Abstract
Objective:
Sepsis remains a costly and prevalent syndrome in hospitals; however, machine learning systems can increase timely sepsis detection using electronic health records. This study validates a gradient boosted ensemble machine learning tool for sepsis detection and prediction, and compares its performance to existing methods.
Materials and Methods:
Retrospective data was drawn from databases at the University of California, San Francisco (UCSF) Medical Center and the Beth Israel Deaconess Medical Center (BIDMC). Adult patient encounters without sepsis on admission, and with at least one recording of each of six vital signs (SpO2, heart rate, respiratory rate, temperature, systolic and diastolic blood pressure) were included. We compared the performance of the machine learning algorithm (MLA) to that of commonly used scoring systems. Area under the receiver operating characteristic (AUROC) curve was our primary measure of accuracy. MLA performance was measured at sepsis onset and 24, and 48 hours prior sepsis onset.
Results:
The MLA achieved AUROC of 0.88, 0.84, and 0.83 for sepsis onset and 24 and 48 hours prior to onset, respectively. These values were superior to those of SIRS (0.66), MEWS (0.61), SOFA (0.72), and qSOFA (0.60) at time of onset. When trained on UCSF data and tested on BIDMC data, sepsis onset AUROC was 0.89.
Discussion and Conclusion:
The MLA predicts sepsis up to 48 hours in advance and identifies sepsis onset more accurately than commonly used tools, maintaining high performance for sepsis detection when trained and tested on separate datasets.
Keywords: Sepsis, Machine Learning, Electronic Health Records, Prediction
Introduction
Sepsis remains one of the most prevalent and costly syndromes in hospitals nationwide. Formerly classified in a three tier system progressing from sepsis to severe sepsis to septic shock, sepsis was recently redefined as a two tier progression from sepsis (encompassing severe sepsis) to septic shock [1]. Sepsis is often life-threatening, with reported mortality rates of 25 to 40 percent in recent literature [2, 3]. Sepsis also places a cost burden of approximately $24 billion on the U.S. healthcare system annually [4]. Promptly diagnosing sepsis and intervening before progression to septic shock has been associated with increased patient survival and reduced hospital length of stay [5, 6]. Sepsis is characterized as organ dysfunction resulting from a systemic inflammatory response to infection; however, the source of infection and the host response can vary greatly by patient, leading to difficulties in rapid detection. Consequently, a recent focus in medical research has concerned automated patient surveillance for timely detection of sepsis in hospitalized patients.
The widespread implementation of electronic health records (EHRs) in hospitals have made automated clinical decision and prediction systems more feasible, potentially improving the surveillance and treatment of complex syndromes [7]. Such systems provide alerts and suggestions by transforming a patient’s medical data into clinically accessible information in order to enhance patient care [8]. Currently, most methods for sepsis detection use rules-based gold standards to generate alerts and guidelines, and few have been developed with predictive capabilities [9–11].
Rules-based sepsis scoring systems commonly used in the clinical setting include the Systemic Inflammatory Response Syndrome (SIRS) criteria [12], the Sequential Organ Failure Assessment (SOFA) score [13], and the Modified Early Warning Score (MEWS) [14]. Several studies have implemented versions of these scoring systems into EHRs [9–11]. While these tools may demonstrate high sensitivity, they provide suboptimal specificity, and are not designed to predict sepsis development. Furthermore, diverse patient populations and the heterogeneity of infection source may not be accurately represented in rules-based scores. In contrast, machine learning-based prediction tools have the potential to provide advance notice of sepsis risk, higher specificity, and more generalizability, potentially enabling clinicians to intervene earlier while also reducing the burden of false alarms.
We have developed a machine learning-based sepsis prediction algorithm using patient EHR data as input, and previously have validated its ability to predict sepsis, severe sepsis, and septic shock up to four hours prior to onset [15–17]. Our prior studies have been conducted on a single center critical care population.
In this study, we define sepsis according to the more recent definition of Singer et al. [1]. We aim to assess the retrospective performance of the machine learning algorithm further in advance, up to two days prior to sepsis onset, on a mixed-ward data set using only vital sign inputs. Further, we aim to evaluate the algorithm’s robustness through cross-population validation, and compare the performance to that of commonly used rules-based scoring systems.
Materials and methods
Ethics approval and consent to participate
Before receipt of datasets, all patient records were de-identified according to the Health Insurance Portability and Accountability Act (HIPAA) standards. Collection of data did not impact patient safety. Approval for this study was granted by the Institutional Review Board (IRB) at UCSF.
