Abstract
Expansion of the genetic code with unnatural amino acids (Uaas) has significantly increased the chemical space available to proteins for exploitation. Due to the inherent limitation of translational machinery and the required compatibility with biological settings, function groups introduced via Uaas to date are restricted to chemically inert, bioorthogonal, or latent bioreactive groups. To break this barrier, here we report a new strategy enabling the specific incorporation of biochemically reactive amino acids into proteins. A latent bioreactive amino acid is genetically encoded at a position proximal to the target natural amino acid; they react via proximity-enabled reactivity, selectively converting the latter into a reactive residue in situ. Using this Genetically Encoded Chemical COnversion (GECCO) strategy and harnessing the sulfur-fluoride exchange (SuFEx) reaction between fluorosulfate-L-tyrosine and serine or threonine, we site-specifically generated the reactive dehydroalanine and dehydrobutyrine into proteins. GECCO works both inter- and intra-molecularly, and is compatible with various proteins. We further labeled the resultant dehydroalanine-containing protein with thiol-saccharide to generate glycoprotein mimetics. GECCO represents a new solution for selectively introducing biochemically reactive amino acids into proteins and is expected to open new avenues for exploiting chemistry in live systems for biological research and engineering.
Graphical Abstract
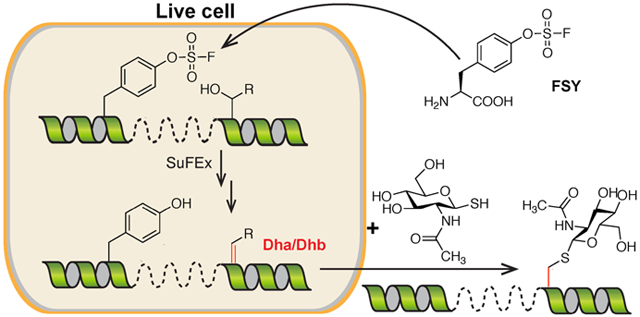
Through engineering orthogonal components for protein translation, unnatural amino acids (Uaas) have been genetically encoded in various cells and model organisms.1-5 To be compatible with biological settings, side chains of these encoded Uaas are mainly chemically inert or bioorthogonal.2,3,6 A recent breakthrough is the encoding of latent bioreactive Uaas, which are unreactive inside cells but once incorporated into proteins able to form covalent bonds with natural amino acid residues in proximity.7-10 Nonetheless, it remains infeasible to selectively introduce biochemically reactive Uaas into proteins in live cells, because the biochemical reactivity of the Uaa may interfere with protein translation and other biological processes. Nature, on the other hand, has installed the reactive dehydroalanine (Dha) and dehydrobutyrine (Dhb) into proteins through enzymatic posttranslational modifications, which are used to create unique intra-protein bridges in lantipeptides and thiopeptides possessing antimicrobial and antitumor activities.11,12 Through chemical conversion Dha and Dhb can also be generated in vitro, and the unique structure and reactivity of α,β-unsaturated carbonyl moiety in Dha have been harnessed for chemical mutagenesis and installation of a broad range of posttranslational modifications, providing an invaluable route for studying proteins.13,21 However, due to the cellular incompatibility of reagents and conditions used for chemical conversion, methods reported to date cannot generate Dha or Dhb in vivo.
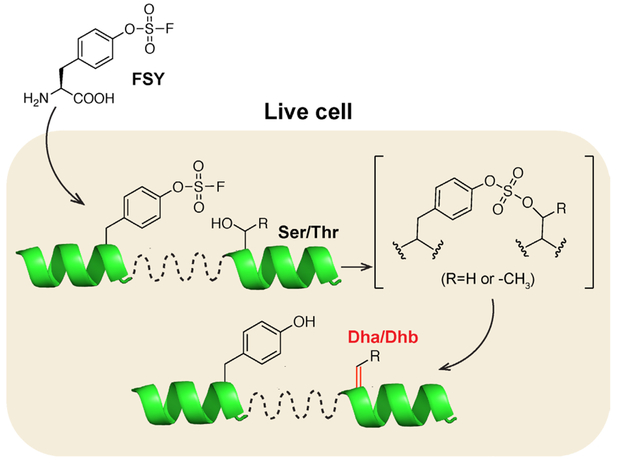
Here we present a novel strategy to selectively introduce biochemically reactive Uaas into proteins. We genetically encode a latent bioreactive Uaa into proteins at a site proximal to the target position; enabled by proximity-enhanced reactivity,10 the latent bioreactive Uaa then reacts with the nearby target natural amino acid residue, selectively converting it into a biochemically reactive Uaa (Fig. 1). Through incorporating the latent bioreactive Uaa fluorosulfate-L-tyrosine (FSY) and harnessing its reaction with Ser or Thr, we demonstrated the generation of Dha and Dhb on proteins. We further showed that the resultant Dha could be used to selectively attach saccharide for generating glycoprotein mimetics. We expect that Dha and Dhb generated in situ will enable versatile chemical means for protein research and engineering in live cells, and that this Genetically Encoded Chemical COnversion (GECCO) strategy will open a new avenue for selective introduction of biochemically reactive amino acids in vivo.
