Abstract
Long non–coding RNAs (lncRNAs) are key molecules that regulate gene expression in a variety of organisms. LncRNAs can drive different transcriptional and post-transcriptional events that impact cellular functions. Recent studies have identified many lncRNAs associated with immune cell development and activation; however, an understanding of their functional role in host immunity to infection is just emerging. Here, we provide a detailed and updated review of the functional roles of lncRNAs in regulating mammalian immune responses during host-pathogen interactions, as these functions may be either beneficial or detrimental to the host. With increased mechanistic insight on the roles of lncRNAs, it may be possible to design and/or improve lncRNA-based therapies to treat a variety of infectious and inflammatory diseases.
LncRNAs: Identification, Characteristics and Classification
It is well known that only < 2% of the mammalian genome encodes proteins, while the rest of the genes encode non-protein coding RNAs [1]. Non-coding RNAs can be divided into small and long non-coding RNAs (lncRNAs). LncRNAs are non-coding RNAs transcribed by RNA polymerase II (RNA Pol II) and are longer than 200 nucleotides. This arbitrary cut-off in size distinguishes them from small non-coding RNAs such as microRNAs (miRNAs) and short interfering RNAs (siRNAs). Similar to miRNAs, lncRNAs have emerged as new regulators of expression of inflammatory response genes in mammalian leukocytes [2]. In addition, similar to messenger RNAs (mRNAs), most lncRNAs are capped, polyadenylated and spliced [3]. Furthermore, a comprehensive analysis of their expression in multiple human organs has indicated that lncRNAs may be more tissue-specific than protein-coding genes, albeit with lower expression than the latter [4]. While some lncRNAs have been reported to encode small peptides [5, 6], the functional significance of such peptides is often unclear; and, for the purpose of this review, these will not be discussed, but readers are directed to other excellent reviews [7, 8].
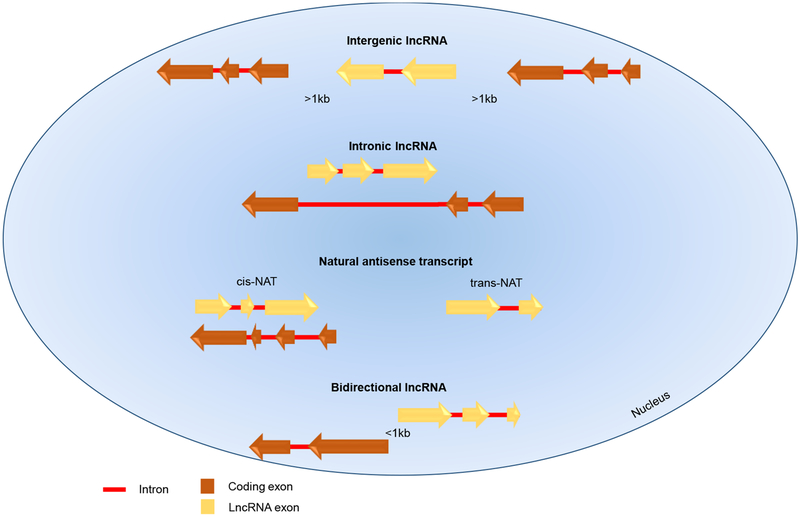
LncRNAs are classified depending on their genomic location (Box1). So far, four classes of lncRNAs are known: long intergenic non-coding RNAs (lincRNAs), bidirectional lncRNAs, intronic non-coding RNAs, and natural antisense transcripts (NATs) [2, 9] (Figure 1). Initially, lncRNAs were considered as “transcriptional noise” without biological function, but emerging evidence has unraveled their important regulatory functions, including controlling both transcriptional and post-transcriptional events, such as DNA methylation, histone modification, splicing, transcription, and translation [2]. Recent research using multiple cutting edge techniques (Box 2) has uncovered functional roles for many lncRNAs in the regulation of cellular events, including cell differentiation [10–14] and malignant transformation [15, 16]. However, the functions of many lncRNAs remain unknown. A role of lncRNAs in pathogenesis of infectious, inflammatory, and autoimmune diseases is just beginning to be understood.
Box 1. Genomic location of lncRNAs.
Based on their genomic location, lncRNAs are distinguished as long intergenic non-coding RNAs (lincRNAs), bidirectional lncRNAs, intronic non-coding RNAs, and natural antisense transcripts (NATs). LincRNAs are situated between two protein-coding genes and are at least 1 kb away from them. Bidirectional lncRNAs are oriented head-to-head with a protein-coding gene within 1kb, and their transcript shows a similar expression pattern to its protein-coding counterpart, suggesting the sharing of a bidirectional promoter [2, 9, 88]. Since lincRNAs and bidirectional lncRNAs do not overlap with other genes, they are referred to as non-overlapping lncRNAs. Intronic RNAs arise from the intronic regions of the genome, whereas NATs are lncRNAs complementary to protein-coding genes. Intronic lncRNAs and NATs are overlapping lncRNAs [9]. NATs are the most common lncRNAs and comprise cis-NATs, complementary to a protein-coding gene located in their same genomic location, and trans-NATs, which arise in a different genomic location compared to the transcript to which they are complementary [89].
Figure 1. Location of Mammalian LncRNAs Relative to Nearby Protein-Coding Genes.
Mammalian LncRNAs are classified depending on their genomic location. Intergenic lncRNAs and bidirectional lncRNAs do not overlap with other genes. Intergenic lncRNAs are situated between two protein-coding genes, at least 1 kb away from them; bidirectional lncRNAs are oriented head to head with a protein-coding gene within 1kb. Intronic lncRNAs and NATs show some degree of overlap with other genes [9]. Intronic lncRNAs arise from the intronic regions of protein-coding genes; NATs are lncRNAs complementary to protein-coding genes and may be categorized as cis-NATs, complementary to a protein-coding gene located in their same genomic location; and trans-NATs, arising in a different genomic location [89, 99]. Arrows show transcriptional direction.
Box 2. New methodologies for studying lncRNAs.
LncRNAs Annotation Tools
Every year, hundreds of lncRNAs are discovered and annotated using cutting-edge transcriptome sequencing techniques, e.g. RNA sequencing (RNA-seq) and rapid amplification of cDNA ends sequencing (RACE-seq), developed to complete lncRNA annotations, albeit with relatively low throughput [90]. The newly-established RNA Capture Long-Seq (CLS) method couples targeted RNA capture with third-generation long-read cDNA sequencing aiming to accelerate progress towards a faster and higher-quality mammalian transcriptome annotation [91, 92]. With CLS, a comprehensive capture library was obtained targeting intergenic GENCODE lncRNAs in human and mouse tissues [92]. In the human survival associated mitochondrial melanoma specific oncogenic non-coding RNA (SAMMSON) oncogene (LINC01212), CLS unveiled previously unannotated exons, splice sites, and transcription termination sites [92]. With CLS, existing gene models might be improved, and putative novel loci identified [92].
Discovery of LncRNA-Interacting Partners
New RNA-based techniques, e.g. Photoactivatable ribonucleotide-enhanced cross-linking and immunoprecipitation (PAR-CLIP) [93], and Capture hybridization analysis of RNA targets (CHART) [94] have led to the identification of several lncRNA-interacting partners.
in vitro RNA antisense purification (RAP) has been used to map the localization of lncRNAs across a genome; the localization of Xist during the initiation and maintenance of XCI in mouse lung fibroblasts was mapped [95]. Coupling RAP with mass spectrometry (RAP-MS) uncovered the Xist-interacting protein SHARP in mice, providing insight into Xist-mediated transcriptional silencing [19]. RAP-MS uses ultraviolet crosslinking to create covalent bonds between directly-interacting RNA and protein, and purifies lncRNAs in denaturing conditions to disrupt non-covalent interactions [19] RAP-MS may constitute an important tool in the lncRNA field, perhaps unveiling lncRNA mechanisms that have thus far proved elusive [19].
Chromosomal looping creates a three dimensional folding that allows distant genes, i.e. promoters and enhancers, to be spatially close [96, 97]. New bioinformatics tools might enable the prediction of lncRNA interactions, genomic targets, and lncRNA secondary structures. Using a combination of different cutting-edge techniques, including Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET), a role for 3D chromatin topology in modulating innate immunity was recently highlighted [98]. Specifically, 3D chromatin architecture could correctly position a subset of immune gene–priming lncRNAs (IPLs) proximal to immune genes prior to their activation. In human cells (HUVEC), the IPL upstream master lncRNA of the inflammatory chemokine locus (UMLILO) could direct the WDR5–mixed lineage leukemia protein 1 (MLL1) complex across CXCL gene promoters, thus facilitating H3K4me3 epigenetic priming [98].
Overall, the development of new computational and/or biochemical tools, as well as improvements in existing technologies are fundamental to the discovery of new lncRNAs, their interacting partners, and their putative functions.
Here, we provide a comprehensive and timely overview of newly-discovered lncRNAs that have been implicated in the regulation of immune responses and host defense against pathogens. Increased knowledge of the functions of lncRNAs might allow us to attain relevant molecular insights into the regulation of immune responses during host-pathogen interactions, with the ultimate aim of developing new putative therapeutic targets to treat a subset of infectious diseases.