Measurements
Sepsis risk scores were generated by analyzing patient measurements of each of the six following vital signs: systolic blood pressure, diastolic blood pressure, heart rate, respiratory rate, peripheral oxygen saturation (SpO2), and temperature. Patient encounters were required to have at least one of each vital sign recorded for inclusion in the study. We used only these vital signs to generate features to determine sepsis risk scores. These measurements were selected because they are relevant to the onset of sepsis, and are routinely collected without a pre-existing clinical suspicion of sepsis.
Data sources
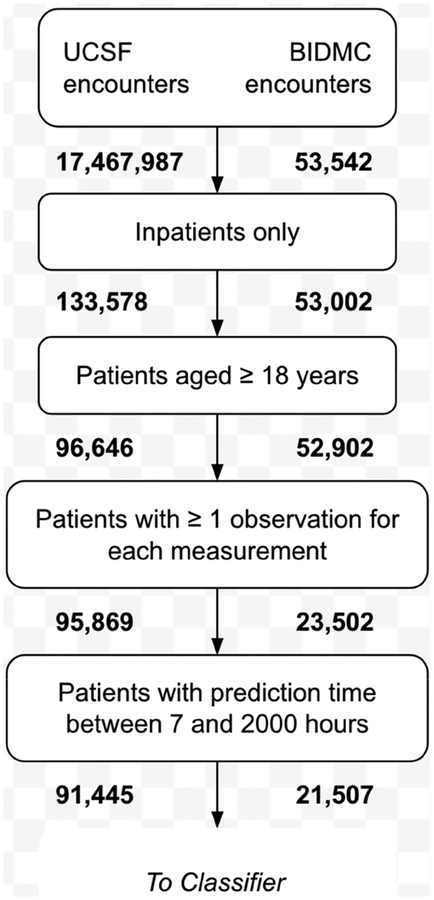
The data used in this study were collected from the University of California, San Francisco Medical Center (UCSF; San Francisco, CA), and the Beth Israel Deaconess Medical Center (BIDMC; Boston, MA). The UCSF data set contained 17,467,987 inpatient and outpatient encounters from the Parnassus Heights, Mission Bay, and Mount Zion campuses from June 2011 to March 2016. Of the 96,646 inpatients over the age of 18, 95,869 had at least one documented recording of each vital sign. From these encounters, we further limited our selection to those patients with hospital stays between 7 and 2,000 hours, which ultimately left 91,445 patients in our final cohort (Figure 1). We used subsets of this final cohort in our 24- and 48-hour lookahead analyses, depending on patient length of stay. The data from UCSF were collected in the intensive care unit (ICU), emergency department (ED), and floor units; thus, different levels of data collection frequency and care provision were captured in this cohort. We were unable to determine the location of a patient at a given point in time, because the data did not include unit transfer timestamps. BIDMC data were obtained from the Medical Information Mart for Intensive Care III (MIMIC-III) v1.3 database, compiled by the MIT Laboratory for Computational Physiology and their associated collaborators [18]. 61,532 BIDMC ICU encounters from 2001 to 2012 are documented in this database. Of the 52,902 inpatient encounters for which the patient was at-or-over the age of 18, 21,507 had at least one recording of each vital sign and were included in the final cohort. Excluded adult inpatient encounters had one or more vital signs for which there were no recorded measurements, and were not necessarily missing measurements of all vital signs.
Figure 1.
Patient encounter inclusion diagram for UCSF and BIDMC data. All patient encounters meeting the sequential inclusion criteria outlined above were included in training and testing sets. Subsets included for each lookahead time varied depending on patient length of stay.
Outcomes and procedures
The primary outcome measured was the algorithm’s ability to identify septic patients at the time of onset and 24 and 48 hours prior to onset. The algorithm’s performance was determined using the area under the receiver operating characteristic (AUROC) curve metric.
PostgreSQL (PostgreSQL Global Development Group; Berkeley, CA) database queries were written to extract and store measurements in comma separated value (CSV) files [19]. The six vital signs (heart rate, respiratory rate, temperature, SpO2, systolic blood pressure, and diastolic blood pressure) were the only inputs used to generate features for sepsis risk predictions. If a patient did not have at least one new measurement for every hour preceding their defined onset time, the missing value was determined with carry-forward imputation using the patient’s previous recorded value. In the case of multiple measurements within an hour, the mean was calculated and used in place of an individual measurement. This procedure helped to minimize non-physiological data regarding measurement frequency that was passed to the classifier.
