Abstract
Rabies virus (RABV) constitutes a major social and economic burden associated with 60,000 deaths annually worldwide. Although pre- and post-exposure treatment options are available, they are efficacious only when initiated prior to the onset of clinical symptoms. Aggravating the problem, the current RABV vaccine does not cross-protect against the emerging zoonotic phylogroup II lyssaviruses. A requirement for an uninterrupted cold chain and high cost of the immunoglobulin component of rabies prophylaxis generate an unmet need for the development of RABV-specific antivirals. We discuss desirable anti-RABV drug profiles, past efforts to address the problem and inhibitor candidates identified, and examine how the rapidly expanding structural insight into RABV protein organization has illuminated novel druggable target candidates and paved the way to structure-aided drug optimization. Special emphasis is given to the viral RNA-dependent RNA polymerase complex as a promising target for direct-acting broad-spectrum RABV inhibitors.
Introduction
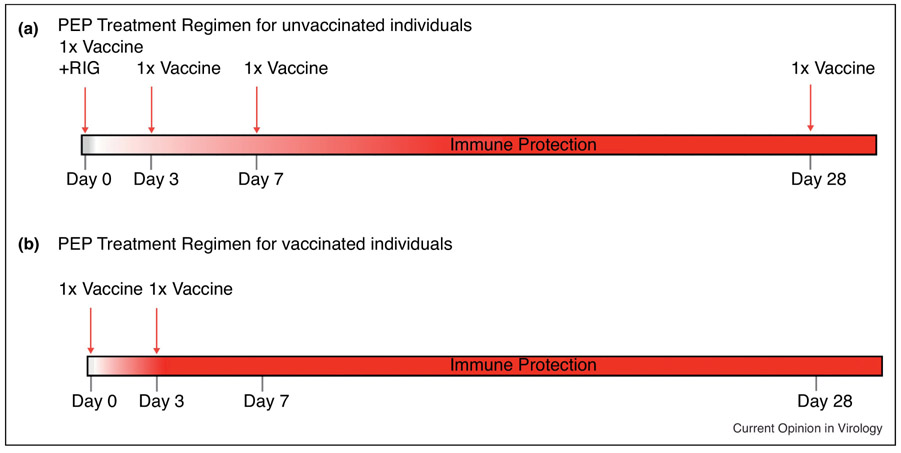
The devastating signs and symptoms of rabies disease have been documented as far back as 2,000 B.C. in the Eshnunna tablets of Mesopotamia [1]. Even now, in the second millennia A.D, rabies disease continues to be a social and economic hardship with approximately 60,000 deaths worldwide, nearly $8.6 billion in economic burden, and $1.5 billion spent on post-exposure prophylaxis treatment (PEP) alone [2]. The causative agents, lyssaviruses, within the Rhabdoviridae family, are characterized as zoonotic, neurotropic negative-sense non-segmented RNA viruses. Transmission of rabies virus (RABV) occurs typically through the transfer of infectious saliva from the percutaneous bite of a mammal, usually a dog [2]. Through axoplasmic transport, RABV enters the central nervous system (CNS) where it begins to replicate, causing severe neuronal dysfunction [3-5]. Rabies is vaccine-preventable as well as treatable early after infection. After the onset of clinical symptoms, however, almost all patients succumb to the infection, progressing toward coma and ultimately death [6]. RABV's ability to effectively subvert the host immune system through evasion of TLR signaling, downregulation of IFN signaling, and prevention of adaptive responses by maintaining lowered blood-brain barrier (BBB) permeability, and induction of T-cell apoptosis exemplifies why early intervention is critical [7-9]. As depicted in figure 1, treatment of rabies consists of rabies immune-globin (RIG) and four doses of the vaccine over a 4-week period. PEP is recommended for previously vaccinated individuals as well, and consists of vaccine doses on days 0 and 3. A single PEP regimen costs at least $3,000 in the United States [2]. This expense of rabies PEP is predominantly due to the high cost of producing human rabies immune-globin HRIG, a human plasma-based product, with a relatively short shelf life and need for extensive quality assurance [10]. A second contributor to the high treatment cost is the requirement of four doses of rabies vaccine, which typically costs $260 per dose in the USA and Europe. In Africa and Asia, where 95% of rabies-related deaths occur, PEP averages $40 and $49, respectively. This expense is often out of reach in areas with a daily family income of approximately $1-2. The number of people worldwide that receive rabies PEP as well as the crippling debt associated with it is estimated to reach a staggering 15 million annually [2]. Furthermore, the current vaccine is likely ineffective against emerging zoonotic lyssaviruses of phylogroup II such as Mokola (MOKV) and Lagos bat viruses [11-15]. The high cost of HRIG and the current vaccine, along with cold-chain requirements for both, present an urgent and unmet clinical need for the development of safe, cost-effective, efficacious, shelf-stable, and cross-protective antivirals against lyssavirus phylogroups associated with human rabies disease. Antiviral compounds could be used to replace the HRIG or other RIG component in current rabies PEP.
Figure 1:
Schematic diagram representing the current post-exposure prophylaxis treatment (PEP) schedule as recommended by the WHO, A) for naïve individuals and B) previously vaccinated individuals. [2]
Lyssavirus Virion Organization
Lyssaviruses contain RNA genomes of approximately 12 kb. The virion of lyssaviruses, as with the other family members of Rhabdoviridae, is characterized by a bullet-shape with a length of 180 nm, and average diameter of 70 nm. Lyssaviruses are enveloped by a lipid protein coat studded with the viral receptor glycoprotein (G). G is the predominant target for the host humoral immune response. The RABV G is a trimeric type-1 membrane protein, capable of major conformational rearrangements upon receptor binding and subsequent endocytotic internalization and acidification of the endosome [6]. Nicotinic acetylcholine receptor (nAChR), the neuronal cell adhesion molecule (NCAM), and the p75 neurotrophin receptor (p75NTR) have all been implicated in serving as host receptors for RABV, and several other membrane components have been proposed to aid in viral entry as well [16]. Matrix protein (M) interacts with the cytoplasmic tails of G, lining the viral envelope [17, 18]. Within the virion, the viral genome is encapsidated by nucleocapsid (N), resulting in a helical ribonucleoprotein complex (RNP). Encapsidation of the viral genome is a protective measure against RNAse digest and it reduces triggering of innate cellular immune response pathways. Recognition of N by the viral RNA-dependent RNA polymerase (vRdRp) is essential for viral replication [19]. N also plays a significant role in evading the innate immune response and enhancing viral pathogenicity through virion cell-to-cell spread [20]. The vRdRp is a hetero-oligomeric protein complex consisting of the large protein (L) and the phosphoprotein (P) [6]. Of these, P, as depicted in figure 2, is the noncatalytic cofactor of the vRdRp that mediates the interaction with RNP to position L for RNA synthesis. P also guides nascently folded RNA-free N (N0) to newly synthesized genomic RNA during replication [19]. Up to five alternative start codons result in five N-terminally truncated P protein variants, which have been implicated as Type-1 IFN antagonists [21]. This is accomplished through direct interaction with STAT1 and STAT2 proteins as well as suppressing IRF-3 [22-24]. L is approximately 250 kDa in mass and provides all enzymatic activity required for viral genome synthesis. Sequence homology among other members of Mononegavirales have revealed 6 highly conserved regions (CRs) within L, as shown in figure 3a [25]. These CRs have been implicated in the different catalytic functions for productive replication. CRII and CRIII are required for phosphodiester bond formation, with III containing a GDN motif starting at residue 729 that is considered to form the catalytic center [26-28]. CRV is implicated in mediating viral mRNA capping through GDP polyribonucleotidyltransferase (PRNTase) activity [29-33]. CRVI contains a K-D-K-E motif that is characteristic for methyltransferase (MTase) activities [34-36]
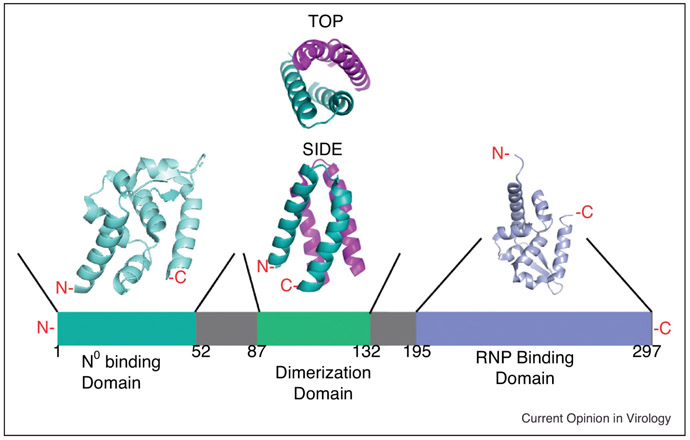
Figure 2:
Schematic representation of the modular organization of the RABV phosphoprotein (P). The N0 binding domain is teal, the dimerization domain is green, and the ribonucleoprotein binding domain (RNP) is periwinkle. The solved crystal structure for the N0 binding domain is depicted in teal (PDB 3OA1). The solved crystal structure for the dimerization domain is depicted in green and pink with both top and side views (PDB 3L32). The solved structure for the RNP is depicted in periwinkle (PDB 1VYI). [19, 127, 135, 137, 138]
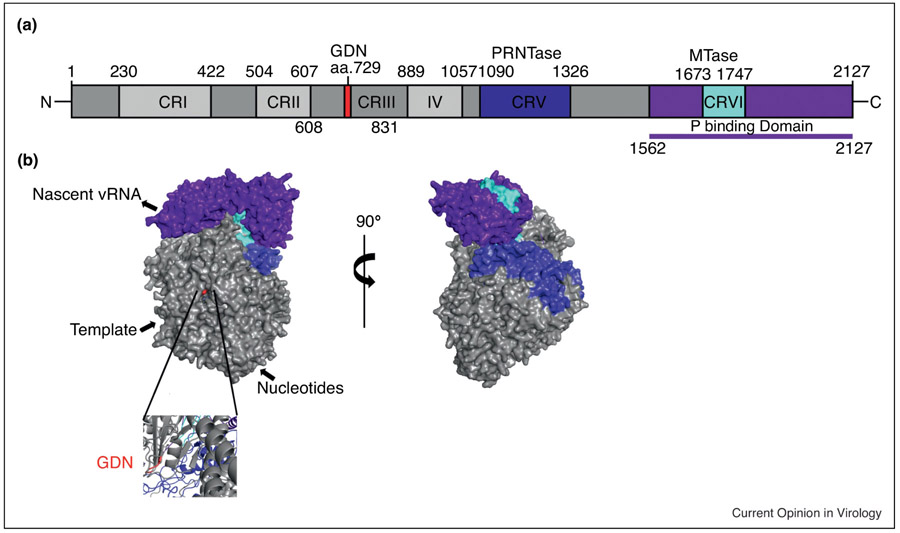
Figure 3:
A) Schematic diagram depicting the domain organization of RABV large protein (L). The GDN polymerization motif is in red. The polyribonucleotidyltransferase (PRNTase) is in blue. The methyltransferase (MTase) is in cyan. The phosphoprotein (P) binding region is purple. Conserved regions (CR) of the non-segmented negative-sense RNA viruses are labelled CR I -VI. B) Surface representation of the RABV L generated by homology modelling based on the coordinates reported for the closely related VSV L structure with the same color scheme as described by 3A. Below is a zoomed in ribbon representation of the GDN motif responsible for polymerase activity. [25, 33, 36, 128]
Current Treatment of RABV Infection
The high cost and cold chain requirement of rabies biologics have revealed an urgent need for the development of alternative antiviral compounds. To date there have been 14 documented survivors of symptomatic rabies disease, of whom all but one received vaccine and PEP. However, all of these survivors exhibit severe neurological sequelae [37]. The single documented survivor, who did not receive PEP nor vaccine, was treated with what is now termed “The Milwaukee Protocol”. This treatment method involves induction of a therapeutic coma accompanied by ketamine and amantadine infusions [38]. This survivor had anti-RABV antibodies, suggesting that she was infected with a untypical RABV strain and that her immune system was responsible for the clearance, though the strain was never identified [39]. Also, this original Milwaukee protocol patient developed neurological side effects that never fully resolved [38]. Subsequent application of the Milwaukee protocol has resulted in 31 deaths and only 1-2 additional survivors, who both developed severe neuronal sequelae, casting considerable doubt on the overall efficacy of the approach [39, 40].
Administered in conjunction with RIG and RABV vaccine, broad-spectrum antiviral therapeutics are used for the treatment of highly aggressive rabies cases. These include ribavirin, interferon-alpha (IFN-α), and ketamine/amantadine [41, 42]. Ribavirin is a broad-spectrum guanosine nucleoside analog with unclear mechanism of action including inhibition of inosine monophosphate dehydrogenase (IMPDH) purine de novo synthesis, direct incorporation into nascent viral genomes causing lethal mutagenesis, and inhibition of mRNA capping [43-45]. Although efficacious in vivo, ribavirin has shown no activity against clinical rabies [41, 46]. This is supported by a 15 year-long study in which 16 RABV infected patients were treated with ribavirin, but no beneficial effect was observed [47]. Also, in two separate cases, one in Thailand, and one in the United States, ribavirin was administered, again demonstrating a lack of efficacy [46, 48]. It is hypothesized that this disappointing performance of ribavirin is due to its interference with the Th1/Th2 immune response, thus hindering production of effective antibodies that are essential for RABV clearance [49].
Inducing an antiviral state through triggering of innate immune response pathways, anti-RABV activity of IFN-α is ameliorated through counteraction by the lyssavirus phosphoprotein (P) [8, 46]. Currently up to five truncated isotypes of P (P1-5) have been identified that play distinct roles in antagonizing the type-1 IFN response. P1 and P2 encode for a nuclear export signal and directly bind to phosphorylated STAT1, thus sequestering it to the cytoplasm [50, 51]. P3 binds directly to microtubules to prevent STAT1 shuttling into the nucleus and it furthermore can interact directly with STAT1 to block DNA binding [52, 53]. The inhibition of STAT1 also renders exogenous IFN-α ineffective for RABV therapy.
Ketamine and amantadine are core components of the Milwaukee protocol anti-RABV approach. Both drugs are non-competitive NMDA receptor antagonists and have been shown to prevent uncoating and release of RABV particles. Ketamine requires prohibitively high concentrations to effectively block viral replication, however, which are not achievable in human therapy [38, 40, 42, 54].
All current antiviral strategies to block RABV replication are thus compromised by substantial limitations, adding little to improve case fatality rates of symptomatic disease. This lack of an effective therapeutic option creates an urgent and currently unmet clinical need for the development of next-generation antivirals that can be used as an additional component of current combinatorial PEP and may improve management of established rabies cases.
Anti-RABV Drug Discovery
Most of the deaths attributed to RABV infection result from socio-economic barriers, reagent shortages, and inability to maintain an uninterrupted cold chain for the transport of biologics for PEP [11]. The discovery of novel antiviral compounds may offer a fresh avenue to address these concerns. Small-molecule antivirals in general have the advantages of cost-effective manufacture – addressing supply concerns – as well as high shelf-stability, enabling developing countries to stock-pile life-saving supplies [42]. Currently, there are no small-molecule inhibitors licensed for therapeutic use, despite several attempts at discovery [41, 54-62]. Based on previous antiviral activity of phenolic compounds against viruses such as HIV, herpes simplex virus and influenza virus, 24 representatives of this class were tested against RABV. Based on visually scoring of viral cytopathic effects (CPE) after infection with the Pasteur virus (PV) RABV strain, 50%-effective concentrations (EC50) were in all cases >50 μM [54]. Although a specific mechanism of activity was not evaluated in this study, it was suggested that the antiviral activities of phenolic compounds may be attributed to their interaction with host cell group-specific antigens (GAGs), blocking viral entry [63]. Another anti-RABV drug discovery campaign employed cell-free translation, using fluorescently tagged mRNA to screen the Prosetta compound library in search of hits that prevented nucleocapsid assembly. This exercise yielded a hit directed against the ABCE1 transporter that showed considerable cytotoxicity with 50% cytotoxic concentrations (CC50) of 2.5-10 μM [55].
These early attempts for anti-RABV drug discovery did not yield viable hit candidates, but exposed some challenges to anti-RABV drug screening approach. A significant deterrent to automated anti-RABV drug discovery, for instance, is the BSL2/3 containment requirements imposed by replication-competent RABVs and mandatory rabies vaccination as well restriction to the laboratory of not vaccinated people.. Based on the original reverse genetics system developed for the SAD-B19 RABV strain [64], however, minireplicon systems and single-cycle reporter RABV viruses were developed that allow study of the RABV polymerase activity and in-cell replication in a BSL2 setting [65-73]. Analogous to the precedent set by successful screens employing single-cycle HIV, hepatitis C virus, and influenza virus reporter strains [74-76], the single-cycle based approach in particular offers an exciting drug discovery perspective. Transient-transfection based minigenome drug screens have furthermore been attempted to identify, for instance, Ebola virus polymerase inhibitors [77-79]. Applied to the RABV problem, single-cycle reporter viruses and/or minireplicon systems may offer a viable solution to the biocontainment challenge to automated large-scale drug discovery.
Host-Directed RABV Inhibitors
The analysis of the host-RABV interactome has revealed cellular proteins that are essential for completion of the RABV life-cycle and may provide exploitable antiviral target opportunities [80, 81]. Targeting of host factors involved in viral replication and pathogenesis offers several advantages over direct-acting antivirals. The frequency of viral escape is typically reduced when host factors are targeted [62, 82-84], due to the fact that the host genetic information is much more stable than that of error-prone RNA viruses and resistance mutations can never become fixed in circulating viral strains. Also, the potential for a broadened indication spectrum is heightened if the targeted host factor is highjacked by related viruses within the same family, preparing the path for targeting the emerging phylogroup II lyssaviruses [55].
