Abstract
A major challenge in developmental biology is unraveling the precise regulation of plant stem cell maintenance and the transition to a fully differentiated cell. In this review, we highlight major themes coordinating the acquisition of cell identity and subsequent differentiation in plants. Plant cells are immobile and establish position-dependent cell lineages that rely heavily on external cues. Central players are the hormones auxin and cytokinin, which balance cell division and differentiation during organogenesis. Transcription factors and miRNAs, many of which are mobile in plants, establish gene regulatory networks that communicate cell position and fate. Small peptide signaling also provides positional cues as new cell types emerge from stem cell division and progress through differentiation. These pathways recruit similar players for patterning different organs, emphasizing the modular nature of gene regulatory networks. Finally, we speculate on the outstanding questions in the field and discuss how they may be addressed by emerging technologies.
Keywords: plant stem cells, asymmetric divisions, meristem, transcriptional regulation, mobile signals, cell identity
INTRODUCTION
In multicellular organisms, the choice to divide or differentiate lies at the core of cellular decision making. The advent of multicellularity required the diversification of cell types to carry out specialized functions. While all cell types share the same genetic information encoded in their DNA, an array of regulatory mechanisms specify distinct programs of gene expression. These changes in gene expression modify the complement of proteins within a given cell, resulting in an altered competence to respond to signals and patterning cues.
Central to development are asymmetric divisions during which one cell divides to produce two daughter cells that go on to acquire different fates (Horvitz & Herskowitz 1992). Stem cells by definition undergo asymmetric divisions to self-renew and produce new cell types (Morrison et al. 1997). Plants maintain a constant pool of stem cells to drive growth throughout their life spans (Heidstra & Sabatini 2014). These pools of stem cells are found in meristems, which are regions of plant tissue composed of a central stem cell niche and actively dividing stem cell progeny (Scheres 2007). This review focuses on the two primary meristems established during embryogenesis, the shoot apical meristem (SAM) and the root apical meristem (RAM). In addition to these primary meristems, other stem cells arise in the establishment of specialized cell lineages during differentiation. Examples include the outgrowth of lateral organs in the shoot and root and the formation of epidermal pores that mediate gas exchange (Nadeau & Sack 2003, Scheres 2007).
Plants exhibit remarkable developmental plasticity—a fundamental feature of the survival of a plant because it is rooted to the ground. Since plant cells are immobile, intercellular communication is vital for the establishment and maintenance of cell identity (Aida et al. 2004, van den Berg et al. 1997, Yadav et al. 2010). In plants, it is well established that positional cues regulate cell identity (Kidner et al. 2000, van den Berg et al. 1995). While there is evidence for lineage-based information encoded in daughter cells, such information can be overridden by extrinsic positional cues. Ablation and regeneration studies have demonstrated that most cell types in the meristem can adopt a new position-dependent fate (Efroni et al. 2016, van den Berg et al. 1997). Thus, most plant cells have the capacity to assume a new cell identity, although maintenance of identity also requires continuous signaling and regulatory feedback (Helariutta et al. 2000; Sabatini et al. 1999, 2003).
Specific combinations of transcriptional regulators can revert the identity of a differentiated cell into a stem cell in both plants and animals (Gallois et al. 2004, Huangfu et al. 2008, Smith et al. 2016). Cell identity is therefore established and maintained by myriad interactions of molecular regulators (Caggiano et al. 2017, Moreno-Risueno et al. 2015). This includes hormonal fluxes and mobile signals from neighboring cells that converge on transcription factor activity (Drisch & Stahl 2015, Galli & Gallavotti 2016). Moreover, the active interplay of transcription factors and other signaling molecules cues cell identity and ensures robust patterning of the plant body (Rahni et al. 2016). Evolution tinkers and reuses molecular regulators to generate diversity, such as the expansion of gene families that take on similar roles in different tissues (Dolzblasz et al. 2016, Zhou et al. 2015). Biology is thus full of repeating patterns that form the foundation of the nuanced information processing that is executed at the molecular level.
SPATIAL ORGANIZATION OF PLANT STEM CELLS
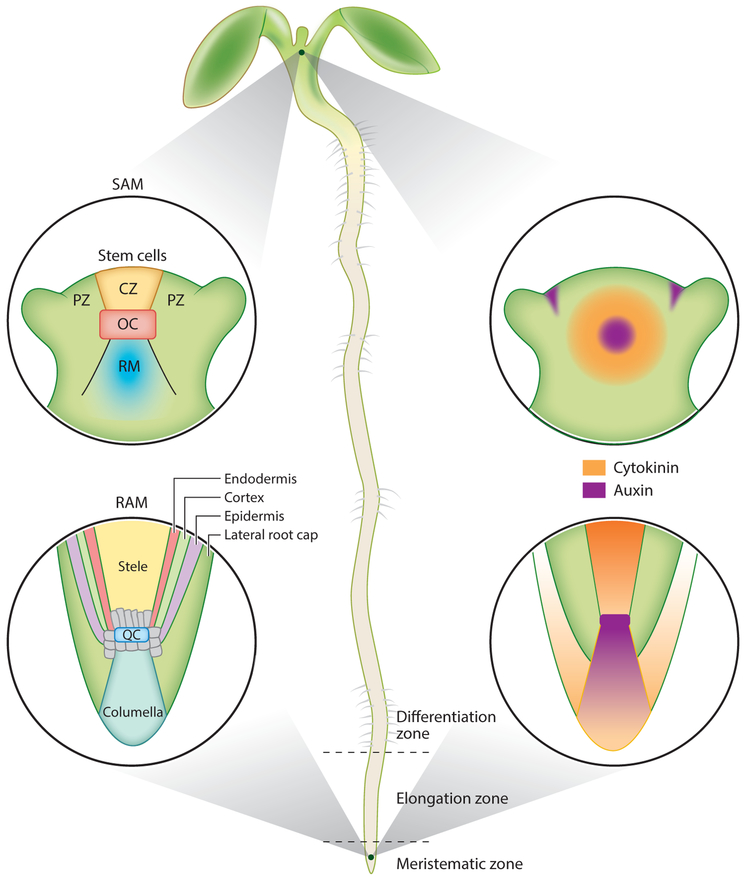
Aspects of regulation in the SAM and the RAM are almost mirror images and follow opposing gradients of differentiation along the primary axis of the plant body (Figure 1).
Figure 1.
An Arabidopsis seedling depicting the developmental organization of (moving counterclockwise from top left) the shoot apical meristem (SAM), root apical meristem (RAM), root developmental zones, and hormone domains of the RAM and SAM. (Top left) The SAM is positioned by the central organizing center (OC), which maintains the stem cells in the overlying central zone (CZ). Cells differentiate into either leaf or shoot cells as they are displaced into the peripheral zone (PZ) or the rib meristem (RM). (Bottom left) The quiescent center (QC) maintains the surrounding stem cells (gray) that give rise to all the tissues in the root. (Bottom center) Actively dividing cells are found in the meristematic zone. The transition to differentiation starts in the elongation zone when cells exit the mitotic cycle and enter a period of endoreduplication. Cells acquire their specialized features, such as root hairs, in the differentiation zone. (Right) The hormones auxin and cytokinin establish juxtaposed signaling domains that regulate division and differentiation in the SAM and RAM.
The SAM produces all the aerial organs of the plant, including the stem, leaves, and flowers. A small group of cells functions as the organizing center (OC), which maintains the stem cell population in the overlying layers of the central zone (CZ) at the very tip of the meristem (Figure 1, top left). Division of stem cells in the CZ can result in displacement of a daughter cell to the peripheral zone (PZ) at the edges of the meristem or to the rib meristem (RM) beneath the CZ. Daughter cell displacement leads to the competence to differentiate into leaf or flower primordia at the PZ or into various cell types of the elongating stem at the RM (Gaillochet & Lohmann 2015).
