Capsule Summary:
We identified a strong association between peanut allergy and the MALT1 locus in LEAP participants in the peanut avoidance group with 58.6% of carriers developing peanut allergy at 60 months as compared to 12.7% of non-carriers.
Keywords: Peanut allergy, MALT1, GWAS, food allergy, immunogenetics, early allergen exposure, IgE
To the Editor:
The Learning Early about Peanut Allergy (LEAP) trial (1, 2) motivated a change in pediatric guidelines for the early introduction of dietary peanut as an effective strategy for the prevention of peanut allergy. LEAP participants were presumed to be at increased risk for peanut allergy(3), and dietary introduction of peanut protein beginning in the first four to eleven months of life significantly decreased the frequency of peanut allergy later in childhood and modulated the immune response to peanuts in this at-risk group (1). To identify the genetic determinants of peanut allergy in the LEAP participants, whole genome sequencing (WGS) was performed (Supplementary Information Sections 1–2, Table S1). Following published standards for WGS data (4) (Supplementary Information Sections 3–4), there were 542 per protocol LEAP participants available for genome-wide genetic association tests, including 49 with peanut allergy, which was defined as a positive result on a double-blind placebo-controlled oral food challenge at 60 months of age (Supplementary Information Section 1). The de-convolution of genetic ancestry aligns well with self-reported race/ethnicity (Supplementary Information Section 5, Fig S1). Given the high success of early introduction of dietary peanut in the LEAP trial, 48 peanut allergy participants were from the peanut avoidance arm and only one from the consumption arm (Table S1). Therefore, genome-wide association was assessed for peanut allergy in the 275 participants from the avoidance arm (N=48 peanut allergic/N=227 non peanut allergic) on a total of 4,444,069 single nucleotide variants (SNVs, Fig 1, Supplementary Information Section 6, Fig S2) in a discovery analysis. Subsequent follow up of the peak genetic signal(s) was extended to include the participants from the consumption arm (N=267) with immunological quantitative traits of importance (Fig 2) to facilitate the examination of the identified genetic loci in the context of the intervention.
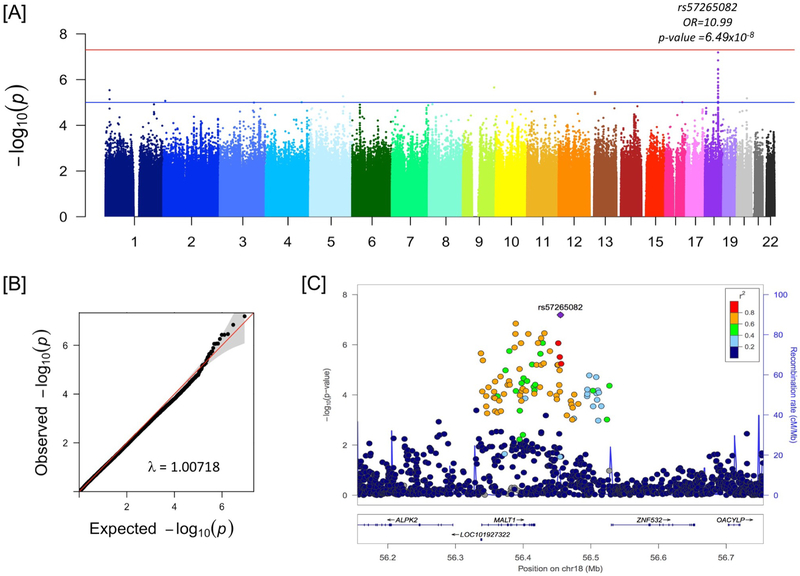
Figure 1:
Genome-wide association with peanut allergy at 60 months in N=275 LEAP participants in the peanut avoidance group. Panel A is the genome-wide Manhattan plot for N=4,444,069 SNVs with MAF ≥ 2%, missingness < 5%, and Hardy-Weinberg Equilibrium p ≥ 10−6. Panel B is the quantile-quantile plot of the same data as Panel A. Panel C shows the peak association region on chromosome 18.