Figure 1.
GECCO to site-selectively introduce biochemically reactive amino acids into proteins. The latent bioreactive Uaa FSY reacts with a nearby Ser or Thr via proximity-enabled reactivity, selectively converting the latter into Dha or Dhb.
We recently evolved an orthogonal tRNAPyl/FSYRS pair that genetically incorporates Uaa FSY into proteins in E. coli and mammalian cells.22 The incorporated FSY was found to react with Lys, His, and Tyr in proximity through sulfur-fluoride exchange (SuFEx) reaction, forming covalent protein crosslinks in vivo. However, we did not detect any crosslinking between FSY and Ser or Thr.22 In contrast, arylfluorosulfate installed on chemical probes was able to react with Lys, Tyr, and Ser within a positively charged binding pocket of the specifically bound protein, and the resultant arylfluorosulfate-Ser adduct was found to partially hydrolyze to Dha,23-25 but what occurred to the arylfluorosulfate warhead remains uncharacterized. Prompted by these findings, we aimed to determine whether proximal FSY and Ser incorporated into proteins (instead of on small molecules) would react, whether a positively charged microenvironment is necessary, and what the products would be. We thus incorporated FSY and Ser at different protein contexts without positively charged residues nearby, and characterized their identity using high resolution tandem MS.
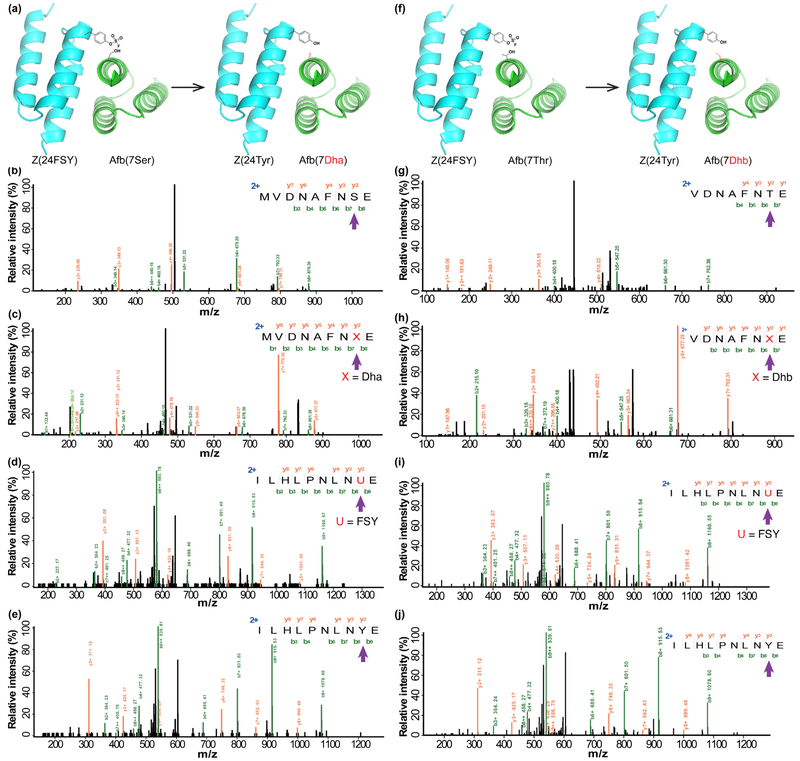
We first examined whether FSY would react with Ser intermolecularly when separately installed on two interacting proteins. Specifically, we incorporated FSY at position E24 of the Z protein, and introduced Ser at position K7 of the Z-binding affibody (Afb) (Fig. 2a), placing them in proximity when Afb binds with Z.26 Proteins Z(24FSY) and Afb(7Ser) were co-expressed in E. coli, allowing them to bind and react in vivo, and then purified and characterized using MS. As expected, tandem MS identified the Ser-containing peptide for the Afb(7Ser) protein (Fig. 2b). To our delight, we also identified this peptide with Dha at the 7Ser position (Fig. 2c). A series of b and y ions in the tandem MS unambiguously indicate the presence of Dha at the 7Ser position, confirming the conversion of Ser to Dha. In addition, we identified the FSY-containing peptide for the Z(24FSY) protein (Fig. 2d), indicating specific incorporation of FSY. Meanwhile, the peptide containing Tyr at the 24FSY position was also identified (Fig. 2e), suggesting the conversion of FSY to Tyr upon reacting with Ser. Based on the peak areas of these peptides in the extracted ion chromatograms (Fig. S1), the conversion rate of Ser to Dha was ca. 3.7% and FSY to Tyr was ca. 4.1%. These results indicate that FSY installed proximal to Ser in proteins could intermolecularly convert Ser into Dha.