In the root, an organizing group of mitotically inactive quiescent center (QC) cells marks the position of the stem cell niche (van den Berg et al. 1997). Surrounding the QC are the stem cells that divide asymmetrically to generate all the tissue types in the root (Figure 1, bottom left). Because plant cells do not move in relation to each other, as the proximal stem cells divide, their daughters are displaced further from the QC. Each division displaces a daughter cell apically, resulting in the downward growth of the root (Cederholm et al. 2012). Each stem cell type generates a cell file of displaced daughters, which go through several additional rounds of division in the transit-amplifying region of the meristematic zone (MZ). In the elongation zone (EZ), cells exit the traditional cell cycle and enter a period of endoreduplication: multiple cycles of DNA replication accompanied by cell elongation, without an intervening cell division (Polyn et al. 2015). The border between the MZ and the EZ therefore defines the transition from cell division to differentiation (Dello Ioio et al. 2007). Finally, the differentiation zone (DZ) is where cells acquire the features of their differentiated fate (Figure 1, bottom center). As a result, the primary axis of the root serves as a linear timeline of development from stem cell to differentiated tissue.
HORMONES AND PATTERNING
Hormones play key roles in patterning both plants and animals. In plants, hormone biosynthesis, trafficking, and signaling establish gradients of accumulation and response that result in distinct cell fates. While several hormones play a role, the hormones auxin and cytokinin are essential for positioning and maintaining meristem activity (Figure 1, right). The balance between division and differentiation relies on a mutual steady state reached by the interactions between these two hormones (Schaller et al. 2015). A cell’s competence to respond to the two hormones guides the gradual acquisition of identity. What establishes this competence to respond?
In the root, auxin regulates stem cell divisions, and cytokinin triggers the transition to differentiation (Blilou et al. 2005, Dello Ioio et al. 2007). These roles are reversed in the shoot, where local auxin accumulation drives organogenesis in the PZ and cytokinins are crucial for cell proliferation in the CZ (Werner et al. 2003, Zhao et al. 2010). Each hormone stimulates a signaling pathway that culminates in the activation of transcription factors, which mediate global changes in gene expression. Auxin signaling has been tied to almost every aspect of plant growth and development. Briefly, an auxin signal promotes the destabilization of its signaling repressors, resulting in the activation of downstream transcription factors (Weijers & Wagner 2016). In contrast, cytokinin signaling relies on a two-component phosphorelay to phosphorylate and activate transcriptional response regulators (Hwang et al. 2012). Both hormones directly activate the expression of their own signaling repressors. They are thus regulated by negative feedback that can determine the duration and extent of response in a given cell.
While these frameworks have provided key insights into the molecular basis of hormone response, there remains much to be fleshed out. Genetic dissection of these signaling pathways is plagued by extensive redundancy among members of large families of transcriptional regulators (Keshishian & Rashotte 2015, Lokerse & Weijers 2009). Transcriptional reporters serve as the principal readout of hormone activity, but sifting through the web of signaling components that relay a specific signal remains a major challenge (Liao et al. 2015, Zürcher et al. 2013). Moreover, how these hormones mediate transcriptional repression remains an open question. Almost every hormone pathway has been connected to TOPLESS (TPL), a transcriptional cofactor related to the Groucho/Tup1 family of corepressors (Causier et al. 2012, Szemenyei et al. 2008). TPL recruitment is associated with a histone deacetylase (HDAC), suggesting that repression involves changes in chromatin state (Long et al. 2006). While several chromatin modifiers have been identified to play a role in hormonal regulation of cell fate (Efroni et al. 2013, Wu et al. 2015, Yang et al. 2015), the path from hormone perception to transcriptional response remains largely a black box.
As early as embryogenesis, complementary patterns of auxin and cytokinin signaling appear to define spatial boundaries, but the precise molecular mechanism has not been determined (Müller & Sheen 2008). Overlapping expression patterns of key cytokinin and auxin regulators have led to the identification of genetic circuits in which auxin signaling converges on cytokinin’s regulators of biosynthesis, accumulation, and signaling and vice versa (Chandler & Werr 2015). Through a combination of modeling and experimentation, DiMambro et al. (2017) recently demonstrated how an auxin minimum at the boundary between the MZ and the EZ in the root provides a positional cue for the switch to differentiation. Previous work demonstrated that cytokinin induces repression of auxin signaling in this domain, resulting in the inhibition of the PIN1 auxin transporter (Dello Ioio et al. 2008). New evidence indicates that cytokinin further enforces this minimum by inducing members of the GRETCHEN HAGEN 3 (GH3) gene family (Di Mambro et al. 2017). GH3 family members function as auxin conjugation enzymes that convert auxin to an inactive state (Staswick et al. 2005). Cytokinin-induced repression of auxin signaling and auxin inactivation by a GH3 result in an auxin minimum positioned where cells exit the MZ and transition to differentiation (Di Mambro et al. 2017). Di Mambro et al. (2017) argue that auxin thus patterns the root in a self-organizing manner, with cytokinin signaling as one of multiple inputs.
The remarkable ability of plants to regenerate is also dependent on auxin and cytokinin (Su & Zhang 2014). Given the right combination of hormones, organogenesis can be induced in explants prepared from almost any tissue. This chemical regulation has been recognized since the 1950s, shortly after the identification of cytokinins (Skoog & Miller 1957). To induce the formation of a callus, explants are grown on media containing a high ratio of auxin to cytokinin. Root induction requires only auxin, while shoot induction requires a high cytokinin-to-auxin ratio (Feldmann & Marks 1986). Thus, the balance of auxin and cytokinin directs organ formation both in vivo and in vitro.
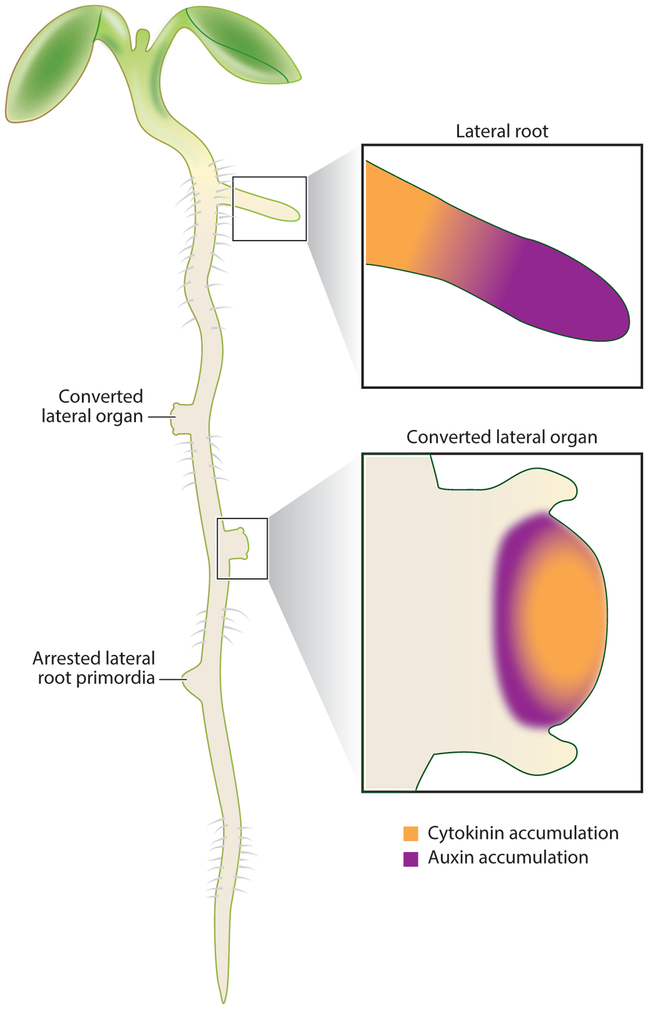
The plasticity of organ identity in plants was recently investigated using alternate auxin and cytokinin treatments to convert lateral root primordia into shoot meristems, without the formation of a dedifferentiated callus (Rosspopoff et al. 2017). Lateral root development has been extensively studied as a model for de novo organogenesis (Malamy & Benfey 1997). Cytokinin treatment resulted in direct conversion of lateral root primordia into a growing shoot in as few as 4 days (Figure 2). This conversion was possible only after the establishment of a stem cell niche in the lateral root primordia. During this stage, there is a characteristic auxin maximum at the emerging lateral root tip and no cytokinin signal. In the organ undergoing conversion, cytokinin accumulates in the central vasculature, and auxin is depleted from the root tip. As the shoot meristem develops, new auxin maxima form at the base of leaf primordia, and a broad cytokinin domain is established in the inner layers of the meristem. Notably, the hormone domains shift from that of root primordia into domains characteristic of a SAM prior to the expression of SAM-specific regulators.