However, host-directed antivirals are more prone to inducing severe adverse effects [85-87]. For example, erlotinib, dasatinib, and ezetimibe are all broad-spectrum entry inhibitors of hepatitis C virus that have no documented escape mutations [88, 89]. However, these drugs have revealed several adverse effects for lung, liver, and kidney functions, as well as cause rash and diarrhea in cancer patients, thus demonstrating the tightrope balance of targeting host proteins that are essential for cellular function [90-92]. Identifying pursuable host targets thus remains to be a challenging endeavor [93, 94]. RABV entry is mediated by clathrin-coated endocytosis which is dependent on actin cytoskeleton reorganization for internalization [95]. A closely related Rhabdovirus, vesicular stomatitis virus (VSV), employs a comparable entry strategy [96, 97]. Dynamin is a critical component of multiple endocytotic pathways and plays a role in actin reassembly and organization [95, 97]. Dynasore, a membrane-permeable inhibitor of dynamin GTPase activity, has been shown to inhibit the entry process for several viruses, including RABV and VSV [95, 98]. A similar dynamin inhibitor, AMBA, has also shown antiviral effects against HSV, but the selectivity index (SI = CC50/EC50) was at a low <21. When tested in vivo, there was only a 50% survival rate of mice given a lethal RABV challenge, greatly compromising therapeutic potential against the RABV indication [99]. Since AMBA was a direct hit compound from a high-throughput screen, synthetic optimization to improve efficacy was suggested. However, greater affinity for dynamic targets must be anticipated to coincide with increased cytotoxicity, since dynamin is an essential cellular protein. Analysis of RABV entry into non-neuronal cells has revealed preference for cholesterol-rich microdomains [16, 100-106]. Depletion of membrane cholesterol, however, did not affect viral entry, suggesting RABV may have another route of entry into these cell types [107].
RABV encephalitic pathogenesis is Raf/MEK/ERK kinase pathway-dependent [108]. Sorafenib is a small-molecule tyrosine kinase inhibitor that curbs angiogenesis in cancer patients [109]. In an attempt of repurposing cancer drugs as antivirals, sorafenib has been highlighted as a broad-range antiviral against adenovirus, mumps virus, chikungunya virus, dengue virus, West Nile virus, Yellow fever virus, and enterovirus 71 [110-116]. When administered at non-cytotoxic levels, however, sorafenib given in combination with IFN-β reduced RABV load by less than one order of magnitude (74% inhibition) [22]. Furthermore, this compound was associated with GI complications and other severe adverse effects in cancer patients, making it an unlikely antiviral candidate [117-123].
The neural precursor cell expressed developmentally down-regulated protein 4 (Nedd4) is a highly conserved and universally expressed ubiquitin ligase that regulates steady state levels of many membrane proteins including ion channels and membrane receptors. The recently characterized WW domain of Nedd4 has been associated with a number of functions that facilitate protein-protein interactions [124]. This domain was shown to provide a binding site for M proteins of rhabdoviruses and filoviruses, promoting viral budding and egress [125]. A hit compound synthesized from an in silico screen based on the structural information of the WW domain showed some initial efficacy against RABV. Further synthetic optimization of the scaffold yielded two hit compounds that specifically targeted the Nedd4 WW domain, competitively inhibited viral M protein binding to the domain, and were only minimally cytotoxic [126]. Further in vivo evaluation is required, however, to determine whether the Nedd4 WW domain will persevere as a viable druggable target.
Direct-Acting Antivirals
Despite the anticipated strengths of host-directed antiviral approaches, unacceptable cytotoxicity often shifts focus towards direct-acting inhibitors that offer the potential for wider therapeutic windows [59]. A co-crystal structure of N with viral RNA was solved (figure 4) that may be exploitable for drug discovery through structure-guided design [127]. The conserved nature of N and its critical interactions with multiple viral and cellular proteins make it overall an attractive druggable candidate [128-130]. Proof-of-concept comes from liposomally delivered siRNAs that were designed to address highly conserved N sequences across different RABV strains. RABV replication was significantly inhibited in vitro through this approach, validating druggability of RABV N. In vivo, however, the siRNAs protected at best 60% of lethally RABV challenged mice [61], which was attributed to inefficient delivery and uptake by the animals [59, 61].
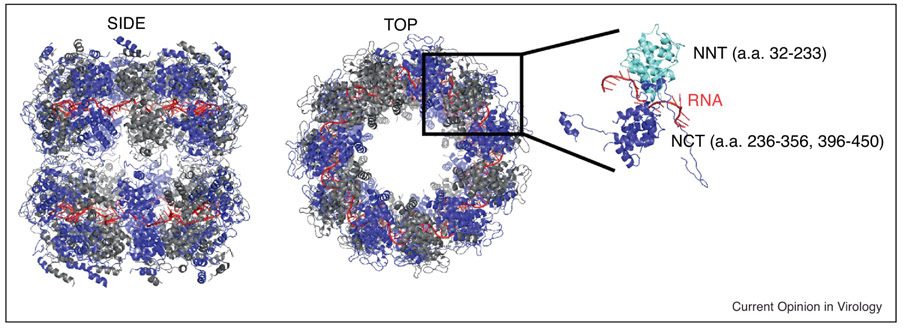
Figure 4:
Structural representation of the ribonucleoprotein complex (PDB 2GTT) with both side (left) and top (center) views. Individual nucleoprotein (N) protomers are depicted as alternating blue and grey with RNA as a red coil. The far right represents a single N protomer with the n-terminal residues (NNT) in cyan and the c-terminal residues (NCT) in blue. [127, 137]
Conformational changes of N are likewise vital for its bioactivity. Phosphorylation at serine389, for instance, is considered to allow N to loosen its interaction with RNA, enabling access of the vRdRp to the encapsidated RNA template [131, 132]. These dynamic changes in nucleoprotein structure may be targeted by allosteric inhibitors that trap N in specific conformations, preventing rearrangements required for polymerization [133]. In addition to targeting N directly, preventing the interaction of the viral P-L polymerase complex with N in the assembled RNP may be a viable antiviral strategy. Replication of the distantly related respiratory syncytial virus, for instance, is potently blocked by the small-molecule compound RSV604 that is considered to interfere with P-RNP binding [134]. Structural models of RNP and the C-terminal domain of P proteins of both RABV and MOKV revealed close structural homology and interaction mechanism similar to these employed by the related paramyxo-and pneumoviruses [99, 135-138]. Through yeast-2-hybrid screening, several peptides were identified that bind directly to both RABV and MOKV P in highly conserved regions, inhibiting viral replication in minigenome assays. Mechanistically, inhibition by these peptides was due to disruption of RNP-P complex formation [139]. Although therapeutic peptides are often highly specific and show low toxicity, they are frequently proteolytically unstable, display poor membrane permeability, and are often immunogenic when repeat administration is required [140]. Replacement of P binding peptides with a small-molecule inhibitor that directly interacts with P should therefore be considered in search of a broad range therapeutic that is efficacious against both phylogroups, cost-effective, and shows superior stability.
M is of critical importance for viral particle formation, based on self-assembly upon interaction with RNP and the cytoplasmic domain of G [141]. M also induces mitochondria-mediated cellular apoptosis in neuronal cells to promote viral dissemination [142, 143], and alters host cell protein biosynthesis through two mechanisms [144]: suppression of mRNA translation by interaction with Rae1 and block of mRNA export from the nucleus [145]; and shut-off of host gene expression by binding to, and/or modulating of, the phosphorylation site of host cell transcription factors [145]. The structures of both VSV and LABV M proteins have been solved [146], fueling the mechanistic appreciation of M assembly and opening a door towards structure-guided drug design against specific M microdomains. Attractive antiviral targets include M domains involved in M homo-oligomerization, interaction with host proteins, or required for binding to the viral G protein [17, 18, 70, 125, 126, 146-151].
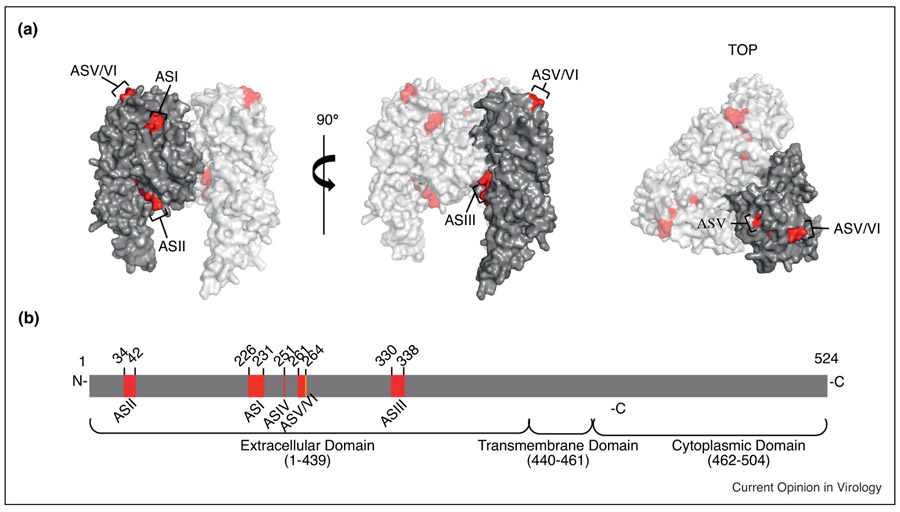
Monoclonal antibodies (mAbs) targeting RABV G have proven antiviral efficacy [15, 152-154]. Broad panels of antibodies against rabies G revealed primary clusters for antibody recognition in distinct regions of the G ectodomain [18, 155-158]. As shown in figure 5, these antigenic sites (AS) include ASI (a.a. 226-231), ASII (a.a.34-42), ASIII (a.a.330-338), ASIV (a.a. 251), ASV (a.a. 261-264), and ASVI (a.a. 264). Considerable efforts have been made to assemble antibody cocktails that target different AS’ to prevent viral escape through antigenic drift and provide cross-protection based on across-strain conservation [14, 42, 159-164]. Despite the initial promise of the approach, antibody therapy does not address the cold-chain problematic or provide cross-protection against the emerging phylogroup II lyssavirus threat. Lifting the cold-chain limitation at least, small molecule fusion inhibitors targeting G should be considered nevertheless to block RABV entry [165]. These can be effective against viral targets, since resistance often coincides with a viral fitness penalty [166-169].
