Abstract
“What I cannot create, I do not understand.” Richard Feynman may have championed reasoning from first principles in his famous blackboard missive, but he could just as well have been referring to the plight of a molecular virologist. What cannot be grown in a controlled laboratory setting, we cannot fully understand. The story of the laboratory domestication of hepatitis C virus (HCV) is now a classic example of virologists applying all manner of inventive skill to create cell-based models of infection in order to clarify prospective drug targets. In this review, we highlight key successes and failures that were instructive in achieving cell based models for HCV studies and drug development. We also emphasize the lessons learned from the ~40 year saga that may be applicable to viruses yet unknown and uncultured.
Introduction
As of late 2018, HCV patients can be prescribed a once-a-day pill that within 8 weeks provides greater than a 95% chance of achieving cure with negligible side effects [1]. This remarkable achievement of HCV direct acting antivirals (DAAs) rests upon three decades of hard work from thousands of basic and clinical scientists, physicians, and drug developers. And while discussions surrounding access to HCV treatment have rightly become urgent [2], it is increasingly clear that the scientific breakthroughs underlying HCV therapy provide a compelling roadmap for drug development against other viruses. In this review, we broadly outline the story of HCV basic research that led to therapeutic success with a focus on cell-based models for drug development (Figure 1). We also highlight key lessons singular to HCV that help emphasize this virus as an exceptional and unique model system to inspire work in 21st century virology and beyond.
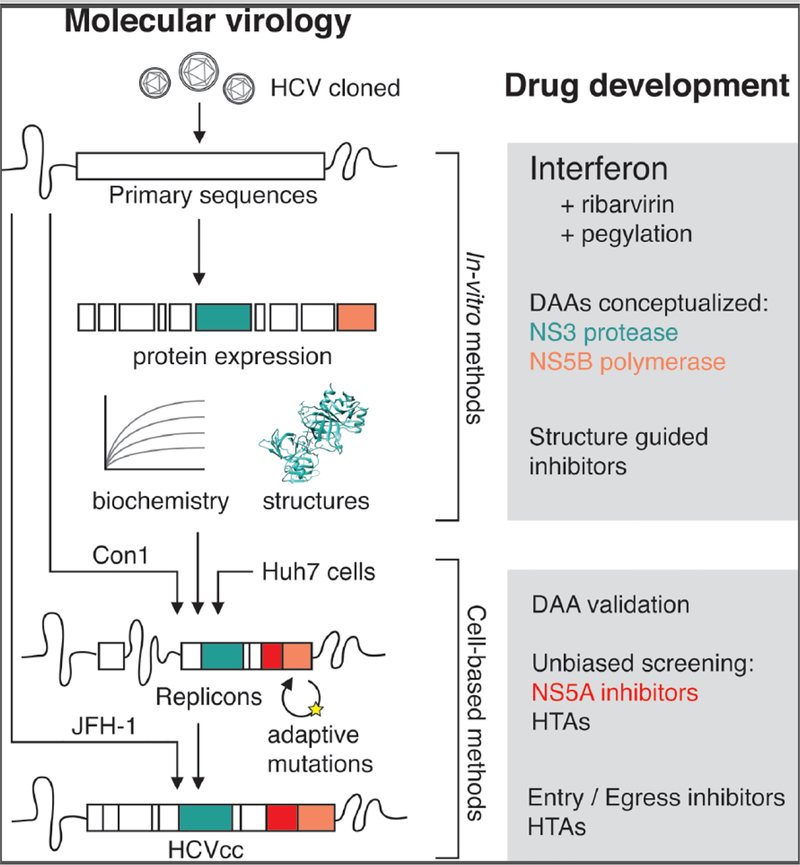
Figure 1: Domesticating hepatitis C virus.
After a decade and a half of observation as NANBH in humans, the cloning of HCV initially permitted in vitro expression, biochemical characterization, and structural studies of viral proteins. These studies subsequently informed the successful creation of the HCV replicon, a major breakthrough for validation, optimization and unbiased screening of DAAs and host-targeting agents (HTAs). This continued with HCVcc, which allowed drug development efforts encompassing the entire virus lifecycle. Major themes in drug development based on biochemical or cell-based models are presented in boxes at right.