Additionally, data were collected to generate the rules-based scoring system comparators and the Sepsis-3 gold standard. We compared the predictive algorithm to the commonly implemented SIRS, SOFA, MEWS, and qSOFA scores. We determined the presence of SIRS criteria as in Jaimes et al. [12]. We computed each patient’s MEWS score [14] as in Fullerton et al. [20]. The qSOFA score was calculated as in Singer et al. [1]. Although the SOFA score is part of the gold-standard sepsis definition, we also evaluated its capacity as a standalone detector of sepsis onset. Calculation of these scores required the creation of CSV files for PaO2, FiO2,Glasgow Coma Scale, vasopressor doses, bilirubin level, platelet count, and white blood cell count.
The Sepsis-3 gold standard was based on the 2016 consensus definition which defined sepsis as “life-threatening organ dysfunction caused by a dysregulated host response to infection,” with organ dysfunction defined as a change in Sequential Organ Failure Assessment (SOFA) score of ≥ 2 points [1]. We followed the definition proposed by Seymour et al. for identifying changes in SOFA score [21]. We identified suspicion of infection as either a culture drawn together with antibiotics administered within 24 hours, or antibiotics administered together with a culture drawn within 72 hours. This method adheres to the retrospective validation of Seymour et al. [21]. We designated the first time that both the SOFA score and infection thresholds were met as the time of sepsis onset.
Of the 91,445 encounters from UCSF satisfying the inclusion criteria of Figure 1, there were 2,649 encounters meeting the Sepsis-3 criteria, for a prevalence of 2.9%. Of the 21,507 BIDMC encounters, 1,024 met the Sepsis-3 criteria, for a prevalence of 4.8%. Together, the 112,952 encounters had an overall Sepsis-3 prevalence of 3.3%. We note that many encounters meeting the Sepsis-3 criteria had an associated time of sepsis onset occurring within the first seven hours of stay and were therefore excluded by the final step of the inclusion criteria.
Patients who never developed sepsis were assigned an “onset time” at random according to a continuous, uniform probability distribution, as negative class examples. Data collected at and before the patient’s onset time were used to generate the algorithm’s risk scores. In order to enable prediction windows, all patient encounters presenting a diagnosis of sepsis on admission, or those for which sepsis was diagnosed within seven hours of admission, were excluded from analysis.
Before training the classifier, patient encounters were divided into subgroups depending on length of stay. For example, if a patient spent 25 hours in the hospital before developing sepsis, the patient encounter was included in 24-hour prediction experiments, but could not be included in the 48-hour experiments. Respectively, of the 20,590 BIDMC and 89,000 UCSF encounters with at least 24 hours of stay data, 107 and 267 encounters had a Sepsis-3 diagnosis, after spending at least 24 hours in the hospital. Of the 20,533 BIDMC and 88,887 UCSF encounters with at least 48 hours of stay data, 50 and 97 encounters had a Sepsis-3 diagnosis after spending 48 hours of more in the hospital, respectively. The few encounters with an onset time exceeding 2,000 hours in the hospital were excluded to limit the size of the computation matrices.
The machine learning prediction algorithm
Gradient boosted trees were used to create the classifier, using scores from an ensemble of trees to calculate total scores. Predictions were generated from the six patient vital signs at prediction time, one hour before prediction time, and two hours before prediction time, as well as the hourly differences in each value. These values for each vital sign were concatenated into a causal feature vector x with 30 elements (five values from each of six measurement channels).
We created the trees in the Python XGBoost package [22]. Each branch of a tree was split by creating two groups of a feature. We limited branching to four, three, and six branches, for 0-hour, 24-hour, and 48-hour prediction, respectively. We did so on the basis of five-fold cross-validation grid search on the training set; we elaborate this below. It was also on the basis of this grid search that we chose the learning rate parameter of XGBoost to 0.05, 0.12, and 0.12, for 0-hour, 24-hour, and 48-hour prediction, respectively. It was not important for us to restrict the maximum number of trees in each ensemble, as this was superseded by the use of early stopping to prevent model overfit. The resulting ensemble was used to iteratively calculate total risk scores; these risk scores were used to determine the presence or likely development of sepsis for each patient.
The UCSF encounters were split into two groups, 80% of encounters were allocated uniformly-at-random to a training set, and 20% of encounters were randomly allocated to an independent, hold-out test set. To perform the hyperparameter grid search, we conducted five- fold cross-validation using only the encounters allocated to the training set. We searched a parameter space around previously reported hyperparameters for a similar prediction task [23,24]. For the maximum number of branchings, we searched integers from 3 to 8 and, for the learning rate, we searched from 0.05 to 0.12 in increments of 0.01. For each lookahead, we selected the combination of number of branchings and learning rate which produced the best average area under the receiver operating characteristic (AUROC) curve across the five folds.