Figure 5:
Schematic representation of the RABV glycoprotein (G) antigenic sites (AS). A) Side view (left) and 90° turn (center) and top view (right) of homology model of RABV G based on VSV G (PDB 2J6J). Residues of AS are highlighted in red for each view. B) Linear schematic showing relative position and amino acid numbering for ASI-VI within the extracellular domain of G. [129, 157, 191, 195].
Harboring all catalytic centers of the viral polymerase complex, L has emerged as a leading drug target for many negative-sense RNA viruses because of its highly conserved nature and the density of candidate target sites including domains required for RNA synthesis, mRNA capping, or mRNA methylation [25, 128, 170-174].
Favipiravir (T-705) is a broad-spectrum RNA virus inhibitor that is currently licensed in Japan for stock-piling against pandemic influenza viruses resistant to oseltamivir. T-705 has been shown to be effective against Ebola virus, also within order Mononegavirales and therefore was considered for use against RABV also [175-181]. When tested in mouse neuroblastoma neural 2a cells, T-705 indeed lowered RABV titers by three to four order of magnitude (EC50 of 32.4 μM against a circulating RABV strain and 44.3 μM against the vaccine strain). However, the survival rate of mice infected with RABV and treated with T-705 was only approximately 50% (5 out of 9 animals examined), and the surviving mice developed limb paralysis, indicating viral circulation within the nervous system [60]. T-705 was also the least effective in a panel of drug combinations with IFN-α, suggesting that it is not an optimal-antiviral agent against RABV [112].
Screening of several ribavirin analogs returned two compounds, EICAR and EICNR, that had superior antiviral potency in human neuroblastoma cells (EC50 0.9 μM, and 3.8 μM, respectively), compared to the ribavirin EC50 of 18.6 μM [57]. However, testing of these compounds was limited to the RABV vaccine strain, necessitating further examination in vivo against clinically-relevant pathogenic RABVs. If effective in vivo, these analogs may have the potential to replace ribavirin in current PEP.
Beyond ribavirin, ribonucleoside analogs present an exciting option for the discovery of broad-spectrum lyssavirus inhibitors. For instance, the ribonucleoside analogy N4-hydroxycytidine reportedly blocks both seasonal and pathogenic influenza virus strains, respiratory syncytial virus, Ebola virus, chikungunya virus, and hepatitis B and C viruses [182-188] with excellent pharmacological properties [182]. If antiviral activity equally extends to RABV and related lyssaviruses, compounds like N4-hydroxycytidine may present an example for a viable next-generation therapeutic option to address the rabies challenge.
Summary
The high cost of PEP for RABV and the lack of cross-protection against the emerging zoonotic lyssaviruses of phylogroup II have underscored the unmet demand of an updated treatment regimen. Ideal alternatives break the cold-chain requirement, be BBB permeable, RABV specific, safe, and efficacious. Advances in molecular biology and a better understanding of RABV pathogenesis have led to new approaches to address the problem. Of all options, we consider the RABV polymerase complex to represent the most promising target for direct-acting antivirals due to the comparably low conservation of G across lyssaviruses [12, 17, 158, 174, 189-192]. The heterologous polymerase complex offers druggable protein-protein interfaces, essential enzymatic centers, and opportunity for allosteric and competitive substrate-analog inhibitors. Crystal structures have been determined for the RABV N-RNA complex and the P-RNP [127, 135, 137] complex, providing an exciting starting point for future structure-guided drug design.
Table 1.
Host-directed anti-RABV compounds
| Compound | Target | EC50 (uM) | CC50 (uM) | SI (CC50/EC50) | Ref. |
|---|---|---|---|---|---|
| Catechin | Host Cell GAGs | 36.50 ± 8.40 | 124.33 ± 33.53 | 3 | [54, 63] |
| Quercetin | Host Cell GAGs | 191.68 ± 24.25 | 670.02 ± 180.18 | 3 | [54, 63] |
| 3,4,5-Trimethoxybenzoic acid | Host Cell GAGs | 2142.74 ± 266.37 | >5042.41 | 2 | [54, 63] |
| Trimethoxyacetophenone | Host Cell GAGs | 1023.98 ± 64.62 | 3738.98 ± 1099.17 | 3 | [54, 63] |
| 3,4,5-Trimethoxybenzoic acid ethyl ester | Host Cell GAGs | 822.23 ± 134.38 | 3204.20 ± 397.87 | 3 | [54, 63] |
| Butyl gallate | Host Cell GAGs | 109.79 | 113.23 ± 52.35 | 1 | [54, 63] |
| PAV-866 | ABCE1 | ~0.15–0.30 | ~2.5–10 | ~100 | [55] |
| Sorafenib | Tyrosine Kinases | 1.463 | >160 | 109 | [22] |
| 2-piperidin-3-yl-benzothiazole analog | Nedd4 | 0.345 | >1 | >3 | [126] |
| 1-acetyl-3-(2,2,2-trifluoroethyl)-urea analog | Nedd4 | 0.210 | >1 | >8 | [126] |
Table 2.
Viral-Directed anti-RABV Compounds
| Compound | Target | EC50 (uM) | CC50 (uM) | SI (CC50/EC50) | Ref. |
|---|---|---|---|---|---|
| Ketamine | vRNA Synthesis | 922.93 ± 68.48 | 3010.69 ± 171.26 | 3.3 | [54, 193] |
| Ribavirin | de novo purine synthesis | 18.55 | >200 | >10 | [57] |
| EICAR | de novo purine synthesis | 0.90 | >200 | >200 | [57] |
| EICNR | de novo purine synthesis | 3.80 | >200 | >50 | [57] |
| Favipiravir | vRdRp | 32.4 | >2500mM | >1000 | [60, 194] |
Acknowledgements
This work was supported, in part, by public health service grant AI127823 from the NIH/NIAID (to R.K.P. and M.J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fales FM, Chapter 2: mesopotamia. Handb Clin Neurol, 2010. 95: p. 15–27. [DOI] [PubMed] [Google Scholar]
- 2.O'Brien KL, Nolan T, and Rabies S.W.o., The WHO position on rabies immunization - 2018 updates. Vaccine, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhingra V, et al. , Proteomic profiling reveals that rabies virus infection results in differential expression of host proteins involved in ion homeostasis and synaptic physiology in the central nervous system. J Neurovirol, 2007. 13(2): p. 107–17. [DOI] [PubMed] [Google Scholar]
- 4.Gillet JP, Derer P, and Tsiang H, Axonal transport of rabies virus in the central nervous system of the rat. J Neuropathol Exp Neurol, 1986. 45(6): p. 619–34. [DOI] [PubMed] [Google Scholar]
- 5.Jackson AC, Diabolical effects of rabies encephalitis. J Neurovirol, 2016. 22(1): p. 8–13. [DOI] [PubMed] [Google Scholar]
- 6*.Davis BM, Rall GF, and Schnell MJ, Everything You Always Wanted to Know About Rabies Virus (But Were Afraid to Ask). Annu Rev Virol, 2015. 2(1): p. 451–71. [DOI] [PMC free article] [PubMed] [Google Scholar]; Excellent review that provides extensive overview of rabies virus disease progression within the human host. The authors also point out unanswered research questions and the current strategies used within the field to solve them.
- 7.Lafon M, Evasive strategies in rabies virus infection. Adv Virus Res, 2011. 79: p. 33–53. [DOI] [PubMed] [Google Scholar]
- 8.Scott TP, and Nel LH, Subversion of the Immune Response by Rabies Virus. Viruses, 2016. 8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9*.Ito N, Moseley GW, and Sugiyama M, The importance of immune evasion in the pathogenesis of rabies virus. J Vet Med Sci, 2016. 78(7): p. 1089–98. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors provide invaluable insight into the current understanding of rabies virus immune evasion strategies. This review points out the importance of keeping in mind such immune evasion strategies for future drug development.