Mechanism of Action of LncRNAs
LncRNAs can regulate gene expression by interacting with genomic DNA, RNA or proteins, and their functions depend on their subcellular localization, which might be either the nucleus or the cytoplasm [17]. LncRNAs may operate in cis (see Glossary) or trans, and may act as a guide, scaffold, or decoy [17]. Xist (X-inactive specific transcript), a nuclear lncRNA that has been studied for decades in relation to its involvement in X chromosome inactivation during mammalian female development, constitutes an example of a lncRNA interacting with genomic DNA [18]. Xist is a large transcript of 20 kb that binds DNA in cis and interacts with a large number of proteins [18]. For instance, RNA antisense purification-mass spectrometry (RAP-MS) has revealed that the protein SMRT and HDAC associated repressor protein (SHARP), interacts with Xist and is necessary for X chromosome silencing in mouse embryonic stem (ES) cells [19]. The knockdown of the co-repressor silencing mediator of retinoic acid and thyroid hormone receptor SMRT or histone deacetylase 3 (HDAC3) in both male and female ES cells abrogated silencing of X chromosome genes upon induction of Xist expression [19]. Furthermore, knockdown of HDAC3 in both male and female ES cells eliminated the exclusion of RNA Pol II from the inactive X chromosome [19]. These findings demonstrate that Xist interacts directly with SHARP, which recruits SMRT and activates HDAC3, thus promoting the exclusion of RNA Pol II from the inactive X chromosome [19].
Another common mechanism of action of lncRNAs is the regulation of histone modifications via their association with ribonucleoprotein (RNPs) complexes, or by interacting with transcription factors. THRIL (TNFα and hnRNPL related immunoregulatory lncRNA), an antisense lncRNA that partially overlaps the 3’ UTR of brain protein I3 binding protein (BRI3BP) coding gene, is an example of a lncRNA that regulates histone modifications via RNPs [20]. In the human monocytic cell line THP-1, pull-down assays and chromatin immunoprecipitation (ChIP) analysis revealed that in response to Pam3CSK4 (synthetic lipoprotein), THRIL interacts with the heterogeneous nuclear RNPL (hnRNPL) at the tumor necrosis factor (TNF)-α (TNFA) promoter and induces TNF-α transcription [20]. Moreover, addition of exogenous human TNF-α resulted in downregulation of THRIL mRNA in THP-1 cells, suggesting that this lncRNA might be part of a protective feedback loop to control TNF-α production [20]. Consistent with this, THRIL and TNF-α expression can be altered in Kawasaki disease, an inflammatory disease of children [20]. Analysis of patient blood samples revealed that compared to the convalescent phase of this disease, THRIL expression was significantly lower in the acute phase, whereas TNF-α was elevated [20].
NKILA (NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) interacting lncRNA) is an example of a lncRNA that regulates histone modifications by interacting with transcription factors. NKILA regulates NF-kB signaling and can repress human breast cancer-associated inflammation [21]. Specifically, in response to lipopolysaccharide (LPS), NKILA is upregulated in several human breast cancer cell lines [21]. However, high-metastatic breast cancer lines with high NF-κB activities have shown much lower NKILA expression than low-metastatic lines, suggesting that NKILA can act as a negative regulator of NF-κB signaling [21]. In in vitro assays, together with gain-of-function/loss-of-function studies in human breast cancer cell lines, have shown that NKILA binds to the p65-NF-kB/IkB (nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor) complex in the cytoplasm, and inhibits NF-kB signaling by masking the phosphorylation sites of IkB, thus stabilizing the NF-kB/IkB complex [21]. Furthermore, NKILA expression and NF-kB activity have been measured in primary breast epithelial cells isolated from normal breast tissues, or from patients with invasive breast cancers with and without metastasis [21]. In these, NKILA expression was abundant in normal breast epithelial cells, but was lower in cells from non-metastatic breast tumors and was further reduced in those with metastasis, indicating that NKILA reduction might potentially predict poor clinical outcome in patients with breast cancer [21]. As expected, NF-kB transcriptional activities were negatively correlated with NKILA expression in these cells [21].
Additionally, lncRNAs can regulate a variety of biological processes at the post-transcriptional level by affecting translation efficiency, mRNA stability, and splicing. For instance, in HEK cells, lncRNA Uchl1 AS (ubiquitin carboxyl-terminal hydrolase L1 antisense) increases UCHL1 protein synthesis, without affecting mRNA quantity, suggesting that lncRNA Uchl1 AS regulates UCHL1 expression at a post-transcriptional level [22]. Furthermore, in a murine dopaminergic cell line, polysome profiles have shown that functional lncRNA Uchl1 AS is necessary for Uchl1 mRNA association to heavy polysomes and translation.[22]. Half Staufen1 (STAU1)-binding site RNAs (½-sbsRNAs) can mediate mRNA decay thereby promoting the degradation of mRNA targets [23]. Indeed, ½-sbsRNA1, one of the seven different cytoplasmic ½-sbsRNAs characterized in HeLa cells, was shown to bind to its mRNA target [23]. Downregulation of ½-sbsRNA1 led to its target upregulation; furthermore, co-immunoprecipitation studies demonstrated that ½-sbsRNA1 was required for STAU1-mediated mRNA decay (SMD) [23]. These examples suggest that nuclear lncRNAs are most likely to participate in transcriptional events either by interacting with genomic DNA [18] or by forming RNA-protein complexes, [20]. By contrast, cytoplasmic lncRNAs might regulate gene expression at post-transcriptional levels [23]. Thus, subcellular localization, as well as the nature of lncRNA interacting partners can influence lncRNA targets and their regulatory functions.
LncRNA-Mediated Modulation of Immune Responses Against Microbial Components
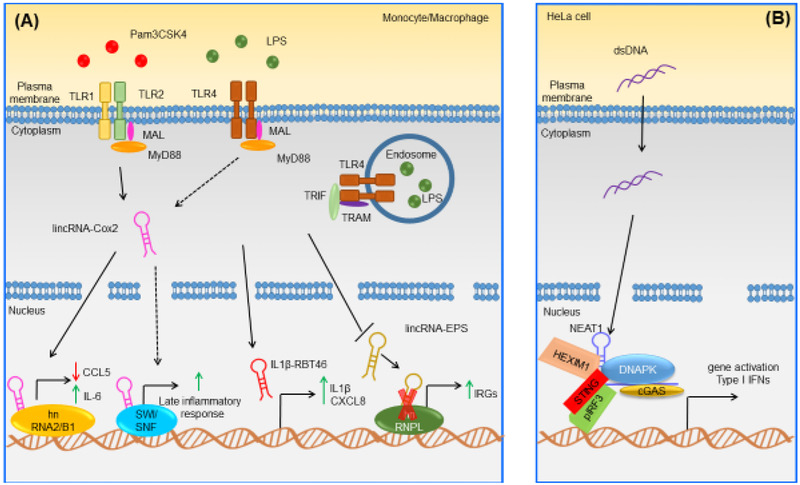
Several species of lncRNAs have been shown to play a key role in modulating immunity against microbial components such as LPS and Pam3CSK4 (Figure 2). For example, lincRNA-Cox2 is a cyclooxygenase 2 (cox2)-neighboring lncRNA, identified in murine dendritic cells (DCs) exposed to LPS [24]. In bone marrow-derived mouse macrophages (BMDMs), lincRNA-Cox2 can be found in the cytoplasm or in the nucleus and is highly upregulated when these cells are stimulated with LPS or Pam3CSK4 [25] relative to control cells. Elevated lincRNA-Cox2 expression has also been noted in Listeria monocytogenes-infected macrophages and in splenocytes isolated from L. monocytogenes-infected mice relative to uninfected cells. [25]. Moreover, in BMDMs stimulated with Pam3CSK4, silencing of lincRNA-Cox2 by short hairpin RNA (shRNA) led to induction and repression of immune response genes C-C motif ligand 5 (Ccl5) and interleukin 6 (Il6), respectively [25]. Mechanistically, hnRNP-A/B and hnRNP-A2/B1 were identified as specific binding partners for lincRNA-Cox2 in both BMDM nuclear and cytosolic fractions [25]. In addition, knockdown of hnRNP-A/B and hnRNP-A2/B1 by shRNA in BMDMs resulted in enhanced Ccl5 protein expression in both unstimulated and Pam3CSK4-stimulated cells; furthermore ChIP assays revealed increased RNA Pol II binding at the Ccl5 promoter when lincRNA-Cox2 -- or either of the hnRNPs -- were silenced in unstimulated cells. This suggesting a critical role for these molecules in the regulation of Ccl5 transcription [25]. It is reasonable to speculate that lincRNA-Cox2 might regulate the transcription of immune genes by interacting with hnRNP-A/B and hnRNP-A2/B1 [25]. Another study showed that siRNA knockdown of lincRNA-Cox2 attenuated the transcription of late-primary genes triggered by LPS in BV2 mouse microglia cells [26] Furthermore, in vitro assays revealed a physical association between NF-κB subunits (RelA and p50) and the SWItch/Sucrose non-fermentable (SWI/SNF) complex in LPS-stimulated RAW264.7 cells; siRNA knockdown of lincRNA-Cox2 decreased the association between NF-κB subunits (RelA and p50) and the SWI/SNF complex [26]. These findings suggest that upon LPS stimulation of murine macrophages, lincRNA-Cox2 can bind the SWI/SNF complex, modulating chromatin remodeling and promoting NF-κB-dependent transcription of late inflammatory genes such as Ccl5 [26] . Thus, it appears that the effect of lincRNA-Cox2 on gene transcription in vitro can be cell type- and immune pathway-dependent. Furthermore, a recent study examined the function of lincRNA-Cox2 during LPS-dependent immune responses in vivo, using CRISPR-Cas9-generated lincRNA-Cox2 intron-less-mutant mice [27]. Intraperitoneal (i.p) LPS injection in these mice led to upregulation of interferon-stimulated genes, including Ccl5 and Ip10, whereas proinflammatory gene expression, including Il5, Lif, and Il17, was reduced compared to wild type (WT) mice [27]. All genes affected in mutant mice were located on different chromosomes relative to lincRNA-Cox2, confirming that lincRNA-Cox2 functions in trans could control immune responses in vivo [27]. These results supported earlier in vitro findings indicating that mouse lincRNA-Cox2 can both promote and inhibit the expression of innate immune genes [25] [27].