We further divided the 80% training set into ten, equally-sized groups for ten-fold cross validation, again allocating the encounters uniformly-at-random. Nine of these groups were used to train the algorithm, and the remaining group was used to test it. We cycled the algorithm through each of the ten possible combinations of training and testing sets, and also tested each combination on the independent test set. Performance metrics for the MLA were obtained as the average of the performance metrics of the ten cross-validation models, including both the figure-based information and tabular results in the Results section. In particular, we report feature importance scores generated by XGBoost, which represent the number of times each feature was used to split the data across the trees; these scores were averaged across the five folds. Additionally, the cross-validation results were used to calculate standard deviation in AUROC.
As described in the previous section, for 24- and 48-hour prediction, we further limited the patient encounter cohorts to those encounters with enough stay data. Consequently, there were fewer septic patient encounters in these cohorts, leading to a degree of class imbalance. As opposed to artificially inflating the number of septic patients through minority oversampling, in a way which may not reflect the prospective setting in which such a MLA would be used, we relied on XGBoost’s built-in handling of imbalanced classes [22].
Cross-population validation methods
The cross-population validation experiments were used to evaluate the algorithm’s sepsis detection performance when trained on one institution’s data set and tested on another with demographic and clinical differences. After training the algorithm on UCSF data, its performance for sepsis detection was tested on BIDMC patient measurements, with no retraining on the target dataset. Testing was performed on the entire dataset. The algorithm was trained as described above, and testing was performed at time of onset only to facilitate comparison with rules-based methods.
Results
The final patient population included 91,445 encounters from UCSF, and 21,507 encounters from BIDMC. Demographic information for both patient populations is presented in Table 1. Significant differences between the data sets include the visited hospital units, sepsis prevalences, and age distributions. The BIDMC database contained only ICU stays, while the UCSF dataset contained inpatient encounters across all wards. These dissimilar datasets were chosen to provide an evaluation of the prediction algorithm’s generalizability across diverse patient populations.
Table 1.
Demographic characteristics for UCSF and BIDMC cohorts.
| Demographic Overview | Characteristic | UCSF (%) | BIDMC (%) |
|---|---|---|---|
| Gender | Female | 54.62 | 43.87 |
| Male | 45.38 | 56.13 | |
| Agea | 18–29 | 11.50 | 4.34 |
| 30–39 | 15.20 | 4.99 | |
| 40–49 | 13.02 | 9.73 | |
| 50–59 | 18.62 | 17.89 | |
| 60–69 | 21.22 | 22.79 | |
| 70+ | 20.44 | 40.26 | |
| Length of Stay (days)b | 0–2 | 30.35 | 49.34 |
| 3–5 | 38.42 | 32.38 | |
| 6–8 | 14.21 | 8.24 | |
| 9+ | 17.02 | 10.05 | |
| Death During Hospital Stay | Yes | 2.19 | 27.08 |
| No | 97.81 | 72.92 | |
| ICD-9 Code | Sepsis | 5.83 | 3.48 |
| Severe Sepsis | 3.69 | 10.19 | |
| Septic Shock | 1.85 | 7.00 |
Note: The UCSF cohort includes 91,445 patients. The BIDMC cohort includes 21,507 patients.
UCSF: median 55, IQR (38, 67); BIDMC: median 65, IQR (53, 77);
UCSF: median 4, IQR (2, 6.32); BIDMC: median 3, IQR (2, 4)
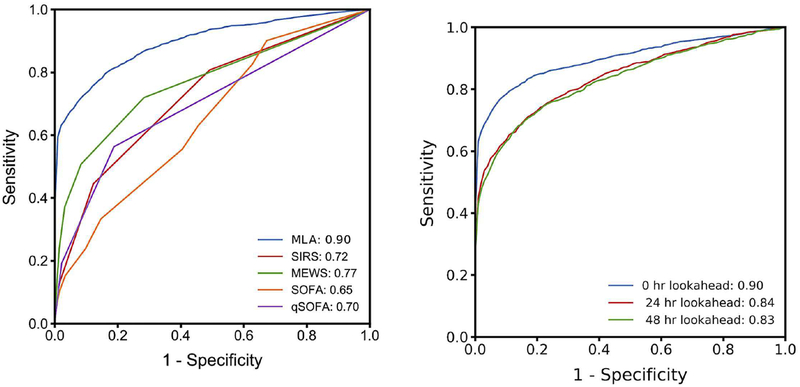
At the time of sepsis onset, the machine learning algorithm trained and tested on the UCSF data set demonstrated a higher AUROC (0.88) than the SIRS (0.66), MEWS (0.61), SOFA (0.72), or qSOFA (0.60) scoring systems for the same data set (Figure 2). A comparison of additional performance measures can be found in Table 2. For these operating points, the specificities of the MLA, SIRS, and SOFA correspond to respective false alarm rates of 0.22, 0.49, and 0.41; in other words, SIRS and SOFA produced 2.22 and 1.86 times as many false alerts as did the MLA.