- 10.Dodet B, et al. , Human rabies deaths in Africa: breaking the cycle of indifference. Int Health, 2015. 7(1): p. 4–6. [DOI] [PubMed] [Google Scholar]
- 11.Rabies vaccines. WHO position paper. Wkly Epidemiol Rec, 2007. 82(49-50): p. 425–35. [PubMed] [Google Scholar]
- 12.Badrane H, et al. , Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J Virol, 2001. 75(7): p. 3268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourhy H, Sureau P, and Tordo N, From rabies to rabies-related viruses. Vet Microbiol, 1990. 23(1-4): p. 115–28. [DOI] [PubMed] [Google Scholar]
- 14.Bussereau F, et al. , Monoclonal antibodies to Mokola virus for identification of rabies and rabies-related viruses. J Clin Microbiol, 1988. 26(12): p. 2489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent J, Bussereau F, and Sureau P, Immunological relationships between rabies virus and rabies-related viruses studied with monoclonal antibodies to Mokola virus. Ann Inst Pasteur Virol, 1988. 139(2): p. 157–73. [DOI] [PubMed] [Google Scholar]
- 16.Lafon M, Rabies virus receptors. J Neurovirol, 2005. 11(1): p. 82–7. [DOI] [PubMed] [Google Scholar]
- 17.Kgaladi J, et al. , Pathogenicity and Immunogenicity of Recombinant Rabies Viruses Expressing the Lagos Bat Virus Matrix and Glycoprotein: Perspectives for a Pan-Lyssavirus Vaccine. Trop Med Infect Dis, 2017. 2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marston DA, et al. , Interspecies protein substitution to investigate the role of the lyssavirus glycoprotein. J Gen Virol, 2013. 94(Pt 2): p. 284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeiro Ede A Jr., et al. , Binding of rabies virus polymerase cofactor to recombinant circular nucleoprotein-RNA complexes. J Mol Biol, 2009. 394(3): p. 558–75. [DOI] [PubMed] [Google Scholar]
- 20.Masatani T, et al. , Importance of rabies virus nucleoprotein in viral evasion of interferon response in the brain. Microbiol Immunol, 2013. 57(7): p. 511–7. [DOI] [PubMed] [Google Scholar]
- 21.Okada K, et al. , Roles of the Rabies Virus Phosphoprotein Isoforms in Pathogenesis. J Virol, 2016. 90(18): p. 8226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Marosi A, et al. , Evaluation of in vitro inhibitory potential of type-I interferons and different antiviral compounds on rabies virus replication. Vaccine, 2018. [DOI] [PubMed] [Google Scholar]; Authors compare the efficacy of interferon treatment in combination with broad-spectrum anti-viral inhibitors. This study showed that IFN-α was did not exhibit synergy when combined with Ribavirin, T-705, nor sorafenib. However, synergy was observed when IFN-β was used in conjunction with Ribavirin, T-705, or sorafenib, suggesting a novel combination therapy to treat rabies disease.
- 23.Brzozka K, Finke S, and Conzelmann KK, Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J Virol, 2006. 80(6): p. 2675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brzozka K, Finke S, and Conzelmann KK, Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol, 2005. 79(12): p. 7673–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poch O, et al. , Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol, 1990. 71 (Pt 5): p. 1153–62. [DOI] [PubMed] [Google Scholar]
- 26.Singh R, et al. , Rabies - epidemiology, pathogenesis, public health concerns and advances in diagnosis and control: a comprehensive review. Vet Q, 2017. 37(1): p. 212–251. [DOI] [PubMed] [Google Scholar]
- 27.Schnell MJ, and Conzelmann KK, Polymerase activity of in vitro mutated rabies virus L protein. Virology, 1995. 214(2): p. 522–30. [DOI] [PubMed] [Google Scholar]
- 28.Sleat DE, and Banerjee AK, Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol, 1993. 67(3): p. 1334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogino M, and Ogino T, 5'-Phospho-RNA Acceptor Specificity of GDP Polyribonucleotidyltransferase of Vesicular Stomatitis Virus in mRNA Capping. J Virol, 2017. 91(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino M, et al. , The Rabies Virus L Protein Catalyzes mRNA Capping with GDP Polyribonucleotidyltransferase Activity. Viruses, 2016. 8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JT, McElvain LE, and Whelan SP, Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J Virol, 2007. 81(20): p. 11499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino T, and Banerjee AK, Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell, 2007. 25(1): p. 85–97. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Wang JT, and Whelan SP, A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A, 2006. 103(22): p. 8493–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y, et al. , mRNA cap methylation influences pathogenesis of vesicular stomatitis virus in vivo. J Virol, 2014. 88(5): p. 2913–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahmeh AA, et al. , Ribose 2'-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J Virol, 2009. 83(21): p. 11043–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Fontaine-Rodriguez EC, and Whelan SP, Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol, 2005. 79(21): p. 13373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fooks AR, et al. , Rabies. Nat Rev Dis Primers, 2017. 3: p. 17091. [DOI] [PubMed] [Google Scholar]
- 38.Willoughby RE Jr., et al. , Survival after treatment of rabies with induction of coma. N Engl J Med, 2005. 352(24): p. 2508–14. [DOI] [PubMed] [Google Scholar]
- 39.Aramburo A, et al. , Failure of the Milwaukee protocol in a child with rabies. Clin Infect Dis, 2011. 53(6): p. 572–4. [DOI] [PubMed] [Google Scholar]
- 40.Rubin J, et al. , Applying the Milwaukee protocol to treat canine rabies in Equatorial Guinea. Scand J Infect Dis, 2009. 41(5): p. 372–5. [DOI] [PubMed] [Google Scholar]
- 41.Bussereau F, et al. , Treatment of rabies in mice and foxes with antiviral compounds. Acta Virol, 1988. 32(1): p. 33–49. [PubMed] [Google Scholar]
- 42.Jackson AC, Current and future approaches to the therapy of human rabies. Antiviral Res, 2013. 99(1): p. 61–7. [DOI] [PubMed] [Google Scholar]
- 43.Balzarini J, et al. , Eicar (5-ethynyl-1-beta-D-ribofuranosylimidazole-4-carboxamide). A novel potent inhibitor of inosinate dehydrogenase activity and guanylate biosynthesis. J Biol Chem, 1993. 268(33): p. 24591–8. [PubMed] [Google Scholar]
- 44.Debing Y, et al. , Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrob Agents Chemother, 2014. 58(1): p. 267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leyssen P, et al. , The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J Virol, 2005. 79(3): p. 1943–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warrell MJ, et al. , Failure of interferon alfa and tribavirin in rabies encephalitis. BMJ, 1989. 299(6703): p. 830–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kureishi A, et al. , Rabies in China: recommendations for control. Bull World Health Organ, 1992. 70(4): p. 443–50. [PMC free article] [PubMed] [Google Scholar]
- 48.Centers for Disease, C., Human rabies--Texas. MMWR Morb Mortal Wkly Rep, 1984. 33(33): p. 469–70. [PubMed] [Google Scholar]
- 49.Powers CN, Peavy DL, and Knight V, Selective inhibition of functional lymphocyte subpopulations by ribavirin. Antimicrob Agents Chemother, 1982. 22(1): p. 108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiltzer L, et al. , Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis, 2014. 209(11): p. 1744–53. [DOI] [PubMed] [Google Scholar]
- 51.Pasdeloup D, et al. , Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology, 2005. 334(2): p. 284–93. [DOI] [PubMed] [Google Scholar]
- 52.Moseley GW, et al. , Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J Cell Sci, 2009. 122(Pt 20): p. 3652–62. [DOI] [PubMed] [Google Scholar]
- 53.Vidy A, et al. , The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J Virol, 2007. 81(8): p. 4255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chavez JH, et al. , Evaluation of antiviral activity of phenolic compounds and derivatives against rabies virus. Vet Microbiol, 2006. 116(1-3): p. 53–9. [DOI] [PubMed] [Google Scholar]
- 55.Lingappa UF, et al. , Host-rabies virus protein-protein interactions as druggable antiviral targets. Proc Natl Acad Sci U S A, 2013. 110(10): p. E861–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogee S, et al. , Pyrimethamine inhibits rabies virus replication in vitro. Antiviral Res, 2018. [DOI] [PubMed] [Google Scholar]
- 57.Anindita PD, et al. , Ribavirin-related compounds exert in vitro inhibitory effects toward rabies virus. Antiviral Res, 2018. 154: p. 1–9. [DOI] [PubMed] [Google Scholar]
- 58.Mechlia MB, et al. , Dermaseptins as potential antirabies compounds. Vaccine, 2018. [DOI] [PubMed] [Google Scholar]
- 59.Zhu S, and Guo C, Rabies Control and Treatment: From Prophylaxis to Strategies with Curative Potential. Viruses, 2016. 8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60*.Yamada K, et al. , Efficacy of Favipiravir (T-705) in Rabies Postexposure Prophylaxis. J Infect Dis, 2016. 213(8): p. 1253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate the efficacy of favipiravir (T-705) against rabies virus. Not only were viral titers dramatically decreased, but lowered viral titers were observed neuronal cell lines. The compound was also just as effective as current PEP treatment and could be a suitable alternative that breaks the cold-chain.
- 61.Yang YJ, et al. , Small interfering RNAs targeting the rabies virus nucleoprotein gene. Virus Res, 2012. 169(1): p. 169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dacheux L, Delmas O, and Bourhy H, Human rabies encephalitis prevention and treatment: progress since Pasteur's discovery. Infect Disord Drug Targets, 2011. 11(3): p. 251–99. [DOI] [PubMed] [Google Scholar]
- 63*.Wu YH, et al. , Structure Properties and Mechanisms of Action of Naturally Originated Phenolic Acids and Their Derivatives against Human Viral Infections. Curr Med Chem, 2017. 24(38): p. 4279–4302. [DOI] [PubMed] [Google Scholar]; The authors compare the anti-rabies effect of several phenolic compounds. The authors identify potential phenolic scaffolds that could be investigated for further structure-activity relationship studies.