Figure 2.
Generation of Dha and Dhb on proteins via intermolecular GECCO in E. coli. (a) Structure of Afb-Z complex (PDB: 1LP1) showing two proximal sites for placing FSY and the target Ser. (b-c) Tandem mass spectra identifying Ser and Dha at site 7 of the Afb protein. (d-e) Tandem mass spectra identifying FSY and Tyr at site 24 of the Z protein. (f) Structure of Afb-Z complex (PDB: 1LP1) showing two proximal sites for placing FSY and the target Thr. (g-h) Tandem mass spectra identifying Thr and Dhb at site 7 of the Afb protein. (i-j) Tandem mass spectra identifying FSY and Tyr at site 24 of the Z protein.
We next examined whether FSY could convert Thr to Dhb in proteins. Similarly, proteins Z(24FSY) and Afb(7Thr) were co-expressed in E. coli (Fig. 2f). MS analyses of the purified proteins identified the Thr-containing peptide (Fig. 2g) as well as the Dhb-containing peptide (Fig. 2h) ofAfb(7Thr), validating the conversion of 7Thr to Dhb. Again, for protein Z(24FSY), the FSY-containing peptide was identified (Fig. 2i), together with this peptide wherein FSY was converted into Tyr (Fig. 2j). Extracted ion chromatograms of these peptides indicate that the conversion rate of Thr to Dhb was ca. 5.5% and FSY to Tyr was ca. 7.2% (Fig. S2). These data suggest that FSY was able to convert both Ser and Thr into the respective Dha and Dhb, while changing itself back to the natural amino acid Tyr.
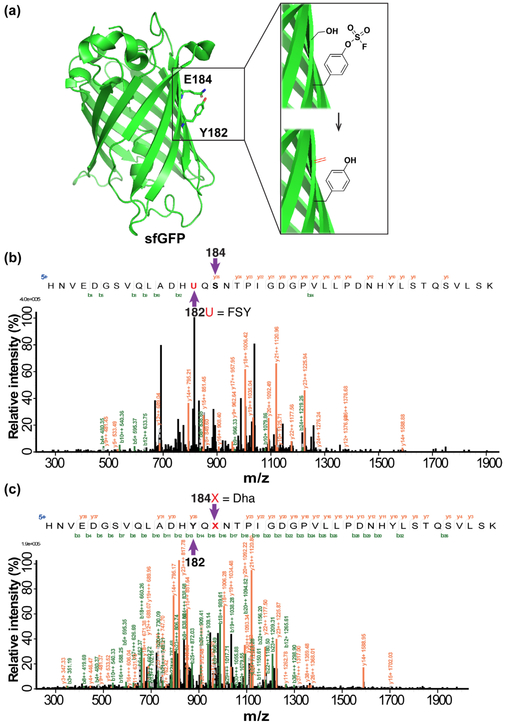
We then determined whether Dha could be generated by FSY through intramolecular conversion. We initially targeted a Ser located on a β-strand of the super-fold green fluorescent protein (sfGFP). FSY was incorporated at the permissive site Tyr182 and a Ser was placed at site 184, the i+2 position so that both side chains pointing to the same side of the β-strand (Fig. 3a). This mutant sfGFP(182FSY/184Ser) was expressed in E. coli and then characterized by MS. The peptide containing FSY at site 182 and Ser at site 184 was identified, again indicating specific incorporation of FSY (Fig. 3b). As expected, this peptide was also identified containing Tyr at site 182 and Dha at site 184 (Fig. 3c). The conversion rate of Ser to Dha was 1.3% (Fig. S3). We note that no peptide containing FSY182/Dha184 was detected, that is, whenever Dha was detected at site 184, site 182 was always found to be Tyr, indicating that conversion of Ser to Dha was consequentially associated with conversion of FSY to Tyr. The Dha conversion rate was low possibly because the rigidity of the sfGFP β-strand does not allow ample contact of FSY with Ser; computational modeling indicates that the side chains of FSY and Ser point away from each other in their sterically allowed rotamers (Fig. S4).