Figure 2.
MLA performance compared to those of competing systems and in hours before onset. A) Comparison of ROC and AUROC for MLA and competing rules-based scoring systems at time of sepsis onset in the UCSF patient population. B) ROC curve and AUROC for MLA at 0, 24, and 48 hours before sepsis onset for UCSF patient data.
Abbreviations: MEWS, modified early warning score; MLA, machine learning algorithm; qSOFA: quick SOFA; SIRS, systemic inflammatory response syndrome; and SOFA, sequential (sepsis-related) organ failure assessment.
Table 2.
MLA performance measures before sepsis onset and competing scores at time of onset.
| MLA (t = 0) |
MLA (t = −24) |
MLA (t = −48) |
SIRS (t = 0) |
MEWS (t = 0) |
SOFA (t = 0) |
qSOFAa (t = 0) |
|
|---|---|---|---|---|---|---|---|
| AUROC (SD) |
0.88 (0.008) |
0.84 (0.04) |
0.83 (0.04) |
0.66 | 0.61 | 0.72 | 0.60 |
| Sensitivity | 0.80 | 0.80 | 0.84 | 0.70 | 0.52 | 0.78 | 0.37 |
| Specificity | 0.78 | 0.72 | 0.66 | 0.51 | 0.72 | 0.59 | 0.81 |
| DOR | 14.79 | 13.69 | 13.15 | 2.44 | 2.81 | 5.20 | 2.60 |
| LR+ | 3.76 | 2.57 | 2.86 | 1.43 | 1.86 | 1.92 | 2.00 |
| LR− | 0.26 | 0.28 | 0.21 | 0.56 | 0.66 | 0.37 | 0.77 |
Note: Performance measures for MLA at 0, 24, and 48 hours before onset, and competing scores at time of onset, measured on UCSF patient data. Operating points were chosen for sensitivities close to 0.80. Standard deviation in AUROC across cross-validation folds was calculated only for the MLA.
Abbreviations: AUROC, area under the receiver operating characteristic; DOR, diagnostic odds ratio; LR+, positive likelihood ratio; LR-, negative likelihood ratio; MEWS, modified early warning score; MLA, machine learning algorithm; qSOFA, quick SOFA; SD, standard deviation; SIRS, systemic inflammatory response syndrome; and SOFA, sequential (sepsis-related) organ failure assessment.
All of qSOFA’s operating points produced sensitivities far from 0.80.
At 24 and 48 hours before onset, the algorithm’s AUROC was 0.84 and 0.83, respectively (Figure 2, Table 2). 89,000 UCSF patients (267 septic) were included to calculate the AUROC at 24 hours, and 88,887 UCSF patients (97 septic) were included to calculate the AUROC at 48 hours before onset. The diagnostic odds ratio (DOR) at both 24 and 48 hours pre-onset was superior to those of the rules-based methods determined at the time of onset (Table 2). When trained on the UCSF database and tested BIDMC data (21,507 patients, 1,024 septic) with no retraining, the AUROC value was 0.890.
For each prediction window, we obtained feature importance scores (Supplementary Table 1). The five features with the highest importance scores are collected in Table 3. Across prediction windows, age was the most important feature, with a combination of temperature, heart rate, and systolic blood pressure measurements obtaining the majority of the other highest scores.
Table 3.
Features with highest feature importance scores for each lookahead.
| # | 0-Hour Detection | 24-Hour Prediction | 48-Hour Prediction |
|---|---|---|---|
| 1 | Age | Age | Age |
| 2 | HR (t = 0) | HR (t = −2) | HR (t = 0) |
| 3 | Temp (t = 0) | HR (t = −1) | SysABP (t = −2) |
| 4 | SysABP (t = 0) | HR (t = 0) | ΔDiasABP (t = −2 to −1) |
| 5 | Temp (t = −2) | Temp (t = 0) | HR (t = −2) & Temp (t = 0) |
Note: The parentheticals indicate that a feature was measured 0, 1, or 2 hours before the beginning of the prediction window. For example, in the context of 24-hour prediction, t = −2 indicates that the measurement was taken 24 + 2 = 26 hours before the onset time. The ΔDiasABP feature in the 48-hour prediction column corresponds to a difference in measurements between two times, t = −2 and t = −1. The HR (t = −2) and Temp (t = 0) features had the same average importance score for 48-hour prediction.
