madan (KUMC) for histopathology review, and Dr. Harsh Pathak (KUCC) for technical guidance with cell line-derived exosome isolations. The authors greatly appreciate the anonymous reviewers for their insightful and inspiring comments. This study was supported by 1R21CA186846, 1R21CA207816, 1R21EB024101, 1R33CA214333, and P20GM103638 from the NIH. P.Z. was supported by the postdoc award from the Kansas IDeA Network of Biomedical Research Excellence (K-INBRE) under the grant P20GM103418 from NIH/NIGMS. A.K.G. is the Chancellors Distinguished Chair in Biomedical Sciences endowed Professor.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are available for research purposes from the corresponding author on reasonable request.
REFERENCES
- 1.Schwarzenbach H, Nishida N, Calin GA & Pantel K Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 11, 145–156 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Haber DA & Velculescu VE Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 4, 650–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He M & Zeng Y Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer. J Lab Autom 21, 599–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Im H et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nature Biotechnology 32, 490–U219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanez-Mo M et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atay S et al. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci U S A 111, 711–716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melo SA et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, He M & Zeng Y Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab on a chip 16, 3033–3042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squires TM, Messinger RJ & Manalis SR Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol 26, 417–426 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Stroock AD et al. Chaotic mixer for microchannels. Science 295, 647–651 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Shang Y, Zeng Y & Zeng Y Integrated Microfluidic Lectin Barcode Platform for High-Performance Focused Glycomic Profiling. Scientific reports 6, 20297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley SO et al. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nature nanotechnology 9, 969–980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soleymani L, Fang Z, Sargent EH & Kelley SO Programming the detection limits of biosensors through controlled nanostructuring. Nature nanotechnology 4, 844–848 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Bin X, Sargent EH & Kelley SO Nanostructuring of sensors determines the efficiency of biomolecular capture. Analytical chemistry 82, 5928–5931 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Wang S et al. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angewandte Chemie 48, 8970–8973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen GD, Fachin F, Fernandez-Suarez M, Wardle BL & Toner M Nanoporous elements in microfluidics for multiscale manipulation of bioparticles. Small 7, 1061–1067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen GD, Fachin F, Colombini E, Wardle BL & Toner M Nanoporous micro-element arrays for particle interception in microfluidic cell separation. Lab on a chip 12, 3159–3167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fachin F, Chen GD, Toner M & Wardle BL Integration of Bulk Nanoporous Elements in Microfluidic Devices With Application to Biomedical Diagnostics. Journal of Microelectromechanical Systems 20, 1428–1438 (2011). [Google Scholar]
- 19.Zeng Y & Harrison DJ Self-assembled colloidal arrays as three-dimensional nanofluidic sieves for separation of biomolecules on microchips. Analytical chemistry 79, 2289–2295 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Stott SL et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences of the United States of America 107, 18392–18397 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angewandte Chemie 50, 3084–3088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh G, Pillai S, Arpanaei A & Kingshott P Multicomponent colloidal crystals that are tunable over large areas. Soft Matter 7, 3290–3294 (2011). [Google Scholar]
- 23.Nazemifard N et al. A systematic evaluation of the role of crystalline order in nanoporous materials on DNA separation. Lab on a chip 12, 146–152 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Adams AA et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. Journal of the American Chemical Society 130, 8633–8641 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes TP & Kralj JG Engineering and analysis of surface interactions in a microfluidic herringbone micromixer. Lab on a chip 12, 2634–2637 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Sheng W et al. Capture, release and culture of circulating tumor cells from pancreatic cancer patients using an enhanced mixing chip. Lab on a chip 14, 89–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philipse AP & Pathmamanoharan C Liquid Permeation (and Sedimentation) of Dense Colloidal Hard-Sphere Packings. J Colloid Interf Sci 159, 96–107 (1993). [Google Scholar]
- 28.Frishfelds V, Hellstrom JGI & Lundstrom TS Flow-Induced Deformations Within Random Packed Beds of Spheres. Transport Porous Med 104, 43–56 (2014). [Google Scholar]