Figure 3.
Generation of Dha on sfGFP via intramolecular GECCO in E. coli. (a) Crystal structure of sfGFP (PDB: 2B3P) showing site Tyr182 for FSY incorporation to target Ser introduced at site Glu184 on the β-strand. (b-c) Tandem MS spectra of sfGFP (182FSY/184Ser) expressed in E. coli identifying 182FSY/184Ser (b) and 182Tyr/184Dha (c).
We next tested intramolecular conversion in a less rigid protein context in ubiquitin. FSY was introduced in site 45, which has contact with Ser65 and Lys63. After expressing ubiquitin(FSY45) in E. coli followed by MS characterization, we found that FSY predominately crosslinked with Lys63 and Ser65 was converted to Dha in 1.7% yield (Fig. S5).
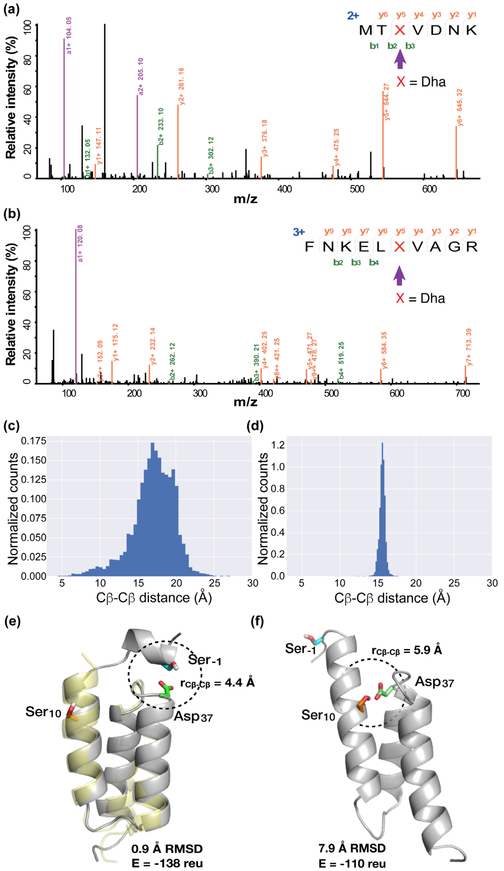
We therefore reasoned that a good contact of FSY with the target Ser without competition from Lys, His, and Tyr, which are known to react with FSY,22 may promote the conversion rate. To test this idea, we incorporated FSY into Afb at site Asp37, which is located in a loop and has no contact with Lys, His or Tyr, and determined how efficiently nearby Ser could be converted into Dha. After expressing Afb (37FSY) in E.coli followed by purification, MS characterization revealed that Ser10 was converted to Dha in 3.9% yield and Ser-1 to Dha in 53% yield (Fig. 4a, 4b, S6). The crystal structure of Afb is available only in complex with the Z protein, and some residues of Afb are missing in this complex structure. To understand the conversion difference, we performed ab initio folding of the Afb sequence containing all residues, including additional residues Met(−3)Thr(−2)Ser(−1) at the N-terminus introduced by cloning. The Cβ-Cβ distances between site 37 and Ser-1 or Ser10 were analyzed in outputted models (Fig. 4c, 4d, S7). Many low energy/low RMSD models contain Ser-1 with a close distance to site 37, whereas very few models contain Ser10 with a close distance to site 37—and these few models are very high energy (Fig. 4c-f). These data support that better contact of FSY with Ser-1 indeed enhances the conversion of Ser to Dha.