Figure 2.
Hormonal treatment can convert lateral root primordia into shoot primordia and switch the characteristic auxin-cytokinin gradient of a root within a short developmental window. Presented is a schematic of how lateral root primordia responded to hormonal treatments by Rosspopoff et al. (2017) at different developmental stages. Early lateral root primordia are aborted. In contrast, intermediate-stage lateral root primordia transition to accumulate cytokinin in the stem cell niche and grow to resemble a shoot apical meristem. Past a certain stage, lateral root primordia maintain their characteristic auxin-cytokinin domains and emerge as lateral roots.
In a recent study of root regeneration, a similar relationship was observed between the formation of complementary auxin and cytokinin domains and the establishment of cell identity in the regenerated root (Efroni et al. 2016). Following removal of the root tip, including the stem cell niche, Efroni et al. (2016) employed lineage tracing and single-cell RNA-seq to follow the gene expression of specific cells during the reestablishment of the stem cell niche. They found that cells from multiple tissues acquired stem cell identity to regenerate the excised root tip. Moreover, manipulation of auxin or cytokinin during regeneration could alter patterning. The authors argue that, similar to embryogenesis, auxin and cytokinin form complementary signaling domains during regeneration that provide spatial cues for positioning the stem cell niche.
REGULATORY FEEDBACK IN THE SHOOT AND ROOT STEM CELL NICHES
Members of the WUSCHEL-RELATED HOMEOBOX (WOX) gene family are required non–cell autonomously to maintain stem cell homeostasis. The best-characterized family members are WUSCHEL (WUS) in the shoot and WOX5 in the root. The onset of WUS expression in the shoot and the onset of WOX5 expression in the root are early markers of stem cell niche establishment. Their expression is restricted to only a few cells, which function as organizers at the center of each niche. Loss of either transcription factor results in the collapse of its stem cell niche (Sarkar et al. 2007). To ensure proper patterning and development, restriction of WUS and WOX5 expression to the organizing cells is tightly regulated by layered feedback mechanisms.
In the SAM, WUS expression is positioned by a gradient of cytokinin (Chickarmane et al. 2012). An activating enzyme of cytokinin is specifically expressed in the uppermost stem cell layer, permitting the diffusion of cytokinin to the lower layers of the SAM (Kurakawa et al. 2007). The underlying WUS domain in the shoot OC contains high levels of a cytokinin receptor, sensitizing the cells to cytokinin and resulting in high levels of cytokinin signaling (Gordon et al. 2009). Cytokinin promotes WUS expression and WUS promotes cytokinin signaling through repression of negative regulators of cytokinin signaling (Leibfried et al. 2005, Meng et al. 2017). Thus, a positive feedback loop between WUS and cytokinin ensures that WUS expression leads to more WUS and high levels of cytokinin signaling (Chickarmane et al. 2012).
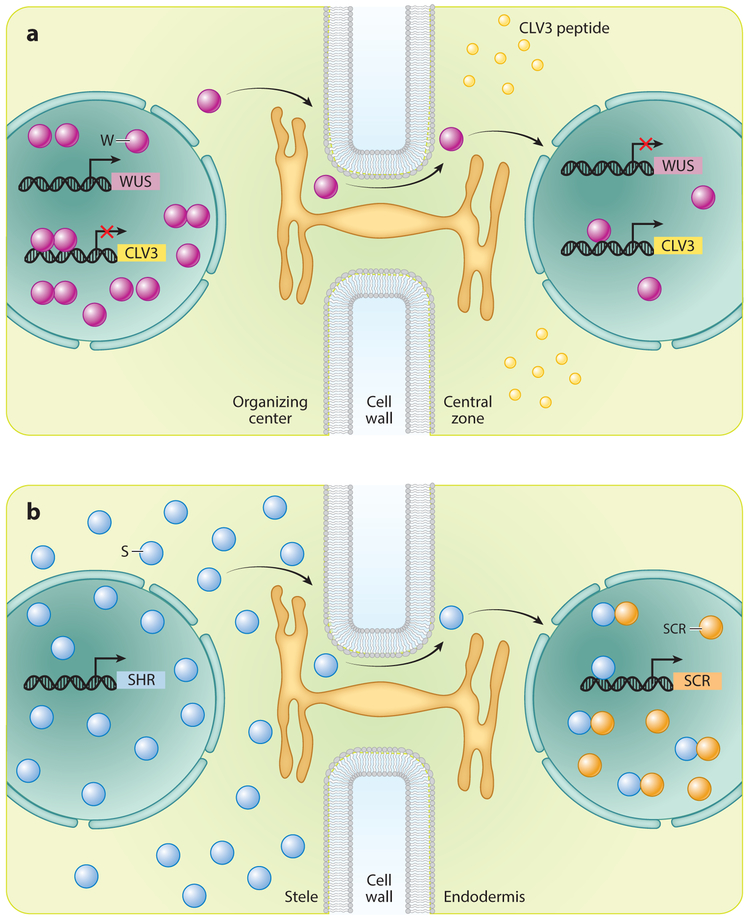
WUS expression in the OC is critical for stem cell fate in the overlying cells of the CZ (Laux et al. 1996, Yadav et al. 2010). Only cells in the OC express WUS, but the WUS protein migrates to the neighboring cell layers of the CZ through plasmodesmata (see sidebar titled Intercellular Trafficking Through Plasmodesmata) (Daum et al. 2014, Yadav et al. 2011). In CZ stem cells, the mobile WUS transcription factor activates expression of its own negative regulator, a small secreted peptide termed CLAVATA3 (CLV3) (Yadav et al. 2011). Recent analysis of WUS binding to cis elements in the regulatory region of CLV3 indicated that the same cis elements mediate repression and activation, depending on the concentration of WUS (Perales et al. 2016). WUS binds to these elements as monomers at low concentrations and as dimers at higher concentrations. These observations provide a plausible explanation for the ability of WUS to activate CLV3 only in the CZ and not in the OC where WUS is expressed. When WUS migrates from its source in the OC to neighboring stem cells, it accumulates at lower levels than at its origin. This lower accumulation results in the activation of CLV3 transcription, limiting the expression domain of WUS (Rodriguez et al. 2016). Conversely, cells transcribing WUS accumulate high levels of WUS protein, leading to repression of CLV3 transcription as well as to the self-sustaining expression of WUS solely in the OC (Figure 3).
INTERCELLULAR TRAFFICKING THROUGH PLASMODESMATA.
Approximately 30% of transcription factors in Arabidopsis are mobile, and there are examples of both transcription factors and small RNAs moving through plasmodesmata to regulate development in plants (Hantke et al. 1995, Lucas et al. 1995, Nakajima et al. 2001, Sessions et al. 2000, Vatén et al. 2011). Passage of signaling molecules can occur through the cell wall matrix or directly through cytoplasmic connections termed plasmodesmata. Plasmodesmata are plasma membrane–lined channels containing a thread of endoplasmic reticulum. Relative to the analogous structure in animals termed gap junctions, plasmodesmata are massive, at a size exclusion limit of 10 kDa and a diameter of 50–60 nm (Kim et al. 2002, Robards & Lucas 1990). There is also evidence that plasmodesmata are selective, allowing passage of some molecules and not others (Hantke et al. 1995, Lucas et al. 1995, Sessions et al. 2000). Recently, a study of how plasmodesmata change during cell maturation showed that plasmodesmata in recently divided cells have almost no space between their plasma membrane and endoplasmic reticulum connections (Nicolas et al. 2017). As cells elongate during maturity, the gap between the plasma membrane and endoplasmic reticulum widens, potentially to accommodate larger molecules (Nicolas et al. 2017).