- 64.Schnell MJ, Mebatsion T, and Conzelmann KK, Infectious rabies viruses from cloned cDNA. EMBO J, 1994. 13(18): p. 4195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conzelmann KK, and Schnell M, Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol, 1994. 68(2): p. 713–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ajorloo M, et al. , Assessment the Efficiency of the Constructed Minigenome of Rabies Virus using PV Strain as Helper Virus. Arch Iran Med, 2016. 19(5): p. 335–41. [PubMed] [Google Scholar]
- 67.Ghanem A, and Conzelmann KK, G gene-deficient single-round rabies viruses for neuronal circuit analysis. Virus Res, 2016. 216: p. 41–54. [DOI] [PubMed] [Google Scholar]
- 68.Cenna J, et al. , Replication-deficient rabies virus-based vaccines are safe and immunogenic in mice and nonhuman primates. J Infect Dis, 2009. 200(8): p. 1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cenna J, et al. , Immune modulating effect by a phosphoprotein-deleted rabies virus vaccine vector expressing two copies of the rabies virus glycoprotein gene. Vaccine, 2008. 26(50): p. 6405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ito N, et al. , Characterization of M gene-deficient rabies virus with advantages of effective immunization and safety as a vaccine strain. Microbiol Immunol, 2005. 49(11): p. 971–9. [DOI] [PubMed] [Google Scholar]
- 71.Morimoto K, Shoji Y, and Inoue S, Characterization of P gene-deficient rabies virus: propagation, pathogenicity and antigenicity. Virus Res, 2005. 111(1): p. 61–7. [DOI] [PubMed] [Google Scholar]
- 72.Shoji Y, et al. , Generation and characterization of P gene-deficient rabies virus. Virology, 2004. 318(1): p. 295–305. [DOI] [PubMed] [Google Scholar]
- 73.Le Mercier P, et al. , A novel expression cassette of lyssavirus shows that the distantly related Mokola virus can rescue a defective rabies virus genome. J Virol, 2002. 76(4): p. 2024–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soezi M, et al. , Toward the development of a single-round infection assay based on EGFP reporting for anti-HIV-1 drug discovery. Rep Biochem Mol Biol, 2015. 4(1): p. 1–9. [PMC free article] [PubMed] [Google Scholar]
- 75.Wichroski MJ, et al. , High-throughput screening and rapid inhibitor triage using an infectious chimeric Hepatitis C virus. PLoS One, 2012. 7(8): p. e42609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nogales A, et al. , Development and applications of single-cycle infectious influenza A virus (sciIAV). Virus Res, 2016. 216: p. 26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelson EV, et al. , An RNA polymerase II-driven Ebola virus minigenome system as an advanced tool for antiviral drug screening. Antiviral Res, 2017. 146: p. 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cressey T, Brauburger K, and Muhlberger E, Modeling Ebola Virus Genome Replication and Transcription with Minigenome Systems. Methods Mol Biol, 2017. 1628: p. 79–92. [DOI] [PubMed] [Google Scholar]
- 79.Hoenen T, and Feldmann H, Reverse Genetics Systems for Filoviruses. Methods Mol Biol, 2017. 1602: p. 159–170. [DOI] [PubMed] [Google Scholar]
- 80*.Mehta S, et al. , Pathway Analysis of Proteomics Profiles in Rabies Infection: Towards Future Biomarkers? OMICS, 2016. 20(2): p. 97–109. [DOI] [PubMed] [Google Scholar]; The authors discover upregulated host proteins during rabies infection. The study finds potential novel pathways that are exploited by rabies virus and suggest that the upregulated proteins can serve as biomarkers for disease recognition as well as potential host-directed drug targets.
- 81.Mehta SM, Banerjee SM, and Chowdhary AS, Postgenomics biomarkers for rabies-the next decade of proteomics. OMICS, 2015. 19(2): p. 67–79. [DOI] [PubMed] [Google Scholar]
- 82.Golz S, and Huser J, Discovery of a new drug--from target identification to ultra-high-throughput screening. Clin Lab, 2007. 53(1-2): p. 77–9. [PubMed] [Google Scholar]
- 83.Seneci P, and Miertus S, Combinatorial chemistry and high-throughput screening in drug discovery: different strategies and formats. Mol Divers, 2000. 5(2): p. 75–89. [DOI] [PubMed] [Google Scholar]
- 84.Krumm SA, et al. , Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS ONE, 2011. 6(5): p. e20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dekant W, Biotransformation and renal processing of nephrotoxic agents. Arch Toxicol Suppl, 1996. 18: p. 163–72. [DOI] [PubMed] [Google Scholar]
- 86.Mutlib AE, et al. , The species-dependent metabolism of efavirenz produces a nephrotoxic glutathione conjugate in rats. Toxicol Appl Pharmacol, 2000. 169(1): p. 102–13. [DOI] [PubMed] [Google Scholar]
- 87**.Plemper RK, and Cox RM, Biology must develop herd immunity against bad-actor molecules. PLoS Pathog, 2018. 14(6): p. e1007038. [DOI] [PMC free article] [PubMed] [Google Scholar]; An exceptional reivew on important considerations for anti-viral drug development. The authors emphasize the importance of filtering out mis-leading compounds that are further revealed as pan-assay interference compounds with questionable chemistry profiles.
- 88.Lupberger J, et al. , EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med, 2011. 17(5): p. 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gerold G, and Pietschmann T, The HCV life cycle: in vitro tissue culture systems and therapeutic targets. Dig Dis, 2014. 32(5): p. 525–37. [DOI] [PubMed] [Google Scholar]
- 90.Hotta K, and Kiura K, Safety profiles of erlotinib therapy in patients with advanced non-small-cell lung cancer. Expert Rev Anticancer Ther, 2011. 11(7): p. 991–7. [DOI] [PubMed] [Google Scholar]
- 91.Castellote J, et al. , Serious drug-induced liver disease secondary to ezetimibe. World J Gastroenterol, 2008. 14(32): p. 5098–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Erba HP, et al. , Improving frontline treatment for chronic myeloid leukemia: emerging evidence for use of nilotinib and dasatinib. Clin Adv Hematol Oncol, 2011. 9(10): p. 734–45. [PubMed] [Google Scholar]
- 93**.Kaufmann SHE, et al. , Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov, 2018. 17(1): p. 35–56. [DOI] [PMC free article] [PubMed] [Google Scholar]; A broad review on the current progress of host-directed therapies. This review provides a well balanced argument for the use of host-directed anti-virals and emphasizes both the pros and cons of such therapies.
- 94.Zumla A, et al. , Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect Dis, 2016. 16(4): p. e47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weir DL, et al. , Host cell virus entry mediated by Australian bat lyssavirus G envelope glycoprotein occurs through a clathrin-mediated endocytic pathway that requires actin and Rab5. Virol J, 2014. 11: p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Piccinotti S, and Whelan SP, Rabies Internalizes into Primary Peripheral Neurons via Clathrin Coated Pits and Requires Fusion at the Cell Body. PLoS Pathog, 2016. 12(7): p. e1005753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Piccinotti S, Kirchhausen T, and Whelan SP, Uptake of rabies virus into epithelial cells by clathrin-mediated endocytosis depends upon actin. J Virol, 2013. 87(21): p. 11637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mues MB, et al. , Dynasore disrupts trafficking of herpes simplex virus proteins. J Virol, 2015. 89(13): p. 6673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dai W, et al. , Antiviral Effects of ABMA against Herpes Simplex Virus Type 2 In Vitro and In Vivo. Viruses, 2018. 10(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ohkubo S, and Nakahata N, [Role of lipid rafts in trimeric G protein-mediated signal transduction]. Yakugaku Zasshi, 2007. 127(1): p. 27–40. [DOI] [PubMed] [Google Scholar]
- 101.Bruses JL, Chauvet N, and Rutishauser U, Membrane lipid rafts are necessary for the maintenance of the (alpha)7 nicotinic acetylcholine receptor in somatic spines of ciliary neurons. J Neurosci, 2001. 21(2): p. 504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roth AL, and Berg DK, Large clusters of alpha7-containing nicotinic acetylcholine receptors on chick spinal cord neurons. J Comp Neurol, 2003. 465(2): p. 195–204. [DOI] [PubMed] [Google Scholar]
- 103.Bilderback TR, Grigsby RJ, and Dobrowsky RT, Association of p75(NTR) with caveolin and localization of neurotrophin-induced sphingomyelin hydrolysis to caveolae. J Biol Chem, 1997. 272(16): p. 10922–7. [DOI] [PubMed] [Google Scholar]
- 104.Conroy WG, Ogden LF, and Berg DK, Cluster formation of alpha7-containing nicotinic receptors at interneuronal interfaces in cell culture. Neuropharmacology, 2000. 39(13): p. 2699–705. [DOI] [PubMed] [Google Scholar]
- 105.Sissoeff L, et al. , Stable trimerization of recombinant rabies virus glycoprotein ectodomain is required for interaction with the p75NTR receptor. J Gen Virol, 2005. 86(Pt 9): p. 2543–52. [DOI] [PubMed] [Google Scholar]
- 106.Shoop RD, Yamada N, and Berg DK, Cytoskeletal links of neuronal acetylcholine receptors containing alpha 7 subunits. J Neurosci, 2000. 20(11): p. 4021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hotta K, et al. , Effect of cellular cholesterol depletion on rabies virus infection. Virus Res, 2009. 139(1): p. 85–90. [DOI] [PubMed] [Google Scholar]
- 108.Manjunatha V, et al. , Inhibition of MEK-ERK1/2-MAP kinase signalling pathway reduces rabies virus induced pathologies in mouse model. Microb Pathog, 2017. 112: p. 38–49. [DOI] [PubMed] [Google Scholar]
- 109.Ivanyi P, et al. , Novel therapies in advanced renal cell carcinoma: management of adverse events from sorafenib and sunitinib. Dtsch Arztebl Int, 2008. 105(13): p. 232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Himmelsbach K, et al. , New aspects of an anti-tumour drug: sorafenib efficiently inhibits HCV replication. Gut, 2009. 58(12): p. 1644–53. [DOI] [PubMed] [Google Scholar]
