Abstract
Transcriptional regulatory elements are typically found in relatively nucleosome-free genomic regions, often referred to as “open chromatin.” DNase I can digest nucleosome-depleted DNA (presumably bound by transcription factors), but DNA in nucleosomes or higher-order chromatin fibers is less accessible to the nuclease. DNase-seq uses high throughput sequencing to permit the interrogation of DNase hypersensitive sites (DHSs) across the entire genome. This protocol details how to perform DNase-seq in vivo using Xenopus embryos.
MATERIALS
Reagents
Agarose gel and reagents for electrophoresis
Buffer A <R>
Deoxyribonuclease I (Sigma-Aldrich D4527)
DNA gel extraction kit (MACHEREY-NAGEL NucleoSpin® Gel and PCR Clean-up 740609.250)
DNaseI 10x, 1x digestion buffer <R>
Iodixanol solutions <R>
OptiPrep™ Density Gradient Medium (Sigma-Aldrich D1556)
Pefabloc® SC PLUS (Roche 11873601001)
Reagents for DNA purification (Phenol:chloroform:isoamyl alcohol)
Protease inhibitor tablets (cOmplete™, EDTA-free, Roche 4693132001)
Protease K (20mg/mL)
RNase A (10mg/mL)
Stock Deoxyribonuclease I (10U/μL) <R>
Stop buffer <R>
Sucrose solution (0.3M) <R>
TE buffer (10mM Tris-HCl, 1mM EDTA, pH 8.0)
Tris-HCl (10mM, pH 8.0)
Triton™ N-101, reduced (Sigma-Aldrich 303135)
Xenopus embryos
Equipment
Low-speed, refrigerated centrifuge (e.g., Sorvall RT-1000B) with a swinging-bucket rotor
Dounce homogenizer (15mL) with 1mm clearance pestle
Nylon mesh, 100-μm pore size (Ted Pella, Inc., 41–12115)
Microcentrifuge tubes (1.5mL Eppendorf tubes, 2mL Sarstept 72.693.005)
Nutator
Polypropylene tubes (15mL)
Syringe (15mL)
Tygon tubing, (Saint-Gobain™ Tygon™ Clear Laboratory Tubing, R-3603)
Ultracentrifuge with swinging-bucket rotor (e.g., Beckman SW32Ti) and appropriate tubes (Polyallomer centrifuge tube, Beckman 326823)
Water baths preset to 37°C, 55°C
Wide-bore P1000 pipette tips (tips clipped with scissors)
METHOD
Isolation of nuclei from Xenopus Embryos
-
1
Obtain dejellied embryos using standard protocols (Ogino et al., 2006). Collect the embryos at the desired stage.
10–20 million cells are required to create a DNase-seq library. This protocol is written based on using 2,000 early gastrulae, which have ~10,000 cells per embryo. These numbers may change with developmental stage, which will require empirical testing of nuclear isolation efficiency and DNase I activity.
During steps 2–9, keep all materials and solutions on ice.
-
2
Transfer the embryos to a pre-chilled 15mL Dounce homogenizer. Gently wash the embryos twice in 10 mL/wash of ice-cold 0.3M sucrose solution. Remove as much of the solution as possible after the second wash.
-
3
Add 4mL of ice-cold 0.3M sucrose solution containing 0.4% (w/v) Triton N-101.
-
4
Homogenize the embryos using 5–7 strokes of the pre-chilled pestle to release intact nuclei.
-
5
Filter the homogenate through a nylon mesh into a pre-chilled 50 mL tube. Wash the mesh with 1mL of 0.3M sucrose solution containing 0.4% (w/v) Triton N-101.
-
6
Mix the filtered homogenate with an equal volume of ice-cold 30% iodixanol to make 15% iodixanol.
-
7
In a 38.5mL polyallomer centrifuge tube, layer 12mL each of 25% and 20% iodixanol solutions and 14mL of 15% iodixanol with the homogenate.
To make sharply separated discontinuous gradients (25%; 20%; 15% iodixanol), first add 20% iodixanol solution to the centrifuge tube. Then, gently add 25% iodixanol solution underneath the 20% iodixanol using a syringe attached to Tygon tubing. The final layer of 15% iodixanol solution containing the homogenized embryos, should be gently pipetted onto the top of 20% gradient.
-
8
Centrifuge at 20,000g using a SW32Ti rotor for 20min at 4°C with maximum braking.