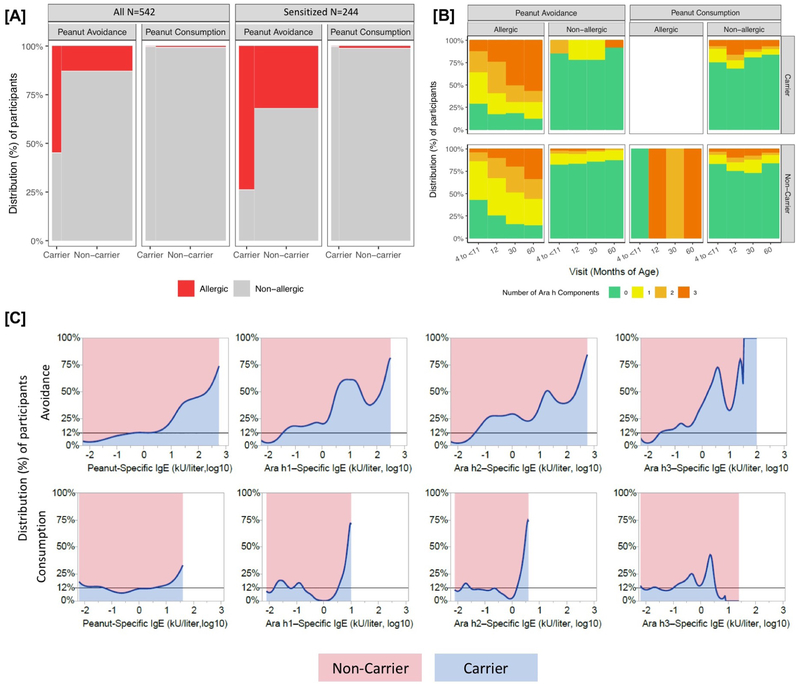
Figure 2:
Panel A shows the proportion of allergic and non-allergic LEAP participants at 60 months of age by treatment group and MALT1 carrier status in all LEAP participants (left) and the sensitized group (right, defined as those with peanut-specific IgE ≥ 0.1 kU/liter at 60 months). Panel B shows the IgE response to Ara h1, Ara h2, and Ara h3 over the course of the LEAP study in all participants (N=542). Ara h status was imputed to 0 for all participants with peanut-specific IgE <0.1. Panel C is the proportion density plots showing the relative distribution of peanut-specific IgE and IgE to Ara h1, Ara h2, and Ara h3 between the MALT1 Carrier and Non-carrier groups at 60 months of age. The horizontal reference line at 12% indicates the proportion of the population with at least one MALT1 risk allele, which illustrates a null distribution with equal proportions of individuals at all titer levels between the carriers and non-carriers. Ara h status was imputed to −2 (log10) for all participants with peanut-specific IgE <0.1 kU/liter. For all panels, imputed genotypes were used for 7 individuals missing allele calls at rs57265082, and Non-carriers were defined as having at least one copy of the T allele (due to the low MAF).
The peak association for peanut allergy in the avoidance group was observed on chromosome 18 (Fig 1A) mapping to the mucosa-associated lymphoid tissue lymphoma translocation (MALT1) gene (Fig 1C, Table S2). The region (Chr18:56337602..56456191) includes strong regulatory signatures for MALT1 expression as well as the expression of the intergenic non-coding RNA (lincRNA), RP11–108P20.1 in the Genotype-Tissue Expression (GTEx) data (Figs S3, S4). However, the specific set of SNVs with p-value <10−5 for peanut allergy only have eQTL signatures for MALT1 (Table S3). The peak associated SNV was rs57265082 with an estimated Odds Ratio (OR) of 10.99, minor allele frequency (MAF) of 5.6%, and p = 6.49×10−8. Gene-based analysis was performed across rare exonic SNVs (MAF ≤5%) using SKAT (Tables S4 and S5). There was nominal association with either all rare exonic SNVs (p = 0.0830) or all rare damaging exonic SNVs (p = 0.0828); however, with the inclusion of the peak WGS variant, rs57265082, the gene-based evidence was very strong (p = 1.89×10−10). Conditioning on the peak SNV, rs57265082, shows that the observed common variant signal is a single genetic locus within the region (Fig S5). There are overall strong differences in the clinical profiles of the MALT1 risk allele carriers compared to non-carriers within the peanut avoidance participants (Table S6). MALT1 is not associated with baseline selection criteria of egg allergy or eczema (p = 0.3241, and p=0.1626 respectively in the avoidance group), and the association between peanut allergy and MALT1 is independent of these baseline selections (Table S7). We observe no association between the key filaggrin variant, R501X, documented to play a role in eczema and peanut allergy (p=0.4014 and MAF of 3.6% in the avoidance group), but recognize that our sample size of N=275 may be underpowered for this.