Non-A, non-B hepatitis
The advent of sensitive serologic tests first for hepatitis B virus (HBV) and then hepatitis A virus (HAV) made it clear by the early 1970’s that a significant transfusion-associated hepatitis was likely caused by an unknown virus(es). Cytomegalovirus and Epstein-Barr virus were quickly ruled out as potential causes of this non-A non-B hepatitis (NANBH) [3,4]. As the hunt for the NANBH virus began, several of its infectious and physical properties were established even before the virus could be identified, mainly from studies using chimpanzees as an experimental model (reviewed in [5]). For example, serum from NANBH patients, when injected into chimpanzees, caused mild but detectable hepatitis, as evidenced by elevated ALT levels, and ultra-structural alterations in the cytoplasm of hepatocytes (reviewed in [6]). Notably, no disease occurred when patient serum was treated with chloroform prior to inoculation into chimpanzees, suggesting that the “mystery” virus likely contained a lipid envelope. [7]. Filtration studies later estimated that the virus was between 30–60 nm in diameter [8]. Based on these physical characteristics, the NANBH virus was tentatively assigned to Togaviridae family of viruses [8]. Many of these propositions were largely vindicated in 1989 when the NANBH virus was finally identified and termed hepatitis C virus (HCV) [9]. In what are now classic experiments, after a decade during which traditional immunological methods to identify the NANBH agent had failed, Michael Houghton and colleagues applied recently developed molecular screening approaches to identify and clone the virus directly. Starting with a recombinant expression library derived from infected chimpanzee plasma, the resulting cDNA library was inserted into λgt11 bacteriophage and expressed in Escherichia coli. The expressed proteins were then screened against serum from NANBH patients to identify reactive clones. One such clone provided the molecular foothold to reveal a large non-host derived RNA molecule that was found predominantly in blinded NANBH samples and thus named HCV [9]. This discovery represented a true first for virology in that the molecular cloning of HCV occurred prior to visualization, growth in cell culture, and serological detection of the virus.
The HCV genome sequence revealed numerous aspects of the virus biology on the basis of analogy with other RNA viruses. HCV is a positive strand RNA virus with a ~10kb genome consisting of single open reading frame of ~3000 amino acids flanked by short non-translated regions. The virus is most closely related to the Flaviviridae family of viruses, particularly pestiviruses such as bovine viral diarrhea virus (BVDV) and classical swine fever virus (CSFV), and it became the founding member of the Hepacivirus genus [10]. Antibody-based diagnostics soon confirmed the presence of HCV in a majority of NANBH samples [11], uncovered widespread contamination of global blood banks [11], and established a clear association of HCV infection with liver disease and hepatocellular carcinoma [12]. These facets quickly underscored the global scale of the HCV threat and the contemporaneous need for preventative measures and therapeutic interventions.
Interferon alpha as NANBH / HCV therapy
That the infectious nature of NANBH could be inferred by the early 1980s led clinicians to consider therapy with interferon alpha (IFNα), by analogy of its successful therapeutic use for chronic HBV [13]. Initially identified in the 1950s as a secreted inhibitory factor for numerous viruses in cell culture [14,15], by the 1980s recombinant IFNα was available to treat HBV and other viruses [16]. IFNα treatment for NANBH resulted in decreased ALT levels that persisted throughout the course of therapy, which if halted resulted in viral rebound [17]. Such responsiveness to interferon became a primary line of evidence in humans that the hepatocellular injury in NANBH was likely the direct or indirect result of a replicating viral agent. After HCV was identified, IFN therapy continued to be the standard of care, with a sustained virologic response (SVR) in 5–25% patients [18]. The addition of the nucleoside analog ribavirin in the late 1990s boosted SVR rates to 30–40% [19], and in the early 21st century, the use of pegylated IFN further improved SVR rates to around 50% [20]. However, IFNα therapy, while effective, was not without complications related to adverse side effects, long (24–48 weeks) treatment regimens, uneven efficacy across viral genotypes, and frequent viral relapse (reviewed in [21]). These limitations of IFN-based treatment continually reinforced the need to cultivate systems and strategies for DAA development.