-
9
Harvest the nuclei above the interface between the 25% iodixanol cushion and the 20% iodixanol layer. Transfer the nuclei to a pre-chilled 15mL polypropylene tube. Dilute the nuclei with two volumes of 0.3M sucrose solution and mix gently by inverting several times. Centrifuge at 1,000g with a swinging-bucket rotor for 5min at 4°C. Remove the solution by pipetting.
-
10
Gently resuspend the pellet in 5mL of ice-cold buffer A. Centrifuge at 1,000g with a swinging-bucket rotor for 5min at 4 °C. Remove as much of the solution as possible by gentle pipetting.
Nuclei can be quantitated by DAPI staining of the nuclei on a hemocytometer.
DNase I Digestion of the Isolated Nuclei
For early gastrulae, nuclei from 500 embryos are used in one digestion reaction in a 2mL microcentrifuge tube. If starting from 2,000 embryos, resuspend the nuclei with 1200μL (4×300μL) of 1x digestion buffer and prepare four reaction tubes at step 12.
-
11
During centrifugation, steps 9 or 10 above, prepare tubes for the DNase I digestion reactions, as follows:
Add 200μL of 1x digestion buffer into each 2mL microcentrifuge tube. Then add needed amounts of 10U/μL of stock DNase I (e.g., 1, 2, 4 and 5μL for final 10, 20, 40 and 50U/rxn respectively). Gently flick to mix.
The efficiency of the DNase release of hypersensitive site fragments can be measured relative to “insensitive” regions using qPCR (see Discussion).
-
12
Resuspend the nuclear pellet from step 10 in N × 300μL (N = number of reactions) of 1x digestion buffer by gentle pipetting with a wide-bore pipette tip.
-
13
Transfer 300μL volumes of nuclear suspension to reaction tubes described in step 11 with a wide-bore pipette tip. Mix the samples by gentle pipetting.
-
14
Incubate the samples at 37 °C for exactly 3 minutes for the DNase I digestion.
-
15
Add 500μL of stop buffer. Mix by gentle inverting the tube. Incubate at 37 °C for 15 minutes.
Recovery of the DNase I hypersensitive fragments
-
16
Transfer samples to 15mL conical tubes and add 2mL of TE. Add 60μL of 10mg/ml of RNase A to each tube.. Incubate at 37 °C for 1 hour.
-
17
Add 40μL of 20mg/mL proteinase K and incubate 2 hours to overnight at 55 °C.
-
18
Perform phenol:chloroform:isoamyl alcohol extractions twice followed by one chloroform extraction, then recover the DNA by precipitation using 1/10 volume of 5M NaCl and two volumes of ethanol.
To prevent shearing of genomic DNA, rock the phenol:chloroform:isoamyl alcohol with sample on a nutator at 4 °C for 30minutes.
-
19
Resuspend DNA pellet in 30μL TE.
-
20
Run a 1% agarose gel. Cut a gel piece containing 50–500bp DHS fragments. Use a gel extraction kit. Elute the DNA in 20–30μL of 10mM Tris-HCl, pH8.0.
Ethidium bromide or other DNA staining dye like SYBR gold can be used for gel staining. Run the gel at low voltage (~10 Volts/cm) to reduce heat generation and permit better size resolution.
Size-fractionated DNA is ready for qPCR and/or for generating a DNase-seq library (see Discussion) and can be stored at −20 °C.
20–30ng of size-fractionated DNase I-digested DNA from 2,000 early gastrulae (10,000 cells/embryo) was quantitated by Qubit. Less than 0.1% of starting total embryonic DNA is expected to be recovered.
TROUBLESHOOTING
Problem (Step 20): After DNase I digestion, the DNA shows a ladder pattern upon electrophoresis, with multiple bands in ~150bp increments (or alternatively all fragments are less than 500bp).