Figure 2. Examples of Known Mammalian LncRNAs Implicated in Immune Responses against Microbial Components.
(A) In murine macrophages, lincRNA-Cox2 is upregulated in response to LPS and Pam3CSK4 [25]. In murine macrophages and in response to Pam3CSK4, lincRNA-Cox2 binds hnRNPA/B and hnRNPA2/B1 leading to both activation and repression of different classes of genes [25]; in murine macrophages and in response to LPS, lincRNA-Cox2 can bind the SWI/SNF complex, leading to late inflammatory gene activation [26]. In human monocytes, IL-1β-RBT46 is upregulated following LPS stimulation, and enhances LPS-induced expression and release of proinflammatory mediators IL-1β and CXCL8 [28]. In murine macrophages, lincRNA-EPS is downregulated following LPS stimulation and at a steady state, lincRNA-EPS restrains IRG activation by interacting with hnRNPL [29]. However, after LPS-induced downregulation of lincRNA-EPS, its inhibitory function no longer appears to be exerted, leading to the upregulation of IRGs both in vitro and in vivo in mice [29]. (B) In HeLa cells, NEAT1 is upregulated in response to ISD and binds HEXIM1, DNAPK and paraspeckle components, thus forming the HDP-RNP complex [33]. Upon stimulation with ISD, the HDP-RNP is remodeled in its composition, with recruitment of STING, release of paraspeckle components, and phosphorylation of IRF3 [33]. Abbreviations: DNAPK, DNA-dependent protein kinase; MAL, MyD88 adapter-like; MyD88, myeloid differentiation primary response 88; pIRF3, phosphorylated interferon regulatory factor 3; STING, stimulator of interferon genes; TLR, Toll-like receptor; TRAM, TRIF-related adaptor molecule; TRIF, TIR-domain-containing adapter-inducing interferon-β.
Another lncRNA involved in the regulation of LPS-mediated immune responses is IL-1β-RBT46. Nuclearly-localized IL-1β-RBT46 originates from a region of bidirectional transcription. Specifically, in human monocytes, genomic association (with the Genomic Association Tester tool) showed a significant enrichment for NF-κB binding sites at genomic locations of IL-1β-RBT46, indincating that IL-1β-RBT46 was a NF-κB-regulated lncRNA [28]. In human monocytic THP-1 cells, IL-1β-RBT46 was upregulated in response to LPS in vitro, and knockdown of IL-1β-RBT46 expression enhanced LPS-induced expression and release of pro-inflammatory factors, IL-1β and CXCL8, relative to control cells [28]. These findings suggested that IL-1β-RBT46 could regulate IL1B transcription in cis, but also appeared to act in trans, regulating the transcription and release of CXCL8, albeit to a lesser extent [28].
However, the underlying molecular mechanism of this regulation is unclear and warrants extensive investigation [28]. In contrast, lincRNA-EPS (erythroid pro survival) is a lncRNA reported as downregulated in BMDMs upon in vitro LPS stimulation and infection with L. monocytogenes or Sendai virus (SeV) compared to control cells [29]. Mechanistically, in vitro RNA-protein binding assays revealed an interaction between lincRNA-EPS and hnRNPL in murine macrophages [29]; furthermore, BMDMs from lincRNA-EPS global knockout (KO) mice stimulated with LPS showed increased expression of immunity-related genes (IRGs) , such as Il6 and Ccl5, compared to unstimulated cells [29]. BMDMs from lincRNA-EPS KO mice also exhibited an enhanced recruitment of RNA Pol II at IRGs (e.g. Ccl5) promoters following LPS-stimulation, suggesting that lincRNA-EPS could regulate the expression of these genes at a transcriptional level [29] In addition, lincRNA-EPS suppressed inflammation in vivo, given that lincRNA-EPS KO mice displayed increased expression of IRGs such as those encoding IL-6, CCL5 and IL1-α relative to controls. The mice were significantly more susceptible to septic shock relative to WT mice [29].
Another lncRNA that has been implicated in the regulation of immune responses to microbial components is NEAT1 (nuclear enriched abundant transcript-1)-- first identified as a lncRNA involved in the assembly of paraspeckles in humans [30]. Two major isoforms of NEAT1 are known in mice and humans: human NEAT1v1, is the 3.7 kb polyadenylated variant, while human NEAT1v2 is the 23 kb non-polyadenylated variant [31, 32]. Recently, in HeLa cells, NEAT1 was found to form a complex with the transcriptional inhibitor hexamethylene bisacetamide inducible-1 (HEXIM1), together with subunits of the DNA-dependent protein kinase (DNAPK) complex (DNAPKc, Ku70, and Ku80), as well as paraspeckles components [33]. This previously unknown complex is termed HDP-RNP (HEXIM1-DNA-PK-paraspeckle components-ribonucleoprotein complex), and NEAT1 is required for its assembly [33]. Functionally, relative to WT HeLa cells, siRNA knockdown of HDP-RNP subunits, including NEAT1, resulted in loss of interferon stimulatory DNA (ISD)-mediated and cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) –mediated interferon regulatory factor-3 (IRF3) phosphorylation, as well as interferon (IFN)-β mRNA expression [33]. These data suggested that HDP-RNP could regulate DNA-mediated innate immune responses upstream of IRF3 phosphorylation and downstream of cGAMP synthesis [33]. Mechanistically, upon stimulation with ISD, the cytosolic DNA receptor, cyclic GMP-AMP synthase (cGAS), along with its partner polyglutamine binding protein 1 (PQBP1), and IRF3, interacted with HDP-RNP. HDP-RNP could then be remodeled with the recruitment of stimulator of interferon genes (STING) and the release of paraspeckle components, thus leading to phosphorylation of DNAPKc and IRF3, and subsequent type I IFN production [33]. Of note, The HDP-RNP complex is also required for Kaposi’s Sarcoma-associated Herpesvirus (KSHV)-mediated activation of the innate immune response, exerting an antiviral function [33]. Indeed, siRNA knockdown of HEXIM1, Ku70, NEAT1, and STING in human umbilical vein endothelial cells (HUVECs) resulted in inhibition of KSHV-mediated IFNβ production, suggesting a reduction in
antiviral response compared to WT [33]. KSHV encodes ORF52, an abundant gamma herpesvirus-specific tegument protein known to target cGAS and inhibit its activation [34]. Ectopic expression of ORF52 in HUVECs cells resulted in loss of binding of cGAS and PQBP1 to HEXIM1, suggesting that targeting HEXIM1-cGAS interactions might constitute a viral strategy to avoid the initiation of innate immunity in the host [33].
Collectively, a number of examples of lncRNAs have been identified as contributing to the regulation of immune responses to microbial components, such as LPS, Pam3CSK4, foreign DNA, among others, particularly using mouse models or cells in vitro. However, further studies are evidently required to fully delineate the role of each of these lncRNAs during live bacterial or viral infections in vivo, and across species.
LncRNAs Functioning in Specific Host-Pathogen Interactions and Defense
Accumulating evidence indicates that a number of functional lncRNAs are differentially regulated during microbial infections. These lncRNAs are either host-derived, or encoded by pathogens, and may play a significant role in controlling host-pathogen interactions. They can function via multiple mechanisms, including the regulation of growth and replication of pathogens, or via cell-autonomous anti-microbial defense mechanisms. Moreover, some lncRNAs can be beneficial in promoting microbial clearance, whereas others can enable pathogen survival [35–37].
Host-derived lncRNAs
While it has been long known that intracellular pathogens can regulate the expression of small noncoding RNAs (e.g. miRNAs) affecting host responses [38–44], the study of interactions between pathogens and lncRNAs has just recently begun. Many lncRNAs have been shown to be differentially expressed in myeloid and non-myeloid cells during infection with pathogens such as Mycobacterium tuberculosis [45, 46], Escherichia coli [47], Rabies virus [48] and fungi [49] in humans, and Enterovirus 71 [50] and Toxoplasma gondii [51] in mouse. However, the signaling pathways or cellular events they regulate remain poorly understood. Newly-identified host-derived lncRNAs involved in immune responses against bacterial (Figure 3) and viral (Figure 4) infections are described below.
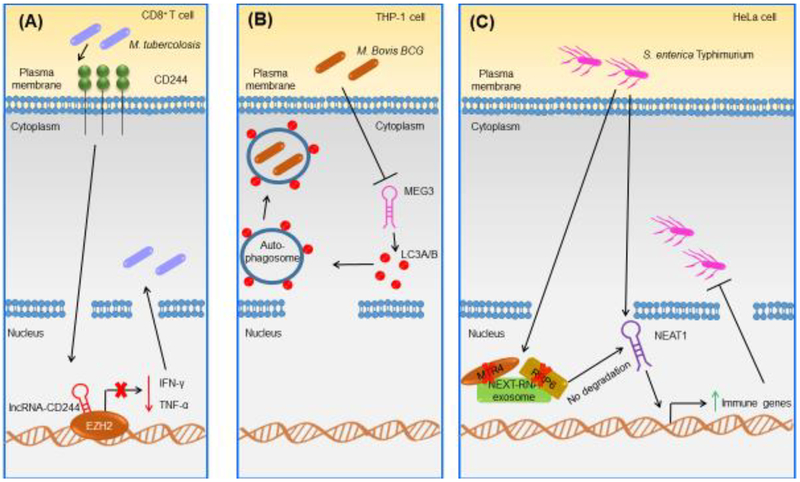
Figure 3. Examples of Known Mammalian Host-Derived LncRNAs Implicated in Host Defense Against Bacterial Infections.