Cell-based models in absentia: early work on HCV
While early basic work on HCV yielded sporadic reports of low level persistent replication, such difficult-to-reproduce findings largely reflected a failure to efficiently propagate the virus in cell culture or animal models beyond chimpanzees (reviewed in [22]). Unlike chimpanzees, cultured human hepatoma cells and primary hepatocytes did not support efficient replication of virus from patient sera. This prompted a series of investigations as to why this might be the case (see next section). Yet, despite the absence of cell culture systems to study the virus, the viral sequence immediately enabled a number of key clinical and experimental observations that were insightful for HCV drug development.
As with other positive-strand RNA viruses, HCV replication is exclusively cytoplasmic and possesses no stable DNA intermediate, as is the case for HIV and HBV. Instead, the virus uses a negative-stranded RNA intermediate, a product of the viral RNA-dependent RNA polymerase (RdRP), to produce positive stranded genomes. In principle, the lack of RdRP activity in host cells presented an obvious and specific viral drug target. Moreover, the lack of a stable intermediate suggested that HCV infection was “curable” in that the virus could be eliminated from an infected individual. One compounding facet of the HCV RdRP activity was the wide sequence heterogeneity within and among infected patients [23]. By 1993, six distinct genotypes of the virus had been identified, differing at the nucleotide level by more than 30%, and unevenly distributed throughout the globe [24,25]. Such genetic diversity in HCV suggested that pan-viral therapies were likely to encounter genotype-specific efficacy problems and that resistance-associated mutations might pre-exist or were likely to develop. This reinforced the need to develop infectious clones from patient isolates, characterize their unique properties, but also strive to define conserved elements and processes crucial to all genotypes.
By analogy with members of the Flaviviridae, the HCV polyprotein was thought to be processed into individual proteins through either viral or cellular protease activity. Different experimental strategies later confirmed this notion and showed that HCV polyprotein was cleaved by the combined action of host and viral proteases to release ten individual proteins [26–30]. Functional analysis of the individual cleavage products identified protease activities for NS2 and NS3 [28–33], helicase and ATPase activities for NS3 [34,35], and RdRP activity for NS5B [36,37]. Meanwhile, contributions largely by pharmaceutical firms yielded the first structures of the NS3 protease [38,39] and NS5B polymerase [40]. These discoveries provided a platform for the rational design of protease and polymerase inhibitors that could be assayed against enzymatic activity in vitro (see below). While promising candidates could be tested in chronically infected chimpanzees, it was clear that a cell based model would be ideal both to test efficacy of candidate drugs and to provide an optimal screening modality for novel therapies.
The dawn of cell culture based HCV replication systems
The failure of patient isolates to infect cells in culture was one of the major problems that occupied HCV researchers during the 1990s. The solution to this problem was provided by progress on two fronts: the development of HCV cDNA clones and the identification of cell lines permissive for HCV RNA replication.
By the mid 1990s, efforts were underway to develop HCV cDNA clones that could be used to launch infection [41]. This approach required accurate knowledge of the terminal ends of the viral genome. While the 5’ end of the genome was determined in 1991 [42], analysis of the 3’ end yielded conflicting reports. This was surprising since positive-strand RNA viruses in this family usually contained highly conserved cis elements at their 3’ ends that played important roles in replication and/or packaging. In the mid 1990’s two research groups independently showed that the HCV 3’ end contained a stretch of highly conserved nucleotides that was missed in the prior studies [43,44]. This sequence, when appended to consensus-derived HCV molecular clones and injected into the liver of chimpanzees, resulted in hepatitis, clearly demonstrating the ability of cloned, full-length viral RNA to initiate infection in vivo. [45,46]. Disappointingly, these cloned genomes failed to launch infection in cultured cells.