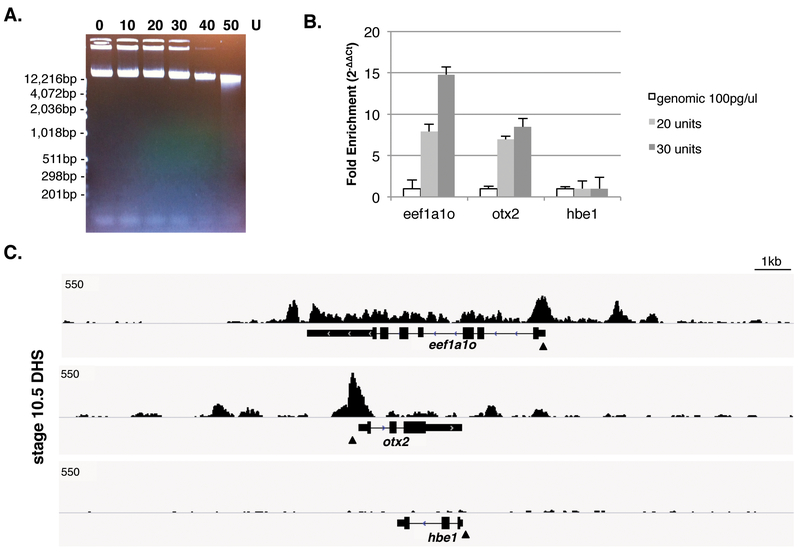
Solution: These observations indicate over-digestion by DNase I, showing either a nucleosomal laddering or more severe DNA degradation. Reducing the amount of DNase I or increasing the number of embryos/nuclei for a digestion reaction should improve the results. Most of the DNA in a DNase I digestion during this short treatment is not digested and the vast majority of DNA is more than 10kb and DNA liberated in the 50–500bp size range will not be visible to the eye (Fig1A).
FIGURE 1.
Methods to detect DHSs using Xenopus embryos A. Gel picture of DNase I digested genomic DNA extracted from isolated nuclei at stage 10.5 Xenopus tropicalis embryos. The amount of high-molecular-weight DNA fragments are gradually decreased as DNase I concentrations increase. B. DNase I hypersensitivity (DHS) monitored by qPCR. DHSs are normalized by Cp value from 100pg of genomic DNA from Xenopus liver using 2−ΔΔCt method. Reference regions are selected from non-expressing gene’s promoters on the desired developmental stage (e.g., hbe1, encoding hemoglobin subunit epsilon 1, which is not expressed until tailbud stages). C. A genome-wide profile of DHSs at gastrula stage by DNase-seq on Xenopus tropicalis genome version 7.1. DHSs are detected on the promoter and enhancer regions of eeflα1ο and otx2 but no DHS is shown around hbe1, which is also used as a reference gene for qPCR at Fig1B. Approximate qPCR regions are marked by arrowheads.
DISCUSSION
The advantage of DNase-seq is that there are no requirements of prior knowledge of histone modifications or transcription factor binding sites and high quality antibodies. This protocol includes modifications of Farzaneh and Pearsons (1978). The use of iso-osmotic iodixanol gradients in place of sucrose not only reduces centrifugation time, but also avoids the removal of water from the nuclei upon being placed in >2M sucrose (Graham, 2002). The DNase I digestion procedure was modified from Neph et al. (2012).
Validation of DHS by qPCR
DNase-seq data generated by the protocol herein shows high correlation with real-time qPCR for regions of expected open chromatin (Fig1B and C). Researchers will need to empirically determine the DNase I digestion conditions that permit the maximal release of DHS regions (without liberating DNase I-insensitive regions of the genome). If other stages are more suited to the experimental question, number of embryos for a desired stage might also need to be modified. The success of DNase I digestion conditions can be monitored by performing qPCR. Well-studied and highly expressed gene’s promoter regions are used as positive controls for qPCR with ~100bp length for amplicons. Negative controls include genes not expressed at the desired stage, serving as reference regions for normalization using 2−ΔΔCt method (Livak and Schmittgen, 2001) to detect DHSs. We have used 100pg of genomic DNA isolated from Xenopus liver as an external standard.
Library generation and data analysis
As input for library construction, 10–30ng of total DNase I digested DNA (size selected in the range of 50–500bp) was used. Libraries are initially quantitated using a Qubit. They are then subjected to quality assessment (size distribution) using a DNA Bioanalyzer, followed by KAPA qPCR quantitation. Using the Illumina HiSeq 2500, 50bp single-end reads were obtained. All sequencing data was aligned to Xenopus tropicalis v7.1 genome (Xenbase, http://www.xenbase.org/, RRID: SCR_003280) using Bowtie v1.0.0 (Langmead et al. 2009) with the following command:
‘bowtie -S -p 16 -v 2 -k 1 -m 1 --best --strata’
Bigwig files were created using deepTool2 bamCoverage (Ramírez et al. 2016) and then visualized using the Broad Institute’s Integrative Genomics Viewer genome browser (Robinson et al. 2011).