(A) In human CD8+ T cells, lncRNA-CD244 is upregulated during Mycobacterium tuberculosis infection in a CD244-dependent manner [52]. LncRNA-CD244 interacts with the chromatin modification enzyme EZH2, leading to trimethylation of H3K27 at the TNFA and IFNG loci [52]. LncRNA-CD244 represses the expression of these two genes and promotes bacterial replication in human CD8+ T cells [52]. (B) In THP-1 cells, MEG3 is downregulated after M. bovis BCG infection [70]. In THP-1 cells, MEG3 affects autophagy by increasing LC3A/B conversion and blocking lysosomal degradation, thus eliminating intracellular M. bovis BCG [70]. (C) In HeLa cells, NEAT1 is upregulated upon Salmonella enterica Typhimurium [37]. The bacterium induces loss of MTR4 and RRP6, where NEAT1 is no longer degraded by the exosome, promoting the transcription of immune genes as well as bacterial clearance [37]. Abbreviations: EZH2, enhancer of zeste homolog 2.
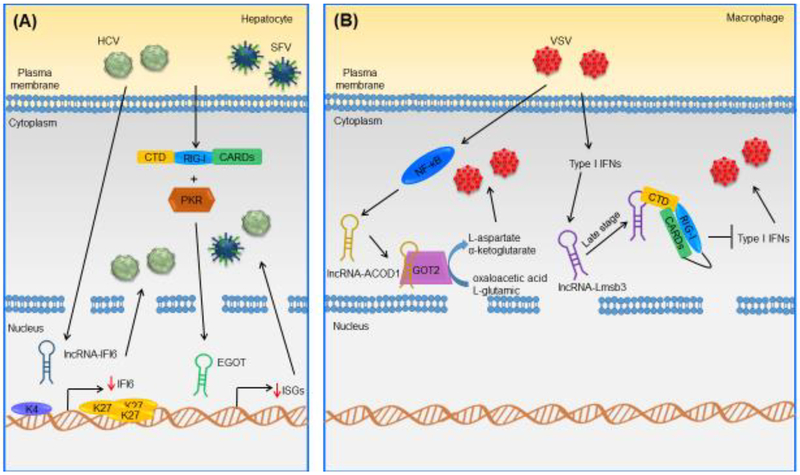
Figure 4. Examples of Known Mammalian Host-Derived LncRNAs Implicated in Host Defense Against Viral Infections.
(A) In human hepatocytes, lncRNA-IFI6 is upregulated in response to HCV. It specifically regulates IFI6 expression, leading to the regulation of H3K4me3 and H3K27me3 marks at the IFI6 promoter, promoting HCV infection in human hepatocytes [64]. In human hepatocytes, EGOT is upregulated upon HCV and SFV infection through the RIG-I and PKR pathways. In human hepatocytes, EGOT increases the expression of several ISGs, negatively affecting the antiviral response [35]. (B) In murine macrophages, lncRNA-ACOD1 is upregulated following VSV infection in an NF-κB-dependent manner. In murine macrophages, lncRNA-ACOD1 binds GOT2, increasing its catalytic activity and production of its metabolites, facilitating viral replication [36]. In murine macrophages, lncRNA-Lmsb3b is a type I IFN-dependent lncRNA induced in response to VSV and SeV [78]. At a late stage of infection, lncRNA-Lms3b binds the RIG-1 CTD, competing with RIG-I ligands; it also maintains RIG-I in a repressed state and prevents RIG-I oligomerization [78]. This inhibits RIG-I signaling and promotes viral replication in murine macrophages [78]. Abbreviations: CARDs, caspase recruitment domains; H3K27me3, trimethylation of lysine 27 on histone H3; H3K4me3, trimethylation of lysine 4 on histone H3; PKR, protein kinase R; RIG-I, retinoic acid-inducible.
Regulation of Gene Transcription via LncRNA-CD244, NeST, NEAT1, IFI6 and EGOT
LncRNA-CD244
LncRNA-CD244 is a lncRNA that overlaps with the 5′ UTR of glutathione S-transferase theta-1 (GST θ1) in human CD8+ T cells [52]. No mouse homolog has been identified for this lncRNA, suggesting that lncRNA-CD244 might not be expressed in all species. A study showed that in vitro, the costimulatory molecule CD244 was upregulated in M. tuberculosis-infected human CD8+ T cells relative to uninfected cells, and this induced the expression of lncRNA-CD244 [52]. Of relevance, CD244+ CD8+ T cells isolated from (peripheral blood mononuclear cells) PBMCs of tuberculosis (TB) patients have been found to express higher lncRNA-CD244 compared with CD244− CD8+ T cells [52], suggesting that CD244 signaling can positively correlate with high expression of lncRNA-CD244; however whether CD244 and lncRNA-CD244 are coregulated, and how CD244 induces lncRNA-CD244 upregulation remain unknown [52]. Furthermore, CD8+ T cells purified from PBMCs of patients with active TB treated with lncRNA-CD244 siRNA or shRNA in vitro, expressed significantly higher IFN-γ and TNF-α proteins relative to control cells, suggesting that lncRNA-CD244 might influence repressive chromatin states at INFG and TNFA loci. [52]. In addition, immunoprecipitation (IP) studies in these cells revealed that lncRNA-CD244 interacted with chromatin modification enzyme enhancer of zeste homolog 2 (EZH2) that catalyzed H3K27me3 trimethylation (repressive mark) at the TNFA and IFNG promoters [52]. In order to confirm these in vitro results in an in vivo model, severe combined immune deficiency mice (SCID) mice were infected with M. tuberculosis, and then infused with LV (lentiviral)-lncRNA–transduced (lncRNA-CD244–depressed) human CD8+ T cells (purified from PBMCs of patients with active TB). These mice exhibited lower bacterial burdens in lungs and blood, as well as less infiltration of red blood cells or damage of pulmonary tissue compared with control mice [52]. This study concluded that lncRNA-CD244 could act as an epigenetic inhibitor of TNFA and IFNG genes by enhancing repressive chromatin marks (and therefore, chromatin state) at the promoter regions of these genes. LncRNA-CD244 could thus inhibit CD8+ T cells immune responses in this humanized mouse model of mycobacterium infection [52]. Presumably, this mechanism might also exist in patients with tuberculosis, but further and robust testing will be needed to establish this. Furthermore, the fact that a mouse homolog has not been identified for this lncRNA constitutes an example of why we need to advance our characterization of lncRNAs sequences and structures.
NeST
NeST (Nettoie Salmonella pas Theiler’s; or Tmevpg1) is an antisense lncRNA encoded in the IFNG/ifng locus and expressed in CD4+ Th1 cells, CD8+ T-cells and NK cells in humans and mice [53–55]. NeST was shown to control manifestations of Theiler’s virus infection in the central nervous system of mice, or in the clearance of Salmonella enterica Thyphimurium murine infection [53]. The ability of inbred mice to clear Theiler’s infection varies greatly from strain to strain [53]. For instance, whereas WT B10.S mice can clear the virus, WT SJL/J mice become persistently infected, and this effect is conferred by different loci [56, 57]. Transgenic B10.S mice expressing either SJL/J- or B10.S-derived NeST RNA have shown increased Theiler’s virus persistence and decreased Salmonella pathogenesis compared with WT B10.S, lacking NeST RNA; this suggested that NeST might be implicated in Theiler’s virus persistence as well as in Salmonella resistance. [53]. In this study, RNA preparations from human 293T cells were co-transfected with ectopic (WD repeat domain 5) WDR5 cDNA (protein involved in histone modifications); when using either B10.S-derived NeST cDNA or SJL/J derived NeST cDNA, NeST interacted with WDR5 [53]. Furthermore, murine activated CD8+ T cells from B10.-expressing SJL/J-derived NeST transgenic mice showed increased H3K4me3 enrichment at the Ifng locus, compared with B10.S mice; this suggested that NeST could epigenetically regulate IFN-γ release [53]. The authors speculated that the disparate effects of NeST might illustrate the role of balanced polymorphisms in susceptibility to infectious diseases [58, 59]. Specifically, NeST polymorphisms might result in increased NeST expression and contribute to differences in T cell responses, altering the magnitude or timing of inflammatory responses, thus conferring susceptibility to Theiler’s virus, but resistance to Salmonella infection [53]. However, whether and how disease-associated SNPs alter human NeST expression remains to be addressed in future studies.
NEAT1
NEAT1, classified as a lncRNA with antiviral functions, is located on human chromosome 11, and its expression is upregulated in response to several viral infections [33, 60–62]. Conversely, the expression of viral DNA, or viral proteins infected cell protein 0 (ICP0) and thymidine kinase (TK), has been shown to be significantly reduced upon NEAT1 knockdown in HeLa cells following Herpes simplex virus (HSV-1) infection, relative to uninfected cells [63]. These data suggest that during HSV-1 infection, NEAT1 might play a significant role in promoting virus replication and viral gene expression [63], an apparent discrepancy with previous findings that might be attributed to cell types and experimental conditions used [33, 60–62]. In the latter study, overexpression of signal transducer and activator of transcription 3 (STAT3) increased HSV-1 viral gene expression significantly relative to controls, whereas STAT3 knockdown dramatically reduced viral gene expression in HeLa cells; this suggested that STAT3 might promote HSV-1 gene expression [63]. Of note, siRNA knockdown of NEAT1 and paraspeckle components in STAT3-overexpressing cells abolished the positive effect of STAT3 on HSV-1 gene expression, suggesting that NEAT1, together with the paraspeckle components, might upregulate STAT3-dependent viral gene transcription [63]. These results were confirmed using an in vivo model, where C57BL/6 mice with skin lesions caused by HSV-1 infection were treated with thermosensitive gels containing either control siRNA, M siSTAT3–2 or M siNEAT1v2 [63]. The last two gels contained siRNAs targeting STAT3 and NEAT1, respectively. The use of these two gels inhibited the development of skin lesions and promoted tissue repair compared to control gel, showing that the inhibition of either STAT3 or NEAT1 resulted in limited HSV-1 replication [63]. Although these results suggest that NEAT1 and STAT3 might be considered as potential therapeutic targets to limit HSV-1 replication, further and robust studies are required to potentially translate these findings to humans [63].