Abbreviations: ΔDiasABP, change in diastolic blood pressure; HR, heart rate; SysABP, systolic blood pressure; Temp, temperature.
Discussion
In this study, the machine learning-based sepsis prediction algorithm demonstrated increased sensitivity and specificity for sepsis over the commonly used SIRS, MEWS, SOFA, and qSOFA scoring systems at the time of sepsis onset (Figure 2, Table 2), and demonstrated robustness to training and testing across separate datasets. Based on the results for the UCSF dataset (Table 2), SIRS and SOFA resulted in 2.22 times and 1.86 times as many false alerts as the MLA, respectively, when identifying a similar proportion of sepsis cases at onset. False alerts are problematic in hospital settings, contributing to alarm fatigue which may lead to compromised patient monitoring and care [25–27]. Therefore, high specificity is desirable in a sepsis alert.
The algorithm also demonstrated high sensitivity and specificity when predicting sepsis onset up to 48 hours in advance. Interestingly, the performance at 48 hours in advance of sepsis onset was roughly equivalent to that 24 hours in advance of onset. This may indicate that changes in vital sign measurements relevant to sepsis development may occur well in advance of the detection of organ failure. Other studies have applied machine learning techniques to predict sepsis onset up to 48 hours prior to organ failure [28], although these algorithms require significantly more data input including patient histories and laboratory test results. This study demonstrates that high sensitivity and specificity for sepsis can be attained up to 48 hours prior to organ failure using only commonly measured vital signs, without requiring laboratory tests which may only be ordered after the initial clinical suspicion of sepsis.
This machine learning approach has potential implications for clinical practice and sepsis management. A recent study has shown that nearly half of patients with severe sepsis (which is similar to the more recent definition of sepsis evaluated in this study), lack sepsis-specific International Classification of Disease (ICD) codes. This study additionally showed that patients with sepsis-specific ICD codes were more likely to receive appropriate sepsis treatment [29]. The significant advance lookahead time provided by this MLA allows for improved risk-stratification of patients, with increased monitoring or more frequent care team assessment for patients with high risk scores. Early recognition of septic patients may facilitate clinical trial enrollment to develop and evaluate new sepsis treatments. Early alerts also provide a window to begin supportive or sepsis bundle treatments before a patient’s condition progresses, potentially improving outcomes.
Implementation of a previous version of this algorithm that predicted sepsis syndromes up to 4 hours in advance led to improved patient outcomes in several prospective settings. In a randomized controlled trial conducted in two adult ICUs at UCSF, use of an MLA was associated with decreases in hospital length of stay, (mean 13.0 days vs. 10.3 days, p = 0.0421) and the in-hospital mortality rate (21.3% vs. 8.96%, p = 0.0176). Use of the previous algorithm was also associated with earlier administration of antibiotics and ordering of blood cultures [30]. The previous algorithm has also improved sepsis-related outcomes at Cape Regional Medical Center [31] and Cabell Huntington Hospital [32], and has been successfully integrated with various EHRs including Epic, Cerner, McKesson, Meditech, Allscripts, Soarian, and Vista. While the present study examines only the UCSF and BIDMC datasets, these prior results are consistent with a broad applicability of MLAs to a variety of datasets and clinical data collection methods.
We note that the algorithm demonstrated high performance on the mixed-ward UCSF dataset. Given the high incidence of sepsis in general wards, and the noted benefits of screening for sepsis on such wards [33], we believe these results are encouraging for future experiments in data-sparse environments outside of the ICU. Indeed, the feature importance scores of Table 3 indicate that the common measurements of heart rate, temperature, and systolic blood pressure, along with age, contributed most to the quality of predictions. We may therefore expect the performance of such a tool to be more robust than tools which rely on less commonly measured information, in environments outside the ICU. The predictive algorithm also performed well in cross-population validation experiments. This may indicate reduced or eliminated need for site-specific data training at a future prospective clinical site implementation of the algorithm.
Limitations
Because this study was a retrospective analysis, we cannot make conclusions about the predictive algorithm’s performance in a live hospital setting. The algorithm may perform differently on real-time data, particularly when run at different time points than those presented in this study. While these data sets had distinct demographic characteristics and represented different hospital units, both settings were urban research hospitals. Thus, data from different environments, such as community hospitals, may not achieve the same cross-population performance. The study population excluded patients who presented with sepsis on admission; the algorithm may perform differently on patients who are admitted with sepsis. Because we did not require the presence of a sepsis related ICD code for a positive classification, it is possible that we positively identified patients with similar acute vital sign trends but without sepsis. Algorithm performance may be lower for such patients than for the general population.