- 111.Lundberg L, et al. , Repurposed FDA-Approved drug sorafenib reduces replication of Venezuelan equine encephalitis virus and other alphaviruses. Antiviral Res, 2018. 157: p. 57–67. [DOI] [PubMed] [Google Scholar]
- 112.Marosi A, et al. , Combination therapy of rabies-infected mice with inhibitors of pro-inflammatory host response, antiviral compounds and human rabies immunoglobulin. Vaccine, 2018. [DOI] [PubMed] [Google Scholar]
- 113.Descamps V, et al. , The kinase-inhibitor sorafenib inhibits multiple steps of the Hepatitis C Virus infectious cycle in vitro. Antiviral Res, 2015. 118: p. 93–102. [DOI] [PubMed] [Google Scholar]
- 114.Gao M, et al. , The multi-targeted kinase inhibitor sorafenib inhibits enterovirus 71 replication by regulating IRES-dependent translation of viral proteins. Antiviral Res, 2014. 106: p. 80–5. [DOI] [PubMed] [Google Scholar]
- 115.Himmelsbach K, and Hildt E, The kinase inhibitor Sorafenib impairs the antiviral effect of interferon alpha on hepatitis C virus replication. Eur J Cell Biol, 2013. 92(1): p. 12–20. [DOI] [PubMed] [Google Scholar]
- 116.Roberts JL, et al. , GRP78/Dna K Is a Target for Nexavar/Stivarga/Votrient in the Treatment of Human Malignancies, Viral Infections and Bacterial Diseases. J Cell Physiol, 2015. 230(10): p. 2552–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rimola J, et al. , Complete response under sorafenib in patients with hepatocellular carcinoma: Relationship with dermatologic adverse events. Hepatology, 2017. [DOI] [PubMed] [Google Scholar]
- 118.Gyawali B, et al. , Risk of serious adverse events and fatal adverse events with sorafenib in patients with solid cancer: a meta-analysis of phase 3 randomized controlled trialsdagger. Ann Oncol, 2017. 28(2): p. 246–253. [DOI] [PubMed] [Google Scholar]
- 119.Granito A, et al. , Prognostic significance of adverse events in patients with hepatocellular carcinoma treated with sorafenib. Therap Adv Gastroenterol, 2016. 9(2): p. 240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Santoni M, et al. , Risk of gastrointestinal events with sorafenib, sunitinib and pazopanib in patients with solid tumors: a systematic review and meta-analysis of clinical trials. Int J Cancer, 2014. 135(4): p. 763–73. [DOI] [PubMed] [Google Scholar]
- 121.Brunocilla PR, et al. , Sorafenib in hepatocellular carcinoma: prospective study on adverse events, quality of life, and related feasibility under daily conditions. Med Oncol, 2013. 30(1): p. 345. [DOI] [PubMed] [Google Scholar]
- 122.Zhao Y, et al. , Drug-related adverse events may predict efficacy in sorafenib therapy for hepatocellular carcinoma. Hepatology, 2012. 56(2): p. 790–1. [DOI] [PubMed] [Google Scholar]
- 123.Choueiri TK, et al. , Risk of arterial thromboembolic events with sunitinib and sorafenib: a systematic review and meta-analysis of clinical trials. J Clin Oncol, 2010. 28(13): p. 2280–5. [DOI] [PubMed] [Google Scholar]
- 124.Goel P, Manning JA, and Kumar S, NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene, 2015. 557(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Harty RN, et al. , A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol, 1999. 73(4): p. 2921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Han Z, et al. , Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol, 2014. 88(13): p. 7294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Albertini AA, et al. , Crystal structure of the rabies virus nucleoprotein-RNA complex. Science, 2006. 313(5785): p. 360–3. [DOI] [PubMed] [Google Scholar]
- 128.Albertini AA, et al. , Structural aspects of rabies virus replication. Cell Mol Life Sci, 2008. 65(2): p. 282–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Holmes EC, et al. , Genetic constraints and the adaptive evolution of rabies virus in nature. Virology, 2002. 292(2): p. 247–57. [DOI] [PubMed] [Google Scholar]
- 130.Kissi B, Tordo N, and Bourhy H, Genetic polymorphism in the rabies virus nucleoprotein gene. Virology, 1995. 209(2): p. 526–37. [DOI] [PubMed] [Google Scholar]
- 131.Wu X, et al. , Both viral transcription and replication are reduced when the rabies virus nucleoprotein is not phosphorylated. J Virol, 2002. 76(9): p. 4153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wu X, Lei X, and Fu ZF, Rabies virus nucleoprotein is phosphorylated by cellular casein kinase II. Biochem Biophys Res Commun, 2003. 304(2): p. 333–8. [DOI] [PubMed] [Google Scholar]
- 133.Smith TG, et al. , Design of future rabies biologics and antiviral drugs. Adv Virus Res, 2011. 79: p. 345–63. [DOI] [PubMed] [Google Scholar]
- 134.Chapman J, et al. , RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob Agents Chemother, 2007. 51(9): p. 3346–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Assenberg R, et al. , Structure of the nucleoprotein binding domain of Mokola virus phosphoprotein. J Virol, 2010. 84(2): p. 1089–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mavrakis M, et al. , Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J Mol Biol, 2004. 343(4): p. 819–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Delmas O, et al. , The structure of the nucleoprotein binding domain of lyssavirus phosphoprotein reveals a structural relationship between the N-RNA binding domains of Rhabdoviridae and Paramyxoviridae. RNA Biol, 2010. 7(3): p. 322–7. [DOI] [PubMed] [Google Scholar]
- 138.Ivanov I, et al. , Structure of the dimerization domain of the rabies virus phosphoprotein. J Virol, 2010. 84(7): p. 3707–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Real E, et al. , Antiviral drug discovery strategy using combinatorial libraries of structurally constrained peptides. J Virol, 2004. 78(14): p. 7410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Otvos L Jr. and Wade JD, Current challenges in peptide-based drug discovery. Front Chem, 2014. 2: p. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mebatsion T, Weiland F, and Conzelmann KK, Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol, 1999. 73(1): p. 242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zan J, et al. , Rabies virus matrix protein induces apoptosis by targeting mitochondria. Exp Cell Res, 2016. 347(1): p. 83–94. [DOI] [PubMed] [Google Scholar]
- 143.Mita T, et al. , Amino acid at position 95 of the matrix protein is a cytopathic determinant of rabies virus. Virus Res, 2008. 137(1): p. 33–9. [DOI] [PubMed] [Google Scholar]
- 144.Komarova AV, et al. , Rabies virus matrix protein interplay with eIF3, new insights into rabies virus pathogenesis. Nucleic Acids Res, 2007. 35(5): p. 1522–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.von Kobbe C, et al. , Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell, 2000. 6(5): p. 1243–52. [DOI] [PubMed] [Google Scholar]
- 146.Graham SC, et al. , Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog, 2008. 4(12): p. e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zentner I, et al. , Discovery of a small-molecule antiviral targeting the HIV-1 matrix protein. Bioorg Med Chem Lett, 2013. 23(4): p. 1132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wirblich C, et al. , PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J Virol, 2008. 82(19): p. 9730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Irie T, et al. , Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J Virol, 2004. 78(6): p. 2657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Besson B, et al. , Regulation of NF-kappaB by the p105-ABIN2-TPL2 complex and RelAp43 during rabies virus infection. PLoS Pathog, 2017. 13(10): p. e1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ben Khalifa Y, et al. , The matrix protein of rabies virus binds to RelAp43 to modulate NF-kappaB-dependent gene expression related to innate immunity. Sci Rep, 2016. 6: p. 39420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zidar M, Kuzman D, and Ravnik M, Characterisation of protein aggregation with the Smoluchowski coagulation approach for use in biopharmaceuticals. Soft Matter, 2018. 14(29): p. 6001–6012. [DOI] [PubMed] [Google Scholar]
- 153.Thongcharoen P, et al. , Monoclonal antibody studies of rabies viruses isolated from Thailand. Southeast Asian J Trop Med Public Health, 1990. 21(1): p. 129–33. [PubMed] [Google Scholar]
- 154.Lafon M, Bourhy H, and Sureau P, Immunity against the European bat rabies (Duvenhage) virus induced by rabies vaccines: an experimental study in mice. Vaccine, 1988. 6(4): p. 362–8. [DOI] [PubMed] [Google Scholar]
- 155.Nolden T, et al. , Comparative studies on the genetic, antigenic and pathogenic characteristics of Bokeloh bat lyssavirus. J Gen Virol, 2014. 95(Pt 8): p. 1647–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Horton DL, et al. , Antigenic and genetic characterization of a divergent African virus, Ikoma lyssavirus. J Gen Virol, 2014. 95(Pt 5): p. 1025–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Papaneri AB, et al. , Alanine scanning of the rabies virus glycoprotein antigenic site III using recombinant rabies virus: implication for post-exposure treatment. Vaccine, 2013. 31(49): p. 5897–902. [DOI] [PubMed] [Google Scholar]
- 158.Bourhy H, et al. , Antigenic and molecular characterization of bat rabies virus in Europe. J Clin Microbiol, 1992. 30(9): p. 2419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159*.Franka R, et al. , In Vivo Efficacy of a Cocktail of Human Monoclonal Antibodies (CL184) Against Diverse North American Bat Rabies Virus Variants. Trop Med Infect Dis, 2017. 2(3). [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors present a monoclonal antibody cocktail, CL184, that introduces a novel and highly conserved antigenic site on the rabies glycoprotein. CL184 is able to neutralize a wide-breadth of clinically relevant rabies viruses.