Once again, knowledge of other RNA viruses provided a route forward. As RNA viruses were explored as potential gene delivery vectors since the late 1980’s, one key observation was that heterologous genes could be stably expressed in place of viral structural proteins [47]. The resulting self-replicating RNAs, or replicons, could be trans-complemented with structural genes to make infectious virus particles [48]. As tools that enabled functional separation of RNA replication from virus assembly, replicons were successfully used to study Sindbis, Semliki Forest, polio- and rhinoviruses [48–51]. The creation of a selectable replicon for the flavivirus Kunjin [52], combined with the identification of naturally occurring self-replicating BVDV RNAs [53] led to the idea that HCV replicons might be achievable.
The breakthrough of a functional HCV replicon arrived in 1999 [54]. Designed from a genotype 1b isolate, and using human hepatoma cells (Huh7), a selectable marker was engineered within the HCV replicon to screen for surviving clones upon antibiotic exposure [54]. Despite low efficiency (1 colony/1011 in vitro transcript RNAs), the resulting cell colonies displayed all the hallmarks of sustained HCV RNA replication [54]. Subsequent identification of adaptive mutations led to vastly improved HCV replicon efficiency, and importantly, showcased feasibility for drug development by demonstrating HCV replicon inhibition by interferons [55,56]. These breakthroughs initiated an iterative process by which adaptive mutations were exploited to increase replication efficiency [57–60], cell lines identified for higher HCV permissiveness [61,62], and patient isolates screened for full infectivity in permissive cellular contexts. By 2005, with the discovery of a genotype 2a isolate called JFH-1, the entire HCV lifecycle could finally be recapitulated in cell culture [63–65]. The advent of the replicon and later fully infectious cell culture systems for HCV has since afforded enormous benefit for basic HCV research, helping characterize essential host factors [66,67] and permitting detailed mechanistic exploration of the viral lifecycle. But perhaps the greatest impact of the replicon was its immediate use to test and refine candidate drugs against HCV enzymes and as a platform to conduct unbiased screens to identify new classes of HCV inhibitors.
HCV DAA development
As just highlighted, HCV proteins with enzymatic activities gained immediate attention as potential drug targets. An unexpected entry into the clinic were highly potent inhibitors of HCV NS5A, a protein with no known enzymatic functions. As current HCV regimens comprise various combinations of protease, polymerase, and NS5A inhibitors, in this section we will provide a brief overview of historical developments leading up to these drugs.
Protease inhibitors: Coincidentally, phase I clinical trials for the first HIV protease inhibitor started in 1989 (reviewed in [68]), the year HCV was discovered. As soon as the existence of serine protease in the HCV polyprotein was predicted and confirmed, this enzyme became an obvious target for drug development. Due to the lack of HCV cell culture systems for compound screening, initial attempts to discover protease inhibitors heavily relied on biochemical or engineered cell-based assays. The inability of known serine protease inhibitors to efficiently inhibit HCV protease [69] and the novel substrate specificity apparent from its polyprotein cleavage sites made it clear that the design of selective and specific HCV protease inhibitors would be greatly aided by determining a high resolution structure of this enzyme. Structural analyses of HCV protease revealed a flat, somewhat hydrophobic, and solvent-exposed active site [38,39]. The lack of attractive pockets where small molecule inhibitors could bind with high affinity posed a formidable challenge, which slowed progress and discouraged many from pursuing HCV protease inhibitors.
The observation that the HCV protease was inhibited by its own cleavage products paved the way for the design of peptidomimetic inhibitors based on natural NS3 substrates [70,71]. These synthetic peptides bind the protease active site in a covalent or non-covalent fashion to block the enzymatic activity either reversibly or irreversibly, depending on the nature of the inhibitor. Validation for this class of protease inhibitors was already provided by the HIV field where several peptidomimetic inhibitors were in clinical use or at different stages of clinical assessment (reviewed in [72]). In 2003, a peptidomimetic HCV protease inhibitor, BILN 2061, demonstrated impressive efficacy in HCV genotype 1a patients, providing the first proof-of-concept for the use of protease inhibitors, and in fact DAAs as a class, for HCV treatment [73]. The development of this inhibitor, however, was halted after cardiac side effects were observed in extended primate toxicity studies.