RECIPES
Buffer A
| Reagent | Final concentration |
|---|---|
| Tris-HCl (1M, pH 8.0) | 15mM |
| NaCl (5M) | 15mM |
| KCl (1M) | 60mM |
| EDTA (0.5M, pH 8.0) | 1mM |
| EGTA (50mM, pH 8.0) | 0.5mM |
| Spermine (500mM) | 0.5mM |
| Pefabloc SC (20mg/mL) | 0.1mg/mL |
| Dithiothreitol (1M) | 2mM |
| Protease inhibitor (Mini) | - |
| Ultra pure water | - |
Store for up to a week at 4°C. Add Pefabloc SC, dithiothreitol and protease inhibitor cocktail immediately before use.
DNase I 10x Digestion Buffer
| Reagent | Final concentration |
|---|---|
| NaCl (5M) | 750mM |
| CaCl2 (1M) | 60mM |
| Ultra pure water | - |
Store at room temperature up to a year
DNaseI 1x Digestion Buffer
Add 500μL of DNase I 10x digestion buffer to 4.5ml of buffer A to make 5ml of DNase I 1x Digestion Buffer.
Make day of use and allow equilibrating to 37 °C for an hour prior to use.
Iodixanol solution (20%; 25%; 30%)
| Reagent | Final concentration |
|---|---|
| Tris-HCl (1M, pH 8.0) | 10mM |
| Sucrose (1M) | 0.3M |
| MgCl2 (1M) | 5mM |
| KCl (1M) | 25mM |
| NaF (500mM) | 10mM |
| β-Glycerophosphate (1M) | 5mM |
| Sodium pyrophosphate (100mM) | 5mM |
| Spermine (500mM) | 0.5mM |
| Spermidine (500mM) | 0.5mM |
| Pefabloc SC (20mg/mL) | 0.1mg/mL |
| Dithiothreitol (1M) | 2mM |
| Protease inhibitor (Mini) | - |
| Iodixanol (60%) | 20%; 25%; 30% |
| Ultra pure water | - |
OptiPrep™ Density Gradient Medium is 60% iodixanol.
Prepare fresh and keep at 4 °C.
Stock DNaseI (10U/μL)
Solubilize the entire bottle of DNase I (10KU in a bottle) on ice with 1ml of following ice-cold storage buffer.
| Reagent | Final concentration |
|---|---|
| Tris-HCl (1M, pH 7.6) | 20mM |
| NaCl (5M) | 50mM |
| MgCl2 (1M) | 2mM |
| CaCl2 (1M) | 2mM |
| Pefabloc SC (20mg/mL) | 0.1mg/mL |
| Dithiothreitol (1M) | 1mM |
| Glycerol (100%) | 50% |
| Ultra pure water | - |
Aliquot to 50–100μL and store at −20 °C for up to several months
Stop Buffer
| Reagent | Final concentration |
|---|---|
| Tris-HCl (1M, pH 8.0) | 50mM |
| NaCl (5M) | 100mM |
| SDS (20%) | 0.10% |
| EDTA (0.5M, pH 8.0) | 100mM |
| Spermine (500mM) | 1mM |
| Spermidine (500mM) | 0.3mM |
| Ultra pure water | - |
Prepare fresh on the day of use and allow to equilibrate to 37°C for an hour prior to use.
Sucrose Buffer (0.3M)
| Reagent | Final concentration |
|---|---|
| Tris-HCl (1M, pH 8.0) | 10mM |
| Sucrose (1M) | 0.3M |
| MgCl2 (1M) | 5mM |
| KCl (1M) | 25mM |
| NaF (500mM) | 10mM |
| β-Glycerophosphate (1M) | 5mM |
| Sodium pyrophosphate (100mM) | 5mM |
| Spermine (500mM) | 0.5mM |
| Spermidine (500mM) | 0.5mM |
| Pefabloc SC (20mg/mL) | 0.1mg/mL |
| Dithiothreitol (1M) | 2mM |
| Protease inhibitor | - |
| Ultra pure water | - |
Store for up to a week at 4 °C. Add Pefabloc SC, dithiothreitol and protease inhibitor cocktail immediately before use.
REFERENCES
- Farzaneh F, Pearson CK. 1978. A method for isolating uncontaminated nuclei from all stages of developing Xenopus laevis embryos. J Embryol Exp Morphol 48: 101–108. [PubMed] [Google Scholar]
- Graham J 2002. Rapid Purification of Nuclei from Animal and Plant Tissues and Cultured Cells. The Scientific World JOURNAL 2: 1551–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H, McConnell WB, Grainger RM. 2006. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mechanisms of Development 123: 103–113. [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research 44: W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nature Biotechnology 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]