- 160*.Chao TY, et al. , SYN023, a novel humanized monoclonal antibody cocktail, for post-exposure prophylaxis of rabies. PLoS Negl Trop Dis, 2017. 11(12): p. e0006133. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors identify two novel monoclonal antibodies that are effective at neutralizing diverse rabies virus strains. Together as a cocktail these antibodies were found to be just as effective as current hRIG PEP treatment.
- 161.Kulkarni PS, et al. , Development of a new purified vero cell rabies vaccine (Rabivax-S) at the serum institute of India Pvt Ltd. Expert Rev Vaccines, 2017. 16(4): p. 303–311. [DOI] [PubMed] [Google Scholar]
- 162.De Benedictis P, et al. , Development of broad-spectrum human monoclonal antibodies for rabies post-exposure prophylaxis. EMBO Mol Med, 2016. 8(4): p. 407–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Arzensek D, Kuzman D, and Podgornik R, Colloidal interactions between monoclonal antibodies in aqueous solutions. J Colloid Interface Sci, 2012. 384(1): p. 207–16. [DOI] [PubMed] [Google Scholar]
- 164.Goudsmit J, et al. , Comparison of an anti-rabies human monoclonal antibody combination with human polyclonal anti-rabies immune globulin. J Infect Dis, 2006. 193(6): p. 796–801. [DOI] [PubMed] [Google Scholar]
- 165.Gaudin Y, Rabies virus-induced membrane fusion pathway. J Cell Biol, 2000. 150(3): p. 601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mackman RL, et al. , Discovery of an oral respiratory syncytial virus (RSV) fusion inhibitor (GS-5806) and clinical proof of concept in a human RSV challenge study. J Med Chem, 2015. 58(4): p. 1630–43. [DOI] [PubMed] [Google Scholar]
- 167.Yan D, et al. , Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc Natl Acad Sci U S A, 2014. 111(33): p. E3441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Douglas JL, et al. , Small molecules VP-14637 and JNJ-2408068 inhibit respiratory syncytial virus fusion by similar mechanisms. Antimicrob Agents Chemother, 2005. 49(6): p. 2460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Albertini AA, et al. , Molecular and cellular aspects of rhabdovirus entry. Viruses, 2012. 4(1): p. 117–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kiso M, et al. , Combination Therapy With Neuraminidase and Polymerase Inhibitors in Nude Mice Infected With Influenza Virus. J Infect Dis, 2018. 217(6): p. 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Andrei G, et al. , The Anti-Human Immunodeficiency Virus Drug Tenofovir, a Reverse Transcriptase Inhibitor, Also Targets the Herpes Simplex Virus DNA Polymerase. J Infect Dis, 2018. 217(5): p. 790–801. [DOI] [PubMed] [Google Scholar]
- 172.Spengler U, Direct antiviral agents (DAAs) - A new age in the treatment of hepatitis C virus infection. Pharmacol Ther, 2018. 183: p. 118–126. [DOI] [PubMed] [Google Scholar]
- 173.Fearns R, and Plemper RK, Polymerases of paramyxoviruses and pneumoviruses. Virus Res, 2017. 234: p. 87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174**.Cox R, and Plemper RK, The paramyxovirus polymerase complex as a target for next-generation anti-paramyxovirus therapeutics. Front Microbiol, 2015. 6: p. 459. [DOI] [PMC free article] [PubMed] [Google Scholar]; An outstanding review that highlights the RNA dependent RNA polymerase as a promising target for anti-virals due its highly conserved nature, and critical importance for the viral lifecycle. This review describes both nucleoside analogs and small-molecule inhibitors as well as current approaches to anti-viral drug discovery.
- 175.Segura Guerrero NA, et al. , Favipiravir inhibits in vitro Usutu virus replication and delays disease progression in an infection model in mice. Antiviral Res, 2018. [DOI] [PubMed] [Google Scholar]
- 176.Goldhill DH, et al. , Determining the Mutation Bias of Favipiravir in Influenza Using Next-generation Sequencing. J Virol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Franco EJ, et al. , The effectiveness of antiviral agents with broad-spectrum activity against chikungunya virus varies between host cell lines. Antivir Chem Chemother, 2018. 26: p. 2040206618807580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Goldhill DH, et al. , The mechanism of resistance to favipiravir in influenza. Proc Natl Acad Sci U S A, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dawes BE, et al. , Favipiravir (T-705) protects against Nipah virus infection in the hamster model. Sci Rep, 2018. 8(1): p. 7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.de la Torre JC, Extending the Antiviral Value of Favipiravir. J Infect Dis, 2018. 218(4): p. 509–511. [DOI] [PubMed] [Google Scholar]
- 181.Delang L, Abdelnabi R, and Neyts J, Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res, 2018. 153: p. 85–94. [DOI] [PubMed] [Google Scholar]
- 182*.Yoon JJ, et al. , Orally Efficacious Broad-Spectrum Ribonucleoside Analog Inhibitor of Influenza and Respiratory Syncytial Viruses. Antimicrob Agents Chemother, 2018. 62(8). [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe a broad-spectrum ribonucleoside analog inhibitor, NHC, that is highly efficacious against both influenza and respiratory syncytial viruses. Further characterization of the compound reveals it to have great bioavailability and a high barrier against viral escape.
- 183.Ehteshami M, et al. , Characterization of beta-d-N(4)-Hydroxycytidine as a Novel Inhibitor of Chikungunya Virus. Antimicrob Agents Chemother, 2017. 61(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Reynard O, et al. , Identification of a New Ribonucleoside Inhibitor of Ebola Virus Replication. Viruses, 2015. 7(12): p. 6233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Matthes E, and Bunger H, Cellular pharmacology of the anti-hepatitis B virus agent beta-L-2',3'-didehydro-2',3'-dideoxy-N4-hydroxycytidine: relevance for activation in HepG2 cells. Antimicrob Agents Chemother, 2010. 54(1): p. 341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Barnard DL, et al. , Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir Chem Chemother, 2006. 17(5): p. 275–84. [DOI] [PubMed] [Google Scholar]
- 187.Shi J, et al. , Synthesis and in vitro anti-HCV activity of beta-D- and 1-2'-deoxy-2'-fluororibonucleosides. Nucleosides Nucleotides Nucleic Acids, 2005. 24(5-7): p. 875–9. [DOI] [PubMed] [Google Scholar]
- 188.Hollecker L, et al. , Synthesis of beta-enantiomers of N4-hydroxy-3'-deoxypyrimidine nucleosides and their evaluation against bovine viral diarrhoea virus and hepatitis C virus in cell culture. Antivir Chem Chemother, 2004. 15(1): p. 43–55. [DOI] [PubMed] [Google Scholar]
- 189.Muller T, et al. , Development of a mouse monoclonal antibody cocktail for post-exposure rabies prophylaxis in humans. PLoS Negl Trop Dis, 2009. 3(11): p. e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Talbi C, et al. , Evolutionary history and dynamics of dog rabies virus in western and central Africa. J Gen Virol, 2009. 90(Pt 4): p. 783–91. [DOI] [PubMed] [Google Scholar]
- 191.Sakamoto S, et al. , Studies on the structures and antigenic properties of rabies virus glycoprotein analogues produced in yeast cells. Vaccine, 1999. 17(3): p. 205–18. [DOI] [PubMed] [Google Scholar]
- 192.Tuchiya K, et al. , Characterization of rabies virus glycoprotein expressed by recombinant baculovirus. Virus Res, 1992. 25(1-2): p. 1–13. [DOI] [PubMed] [Google Scholar]
- 193.Hemachudha T, et al. , Failure of therapeutic coma and ketamine for therapy of human rabies. J Neurovirol, 2006. 12(5): p. 407–9. [DOI] [PubMed] [Google Scholar]
- 194*.Banyard AC, et al. , Re-evaluating the effect of Favipiravir treatment on rabies virus infection. Vaccine, 2017. [DOI] [PubMed] [Google Scholar]; The authors administer favipiravir to rabies infected mice to determine its efficacy in vivo. Despite promising efficacy in vitro, favipiravir was shown to have limited protective effect on the mice and was shown to only slow the onset of clinical symptoms rather than clear viral infection.
- 195.Roche S, et al. , Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited. Cell Mol Life Sci, 2008. 65(11): p. 1716–28. [DOI] [PMC free article] [PubMed] [Google Scholar]