While termination of BILN 2061 initially dampened enthusiasm for HCV protease inhibitors, several groups continued their efforts, leading ultimately to the approval of two first-generation protease inhibitors in 2011 (reviewed in [69,74]). These protease inhibitors were only approved for genotype 1a patients as part of IFN/ribavirin combination therapy, and improved the treatment success rate from less than 40% in 2001 to above 75% (reviewed in [75]). However, these inhibitors had narrow genotype coverage, a low barrier to resistance, and added adverse side effects. Fortunately these problems were quickly overcome by next generation protease inhibitors which now make up an important part of IFN-free regimens (reviewed in [76]). In addition, this class of inhibitors has the added bonus that the HCV serine protease also cleaves and inactivates MAVS, an essential player in a pathway leading to the induction of interferon (reviewed in [77]).
Polymerase inhibitors: Investigation of HCV polymerase inhibition as a therapeutic strategy lagged behind protease inhibitors partly due to the lack of cell-based assays to validate compounds and partly because of the comparatively slower progress on biochemical and structural characterization of HCV polymerase. It was only in 1996, seven years after HCV discovery, that the RdRP activity was unequivocally assigned to the NS5B protein [36]. It took another three years before the first crystal structure of NS5B was reported [40]. This and the lack of HCV cell culture models slowed down the search for clinically relevant compounds.
As HCV cell culture systems became available in early 2000s, various compounds were tested for their ability to inhibit HCV polymerase. Typically, viral polymerase inhibitors can be divided into two categories—nucleos(t)ide analog active site inhibitors that are incorporated into the growing RNA chain and block further incorporation of incoming nucleotides, and allosteric inhibitors that engage sites elsewhere on the enzyme to inhibit enzymatic function. Early compound screens identified HCV polymerase inhibitors of both classes, and some of these readily reduced viral loads in HCV patients (reviewed in [78]). Interestingly, however, just like protease inhibitors, none of these initial compounds reached the clinic mainly due to unacceptable toxicity during pre-clinical evaluation.
In the meantime, the search for better NS5B inhibitors continued, and finally resulted in 2013 in the clinical approval of the first HCV polymerase inhibitor [79]. This nucleotide pro-drug has broad genotype coverage, a high genetic barrier to resistance, minimal side effects, and now forms the backbone of many of the clinically available HCV therapeutic regimens. Only a year after this first approval, a non-nucleoside polymerase inhibitor was approved as part of an HCV combination therapy. The past few years have seen identification of additional HCV polymerase inhibitors, some of which are in phase III clinical trials (reviewed in [80]).
HCV NS5A inhibitors: One of the most unexpected breakthroughs in HCV drug discovery was the development of NS5A inhibitors. This protein, devoid of known enzymatic activity, was never actively pursued as a drug target. It was only from unbiased compound screening using HCV replicons that NS5A targeting candidates emerged [81,82]. NS5A is a phosphoprotein with several critical functions in HCV replication (reviewed in [83]), which may explain why NS5A targeting compounds are remarkably potent. The first NS5A inhibitor was reported in 2010 using a high throughput screen of over one million compounds on HCV replicon cells [81]. A chemically refined version of this compound, which later received FDA approval in 2015, showed incredible picomolar potency, broad genotype coverage, and high safety profile [82]. Several compounds of this class are now in clinical use in various combinations with HCV protease and polymerase inhibitors to form the mainstay of combination DAA therapy [84].