Recent studies suggest that NEAT1 may play a role in regulating host responses during bacterial infections as well [37]. Specifically, whole-transcriptome analysis of HeLa cells infected with Salmonella enterica Typhimurium, revealed that NEAT1v2, the non-polyadenylated variant of NEAT1, was one of the most upregulated unstable nuclear non-coding RNAs (ncRNAs) relative to uninfected cells [37]. In addition, this upregulation was induced by live Salmonella only, but not by heat-killed Salmonella or its bacterial components, suggesting that bacterial intracellular replication was necessary to achieve increased NEAT1v2 RNA expression [37]. The study further dissected the mechanism of upregulation of NEAT1v2 RNA expression during Salmonella infection. Knockdown of components of different degradation complexes in HeLa cells demonstrated that in uninfected cells, unstable nuclear ncRNAs were degraded by the nuclear exosome targeting (NEXT)-RNA exosome pathway; by contrast, upon Salmonella infection, two RNA decay factors belonging to this complex, termed MTR4 and RRP6, were expressed less in several human and mouse cells relative to uninfected cells. Of note, loss of MTR4 and RRP6 resulted in NEAT1v2 transcript stabilization [37]. CRISPR-Cas9-mediated knockout of NEAT1v2 in HeLa cells reduced immune-related gene expression (e.g. TNF superfamily member 9 (TNFSF9) and CCL2), during Salmonella infection compared with uninfected cells [37]. These results suggested that infection led to MTR4 and RRP6 loss, decreasing unstable nuclear ncRNA turnover, stabilizing NEAT1v2 transcripts, and thus leading to the transcriptional activation of immune genes such as TNFSF9 and CCL2 [37]. However, the molecular mechanism responsible for the decrease in MTR4 and RRP upon infection is still unclear. Consequently, NEAT1 seems to exert varying immune-related functions in a context- and pathogen-dependent manner; however, the mechanisms of function under these different contexts remain to be elucidated.
LncRNA-IFI6
LncRNA-IFI6 is located on human chromosome 1 and overlaps with the antisense strain of interferon alpha-inducible protein 6 (IFI6) within intron 1 [64]. It is the most upregulated gene following IFN-α treatment in several human hepatocyte and hepatic stellate cells, such as Huh7.5.1 (hepatocarcinoma cell line) and primary human hepatocytes (PHHs) [64]. In both Huh7.5.1- and PHH-JFH1-infected cells, lncRNA-IFI6 deficiency significantly enhances lncRNA-IFI6 mRNA and protein expression, and reduces Hepatitis C Virus (HCV) RNA titers relative to control cells[64]. Furthermore, IFI6 can exert critical antiviral activity during HVC infection, where its downregulation by siRNA and by short palindromic repeats/Cas9 guide RNA (gRNA) significantly increases HCV RNA and core protein expression in JFH1‐infected Huh7.5.1 cells, compared with controls [64]. During HCV infection, lncRNA-IFI6 has not been found to affect the expression of other classical interferon-stimulated genes (ISGs), indicating that lncRNA-IFI6 might specifically regulate IFI6 expression under these infection conditions [64]. These results have been confirmed in vivo, where higher HCV RNA titers were associated with lower IFN-induced IFI6 expression in liver biopsies from patients with chronic HCV infection, compared with healthy individuals [64]. Moreover, ChIP assays revealed that in unstimulated Huh7.5.1 cells, lncRNA-IFI6 gRNA significantly increased the enrichment of H3K4me3 at IFI6 transcription start sites but significantly reduced the enrichment of H3K27me3 at the IFI6 gene locus compared with control cells, indicating that lncRNA-IFI6 could affect the transcription of IFI6 through histone modifications [64] . Other studies using multiple lncRNA-IFI6 deletion mutants lacking various domains of this molecule have demonstrated that only the overexpression of the mutant containing the large right arm structure of lncRNA‐IFI6 could significantly increase HCV infection and decreased IFI6 mRNA and protein in Huh7.5.1 cells compared with an empty vector [64]. This finding suggests that this spatial domain might be necessary for the regulatory function of lncRNA-IFI6 [64]. Taken together, these results indicate that IFN‐induced lncRNA-IFI6 is upregulated in response to HCV infection, and lncRNA-IFI6 can specifically downregulate the antiviral gene IFI6 via histone modifications, to promote HCV infection in hepatocytes [64]. Although further studies, i.e. in vivo, are necessary, this finding could assist in the development of applications to prevent persistent HCV infection, and possibly, other viral infections [64].
EGOT
EGOT (eosinophil granule ontogeny transcript) is located on human genome antisense to intronic sequences of the inositol 1,4,5-trisphosphate receptor (ITPR1) gene. EGOT was initially discovered as a lncRNA expressed during human eosinophil development [65]. In mature human eosinophils, EGOT regulates the expression of toxic eosinophil proteins [65]. Recent transcriptome analysis in HuH7 cells identified EGOT as one of the most induced genes in response to HCV infection relative to uninfected cells. [35]. EGOT appears to have homologs in primates and rodents, indicating a possible conserved function [35]. Moreover, inactivation of retinoic acid-inducible gene I (RIG-I) and protein kinase R (PKR) pathways by siRNAs in these cells in vitro reduced the expression of EGOT, whereas cells transfected with ectopic PKR led to EGOT upregulation, demonstrating that EGOT could be induced by both RIG-I and PKR activation in HCV-infected cells relative to uninfected controls. [35]. EGOT was also induced in the liver of HCV-infected patients, but not in patients with hepatocellular carcinoma (HCC) [35]. Moreover, depleting EGOT from HCV-infected HuH7 cells with two independent gapmers led to a marked decrease in viral replication relative to cells transfected with control gapmer. Similar results were obtained in response to Semliki forest virus (SFV) infection, suggesting that EGOT expression might be required for the efficient replication of several viruses, but this remains to be further studied [35]. Genome-wide studies have indicated that EGOT expression in these cell lines negatively correlates with innate immune response genes such as those encoding Toll-like receptor 3 (TLR3) and NF-kB, suggesting possible negative regulation of immune responses by this lncRNA [35]. Accordingly, in HCV-infected Huh7 cells, EGOT inhibition by gapmers has led to increased expression of several ISGs such as ISG15 and ISG56, as well as reduced HCV replication, relative to cells transfected with a gapmer control [35]. These results suggested that EGOT can negatively affect certain aspects of host antiviral responses, presumably in favor of HCV replication; however, the latter remains to be directly shown in vivo, and in addition, the molecular mechanisms by which EGOT can affect the expression of antiviral genes remains unknown [35].
MEG3 in Autophagy Activation
MEG3 (maternally expressed 3) is a tumor suppressor gene that has been associated with carcinogenesis in several types of cancer, including multiple myeloma and pituitary adenomas [66, 67]. It has also been implicated in the activation of autophagy in bladder cancer cell lines [68]. In these cancer cell lines, MEG3 expression was downregulated compared to control cells [66, 69]. One study identified MEG3 as one of the few downregulated lncRNAs following Mycobacterium bovis bacillus Calmette-Guerin (BCG) infection in THP-1 cells, associating for the first time, MEG3 with an infection [70]. Moreover, MEG3 knockdown by siRNA induced the conversion of the autophagosomal marker LC3A/B from the cytosolic form LC3A/B I, to the membrane-bound form LC3A/B II, in both M. bovis BCG infected and non-infected THP-1 derived macrophages [70]. However, specific knockdown of MEG3 in these cells had no effect on LC3B transcription, excluding its potential involvement in modulating LC3A/B expression [70]. The activation of the serine/threonine protein kinase mammalian target of rapamycin (mTOR) is known to be negatively correlated to autophagy induction in human cells [71]. Induction of autophagy by rapamycin-mediated inhibition of mTOR signaling can unblock phagosomal maturation in M. tuberculosis and BCG infection [72]. Thus, upon MEG3 knockdown in THP-1 cells infected with BCG, mTOR-mediated phosphorylation of ribosomal protein S6 kinase beta-1 (p70-S6K) (Thr389) was reduced (slightly) relative to uninfected cells, which might suggest a possible decrease in mTOR activity upon induction of autophagy [73]. By contrast, following BCG infection, the protein p62 seemed to increase relative to uninfected THP-1 cells; p62 is known to be selectively degraded by autophagy [73], and an increase in its expression in MEG3 siRNA-treated THP-1 cells suggested inhibition of its own degradation by autophagy, (inhibiting lysosomal degradation) [70]. Thus, these results support the notion that MEG3 downregulation might induce a block in lysosomal degradation rather than an increase in autophagosome formation, but this has yet to be directly demonstrated [70]. Consistent with a positive correlation between MEG3 downregulation and increased autophagy activity, compared to control cells, infected THP-1-cells treated with MEG3 siRNA have shown enhanced eradication of intracellular BCG and accumulation of a LC3A/B signal co-localizing with BCG [70]. Thus, this suggests that M. bovis BCG-dependent downregulation of MEG3 might eliminate mycobacteria in human macrophages via autophagy, although further studies are necessary.