We observe a weaker association with rs57265082 to sensitization (at 60 months, sensitization is defined as those with peanut-specific IgE ≥ 0.1 kU/liter) in the peanut avoidance group (OR = 4.55, p = 0.0011). Additionally, the MALT1 locus remains significantly associated with peanut allergy (p = 0.0003), even within the subset of sensitized participants in the peanut avoidance group (Fig 2A), supporting its role as a genetic risk factor for allergy and not only sensitization. With the inclusion of the LEAP participants from the consumption arm (N=267), MALT1 was found to be significantly associated with an IgE response to multiple specific peanut allergenic protein components Ara h1, Ara h2, and Ara h3 at 60 months (p = 1.11×10−5, Fig 2B, Supplementary Section 7) in the full set of LEAP participants adjusting for intervention.
When examining specific IgE to peanut, as well as the three major allergenic components of peanut, we observe a progressive divergence in the upper end of the IgE distributions in MALT1 carriers (Fig 2C) with two key observations to note. First, the intervention with peanut exposure effectively reduced peanut-specific IgE irrespective of carrier status (truncated distributions in Fig 2C, bottom panel). Second, within the avoidance group, the levels of peanut-specific IgE between the carriers and non-carriers is markedly different; rs57265082 carriers within the peanut avoidance group had the highest peanut-specific IgE levels as compared to non-carriers (Fig 2C, upper panel). The mean titers of peanut-specific IgE were significantly different between carriers vs non-carriers and by treatment group (interaction p = 1.86×10−5), even after adjusting for the baseline differences in peanut-specific IgE (Fig S6A). Importantly, this effect of MALT1 on peanut-specific IgE in the peanut avoidance group is independent of total IgE (Fig S6B, p=2.03×10−5 for peanut-specific IgE and p=0.366 for total IgE). Finally, the additional value of knowing rs57265082 carrier status in predicting an individual’s likelihood of allergy was evaluated, and rs57265082 was found to be an independent predictor of allergy in the avoidance group (Fig S7).
In this first report of the genetics of peanut allergy within the LEAP study, a key biological candidate, the MALT1 gene, is implicated as an independent risk factor for peanut allergy in the context of peanut avoidance. These associations are irrespective of sensitization status (in Fig S7, sensitization at baseline is defined by skin prick positivity, and in Fig 2A, sensitization at 60 months sensitization is defined as peanut-specific IgE ≥ 0.1 kU/liter), supporting a relationship with progression to symptomatic allergy after peanut sensitization, a disease pattern that is inhibited by early and continuous consumption of peanuts. MALT1 encodes a paracaspase that functions as a critical part of the CARMA1-BCL10-MALT1 (CBM) complex, causing NF-KappaB activation in B and T cells in response to an antigen binding to the B or T cell receptor(5). In T cells, this forms part of the signaling cascade leading to T cell activation(6) and involves the two MALT1 isoforms, MALT1A and MALT1B(7). Given that our top SNVs affect MALT1 expression, it is possible that these variants may predispose an individual to greater allergic disease by altering MALT1 expression or affecting the ratio of MALT1A to MALT1B, thus increasing Th2 differentiation after antigen presentation. Additional genes encoding other members of the CBM complex do not show evidence for association within our discovery data (Fig S8).