Figure 3.
Transcription factor movement through plasmodesmata. (a) WUSCHEL (WUS) is expressed in the organizing center, and WUS protein (W) moves into central zone stem cells. In those cells, W activates the expression of CLAVATA3 (CLV3), a small signaling peptide that prevents WUS expression outside the organizing center. New evidence suggests that WUS dimers repress CLV3 expression in the organizing center. (b) SHORTROOT (SHR) is expressed in the stele, where it localizes to both the nucleus and cytoplasm. SHR protein (S) moves into the endodermis, where it activates the expression of SCARECROW (SCR). Physical interaction with SCR sequesters S to the nucleus, preventing further movement.
Similar to WUS expression in the OC, WOX5 expression is confined to the QC cells in the center of the root stem cell niche (Fiers et al. 2005, Stahl et al. 2009). Yet, WOX5 is also required for maintenance of the surrounding stem cells, particularly for the stem cells of the columella (Sarkar et al. 2007). The columella serves as a gravity-sensing and protective tissue at the very tip of the root. Columella cells rapidly differentiate and originate from a single stem cell layer positioned immediately beneath the QC. Expression of WOX5 driven by the WUS promoter demonstrated that WOX5 is capable of intercellular movement in the SAM (Daum et al. 2014). Recent evidence indicates endogenous WOX5 is mobile in the RAM (Pi et al. 2015). There, WOX5 migrates to the columella stem cells to repress differentiation (Pi et al. 2015).
Similar to the relationship between WUS and cytokinin, WOX5 also has numerous ties with auxin signaling. The auxin gradient in the root meristem peaks in the QC, where WOX5 is specifically expressed (Sabatini et al. 1999). This peak of auxin activity is sustained and focused by auxin regulation of the PIN transporters and by local auxin biosynthesis (Blilou et al. 2005, Ding & Friml 2010). The regulatory relationship between WOX5 and auxin also relies on feedback from each other to maintain robust expression and activity in the QC (Tian et al. 2014). Outside the QC, specific auxin signaling repressors and transcription factors repress WOX5 expression (Ding & Friml 2010).
Chromatin regulation is also emerging as a mechanism for restricting expression of WUS and WOX5 (Ikeuchi et al. 2015). SPLAYED (SYD), an SNF2 chromatin remodeling complex, regulates the transcriptional expression of WUS in the SAM (Kwon et al. 2005). REPRESSOR OF WUSCHEL1 (ROW1) was originally identified for its role in confining WUS expression to the OC of the SAM (Han et al. 2008). Recently, ROW1 was also demonstrated to repress WOX5 expression in the proximal meristem. Loss of ROW1 results in the expansion of WOX5 expression into the proximal meristem, and misexpression of ROW1 in the QC extinguishes WOX5 expression (Zhang et al. 2015). ROW1 represses WOX5 expression by binding to H3K4me3 chromatin marks in the WOX5 promoter. The WOX5 promoter is characterized by these H3K4me3 chromatin marks, which remain unchanged regardless of promoter activity (Aichinger et al. 2011, Zhang et al. 2015). Moreover, both WUS and WOX5 interact with TPL corepressors to mediate repression of their targets (Kieffer et al. 2006, Pi et al. 2015). Many examples of transcriptional repression implicate chromatin modifiers, but the molecular mechanisms remain elusive.
WUS and WOX5 are mostly interchangeable between the root and shoot, indicating that they play similar roles in stem cell maintenance (Sarkar et al. 2007). A recent study tested how well other WOX family members could rescue the wus mutant phenotype (Dolzblasz et al. 2016). Only a few of the closely related family members, including WOX5, were capable of various degrees of rescue. Notably, WUS is capable of complete rescue of the wox5 mutant (Sarkar et al. 2007). Thus, while WUS and WOX5 may differ in some aspects of their functionality, they overlap in many others. HAIRYMERISTEM (HAM) functions as a WUS cofactor in regulating a number of WUS target genes (Zhou et al. 2015). Both the WOX and HAM families are highly conserved and include multiple members whose expression domains overlap, suggesting that the interaction between different pairs of WOX and HAM family members plays a key role in how WOX family members maintain different stem cell niches.
FEEDBACK BETWEEN MOBILE TRANSCRIPTION FACTORS AND miRNAS
Small RNAs have an important role in transcriptional feedback. Two miRNAs that together form a hairpin structure, miR165 and miR166, have an important role in regulating dividing and differentiating tissues (Aukerman 2003, Carlsbecker et al. 2010; reviewed in Couzigou & Combier 2016). In the SAM, the RAM, and other meristems, miR165 and miR166 promote differentiation by repressing members of the HD-ZIP III family of transcription factors (Bao et al. 2004, Williams 2005). These factors include PHAVOLUTA, PHABULOSA, and REVOLUTA, which have important roles in maintaining stem cells in both the SAM and RAM (Emery et al. 2003, Hawker 2004, McConnell et al. 2001, Prigge 2005). In each of these settings, different argonaute (AGO) proteins act in meristematic cells to sequester miR165 and miR166 away from targets or to promote differentiation by relieving repression of HD-ZIP transcription factors (Ji et al. 2011, Kidner & Martienssen 2004, McHale 2004, Zhang & Zhang 2012; reviewed in Zhong & Ye 2007). While the key players in this pathway seem to be recycled in meristematic tissues, the mechanisms by which these pathways intersect with those described above to define stem cells are still largely unknown.
SMALL PEPTIDES IN MERISTEM DEVELOPMENT
Similar to the case for animal development, small peptides such as those of the CLAVATA3/ESR-related (CLE) family are important for signaling in the development and differentiation of plants (Matsubayashi 2014). While the relationship between WUS and CLV3 is well established, much less is understood about the downstream mechanism of CLV3 signaling (Clark et al. 1995, Fletcher et al. 1999, Kondo et al. 2006). In a wild-type SAM, CLV3 represses WUS, allowing cells in the PZ to differentiate (Brand et al. 2000, Laux et al. 1996, Mayer et al. 1998, Schoof et al. 2000). Loss of CLV3 results in enlarged meristems due to expansion of the WUS expression domain (Brand et al. 2000, Schoof et al. 2000). Careful genetic, ectopic expression, and biochemical experiments have demonstrated that CLV3 peptide diffuses to the peripheral meristem, where it activates a signaling cascade through CLAVATA1 (CLV1), a leucine-rich receptor kinase (Clark et al. 1997, Lenhard & Laux 2003, Nimchuk et al. 2011, Ogawa et al. 2008, Rojo 2002). While WUS is a transcriptional target of CLV3 signaling, the molecular events leading to transcriptional repression remain elusive. Multiple receptor complexes are involved in CLV3 signaling, and there is mounting evidence that each complex plays a unique role in stem cell regulation (DeYoung & Clark 2008, DeYoung et al. 2005, Nimchuk 2017).
In the root, a small peptide related to CLV3, CLE40, represses WOX5 expression in stem cells surrounding the QC (Stahl et al. 2009). Expression of CLE40 in the SAM can rescue loss of CLV3 signaling in the shoot, indicating these two peptides are functionally equivalent and differ mainly in expression pattern (Hobe et al. 2003). Unlike CLV3, which is expressed in shoot stem cells, CLE40 is expressed in differentiated columella cells and binds to receptors that are expressed in the overlying columella stem cells (Stahl et al. 2013). Loss of CLE40 results in additional layers of columella stem cells (Stahl et al. 2009). In the root, the CLV1 receptor interacts with ARABIDOPSIS CRINKLY 4 (ACR4), another receptor-like kinase, to detect CLE40 and initiate downstream signaling (De Smet et al. 2008; Stahl et al. 2009, 2013). There is also evidence that other receptors involved in CLV3 sensing in the SAM can detect CLE peptides in the RAM (Kinoshita et al. 2010, Muller et al. 2008, Shimizu et al. 2015). Other CLE family peptides have important roles in root development, such as regulating differentiation in root vascular tissue and in several stages of lateral root development (Depuydt et al. 2013; Fernandez et al. 2013, 2015; Kumpf et al. 2013; Meng et al. 2012).