In another model of infection, a recent report indicated that MEG3 was downregulated in nasopharyngeal (NPA) samples of respiratory syncytial virus (RSV)-infected patients and in BEAS-2B cells infected with RSV, compared with controls, suggesting that MEG3 might be involved in the progression of RSV-associated disease [74]. MEG3 overexpression in RSV-infected BEAS-2B cells led to reduced expression of TLR4 relative to control cells, subsequently leading to suppressed TLR4-dependent p38 mitogen activated protein kinase (MAPK) and NF-κB signaling [74]. This result is relevant in that NF-κB and p38 MAPK signaling pathways have been previously shown to be required for the activation of RSV internalization and replication in several human cell lines [75, 76]. Thus, by suppressing NF-κB and p38 MAPK pathways, MEG3 might play a protective role against the progression of RSV infection in human airway epithelial cells [74]. Future investigations should further elucidate this possibility. Collectively, these results indicate that MEG3 expression may have opposite effects in humans depending on the type of infection; being detrimental for the host in response to M. tuberculosis, while presumably playing a protective role in response to RSV infection. These different effects may also be cell type-dependent; however, further studies are required to elucidate the molecular mechanisms underlying this seemingly contrasting behavior.
LncRNA-ACOD1 as a Regulator of Host Metabolism
LncRNA-ACOD1 is located in proximity of the protein coding gene aconitate decarboxylase 1 (ACOD1/Acod1) in both humans and mice [36]. It was originally identified as a Vesicular stomatitis virus (VSV)-induced, IFN-I–independent, and NF-κB-dependent lncRNA [36]. One study showed that in vitro siRNA-mediated knockdown of lncRNA-ACOD1 in murine macrophages significantly downregulated VSV replication relative to control cells [36]. In vivo, lncRNA-ACOD1 KO mice infected with a sub-lethal dose of VSV, exhibited decreased viral burdens and milder pathology compared with WT mice [36]. Moreover, the effect of lncRNA-ACOD1 on viral load was independent of the IRF3-IFN-β axis, since both lncRNA-ACOD1/Irf3 double KO and lncRNA-ACOD1/interferon alpha/beta receptor subunit 1(Ifnar1) double KO mice infected with VSV showed a dramatic suppression of VSV proliferation, alleviated immune responses, and significantly lower mortality compared with lncRNA-ACOD1 KO mice [36]. This suggested that lncRNA-ACOD1 could promote viral replication -- possibly through a previously unidentified mechanism [36]. At steady state, lncRNA-ACOD1 mainly localized to the cytoplasm, and during infection it bound glutamic-oxaloacetic transaminase (GOT2) -- a key metabolic enzyme catalyzing reversible transamination between metabolites such as oxaloacetic acid and L-glutamic acid into L-aspartate and α-ketoglutarate [77]. GOT2 knockdown and overexpression experiments in mouse peritoneal macrophages in vitro demonstrated that GOT2 was essential for lncRNA-ACOD1 function upon viral infection [36]. Specifically, lncRNA-ACOD1 interacted with the small domain of GOT2 through its 5’ segment, as evidenced from RNA sequencing of the GOT2 complex [36].In addition, the use of an aminotransferase activity assay demonstrated that the binding of lncRNA-ACOD1 to GOT2 enhanced enzymatic activity of this enzyme in vitro [36]. Furthermore, upon VSV infection, lncRNA-ACOD1 KO mice showed decreased L-aspartate and α-ketoglutarate expression (metabolic products of GOT2) compared with WT mice [36]. This suggested that upon VSV infection, lncRNA-ACOD1 might enhance GOT2 catalytic activity and production of its metabolites, facilitating viral replication [36]. In response to viral infection with influenza A/PR/8/34 virus, the human lncRNA-ACOD1 ortholog was upregulated, suggesting that this lncRNA might play a role during viral infections in humans. Moreover, a lower viral load was observed in A549 human alveolar basal epithelial cells following deletion of human lncRNA-ACOD1 by RNA interference [36]. GOT2 also bound human lncRNA-ACOD1 upon influenza A/PR/8/34 viral infection in A549 cells [36]. This suggested that the function of lncRNA-ACOD1 in promoting viral replication might be conserved, at least in humans and mice [36]. Moreover this study proposed a new mechanism by which VSV might exploit host metabolic networks via a lncRNA for its replication. Whether this type of lncRNA mechanism can be extended to other viral infections remains to be investigated.
Lnc-Lsm3b in the Regulation of RIG-I Signaling Pathway
Lnc-Lsm3b is located on chromosome 6 in mice, and contains partial sequences of Lsm3 introns and exons [78]. It was identified as one of the most abundant RIG-I-binding lncRNAs in VSV-infected mouse RAW264.7 macrophages [78]. It is significantly induced upon RNA virus infection (VSV or Sendai virus) in a time-dependent manner; however, its expression is completely abolished in mouse peritoneal macrophages from Ifnar KO mice, suggesting that lnc-Lsm3b could be an IFN-induced lncRNA [78]. VSV infection of CRISPR-Cas9-generated lnc-Lsm3b-deficient RAW264.7 cells resulted in increased production of type I IFNs and IL-6 in cell supernatants relative to control cells, with an evident difference 12 hours post-infection [78]. This suggested that lnc-Lsm3b might be a potent negative regulator of RIG-I-mediated type I IFN and proinflammatory cytokine production at later time points of viral infection [78]. Consistent with this, compared to WT mice, lnc-Lsm3b KO mice intravenously-infected with VSV showed less susceptibility to the virus, higher type I IFN serum concentrations at later time points of infection, and lower organ viral burden [78]. Furthermore, in vitro RNA immunoprecipitation (RIP)-qPCR analysis in HEK293T cells co-transfected with plasmids expressing lnc-Lsm3b and RIG-I showed that lnc-Lsm3b bound the carboxy terminal domain (CTD) of RIG-I [78]. RIG-I binding of lnc-Lsm3b was decreased (slightly) with increasing amounts of viral RNA (extracted from VSV-infected HEK293T cells; this suggested that lnc-Lsm3b might restrict immune responses by competing with RIG-I ligands for binding to the CTD of RIG-I [78]. in vitro gel filtration analysis of peritoneal macrophages isolated from lnc-Lsm3b KO mice showed that lnc-Lsm3b deficiency led to increased formation of RIG-I oligomers compared to cells isolated from WT mice; this suggested that lnc-Lms3b prevented oligomerization of RIG-I, and hence, its activation, resulting in decreased type I IFN response [78]. The authors concluded that while lnc-Lsm3b-mediated inhibition of RIG-I activation could prevent overproduction of type I IFNs in a feedback manner to maintain immune homeostasis, the mechanism also facilitated viral replication at a late stage of infection due to decreased type I IFN production [78]. However, since there is no human equivalent of lnc-Lsm3b, it would be interesting to define whether there are any endogenous lncRNAs that might be recognized by RIG-I in a similar fashion, in humans [78].
Pathogen-derived lncRNAs
Some pathogens are known to synthesize their own lncRNAs to regulate their life cycle or facilitate survival in the host, interfering with host innate immune pathways. Functional lncRNAs have been identified in the genome of several pathogenic microorganisms such as the parasite Schistosoma mansoni [79] and the yeast Saccharomyces cerevisiae [80]; however, whether these lncRNAs play a critical role in infectious processes is not always clear. Pathogen-encoded lncRNAs that might be involved in host-pathogen interactions are described below.
PAN
PAN (polyadenylated nuclear RNA) is encoded by Kaposi’s sarcoma herpesvirus (KSHV) [81] and has been shown to associate with host demethylases, ubiquitously transcribed tetratricopeptide repeat, X chromosome (UTX), and JmjC domain-containing protein 3 (JMJD3), promoting lytic viral replication [82]. Specifically, ChiP assays performed on 293L cells transfected with the KSHV genome revealed UTX and JMJD3 binding to KSHV genomic DNA, only when PAN was expressed; conversely, the binding did not occur when cells contained the KSHV genome but where the PAN gene was removed by homologous recombination [82]. Furthermore, qPCR analysis showed reduced viral copy numbers in 293L cells transfected with the KSHV genome lacking PAN, compared to cells expressing the full KSHV genome; this suggested that PAN expression was essential for virus growth [82]. In addition, human Burkitt lymphoma B cells (BJAB cells) transfected with a plasmid containing PAN, exhibited decreased expression of antiviral and pro-inflammatory genes including IL18, IFNA and IFNG, compared to control cells, suggesting that PAN could suppress the expression of host immune response genes [83]. Consequently, the differential association of PAN with specific chromatin modifying complexes might allow this lncRNA to activate and repress gene expression to regulate the KSHV life cycle. However, further investigations are necessary to clarify the molecular mechanisms by which PAN can modulate host immune responses.
HIV-Encoded Antisense Long Noncoding RNA
The human immunodeficiency virus (HIV)-encoded antisense long noncoding RNA was reported to recruit host chromatin remodeling proteins such as DNA methyltransferase 3a (Dnmt3a), HDAC1 and EZH2 to the 5’ long terminal repeat viral promoter (5′ LTR) of the virus, epigenetically suppressing viral transcription [84]. In this study, ChIP analysis showed that the localization of these chromatin remodeling proteins to the HIV 5′ LTR was diminished in ACH2 cells when the HIV-encoded antisense long noncoding RNA was stably knocked down relative to control cells [84]. Thus, the HIV-encoded antisense long noncoding RNA could reduce viral transcription, leaving the virus in a latent state. How and when HIV latency emerges remains unclear; thus a better understanding of HIV-expressed antisense noncoding RNAs in the HIV viral lifecycle may shed light on viral latency [84].