The sepsis gold standard used in this study is necessarily an imperfect characterization of sepsis. We nevertheless believe it to be useful in developing a sepsis prediction tool, as demonstrated by the improvements in sepsis-related clinical outcomes seen using sepsis prediction algorithms trained with the same gold standard [31,32].
There are also limitations with respect to the experimental design. The AUROC, sensitivity, and specificity calculated for longer prediction windows were calculated using a smaller patient pool than the shorter prediction window metrics. These results could have been skewed by the smaller patient populations. To account for this uncertainty, we presented confidence intervals alongside the predictive AUROCs.
Conclusions
The machine learning algorithm assessed in this study is capable of predicting sepsis up to 48 hours in advance of onset with an AUROC of 0.83. This performance exceeds that of commonly used detection methods at time of onset, and may in turn lead to improved patient outcomes through early detection and clinical intervention.
Supplementary Material
Highlights.
Machine learning algorithm accurately predicts sepsis up to 48 hours in advance
Algorithm performance is superior to commonly-used disease severity scoring systems
Algorithm performs well even when trained and tested on different patient populations
May lead to improved patient outcomes through early detection and intervention
Acknowledgements
We gratefully acknowledge Melissa Jay, Jana Hoffman, Touran Fardeen, and Emily Huynh for their assistance in editing this manuscript. We would like to thank Hamid Mohamadlou, Qingqing Mao, and Thomas Desautels for their work in developing the algorithm.
Funding
Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award numbers R43TR002221 and R43TR002309. The funder had no role in the conduct of the study; collection, management, analysis, and interpretation of data; preparation, review, and approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: CB, YZ, ZJ, ALP, SL, JC, and RD are employees of Dascena, Inc. UC has no conflicts to disclose.
References
- [1].Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8): 801–10. doi: 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Angus DC, Linde-Zwirble WT, Lidicker K, Clermont G, Carcillo J, Pinsky MR.. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29(7): 1303–10. PMID: 11445675. [DOI] [PubMed] [Google Scholar]
- [3].Moss M Epidemiology of sepsis: race, sex, and chronic alcohol abuse. Clin Infect Dis. 2005. November 15;41(7): 490–7. [DOI] [PubMed] [Google Scholar]
- [4].Lagu T, Rothberg MB, Shieh MS, Pekow PS, Steingrub JS, Lindenauer PK. Hospitalizations, costs, and outcomes of severe sepsis in the United States 2003 to 2007. Crit Care Med. 2012. March; 40(3): 754–61. [DOI] [PubMed] [Google Scholar]
- [5].Nguyen HB, Corbett SW, Steele R, Banta J, Clark RT, Hayes SR, et al. Implementation of a bundle of quality indicators for the early management of severe sepsis and septic shock is associated with decreased mortality. Crit Care Med. 2007. April 1;35(4): 1105–12. [DOI] [PubMed] [Google Scholar]
- [6].Shorr AF, Micek ST, Jackson WL Jr, Kollef MH. Economic implications of an evidence-based sepsis protocol: Can we improve outcomes and lower costs?* Crit Care Med. 2007;35(5): 1257–62. [DOI] [PubMed] [Google Scholar]
- [7].Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005. March 14. doi: 10.1136/bmj.38398.500764.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sittig DF, Wright A, Osheroff JA, Middleton B, Teich JM, Ash JS, et al. Grand challenges in clinical decision support. J Biomed Inform. 2008;41: 387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Berger T, Birnbaum A, Bijur P, Kuperman G, Gennis P. A computerized alert screening for severe sepsis in emergency department patients increases lactate testing but does not improve inpatient mortality. Appl Clin Inform. 2010;1(4): 394–407. doi: 10.4338/ACI-2010-09-RA-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hooper MH, Weavind L, Wheeler AP, Martin JB, Gowda SS, Semler MW, et al. Randomized trial of automated, electronic monitoring to facilitate early detection of sepsis in the intensive care unit. Crit Care Med. 2012;40(7): 2096. doi: 10.1097/CCM.0b013e318250a887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Semler MW, Weavind L, Hooper MH, Rice TW, Gowda SS, Nadas A, et al. An electronic tool for the evaluation and treatment of sepsis in the ICU: a randomized controlled trial. Crit Care Med. 2015;43(8): 1595. doi: 10.1097/CCM.0000000000001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jaimes F, Garcés J, Cuervo J, Ramirez F, Ramirez J, Vargas A, et al. The systemic inflammatory response syndrome (SIRS) to identify infected patients in the emergency room. Intensive Care Med. 2003;29: 1368–71. doi: 10.1007/s00134-003-1874-0. [DOI] [PubMed] [Google Scholar]