SMALL PEPTIDES IN DIFFERENTIATION
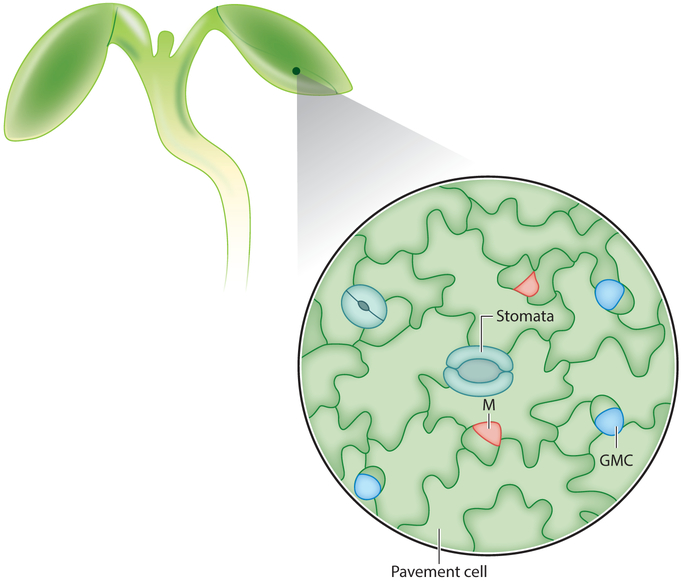
Outside of the meristem, divisions of specialized stem cell lineages are also regulated by small peptides. Members of the epidermal patterning factor (EPF) family of peptides play important roles in leaf patterning (reviewed in Katsir et al. 2011). Stomata are pores in the epidermis that mediate gas exchange (Pillitteri & Torii 2012). They arise from a defined series of asymmetric divisions and one symmetric division that form guard cells, the two cells that regulate stomatal opening (Figure 4). The stereotypical division pattern begins with asymmetric division of a cell termed the meristemoid mother cell, which divides to give rise to a meristemoid cell and a sister cell type. The two daughter cells have different sizes, with the sister cell being much larger than the meristemoid cell. Both progeny continue to asymmetrically divide in a series of amplifying divisions, with cells regenerated as meristemoids transitioning into a fate termed guard mother cells. The guard mother cells finally symmetrically divide to make rounded guard cells, which form a pore. Once the asymmetric divisions begin in one cell, the surrounding cells become incapable of forming stomata and instead differentiate into puzzle piece–shaped pavement cells. This process results in a defined pattern whereby each stoma is surrounded by at least one pavement cell. Although auxin distribution between mother and daughter cells has been implicated in the transition to the final symmetric division that forms the guard cells (Le et al. 2014, Pillitteri & Torii 2012), it is largely unknown how hormone signaling is integrated with other factors to regulate stomatal spacing (Saibo et al. 2003).
Figure 4.
Leaf epidermal cells in various stages of stomatal development. A meristemoid mother cell undergoes the first asymmetric division to produce a meristemoid (M), which continues to divide asymmetrically until it converts into a guard mother cell (GMC). The GMC divides symmetrically to form the two guard cells of the stomatal pore. Each stoma is separated by puzzle piece–shaped pavement cells.
EPF1 and EPF2 have some overlapping effects in promoting division and restricting differentiation at different stages along the path to stomata or pavement cell development (Hara et al. 2009, Hunt & Gray 2009, Pillitteri & Torii 2012). EPF1 inhibits the transition to guard mother cells. EPF2 inhibits the initial asymmetric cell division of meristemoid mother cells and has an important role in ensuring the correct spacing of guard cells (Hara et al. 2009, Hunt & Gray 2009). Overexpression of either EPF1 or EPF2 results in fewer stomatal clusters (Hara et al. 2007, 2009; Hunt & Gray 2009; Pillitteri & Torii 2012). These peptides activate members of the ERECTA (ER) family of receptor kinases and the coreceptor TOO MANY MOUTHS (TMM). ER and TMM can form complexes with the somatic embryogenesis receptor kinase (SERK) family of coreceptors to activate downstream signaling through the mitogen-activated protein kinase (MAPK) pathway (Meng et al. 2015, Pillitteri & Torii 2012). An EPF-like (EPFL) peptide termed STOMAGEN promotes stomatal development by inducing guard cell differentiation. One intriguing question is how such related peptides can exert such different effects. It was recently shown that STOMAGEN also acts through ER family receptors competing with EPF2 for binding to ER (Lee et al. 2015). By contrast, STOMAGEN does not activate MAPK signaling and may actually prevent it (Lee et al. 2015). These findings show how finely tuned this system is, with both inhibiting and activating signals spatially organizing the leaf epidermis. Mechanistically, it is still unknown whether these changes in sensitivity are due to changes in receptor sensitivity or changes in peptide concentration.
EPFL2, another EPFL peptide, has an important role in other aspects of leaf morphogenesis and has an established connection with auxin. Arabidopsis leaves have serrations, the sawlike projections or teeth found at the edges of leaves. Serrations are initiated from small primordia along the leaf margin (Tameshige et al. 2016). In epfl2 mutants, serrations are missing, and there is an expansion of tooth primordia. Auxin is normally restricted to the tips of growing teeth and is important for tooth development. The auxin pathway intersects with EPFL signaling, as auxin has a mutually repressive relationship with EPL2 and ER (Tameshige et al. 2016). These studies demonstrate the importance of the intersection of small peptide signaling and hormonal regulation in defining organ boundaries.
Small peptides also cue the acquisition of specialized features during differentiation. Two recently discovered peptides are required for the proper formation of the root endodermis (Doblas et al. 2017, Nakayama et al. 2017). One role of the endodermis is to provide for selective uptake of solutes. Water and dissolved compounds can pass between cells of the epidermis and cortex. The endodermis generates a waterproof seal made of lignin termed the Casparian strip, which prevents unrestricted flow to the vascular tissue and allows membrane channels to selectively uptake ions (Geldner 2013). The small signaling peptides CASPARIAN STRIP INTEGRITY FACTOR 1 (CIF1) and CIF2 are required for proper sealing of this structure. Mutations in SCHENGEN2 (SGN2), the enzyme required for CIF1 and CIF2 activity, and mutations in both cif1 and cif2 result in defective Casparian strips (Doblas et al. 2017, Nakayama et al. 2017). These peptides act on the endodermis but are derived from the neighboring cells of the vascular tissue. Both genetic and biochemical evidence identified the receptor downstream of CIF1 and CIF2 as SCHENGEN3 (SGN3), a receptor-like kinase with enriched expression in the endodermis (Pfister et al. 2014). The sgn3 mutant also resembles the cif1cif2 double mutant (Doblas et al. 2017, Nakayama et al. 2017, Pfister et al. 2014). Both the CIF peptides and SHORTROOT (SHR) protein (described below) are mobile signals derived from the vasculature, demonstrating the importance of signaling from neighboring cell layers for proper differentiation. This mechanism is similar to induction in animal cell development, such as the well-studied examples of mesoderm induction and EPIDERMAL GROWTH FACTOR signaling in Drosophila eye development (Kimelman 2006, Schweitzer & Shilo 1997).