MALT1 has not been implicated in prior genetic studies, and we are also unable to replicate prior published associations (Table S8) (8, 9). It is important to note that the prior studies compare non-allergic controls to peanut-allergic subjects (8, 9), and the genetic associations identified in these likely represent risk of allergic sensitization and not specifically peanut allergy. In contrast, the LEAP study included only participants who were at high risk for peanut allergy, many of whom were sensitized at baseline, and this unique ascertainment of the LEAP study facilitates our ability to test specifically for the risk of peanut allergy. Yet another singular advantage of the LEAP study is that we are able to interrogate the avoidance group (high incidence of peanut allergy) and contrast this to the consumption group (low incidence of peanut allergy) using quantitative immunological markers to identify the genetic determinants of peanut allergy that are relevant in the absence of peanut exposure. This homogeneity of exposure (i.e. avoidance) and ascertainment (i.e. baseline risk factors) within LEAP account for the ability to detect a strong association with MALT1 despite the limited sample size of N=275 in the discovery analysis; in fact the p-value of 6.49×10−8 for the single variant tests is near the Bonferroni threshold for GWAS significance (5×10−8), and our gene-based analysis results in a p = 1.89×10−10. Targeted genotyping of rs57265082 on additional LEAP participants, including the non per protocol participants, does not change the results from the discovery sample (Supplementary Section 8, Table S9). Furthermore, of the seven participants within the consumption arm that had peanut allergy at baseline, three were MALT1 carriers (unadjusted OR for peanut allergy at baseline in the LEAP consumption group = 5.3, p = 0.0188 using a Pearson chi-square test). However, the lack of a suitable population to use as a replication group is a major limitation of this study, and additional replication will be important to follow up on these associations observed within LEAP.
One striking observation is the differing effect of MALT1 carrier status on peanut-specific IgE patterns between the two intervention arms in LEAP. The introduction of dietary peanut as a strategy for the prevention of peanut allergy is equally effective within carriers and non-carriers. However, our results indicate that within the LEAP participants, MALT1 carriers from the peanut avoidance group have the highest risk for peanut allergy (58.6% of carriers of the MALT1 variant in the avoidance group go on to get peanut allergy in contrast to only 12.7% of the non-carriers, Table S6). Coupled with the observations that 1) the acquisition of additional peanut antigen target specificities in the IgE response is markedly increased in the MALT1 carriers and 2) this peanut-specific IgE response is independent of total IgE, our findings support a genotypephenotype relationship that implicates the MALT1 pathway in the allergic immune pathogenesis of peanut allergy.
Supplementary Material
Acknowledgements
This research was performed as a project of the Immune Tolerance Network, an international clinical research consortium headquartered at the Benaroya Research Institute, and supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under Award Number UM1AI109565. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Drs. Daniel Rotrosen and Alkis Togias from the NIAID for their review of the manuscript, Ms. Monica Campbell from the University of Colorado for her help preparing samples for genotyping, and all the LEAP participants who took part in the study. Dr. Lack reports holding stock and stock options in DBV Technologies. No other potential conflict of interest relevant to this article was reported.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372(9):803–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Togias A, Cooper SF, Acebal ML, Assa’ad A, Baker JR Jr., Beck LA, et al. Addendum guidelines for the prevention of peanut allergy in the United States: Report of the National Institute of Allergy and Infectious Diseases-sponsored expert panel. Ann Allergy Asthma Immunol. 2017;118(2):166–73 e7. [DOI] [PubMed] [Google Scholar]
- 3.Du Toit G, Roberts G, Sayre PH, Plaut M, Bahnson HT, Mitchell H, et al. Identifying infants at high risk of peanut allergy: the Learning Early About Peanut Allergy (LEAP) screening study. J Allergy Clin Immunol. 2013;131(1):135–43 e1–12. [DOI] [PubMed] [Google Scholar]
- 4.Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, O’Connor TD, et al. A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun. 2016;7:12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thome M CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4(5):348–59. [DOI] [PubMed] [Google Scholar]
- 6.Vickery BP, Chin S, Burks AW. Pathophysiology of food allergy. Pediatr Clin North Am. 2011;58(2):363–76, ix-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meininger I, Griesbach RA, Hu D, Gehring T, Seeholzer T, Bertossi A, et al. Alternative splicing of MALT1 controls signalling and activation of CD4(+) T cells. Nat Commun. 2016;7:11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asai Y, Eslami A, van Ginkel CD, Akhabir L, Wan M, Yin D, et al. A Canadian genome-wide association study and meta-analysis confirm HLA as a risk factor for peanut allergy independent of asthma. J Allergy Clin Immunol. 2018;141(4):1513–6. [DOI] [PubMed] [Google Scholar]
- 9.Asai Y, Eslami A, van Ginkel CD, Akhabir L, Wan M, Ellis G, et al. Genome-wide association study and meta-analysis in multiple populations identifies new loci for peanut allergy and establishes C11orf30/EMSY as a genetic risk factor for food allergy. J Allergy Clin Immunol. 2018;141(3):991–1001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.