- 29.Shao H et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nature communications 6, 6999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanwar SS, Dunlay CJ, Simeone DM & Nagrath S Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab on a chip 14, 1891–1900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He M, Crow J, Roth M, Zeng Y & Godwin AK Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab on a chip 14, 3773–3780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reategui E et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nature communications 9, 175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y et al. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat Biomed Eng 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobb RJ et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles 4, 27031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domcke S, Sinha R, Levine DA, Sander C & Schultz N Evaluating cell lines as tumour models by comparison of genomic profiles. 4, 2126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skog J et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature cell biology 10, 1470–1476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies RT et al. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab on a chip 12, 5202–5210 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Li S, Zhao J, Lu P & Xie Y Maximum packing densities of basic 3D objects. Chinese Science Bulletin 55, 114–119 (2010). [Google Scholar]
- 39.Zhao Z, Yang Y, Zeng Y & He M A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab on a chip 16, 489–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chevillet JR et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A 111, 14888–14893 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toffoli G et al. Overexpression of folate binding protein in ovarian cancers. Int J Cancer 74, 193–198 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Kurosaki A et al. Serum folate receptor alpha as a biomarker for ovarian cancer: Implications for diagnosis, prognosis and predicting its local tumor expression. International Journal of Cancer 138, 1994–2002 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Leung F, Dimitromanolakis A, Kobayashi H, Diamandis EP & Kulasingam V Folate-receptor 1 (FOLR1) protein is elevated in the serum of ovarian cancer patients. Clinical Biochemistry 46, 1462–1468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Choi MK, Kim DH & Hyeon T Designed Assembly and Integration of Colloidal Nanocrystals for Device Applications. Advanced materials 28, 1176–1207 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Li Z & Park I Direct synthesis and integration of functional nanostructures in microfluidic devices. Lab on a chip 11, 1946–1951 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Jeon S, Malyarchuk V, White JO & Rogers JA Optically fabricated three dimensional nanofluidic mixers for microfluidic devices. Nano letters 5, 1351–1356 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Park SG, Lee SK, Moon JH & Yang SM Holographic fabrication of three-dimensional nanostructures for microfluidic passive mixing. Lab on a chip 9, 3144–3150 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Hu Y et al. Laser printing hierarchical structures with the aid of controlled capillary-driven self-assembly. Proc Natl Acad Sci U S A 112, 6876–6881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cederquist KB & Kelley SO Nanostructured biomolecular detectors: pushing performance at the nanoscale. Current opinion in chemical biology 16, 415–421 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Vogel N, Retsch M, Fustin CA, Del Campo A & Jonas U Advances in colloidal assembly: the design of structure and hierarchy in two and three dimensions. Chemical reviews 115, 6265–6311 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Bast RC Jr., Hennessy B & Mills GB The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 9, 415–428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodurka DC et al. Reclassification of serous ovarian carcinoma by a 2-tier system: a Gynecologic Oncology Group Study. Cancer 118, 3087–3094 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung A et al. Targeting folate receptor alpha for cancer treatment. Oncotarget 7, 52553–52574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuijk A, van Blaaderen A & Imhof A Synthesis of monodisperse, rodlike silica colloids with tunable aspect ratio. Journal of the American Chemical Society 133, 2346–2349 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Kastelowitz N & Yin H Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. Chembiochem : a European journal of chemical biology 15, 923–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Müllner T, Unger KK & Tallarek U Characterization of microscopic disorder in reconstructed porous materials and assessment of mass transport-relevant structural descriptors. New Journal of Chemistry 40, 3993–4015 (2016). [Google Scholar]
- 57.Frishfelds V, Hellström JGI & Lundström TS Flow-Induced Deformations Within Random Packed Beds of Spheres. Transport Porous Med 104, 43–56 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.