MOBILE TRANSCRIPTION FACTORS REGULATE CELL FATE
The sustained accumulation of auxin induces transcription of PLETHORA (PLT) transcription factors, a family of mobile transcription factors (Mähönen et al. 2014). PLT expression is highest in stem cells and gradually decreases in the shootward direction (Aida et al. 2004, Galinha et al. 2007). PLTs regulate QC and stem cell maintenance and differentiation in what is thought to be a dose-dependent manner (Galinha et al. 2007). Another small family of peptides termed root growth factors (RGFs) maintain the root meristem by regulating the gradient of PLT expression (Matsuzaki et al. 2010, Zhou et al. 2010). The enzyme that modifies the RGFs into their mature form is also regulated by auxin, suggesting convergence of hormonal and small peptide regulation of the PLT gradient (Zhou et al. 2010). Only recently has the clade of receptors for this pathway been identified, highlighting again the complexity of studying small peptides (Ou et al. 2016, Shinohara et al. 2016, Song et al. 2016).
Perhaps the best-characterized mobile transcription factor in Arabidopsis is SHR, which plays multiple roles in root patterning. SHR is a member of the GRAS family of transcription factors, a plant-specific family that regulates various aspects of plant growth and development (Hirsch & Oldroyd 2009). SHR is expressed and translated in the central vascular tissue and moves into the adjacent cell layer (Figure 3b) (Helariutta et al. 2000, Nakajima et al. 2001). This layer includes the root endodermis, the stem cell progenitors of the endodermis, and the QC. As implied by the name, shr mutants have short roots (Benfey et al. 1993), largely due to the QC and stem cell defects that occur in the absence of SHR protein (Helariutta et al. 2000). The primary role of SHR movement, however, is the specification and differentiation of the endodermis. Without SHR, there is no endodermis.
Without movement of SHR into the stem cell progenitors, asymmetric division of the cortex and endodermis stem cell is compromised (Helariutta et al. 2000). A decade of research on this division has uncovered the integral factors that contribute to the timing and decision to divide. Transcription factors, including SHR, can induce cell cycle regulators and phosphorylation of the plant homolog of RETINOBLASTOMA (RB) protein (Cruz-Ramírez et al. 2012, Di Laurenzio et al. 1996, Helariutta et al. 2000, Sozzani et al. 2010). One major question is how auxin signaling is integrated in this process. In modeling of this division, one plausible explanation is that the concentration of auxin at the niche triggers cells with the right levels of transcription factors and RB to divide (Cruz-Ramírez et al. 2012).
Regulating transcription factor movement is also important for robust patterning. SHR expression in the vascular tissue is both nuclear and cytoplasmic, while in the neighboring endodermis SHR protein is mostly nuclear. Available evidence indicates that SHR moves through plasmo-desmata and, unexpectedly, requires nuclear localization for movement. (Gallagher et al. 2004, Vatén et al. 2011). Once in the endodermis, SHR is sequestered to the nucleus. There, SHR induces several transcriptional coregulators that physically interact with SHR and keep it in the nucleus. These cofactors include SCARECROW (SCR), another member of the GRAS family, and several members of the INDETERMINATE DOMAIN (IDD) family of zinc-finger transcription factors, commonly referred to as the BIRDs (Long et al. 2015). Loss of SCR or members of the BIRD family results in the expansion of SHR mobility (Cui et al. 2007, Long et al. 2015). SHR activity in cells external to the endodermis generates additional layers of cortex, the tissue that separates the inner endodermis from the outer epidermis (Cui et al. 2007, Wu et al. 2014). That additional SHR movement results in cortex rather than endodermis formation suggests that additional stele-derived factors are required for endodermis identity.
The transcription factor SHOOT MERISTEMLESS (STM) is mobile and required for SAM maintenance in Arabidopsis (Long et al. 1996). STM’s maize homolog, KNOTTED1 (KN1), was the first mobile transcription factor discovered (Jackson 2002, Jackson et al. 1994, Lucas et al. 1995). In contrast to STM, KN1 has distinct domains of expression and protein localization (Jackson 2002). Both maize KN1 and Arabidopsis STM require their mobility to regulate SAM maintenance (Balkunde et al. 2017, Xu et al. 2011).
While some mobile transcription factors, such as STM, are mobile across large evolutionary distances, others are not. The transcription factor MUTE, which plays a key role in differentiation of stomata (Pillitteri & Torii 2012, Pillitteri et al. 2007), was recently shown to be mobile in the grass species Brachypodium (Raissig et al. 2017). MUTE is a key regulator of cell divisions in leaf patterning, but it is not mobile in Arabidopsis (Pillitteri et al. 2007, 2008). The mobility of MUTE in Brachypodium plausibly explains the presence of additional supporting cells in stomata, which Arabidopsis lacks (Raissig et al. 2017). Modified mobility could explain the patterning differences seen between dicots and monocots.
DISCUSSION
Developmental patterning in time and space emerges from dynamic interactions at the cellular and molecular levels. In plants, intercellular communication is vital for ensuring robust patterning throughout development. Plants utilize recurring modules of intercellular interactions to communicate position and identity. Hormones, mobile transcription factors, small RNAs, and peptides form complementary positional cues to establish and reinforce cell identity. In many cases, duplication and evolution of transcriptional regulators not only confer robustness through redundancy but also generate diversity and specialization. Spatiotemporal development thus relies on the contribution of individual cells and their competence to respond to specific signaling cues.
Signaling between neighboring cells is dynamic and involves multiple feedback loops. This back and forth between cells is essentially an ongoing call and response to both specify and reinforce cell fate. Non–cell autonomous gene regulation by mobile signals provides an elegant solution for robust patterning. The path from signal to gene regulation, however, remains largely obscure. Hormones have long been recognized as chemical messengers, but how the same hormone delivers different messages continues to be an active area of research. The molecular underpinnings of small peptide perception and signaling are complicated by peptide promiscuity and by the range of receptor complexes that propagate the signal. Finally, the mobility of miRNAs and transcription factors suggests that they have different effects in their origin and their terminus. How is spatiotemporal specificity achieved by these factors?
Cell type–specific expression maps and single-cell sequencing have revealed key markers of identity (Birnbaum et al. 2005, Brady et al. 2007, Efroni & Birnbaum 2016, Efroni et al. 2016, Li et al. 2016, Slane et al. 2014). As precision in these technologies continues to improve, it will be possible to track the gene expression program of a cell as it transitions to a new state (Efroni et al. 2016). A number of transcription factors in the root meristem have a graded expression, which could establish a dose-dependent progression of differentiation (Wendrich et al. 2017). This graded expression could also be distance dependent, communicating position within a cell file. In the case of mobile transcription factors, protein levels can differ between the cells in which they are actively expressed and the cells into which they move. Recent advances in in vivo microscopy have enabled the measurement of diffusion and protein binding dynamics of SHR as it moves into the endodermis stem cell to trigger asymmetric cell division (Clark et al. 2016, Long et al. 2017). As described above, SHR plays numerous roles in root development and is active in several cell types. The big outstanding question is how the same transcription factor differs in form or activity across cell types. As our questions continue to become more specific, we will gain new mechanistic insights into the fundamentals of cell identity.