LncRNA-TARE
The malaria pathogen Plasmodium falciparum encodes a family of twenty two telomere-associated lncRNAs called lncRNA-TARE (telomere-associated repetitive element) [85]. P. falciparum genome analysis has shown that lncRNA-TARE neighbor essential genes and factors involved in parasite pathogenesis, such as upsB-type var gene , and are induced after parasite DNA replication; this suggests that lncRNA-TARE may be involved in virulence gene regulation [85]. Furthermore, lncRNA-TARE sequences are enriched with subtelomeric var promoter element 2 (SPE2) motifs, known to function as binding sites for transcription factors such as Plasmodium falciparum ApiAP2 Protein 2 (PfSip2) [86]. In addition, qRT-PCR analysis has demonstrated that during peak parasite DNA replication, the expression of a specific lncRNA-TARE, lncRNA-TARE-4L and PfSip2 is highly correlated with late-stage temporal profiles [85]. Furthermore, PfSip2 is expressed prior to maximal lncRNA-TARE-4L expression, which may indicate PfSip2-mediated induction of lncRNA-TARE and/or co-activation of lncRNA-TARE [85]. lncRNA-TARE might thus play a key role in the transcriptional and/or epigenetic regulation of P. falciparum telomeric and subtelomeric regions, perhaps even in telomere maintenance and virulence gene regulation [85]. However, the precise functions remain unknown and more investigations are evidently necessary [85].
Concluding Remarks
LncRNAs are expressed in all mammalian cell types acting as key regulators of different biological processes. As discussed here, many lncRNAs are emerging as regulators of inflammatory responses of immune cells and host-pathogen interactions. Even though our understanding of the role of lncRNAs in immunity is still in its infancy (see Outstanding questions), during the last few years, several studies have provided new insights into this field. Clear examples are NEAT1 -- shown to play a protective role in humans during Salmonella enterica Thyphimurium infection by triggering the loss of nuclear RNA decay factors [37]. Moreover, lncRNA-ACOD1 by targeting host metabolism, can promote VSV infection in mice, and perhaps modulate influenza A/PR/8/34 virus in humans, although thie latter has not been demonstrated [36]. In addition, lnc-Lsm3b may constitute a molecular decoy involved in saturation of RIG-I binding sites, possibly by restricting the duration of “non-self” RNA-induced innate immune responses during VSV infection in mice [78]. Our knowledge of pathogen-encoded lncRNAs is significantly limited and the possible mechanisms by which these transcripts actively participate in the infectious process remain to be elucidated. For instance, in human cell lines, KSHV-derived PAN has been shown to be associated with host demethylases -- decreasing the expression of certain antiviral and pro-inflammatory genes, and to be fundamental for virus growth. However, how PAN modulates host immune responses and whether its virulent function is confirmed in vivo remain to be addressed. Although significant advances have been made in identifying putative functional roles of various lncRNAs, major limitations remain. Since lncRNAs are not highly conserved across species, it will be extremely challenging to extrapolate results between human and murine experimental systems. Moreover, lncRNAs tend to be tissue- or even cell-specific, thus rendering their functions more difficult to discern at the organismal level. For instance, lincRNA-Cox2 was recently deemed to potentially play differential roles in diverse organs [27]. The in vivo role of many lncRNAs (originally characterized in vitro), remains unknown, particularly in the context of actual host responses to infections. In certain cases (e.g. NEAT1 and MEG3), findings from multiple studies have not been consistent, indicating that further studies are needed to assess a clear role of lncRNAs in these scenarios. Over recent years, humanized mice have been used as pre-clinical models to evaluate new drugs and their safety in a broad array of diseases [87]. Their use in identifying and characterizing new putative lncRNAs would also be critical. For instance, SCID mice engrafted with lncRNA-CD244-depressed human CD8+ T cells have been fundamental in dissecting the role of lncRNA-CD244 during mycobacterial infection. Humanized mice might help overcome certain limitations in lncRNAs studies, facilitating a better functional understanding of human lncRNAs in various disease models. The development of more sophisticated bioinformatics tools will also be fundamental in examining lncRNA sequences and structures; possibly enabling the identification of lncRNA homologs across species and the predictions of lncRNA-interacting partners. Moreover, new high-throughput sequencing methodologies (e.g. RNA Capture Long Seq) and other RNA-based technologies may help to uncover novel putative lncRNAs, lncRNA-interacting molecules and lncRNA functions. Overall, a better understanding of how lncRNAs regulate immune responses and host-pathogen interactions in different contexts might assist the development of lncRNA-based therapeutics to mitigate infections and inflammation-mediated diseases.
Outstanding Questions Box.
Do specific lncRNAs play key roles in the modulation of immune responses and host-pathogen interactions in vivo? In some cases, lncRNAs (e.g. MEG3 and EGOT) have been shown to exert these functions in vitro, whereas their role in vivo has not yet been addressed
Do lncRNAs that have been implicated in immune responses against microbial components (e.g. lincRNA-COX2 and lincRNA-EPS) exert a critical function in host response to infections in vivo as well?
How do pathogens regulate the expression of lncRNAs in a host? Do they secrete particular proteins targeting host machineries? What pathways do they affect?
Do host-derived lncRNAs play a widespread role during infections? So far, only certain host-encoded lncRNAs have been reported to regulate immune responses to viral and/or bacterial infections (e.g. lncRNA-CD244, MEG3, NEAT1 and lncRNA-ACOD1). Whether other host-derived lncRNAs can exert the same function and whether these lncRNAs are involved during other pathogen-induced infections is still unclear.
What are some of the molecular mechanisms that lncRNAs use to exert their specific functions? In many of the examples reported so far, these mechanisms remain unknown or are unclear.
What are some of the functions of pathogen-derived lncRNAs? Do they function to sustain pathogen survival, or are they able to enhance host immune responses? To date, very little is known about this class of lncRNAs.
Can certain lncRNAs be used for the treatment of specific infectious diseases?
Highlights.
LncRNAs are non-coding transcripts longer than 200 nucleotides that bind DNA,
RNA, and proteins, and can regulate gene expression via diverse mechanisms.
Mammalian hosts and pathogens encode lncRNAs that can regulate host-pathogen interactions; these play either beneficial or detrimental roles for host survival.
In human, the host-encoded lncRNA NEAT1 can exert an antibacterial function during Salmonella infection by enhancing host immune gene expression.
The host-encoded lncRNA-ACOD1 can promote viral replication by modulating cellular metabolism in both mouse (VSV infection) and humans (influenza A/PR/8/34 virus infection).
In mice, the host-encoded lncRNA-Lms3b can restrict VSV-induced innate immune responses by inactivating the RIG-I signaling pathway.
The KSHV-encoded lncRNA PAN is critical for virus growth in human cell lines, associating with host demethylases, and decreasing the expression of certain antiviral and pro-inflammatory genes.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R21AI130963. The authors would like to dedicate this publication to the memory of Dr. Andrei Medvedev.
Glossary
- ½-sbsRNAs
lncRNAs that bind mRNAs with the 3’ UTR Alu element, leading to their degradation through SMD
- 3’ UTR
3’ untranslated region of a mRNA immediately following the translation termination site
- 5’ UTR
5’ untranslated region of a mRNA directly upstream from the initiation codon
- Autophagy
process through which cellular material and dysfunctional organelles are degraded and recycled. It also targets intracellular bacteria for lysosomal degradation
- bacillus Calmette-Guerin (BCG)
live attenuated strain of Mycobacterium bovis
- cGAMP
second messenger inducing e.g. a STING-dependent type I IFN response
- cGAS
cytosolic DNA sensor that activates a type-I iFN response
- ChIA-PET
combines ChIP with chromatin conformation capture (3C) technology, detecting when distant DNA regions interact with each other via a protein of interest
- cis-regulatory element
DNA region that regulates the transcription of nearby genes
- CRISPR-Cas9
next-generation genome editing tool
- Decoy
molecule that binds and sequesters other molecules, inhibiting their functions
- DNAPK complex
DNA sensor and inducer of innate immune activation through IRF3, TBK1, and STING
- EZH2
catalytic subunit of the PRC2 complex; can promote transcriptional repression of a target gene
- Gapmers
antisense oligonucleotides used for efficient inhibition of mRNAs and lncRNAs
- Guide
can be a molecule that interacts with protein complexes directing them to specific target genes
- H3K27me3
epigenetic modification that indicates trimethylation of lysine 27 on the histone H3 protein subunit, leading to transcriptional repression
- H3K4me3
epigenetic modification that indicates trimethylation of lysine 4 on the histone H3 protein subunit, leading to transcriptional activation
- HEXIM1
transcriptional regulator acting as an RNA polymerase II transcription inhibitor
- ISD
non-CpG oligomer; strongly enhances IFN-β expression
- JFH1
strain of HCV
- NEXT-RNA exosome pathway
protein complex required for the exosomal degradation of upstream promoter transcripts
- p62
autophagy substrate and adaptor for intracellular bacteria; used as a marker to study autophagic flux
- p70-S6K
hallmark of mTOR activation
- Paraspeckles
subnuclear RNA-protein bodies found in the interchromatin space of mammalian cells
- PKR
upon binding viral dsRNA, undergoes autophosphorylation, regulating translation and multiple signaling pathways
- RACE-seq
method that characterizes cDNA molecules generated by rapid amplification of cDNA ends
- RIG-I
major sensor protein of cytosolic RNA
- RNA decay
process by which RNA molecules are enzymatically degraded
- RNPs
complexes of RNA and protein present in the nucleus during gene transcription, and exerting different functions
- Scaffold
molecule that supports other molecules assembled in a complex
- SCID mice
mice severely deficient in functional B and T lymphocytes
- Septic shock
severe, generalized inflammatory response induced by bloodstream infection with gram-negative bacteria
- STING
molecule that mediates cytosolic DNA-induced signaling events
- SWI/SNF complex
complex that uses adenosine triphosphate-hydrolysis to alter histone–DNA interactions
- Theiler’s virus
single-stranded RNA murine cardiovirus; used as a mouse model for studying virally-induced paralysis, or encephalomyelitis comparable to Multiple sclerosis
- TLR4
receptor that mediates the innate immune response to LPS. Also involved in LPS-independent inflammatory responses triggered by free fatty acids
- trans-regulatory element
DNA region that regulates the transcription of distant genes
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carninci P et al. (2005) The transcriptional landscape of the mammalian genome. Science 309 (5740), 1559–63. [DOI] [PubMed] [Google Scholar]
- 2.Atianand MK et al. (2017) Immunobiology of Long Noncoding RNAs. Annu Rev Immunol 35, 177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S et al. (2012) Landscape of transcription in human cells. Nature 489 (7414), 101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T et al. (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22 (9), 1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson DM et al. (2015) A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160 (4), 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto A et al. (2017) SPAR, a lncRNA encoded mTORC1 inhibitor. Cell Cycle 16 (9), 815–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pang Y et al. (2018) Encoding activities of non-coding RNAs. Theranostics 8 (9), 2496–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi SW et al. (2018) The small peptide world in long noncoding RNAs. Brief Bioinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy MB and Medvedev AE (2016) Long noncoding RNAs as regulators of Toll-like receptor signaling and innate immunity. J Leukoc Biol 99 (6), 839–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotzin JJ et al. (2016) The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537 (7619), 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P et al. (2014) The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344 (6181), 310–3. [DOI] [PubMed] [Google Scholar]