Figure 4.
Generation of Dha on Afb via intramolecular GECCO in E. coli. (a-b) Tandem mass spectra identifying Dha at Ser-1 (a) and Ser10 (b) of the Afb protein. (c-d) Histogram of Cβ-Cβ distances of Ser-1 and Asp37 (c) and of Ser10 and Asp37 (d) in 4,525 low energy models of ab initio folded Afb. (e-f) Representative models from ab initio folding of Afb showing Asp37 close to Ser-1 (e) and close to Ser10 (f). The gold structure in (e) is the aligned Afb backbone of 1LP1.
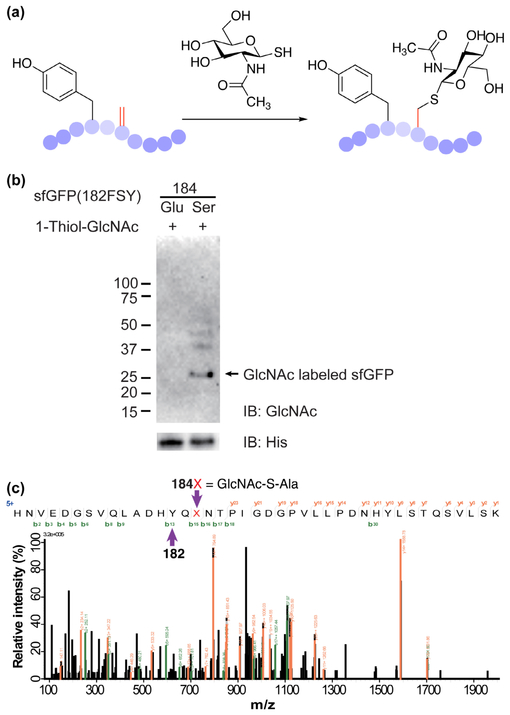
To further demonstrate the chemical identity of Dha generated by FSY conversion, we labeled Dha with a thiol-derivatized saccharide to generate glycoprotein mimetics (Fig. 5a). Methods for preparing site-selectively glycosylated proteins and mimetics are valuable for studying protein glycosylation.17,27-30 We synthesized 2-acetamido-2-deoxy-1-thio-β-D-glucopyranose (1-thiol-GlcNAc) in 3 steps (68% overall yield, Supplementary methods), and incubated 1-thiol-Glc-NAc with sfGFP(182FSY/184Ser) (expressed from E.coli as described above and containing 182Tyr/184Dha) under mild conditions. A similarly expressed and purified sfGFP(182FSY/184Glu) protein was used as the negative control. Western blot analysis of reaction product using an antibody specific for GlcNAc showed that only the Dha-containing sfGFP was labeled by 1-thiol-GlcNAc (Fig. 5b). The reaction product was further analyzed with tandem MS, which clearly confirmed the attachment of 1-thiol-GlcNAc onto Dha at site 184 (Fig. 5c). These results further validate the chemical identity of Dha generated by FSY conversion and its value for selective protein modification.20
Figure 5.
Labeling Dha-containing sfGFP with 1-thiol-GlcNAc. (a) Scheme showing the structure of 1-thiol-GlcNAc and its reaction with Dha. (b) Western blot and (c) tandem MS analysis of reaction product confirming successful labeling of Dha with GlcNAc.
In summary, we developed a new method, GECCO, which enables the genetically introducing of biochemically reactive amino acids into proteins. Harnessing proximity-enabled reactivity, a genetically incorporated latent bioreactive Uaa converts a nearby target natural residue into a reactive amino acid in situ. We demonstrated the conversion of Ser and Thr into the reactive Dha and Dhb, respectively, and the labeling of the Dha-containing protein with a thiol-saccharide to generate glycoprotein mimetics. The conversion occurred both inter-and intra-molecularly on various proteins, with the conversion rate dependent on the contact between FSY and the target Ser/Thr. Dha and Dhb also represent two smallest Uaas introduced into proteins via genetic code expansion to date. Compared with existing methods for introducing Dha to proteins via chemical transformation,20 this work presents a recombinant approach to produce Dha/Dhb-containing proteins without extra chemical treatments. We expect that GECCO will enable the genetically introducing of additional biochemically reactive amino acids in proteins, thus expanding new avenues for exploiting chemistry in live systems for biological research and engineering.