ACKNOWLEDGMENTS
Support for this work is provided by the NIH (R01-GM043778) and by the Howard Hughes Medical Institute. Edith Pierre-Jerome is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Aichinger E, Villar CBR, Di Mambro R, Sabatini S, Köhler C. 2011. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23(3):1047–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, et al. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119(1):109–20 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ. 2003. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell Online 15(11):2730–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkunde R, Kitagawa M, Xu XM, Wang J, Jackson D. 2017. SHOOT MERISTEMLESS trafficking controls axillary meristem formation, meristem size and organ boundaries in Arabidopsis. Plant J. 90(3):435–46. [DOI] [PubMed] [Google Scholar]
- Bao N, Lye K-W, Barton MK. 2004. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7(5):653–62 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA. 1993. Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119(1):57–70 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, et al. 2005. Cell type–specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat. Methods 2(8):615–19 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, et al. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433(7021):39–44 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, et al. 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318(5851):801–6 [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. 2000. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289(5479):617–19 [DOI] [PubMed] [Google Scholar]
- Caggiano MP, Yu X, Bhatia N, Larsson A, Ram H, et al. 2017. Cell type boundaries organize plant development. eLife 6:e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A, Lee J-Y, Roberts CJ, Dettmer J, Lehesranta S, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465(7296):316–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158(1):423–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederholm HM, Iyer-Pascuzzi AS, Benfey PN. 2012. Patterning the primary root in Arabidopsis. Wiley Inter-discip. Rev. Dev. Biol 1(5):675–91 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Werr W 2015. Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci. 20(5):291–300 [DOI] [PubMed] [Google Scholar]
- Chickarmane VS, Gordon SP, Tarr PT, Heisler MG, Meyerowitz EM. 2012. Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. PNAS 109(10):4002–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Hinde E, Winter CM, Fisher AP, Crosti G. 2016. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5:e14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM. 1995. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121:2057–67 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89(4):575–85 [DOI] [PubMed] [Google Scholar]
- Couzigou J-M, Combier J-P. 2016. Plant microRNAs: key regulators of root architecture and biotic interactions. New Phytol. 212(1):22–35 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, et al. 2012. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150(5):1002–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, et al. 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316(5823):421–25 [DOI] [PubMed] [Google Scholar]
- Daum G, Medzihradszky A, Suzaki T, Lohmann JU. 2014. A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. PNAS 111(40):14619–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, et al. 2008. Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322(5901):594–97 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, et al. 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol 17(8):678–82 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, et al. 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322(5906):1380–84 [DOI] [PubMed] [Google Scholar]
- Depuydt S, Rodriguez-Villalon A, Santuari L, Wyser-Rmili C, Ragni L, Hardtke CS. 2013. Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. PNAS 110(17):7074–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE. 2005. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45(1):1–16. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Clark SE. 2008. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 180(2):895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, et al. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86(3):423–33 [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. PNAS 114(36):E7641–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. PNAS 107(26):12046–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, et al. 2017. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355(6322):280–84 [DOI] [PubMed] [Google Scholar]
- Dolzblasz A, Nardmann J, Clerici E, Causier B, van der Graaff E, et al. 2016. Stem cell regulation by Arabidopsis WOX genes. Mol. Plant 9(7):1–41 [DOI] [PubMed] [Google Scholar]
- Drisch RC, Stahl Y. 2015. Function and regulation of transcription factors involved in root apical meristem and stem cell maintenance. Front. Plant Sci 6:505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Birnbaum KD. 2016. The potential of single-cell profiling in plants. Genome Biol. 17(1):1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han S-K, Kim HJ, Wu M-F, Steiner E, et al. 2013. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24(4):438–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip P-L, Rahni R, et al. 2016. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165(7):1721–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, et al. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol 13(20):1768–74 [DOI] [PubMed] [Google Scholar]
- Feldmann KA, Marks MD. 1986. Rapid and efficient regeneration of plants from explants of Arabidopsis thaliana. Plant Sci. 47(1):63–69 [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, et al. 2013. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol. 161(2):954–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, et al. 2015. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. J. Exp. Bot 66(17):5245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, et al. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17(9):2542–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. 1999. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283(5409):1911–14 [DOI] [PubMed] [Google Scholar]
- Gaillochet C, Lohmann JU. 2015. The never-ending story: from pluripotency to plant developmental plasticity. Development 142(13):2237–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, et al. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449(7165):1053–57 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. 2004. Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol 14(20):1847–51 [DOI] [PubMed] [Google Scholar]
- Galli M, Gallavotti A. 2016. Expanding the regulatory network for meristem size in plants. Trends Genet. 32(6):372–83 [DOI] [PubMed] [Google Scholar]
- Gallois J-L, Nora FR, Mizukami Y, Sablowski R. 2004. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18(4):375–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N 2013. The endodermis. Annu. Rev. Plant Biol 64:531–58 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. 2009. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. PNAS 106(38):16529–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li Q, Zhu YX. 2008. Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell 20(6):1482–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke SS, Carpenter R, Coen ES. 1995. Expression of floricaula in single cell layers of periclinal chimeras activates downstream homeotic genes in all layers of floral meristems. Development 121(1):27–35 [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. 2007. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21(14):1720–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, et al. 2009. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 50(6):1019–31 [DOI] [PubMed] [Google Scholar]
- Hawker NP. 2004. Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135(4):2261–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S. 2014. Plant and animal stem cells: similar yet different. Nature Rev. Mol. Cell Biol 15(5):301–12 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, et al. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101(5):555–67 [DOI] [PubMed] [Google Scholar]
- Hirsch S, Oldroyd GED. 2009. GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav 4(8):698–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe M, Müller R, Grünewald M, Brand U, Simon RD. 2003. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol 213(8):371–81 [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Herskowitz I. 1992. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell 68(2):237–55 [DOI] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, et al. 2008. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol 26(11):1269–75 [DOI] [PubMed] [Google Scholar]
- Hunt L, Gray JE. 2009. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol 19(10):864–69 [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. 2012. Cytokinin signaling networks. Annu. Rev. Plant Biol 63:353–80 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Sugimoto K. 2015. Control of plant cell differentiation by histone modification and DNA methylation. Curr. Opin. Plant Biol 28:60–67 [DOI] [PubMed] [Google Scholar]
- Jackson D 2002. Double labeling of KNOTTED1 mRNA and protein reveals multiple potential sites of protein trafficking in the shoot apex. Plant Physiol. 129(4):1423–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. 1994. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120(2):405–13 [Google Scholar]
- Ji L, Liu X, Yan J, Wang W, Yumul RE, et al. 2011. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLOS Genet. 7(3):e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Davies KA, Bergmann DC, Laux T. 2011. Peptide signaling in plant development. Curr. Biol 21(9):R356–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshishian EA, Rashotte AM. 2015. Plant cytokinin signalling. Essays Biochem. 58(0):13–27 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. 2004. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428:81–84 [DOI] [PubMed] [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L. 2000. Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211(2):191–99 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, et al. 2006. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18(3):560–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Hempel FD, Sha K, Pfluger J, Zambryski PC. 2002. Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129(5):1261–72 [DOI] [PubMed] [Google Scholar]
- Kimelman D 2006. Mesoderm induction: from caps to chips. Nat. Rev. Genet 7(5):360–72 [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, et al. 2010. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137(24):4327–27 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, et al. 2006. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313(5788):845–48 [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi C-L, Larrieu A, Stø IM, Butenko MA, et al. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. PNAS 110(13):5235–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, et al. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445(7128):652–55 [DOI] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D. 2005. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19(8):992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jurgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122(1):87–96 [DOI] [PubMed] [Google Scholar]
- Le J, Liu X-G, Yang K-Z, Chen X-L, Zou J-J, et al. 2014. Auxin transport and activity regulate stomatal patterning and development. Nat. Commun 5:3090. [DOI] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin Y-CL, Putarjunan A, et al. 2015. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522(7557):439–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JPC, Busch W, Stehling S, Kehle A, et al. 2005. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438(7071):1172–75 [DOI] [PubMed] [Google Scholar]
- Lenhard M, Laux T. 2003. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130(14):3163–73 [DOI] [PubMed] [Google Scholar]