- [13].Vincent JL, Moreno R, Takala J, Willatts S, De Mendonca A, Bruining H, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22(7): 707–10. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- [14].Subbe CP, Slater A, Menon D, Gemmell L. Validation of physiological scoring systems in the accident and emergency department. Emerg Med J. 2006. November;23(11): 841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Calvert JS, Price DA, Chettipally UK, Barton CW, Feldman MD, Hoffman JL, et al. A computational approach to early sepsis detection. Comput Biol Med. 2016;74(C): 69–73. doi: 10.1016/j.compbiomed.2016.05.003. [DOI] [PubMed] [Google Scholar]
- [16].Calvert JS, Price DA, Chettipally UK, Barton CW, Hoffman JL, Jay M, et al. High-performance detection and early prediction of septic shock for alcohol-use disorder patients. Ann Med Surg. June 2016;8: 50–5. doi: 10.1016/j.amsu.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Desautels T, Calvert J, Hoffman J, Jay M, Kerem Y, Shieh L, et al. Prediction of Sepsis in the Intensive Care Unit With Minimal Electronic Health Record Data: A Machine Learning Approach. JMIR Med Inform. 2016;4(3): 28. doi: 10.2196/medinform.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Johnson AEW, Pollard TJ, Shen L, Lehman LH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].PostgreSQL Global Development Group, https://www.postgresql.org/.
- [20].Fullerton JN, Price CL, Silvey NE, Brace SJ, Perkins GD. Is the Modified Early Warning Score (MEWS) superior to clinician judgement in detecting critical illness in the pre-hospital environment? Resuscitation. 2012;83: 557–62. [DOI] [PubMed] [Google Scholar]
- [21].Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A, et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016. February 23;315(8): 762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen T, Guestrin C. XGBoost: A scalable tree boosting system. Paper presented at the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2016 Aug. [Google Scholar]
- [23].Desautels T, Das R, Calvert J, Trivedi M, Summers C, Wales DJ, Ercole A. Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: a cross-sectional machine learning approach. BMJ open. 2017. September 1;7(9):e017199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mao Q, Jay M, Hoffman JL, Calvert J, Barton C, Shimabukuro D, Shieh L, Chettipally U, Fletcher G, Kerem Y, Zhou Y. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ open. 2018. January 1;8(1):e017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sendelbach S, Funk M. Alarm Fatigue A Patient Safety Concern. AACN advanced critical care. 2013. October 1;24(4):378–86. [DOI] [PubMed] [Google Scholar]
- [26].Cvach M Monitor alarm fatigue: an integrative review. Biomedical instrumentation & technology. 2012. July;46(4):268–77. [DOI] [PubMed] [Google Scholar]
- [27].Mitka M Joint commission warns of alarm fatigue: multitude of alarms from monitoring devices problematic. Jama. 2013. June 12;309(22):2315–6. [DOI] [PubMed] [Google Scholar]
- [28].Henry KE, Hager DN, Pronovost PJ, Saria S. A targeted real-time early warning score (TREWScore) for septic shock. Science translational medicine. 2015. August 5;7(299):299ra122-. [DOI] [PubMed] [Google Scholar]
- [29].Dies AS, Whiles BB, Brown AR, Satterwhite CL, Simpson SQ. Three-Hour Bundle Compliance and Outcomes in Patients With Undiagnosed Severe Sepsis. Chest. 2017. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shimabukuro D, Barton CW, Feldman MD, Mataraso SJ, Das R. Effect of a Machine Learning-Based Severe Sepsis Prediction Algorithm on Patient Survival and Hospital Length of Stay: A Randomized Clinical Trial. BMJ Open Respir Res, In Press. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McCoy A, Das R. Reducing patient mortality, length of stay and readmissions through machine learning-based sepsis prediction in the emergency department, intensive care unit and hospital floor units. BMJ Open Qual. 2017;6: e000158. doi: 10.1136/bmjoq-2017-000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Burdick H, Pino E, Gabel-Comeau D, Gu C, Huang H, Lynn-Palevsky A, Das R. Evaluating a sepsis prediction machine learning algorithm using minimal electronic health record data in the emergency department and intensive care unit. bioRxiv 224014; doi: 10.1101/224014. [DOI] [Google Scholar]
- [33].Bhattacharjee P, Edelson DP, Churpek MM. Identifying Patients With Sepsis on the Hospital Wards. Chest. 2017;151: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.