Supplementary Material
ACKNOWLEDGMENT
W.F.D acknowledges NIH support (R35 GM122603). L.W. acknowledges NIH support (R01GM118384, RF1MH114079).
Footnotes
Supporting Information.
The Supporting Information is available free of charge on the ACS Publications website.
Experimental details and supporting Figures Sl-7 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Wang L; Brock A; Herberich B; Schultz PG Expanding the Genetic Code of Escherichia Coli. Science 2001, 292 (5516), 498–500. [DOI] [PubMed] [Google Scholar]
- (2).Wang L; Schultz PG Expanding the Genetic Code. Angew. Chem. Int. Ed. Engl 2005, 44 (1), 34–66. [DOI] [PubMed] [Google Scholar]
- (3).Liu CC; Schultz PG Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem 2010, 79 (1), 413–444. [DOI] [PubMed] [Google Scholar]
- (4).Wang L Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts. Acc. Chem. Res 2017, 50 (11), 2767–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chen Y; Ma J; Lu W; Tian M; Thauvin M; Yuan C; Volovitch M; Wang Q; Holst J; Liu M; Vriz S; Ye S; Wang L; Li D Heritable Expansion of the Genetic Code in Mouse and Zebrafish. Cell Res 2017,27 (2), 294–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang L; Xie J; Schultz PG Expanding the Genetic Code. Annu. Rev. Biophys. Biomol. Struct 2006, 35 (l), 225–249. [DOI] [PubMed] [Google Scholar]
- (7).Xiang Z; Ren H; Hu YS; Coin I; Wei J; Cang H; Wang L Adding an Unnatural Covalent Bond to Proteins Through Proximity-Enhanced Bioreactivity. Nat. Methods 2013,10 (9), 885–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Xiang Z; Lacey VK; Ren H; Xu J; Burban DJ; Jennings PA; Wang L Proximity-Enabled Protein Crosslinking Through Genetically Encoding Haloalkane Unnatural Amino Acids. Angew. Chem. Int. Ed. Engl 2014,53,2190–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Furman JL; Kang M; Choi S; Cao Y; Wold ED; Sun SB; Smider VV; Schultz PG; Kim CH A Genetically Encoded Aza-Michael Acceptor for Covalent Cross-Linking of Protein-Receptor Complexes. J. Am. Chem. Soc 2014,136 (23), 8411–8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wang L Genetically Encoding New Bioreactivity.N. Biotechnol 2017,38 (Pt A), 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Li H; Xu H; Zhou Y; Zhang J; Long C; Li S; Chen S; Zhou J-M; Shao F The Phosphothreonine Lyase Activity of a Bacterial Type III Effector Family. Science 2007, 315 (5814), 1000–1003. [DOI] [PubMed] [Google Scholar]
- (12).Repka LM; Chekan JR; Nair SK; Van der Donk WA Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem. Rev 2017,117 (8), 5457–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Seebeck FP; Szostak JW Ribosomal Synthesis of Dehydroalanine-Containing Peptides./. J. Am. Chem. Soc 2006,128 (22), 7150–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang J; Schiller SM; Schultz PG A Biosynthetic Route to Dehydroalanine-Containing Proteins. Angew. Chem. Int. Ed 2007, 46 (36), 6849–6851. [DOI] [PubMed] [Google Scholar]
- (15).Guo J; Wang J; Lee JS; Schultz PG Site-Specific Incorporation of Methyl- and Acetyl-Lysine Analogues Into Recombinant Proteins. Angew. Chem. Int. Ed. Engl 2008, 47 (34), 6399–6401. [DOI] [PubMed] [Google Scholar]