- Li S, Yamada M, Han X, Ohler U, Benfey PN. 2016. High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell 39(4):508–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C-Y, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12(3):207–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokerse AS, Weijers D. 2009. Auxin enters the matrix—assembly of response machineries for specific outputs. Curr. Opin. Plant Biol 12(5):520–26 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379(6560):66–69 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312(5779):1520–23 [DOI] [PubMed] [Google Scholar]
- Long Y, Smet W, Cruz-Ramírez A, Castelijns B, de Jonge W, et al. 2015. Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27(4):1185–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, Stahl Y, Weidtkamp-Peters S, Postma M, Zhou W, et al. 2017. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548(7665):97–102 [DOI] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, et al. 1995. Selective trafficking ofKNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270(5244):1980–83 [DOI] [PubMed] [Google Scholar]
- Mähönen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, et al. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515(7525):125–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124(1):33–44 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y 2014. Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol 65:385–413 [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329(5995):1065–67 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95(6):805–15 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411(6838):709–13 [DOI] [PubMed] [Google Scholar]
- McHale NA. 2004. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell Online 16(7):1730–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. 2012. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. PNAS 109(5):1760–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, et al. 2017. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29(6):1357–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chen X, Mang H, Liu C, Yu X, et al. 2015. Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol 25(18):2361–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Sozzani R, Yardimci GG, Petricka JJ, Vernoux T, et al. 2015. Transcriptional control of tissue formation throughout root development. Science 350(6259):426–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Shah NM, Anderson DJ. 1997. Regulatory mechanisms in stem cell biology. Cell. 88(3):287–98 [DOI] [PubMed] [Google Scholar]
- Müller B, Sheen J. 2008. Cytokinin and auxin interaction in root stem-cell specification during early embryo-genesis. Nature 453(7198):1094–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R, Bleckmann A, Simon R. 2008. The receptor kinase CORYNE of Arabidopsis transmits the stem cell–limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell Online 20(4):934–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. 2003. Stomatal development: Cross talk puts mouths in place. Trends Plant Sci. 8(6):294–99 [DOI] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413(6853):307–11 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y. 2017. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355(6322):284–86 [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, Trépout S, Gaston A, Fouché M, et al. 2017. Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants 3:17082. [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL. 2017. CLAVATA1 controls distinct signaling outputs that buffer shoot stem cell proliferation through a two-step transcriptional compensation loop. PLOS Genet. 13(3):e1006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM. 2011. Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol 21(5):345–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y. 2008. Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319(5861):294–94 [DOI] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, et al. 2016. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res. 26(6):686–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Rodriguez K, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV. 2016. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. PNAS 113(41):E6298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister A, Barberon M, Alassimone J, Kalmbach L, Lee Y, et al. 2014. A receptor-like kinase mutant with absent endodermal diffusion barrier displays selective nutrient homeostasis defects. eLife 3:e03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, et al. 2015. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33(5):576–88 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Bogenschutz NL, Torii KU. 2008. The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 49(6):934–43 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. 2007. Termination of asymmetric cell division and differentiation of stomata. Nature 445(7127):501–5 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. 2012. Mechanisms of stomatal development. Annu. Rev. Plant Biol 63:591–614 [DOI] [PubMed] [Google Scholar]
- Polyn S, Willems A, De Veylder L. 2015. Cell cycle entry, maintenance, and exit during plant development. Curr. Opin. Plant Biol 23:1–7 [DOI] [PubMed] [Google Scholar]
- Prigge MJ. 2005. Class III homeodomain–leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17(1):61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahni R, Efroni I, Birnbaum KD. 2016. A case for distributed control of local stem cell behavior in plants. Dev. Cell 38(6):635–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig MT, Matos JL, Anleu Gil MX, Kornfeld A, Bettadapur A, et al. 2017. Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355(6330):1215–18 [DOI] [PubMed] [Google Scholar]
- Robards AW, Lucas WJ. 1990. Plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol 41:369–419 [Google Scholar]
- Rodriguez K, Perales M, Snipes S, Yadav RK, Diaz-Mendoza M, Reddy GV. 2016. DNA-dependenthomodi-merization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. PNAS 113(41):E6307–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E 2002. CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14(5):969–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosspopoff O, Chelysheva L, Saffar J, Lecorgne L, Gey D, et al. 2017. Direct conversion of root primordium into shoot meristem relies on timing of stem cell niche development. Development 144(7):1187–200 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99(5):463–72 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17(3):354–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo NJM, Vriezen WH, Beemster GTS, Van Der Straeten D. 2003. Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 33(6):989–1000 [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, et al. 2007. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446(7137):811–14 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ. 2015. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27(1):44–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B 2007. Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol 8(5):345–54 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100(6):635–44 [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shilo BZ. 1997. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 13(5):191–96 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D. 2000. Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289(5480):779–82 [DOI] [PubMed] [Google Scholar]
- Shimizu N, Ishida T, Yamada M, Shigenobu S, Tabata R, et al. 2015. BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 208(4):1104–13 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y. 2016. Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. PNAS 113(14):3897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Miller CO. 1957. Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol 11:118–30 [PubMed] [Google Scholar]
- Slane D, Kong J, Berendzen KW, Kilian J, Henschen A, et al. 2014. Cell type–specific transcriptome analysis in the early Arabidopsis thaliana embryo. Development 141(24):4831–40 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Sindhu C, Meissner A. 2016. Molecular features of cellular reprogramming and development. Nat. Rev. Mol. Cell Biol 17(3):1–16 [DOI] [PubMed] [Google Scholar]
- Song W, Liu L, Wang J, Wu Z, Zhang H, et al. 2016. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26(6):674–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, et al. 2010. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466(7302):128–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y, Grabowski S, Bleckmann A, Köhnemuth R, Weidtkamp-Peters S, et al. 2013. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol 23(5):362–71 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink RH, Ingram GC, Simon R. 2009. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol 19(11):909–14 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, et al. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17(2):616–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Zhang XS. 2014. The hormonal control of regeneration in plants. Curr. Top. Dev. Biol 108:35–69 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319(5868):1384–86 [DOI] [PubMed] [Google Scholar]
- Tameshige T, Okamoto S, Lee JS, Aida M, Tasaka M, et al. 2016. A secreted peptide and its receptors shape the auxin response pattern and leaf margin morphogenesis. Curr. Biol 26(18):2478–85 [DOI] [PubMed] [Google Scholar]
- Tian H, Wabnik K, Niu T, Li H, Yu Q, et al. 2014. WOX5-IAA17 feedback circuit–mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol. Plant 7(2):277–89 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbek P, Scheres B. 1995. Cell fate in the Arabidopsis root meristem determined by directional signaling. Nature 378(6552):62–65 [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390(6657):287–89 [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof Y-D, Miyashima S, et al. 2011. Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21(6):1144–55 [DOI] [PubMed] [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol 67:539–74 [DOI] [PubMed] [Google Scholar]
- Wendrich JR, Möller BK, Li S, Saiga S, Sozzani R, et al. 2017. Framework for gradual progression of cell ontogeny in the Arabidopsisroot meristem. PNAS 114(42):E8922–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmölling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15(11):2532–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L 2005. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132(16):3657–68 [DOI] [PubMed] [Google Scholar]
- Wu M-F, Yamaguchi N, Xiao J, Bargmann B, Estelle M, et al. 2015. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4:e09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lee C-M, Hayashi T, Price S, Divol F, et al. 2014. A plausible mechanism, based upon SHORT-ROOT movement, for regulating the number of cortex cell layers in roots. PNAS 111(45):16184–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PGM, et al. 2011. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333(6046):1141–44 [DOI] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jonsson H, Reddy GV. 2011. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25(19):2025–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Tavakkoli M, Reddy GV. 2010. WUSCHEL mediates stem cell homeostasis by regulating stem cell number and patterns of cell division and differentiation of stem cell progenitors. Development 137(21):3581–89 [DOI] [PubMed] [Google Scholar]
- Yang S, Li C, Zhao L, Gao S, Lu J, et al. 2015. The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27(6):1670–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiao Y, Liu Z, Zhu Y-X. 2015. ROW1 maintains quiescent centre identity by confining WOX5 expression to specific cells. Nat. Commun 6:6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang X. 2012. Argonautes compete formiR165/166 toregulate shootapical meristem development. Curr. Opin. Plant Biol 15(6):652–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Andersen SU, Ljung K, Dolezal K, Miotk A, et al. 2010. Hormonal control of the shoot stem-cell niche. Nature 465(7301):1089–92 [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H. 2007. Regulation of HD-ZIP III genes by mcroRNA 165. Plant Signal. Behav 2(5):351–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, Zhai Q, Jiang H, et al. 2010. Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22(11):3692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, et al. 2015. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517(7534):377–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zürcher E, Tavor-Deslex D, Lituiev D, Enkerli K, Tarr PT, Müller B. 2013. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161(3):1066–75 [DOI] [PMC free article] [PubMed] [Google Scholar]