- 12.Spurlock CF 3rd et al. (2015) Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat Commun 6, 6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grote P et al. (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24 (2), 206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretz M et al. (2013) Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493 (7431), 231–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arun G et al. (2016) Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30 (1), 34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner JR et al. (2014) PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res 74 (6), 1651–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang KC and Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43 (6), 904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Rocha ST and Heard E (2017) Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol 24 (3),197–204. [DOI] [PubMed] [Google Scholar]
- 19.McHugh CA et al. (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521 (7551), 232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z et al. (2014) The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 111 (3), 1002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B et al. (2015) A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27 (3), 370–81. [DOI] [PubMed] [Google Scholar]
- 22.Carrieri C et al. (2012) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491 (7424), 454–7. [DOI] [PubMed] [Google Scholar]
- 23.Gong C and Maquat LE (2011) lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470 (7333), 284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458 (7235), 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter S et al. (2013) A long noncoding RNA mediates both activation and repression of immune response genes. Science 341 (6147), 789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu G et al. (2016) LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J Immunol 196 (6), 2799–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elling R et al. (2018) Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep 25 (6), 1511–1524 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilott NE et al. (2015) Corrigendum: Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat Commun 6, 6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atianand MK et al. (2016) A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 165 (7), 1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemson CM et al. (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33 (6), 717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchinson JN et al. (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunwoo H et al. (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19 (3), 347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morchikh M et al. (2017) HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol Cell 67 (3), 387–399 e5. [DOI] [PubMed] [Google Scholar]
- 34.Wu JJ et al. (2015) Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 18 (3), 333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carnero E et al. (2016) Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep 17 (7), 1013–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang P et al. (2017) An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science 358 (6366), 1051–1055. [DOI] [PubMed] [Google Scholar]
- 37.Imamura K et al. (2018) Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J 37 (13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez A et al. (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316 (5824), 608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharbati S et al. (2012) Quantification and accurate normalisation of small RNAs through new custom RT-qPCR arrays demonstrates Salmonella-induced microRNAs in human monocytes. BMC Genomics 13, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoeke L et al. (2013) Intestinal Salmonella typhimurium infection leads to miR-29a induced caveolin 2 regulation. PLoS One 8 (6), e67300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pawar K et al. (2016) Mycobacterium bovis BCG Interferes with miR-3619–5p Control of Cathepsin S in the Process of Autophagy. Front Cell Infect Microbiol 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zur Bruegge J et al. (2016) MicroRNA Response of Primary Human Macrophages to Arcobacter Butzleri Infection. Eur J Microbiol Immunol (Bp) 6 (2), 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazra B et al. (2017) The host microRNA miR-301a blocks the IRF1-mediated neuronal innate immune response to Japanese encephalitis virus infection. Sci Signal 10 (466), eaaf5185. [DOI] [PubMed] [Google Scholar]
- 44.Ho BC et al. (2011) Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9 (1), 58–69. [DOI] [PubMed] [Google Scholar]
- 45.Yi Z et al. (2014) Identifcation of differentially expressed long non-coding RNAs in CD4+ T cells response to latent tuberculosis infection. J Infect 69 (6), 558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ZL et al. (2017) Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci Rep 7 (1), 16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang R et al. (2016) Differential transcription profiles of long non-coding RNAs in primary human brain microvascular endothelial cells in response to meningitic Escherichia coli. Sci Rep 6, 38903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji S et al. (2018) Microarray analysis of lncRNA expression in rabies virus infected human neuroblastoma cells. Infect Genet Evol. [DOI] [PubMed] [Google Scholar]
- 49.Riege K et al. (2017) Massive Effect on LncRNAs in Human Monocytes During Fungal and Bacterial Infections and in Response to Vitamins A and D. Sci Rep 7, 40598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y et al. (2018) Characterization of Critical Functions of Long Non-Coding RNAs and mRNAs in Rhabdomyosarcoma Cells and Mouse Skeletal Muscle Infected by Enterovirus 71 Using RNA-Seq. Viruses 10 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menard KL et al. (2018) Toxoplasma gondii Manipulates Expression of Host Long Noncoding RNA during Intracellular Infection. Sci Rep 8 (1), 15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y et al. (2015) Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A 112 (29), E3883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomez JA et al. (2013) The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 152 (4), 743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vigneau S et al. (2003) Tmevpg1, a candidate gene for the control of Theiler’s virus persistence, could be implicated in the regulation of gamma interferon. J Virol 77 (10), 5632–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collier SP et al. (2012) Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol 189 (5), 2084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aubagnac S et al. (2002) Bone marrow chimeras reveal non-H-2 hematopoietic control of susceptibility to Theiler’s virus persistent infection. J Virol 76 (11), 5807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bureau JF et al. (1992) The interaction of two groups of murine genes determines the persistence of Theiler’s virus in the central nervous system. J Virol 66 (8), 4698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cagliani R et al. (2011) Balancing selection is common in the extended MHC region but most alleles with opposite risk profile for autoimmune diseases are neutrally evolving. BMC Evol Biol 11, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang K et al. (2010) Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet 19 (10), 2059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q et al. (2013) NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio 4 (1), e00596–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Imamura K et al. (2014) Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell 53 (3), 393–406. [DOI] [PubMed] [Google Scholar]
- 62.Ma H et al. (2017) The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. J Virol 91 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z et al. (2017) NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell Mol Life Sci 74 (6), 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X et al. (2018) A novel lncRNA regulates HCV infection through IFI6. Hepatology. [Google Scholar]
- 65.Wagner LA et al. (2007) EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 109 (12), 5191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benetatos L et al. (2011) MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer 129 (4), 773–9. [DOI] [PubMed] [Google Scholar]
- 67.Zhou Y et al. (2012) MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol 48 (3), R45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ying L et al. (2013) Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst 9 (3), 407–11. [DOI] [PubMed] [Google Scholar]
- 69.Anwar SL et al. (2012) Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma. PLoS One 7 (11), e49462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pawar K et al. (2016) Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci Rep 6, 19416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jung CH et al. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20 (7), 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gutierrez MG et al. (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119 (6), 753–66. [DOI] [PubMed] [Google Scholar]
- 73.Bjorkoy G et al. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171 (4), 603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tao XW et al. (2018) LncRNA MEG3 ameliorates respiratory syncytial virus infection by suppressing TLR4 signaling. Mol Med Rep 17 (3), 4138–4144. [DOI] [PubMed] [Google Scholar]
- 75.Marchant D et al. (2010) Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. J Virol 84 (21), 11359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marr N and Turvey SE (2012) Role of human TLR4 in respiratory syncytial virus-induced NF-kappaB activation, viral entry and replication. Innate Immun 18 (6), 856–65. [DOI] [PubMed] [Google Scholar]
- 77.Toney MD (2014) Aspartate aminotransferase: an old dog teaches new tricks. Arch Biochem Biophys 544, 119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang M et al. (2018) Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 173 (4), 906–919 e13. [DOI] [PubMed] [Google Scholar]
- 79.Vasconcelos EJR et al. (2017) The Schistosoma mansoni genome encodes thousands of long non-coding RNAs predicted to be functional at different parasite life-cycle stages. Sci Rep 7 (1), 10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niederer RO et al. (2017) Long Noncoding RNAs in the Yeast S. cerevisiae. Adv Exp Med Biol 1008, 119–132. [DOI] [PubMed] [Google Scholar]
- 81.Sun R et al. (1996) Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 93 (21), 11883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rossetto CC and Pari G (2012) KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 8 (5), e1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rossetto CC and Pari GS (2011) Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J Virol 85 (24), 13290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saayman S et al. (2014) An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 22 (6), 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Broadbent KM et al. (2011) A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol 12 (6), R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flueck C et al. (2010) A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog 6 (2), e1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walsh NC et al. (2017) Humanized Mouse Models of Clinical Disease. Annu Rev Pathol 12, 187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mercer TR et al. (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 105 (2), 716–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Werner A et al. (2009) What do natural antisense transcripts regulate? RNA Biol 6 (1), 43–8. [DOI] [PubMed] [Google Scholar]
- 90.Lagarde J et al. (2016) Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq). Nat Commun 7, 12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frankish A et al. (2018) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lagarde J et al. (2017) High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet 49 (12), 1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaneko S et al. (2013) PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20 (11), 1258–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simon MD et al. (2013) High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504 (7480), 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Engreitz JM et al. (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341 (6147), 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128 (4), 787–800. [DOI] [PubMed] [Google Scholar]
- 97.Dekker J (2008) Gene regulation in the third dimension. Science 319 (5871), 1793–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fanucchi S et al. (2019) Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat Genet 51 (1), 138–150. [DOI] [PubMed] [Google Scholar]
- 99.Werner A and Sayer JA (2009) Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens 18 (4), 343–9. [DOI] [PubMed] [Google Scholar]