- (16).Wang ZU; Wang Y-S; Pai P-J; Russell WK; Russell DH; Liu WR A Facile Method to Synthesize Histones with Posttranslational Modification Mimics. Biochemistry 2012, 51 (26), 5232–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wright TH; Bower BJ; Chalker JM; Bernardes GJL; Wiewiora R; Ng WL; Raj R; Faulkner S; Vallee MRJ; Phanumartwiwath A; Coleman OD; Thezenas ML; Khan M; Galan SRG; Lercher L; Schombs MW; Gerstberger S; Palm-Espling ME; Baldwin AJ; Kessler BM; Claridge TDW; Mohammed S; Davis BG Posttranslational Mutagenesis: a Chemical Strategy for Exploring Protein Side-Chain Diversity. Science 2016, 354 (6312), aag1465–aag1465. [DOI] [PubMed] [Google Scholar]
- (18).Yang A; Ha S; Ahn J; Kim R; Kim S; Lee Y; Kim J; Soil D; Lee H-Y; Park H-S A Chemical Biology Route to Site-Specific Authentic Protein Modifications. Science 2016, 354 (6312), 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Freedy AM; Matos MJ; Boutureira O; Corzana F; Guerreiro A; Akkapeddi P; Somovilla VJ; Rodrigues T; Nicholls K; Xie B; Jiméanez-Osés G; Brindle KM; Neves AA; Bernardes GJL Chemoselective Installation of Amine Bonds on Proteins Through Aza-Michael Ligation. J. Am. Chem. Soc 2017, 139 (50), 18365–18375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Dadová J; Galan SR; Davis BG Synthesis of Modified Proteins via Functionalization of Dehydroalanine. Curr. Opin. Chem. Biol 2018, 46, 71–81. [DOI] [PubMed] [Google Scholar]
- (21).de Bruijn AD; Roelfes G Catalytic Modification of Dehydroalanine in Peptides and Proteins by Palladium-Mediated Cross-Coupling. Chemistry 2018, 24 (48), 12728–12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wang N; Yang B; Fu C; Zhu H; Zheng F; Kobayashi T; Liu J; Li S; Ma C; Wang PG; Wang Q; Wang L Genetically Encoding Fluorosulfate-L-Tyrosine to React with Lysine, Histidine, and Tyrosine via SuFEx in Proteins in Vivo. J. Am. Chem. Soc 2018, 140, 4995–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chen W; Dong J; Plate L; Mortenson DE; Brighty GJ; Li S; Liu Y; Galmozzi A; Lee PS; Hulce JJ; Cravatt BF; Saez E; Powers ET; Wilson IA; Sharpless KB; Kelly JW Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(S) Through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue. J. Am. Chem. Soc 2016, 138 (23), 7353–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mortenson DE; Brighty GJ; Plate L; Bare G; Chen W; Li S; Wang H; Cravatt BF; Forli S; Powers ET; Sharpless KB; Wilson IA; Kelly JW “Inverse Drug Discovery” Strategy to Identify Proteins That Are Targeted by Latent Electrophiles as Exemplified by Aryl Fluorosulfates. J. Am. Chem. Soc 2018, 140 (1), 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Fadeyi OO; Hoth LR; Choi C; Feng X; Gopalsamy A; Hett EC; Kyne RE; Robinson RP; Jones LH Covalent Enzyme Inhibition Through Fluorosulfate Modification of a Noncatalytic Serine Residue. ACS Chem. Biol 2017, 12 (8), 2015–2020. [DOI] [PubMed] [Google Scholar]
- (26).Hogbom M; Eklund M; Nygren PA; Nordlund P Structural Basis for Recognition by an in Vitro Evolved Affibody. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (6), 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu H; Wang L; Brock A; Wong C-H; Schultz PG A Method for the Generation of Glycoprotein Mimetics. J. Am. Chem. Soc 2003, 125 (7), 1702–1703. [DOI] [PubMed] [Google Scholar]
- (28).Kiessling LL; Grim JC Glycopolymer Probes of Signal Transduction. Chem. Soc. Rev 2013, 42 (10), 4476–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tiwari VK; Mishra BB; Mishra KB; Mishra N; Singh AS; Chen X Cu-Catalyzed Click Reaction in Carbohydrate Chemistry. Chem. Rev 2016, 116 (5), 3086–3240. [DOI] [PubMed] [Google Scholar]
- (30).Li C; Wang L-X Chemoenzymatic Methods for the Synthesis of Glycoproteins. Chem. Rev 2018, 118 (17), 8359–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.