Some inspiring lessons from basic HCV research
HCV has been instructive to multiple generations of researchers across the globe. While certainly not exhaustive, we group the lessons of HCV research into four broad themes. The first is motivation by analogy. Throughout the history of HCV research, knowledge of other viruses, both well studied and obscure, informed virtually all aspects of hypothesis formulation, experimental design, assay development and selection of drug targets. For example, the known biology of flaviviruses was pivotal in pointing to likely functions of HCV viral gene products [85]. Likewise, the modularity of numerous virus-derived molecular biology tools, such as IRESes and phage promoters, were essential for basic HCV research. These components enabled development of tools such as HCV replicons, which were directly inspired from work with Kunjin virus and BVDV [86]. Similarly, development of protease and polymerase inhibitors for HIV and HBV blazed a path for HCV drug development. It is our view that the plethora of knowledge gained from HCV studies will likely inform future work on viruses yet unknown.
The second theme is that every virus has surprises and HCV has been no exception. The requirement by HCV of the liver specific microRNA-122 [87] is an exceptional example in this vein, that in short course inspired a first-in-class RNA based therapeutic with remarkable potency [88]. While beyond the scope of this review, basic HCV research has furnished unanticipated discoveries on immune mechanisms of viral control [89,90], host polymorphisms that regulate viral clearance [77,91], and cell biological understanding of viral replication factories [92,93]; all of which have and will continue to motivate host-targeting agents (HTAs) as additional therapeutic options [94].
The third theme is that animal models can be simultaneously required and completely optional for investigation of a given virus. For HCV, it is evident that the chimpanzee was essential in the early days to establish the basic properties of the virus. The chimpanzee model was also instrumental in confirming the infectivity of RNA clones, convincing the community that all necessary information for fully infectious virus was present in cloned viral sequences, and that cell based systems could be developed if the correct cellular context was identified [86]. As soon as the cell based models for HCV research became available, the importance of chimpanzees as an animal model diminished, although monetary and ethical issues also contributed [95]. While a better animal model for HCV was and continues to be sought, particularly for questions of viral pathogenesis, carcinogenesis, and vaccine development, the creation of the HCV replicon was sufficient for effective drug development in the absence of a scalable animal model. One may wonder if this will in hindsight be an accident of history, or perhaps applicable to other virus infections and additional diseases.
The fourth theme is that technology development has iteratively shortened the timeline for HCV discoveries, as is generally true now for most of virology. The impacts of this timeline compaction are only beginning to be felt, but are perhaps reflected in the stunning pace of recent work on Zika virus, from molecular virology and pathogenesis to vaccine development (reviewed in [93,96,97]). It is our view that virology is at a unique and exciting moment, where researchers benefit from enormous technological advances to study and tame viruses that wreak havoc on human health.
Acknowledgements
We apologize to colleagues whose work was not referenced due to space constraints. The authors’ work on innate immunity, rodent hepacivairuses, and RNA regulation is supported by NIAID/NIH grants R01AI091707, R01AI131688, and R01AI116943, respectively (C.M.R.); HCV work was supported by NCI grant R01CA057973. Further support comes from a Charles H. Revson Senior Fellowship in Biomedical Science (J.M.L), and a Helmsley Postdoctoral Fellowship for Basic and Translational Research at The Rockefeller University (M.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baumert TF, Berg T, Lim JK, Nelson DR: Status of Direct-acting Antiviral Therapy for HCV Infection and Remaining Challenges. Gastroenterology 2018, doi: 10.1053/j.gastro.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization: Global report on access to hepatitis C treatment. Focus on overcoming barriers. 2016, [no volume].
- •3. Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV: Transfusion-associated hepatitis not due to viral hepatitis type A or B. New England Journal of Medicine 1975, 292:767–770. [DOI] [PubMed] [Google Scholar]
- •4. Alter HJ, Holland PV, Morrow AG, Purcell RH, Feinstone SM, Moritsugu Y: Clinical and serological analysis of transfusion-associated hepatitis. Lancet 1975, 2:838–841. [DOI] [PubMed] [Google Scholar]
- 5.Alter HJ: Clinical, virological and epidemiological basis for the treatment of chronic non-A, non-B hepatitis. J Hepatol 1990, 11 Suppl 1:S19–25. [DOI] [PubMed] [Google Scholar]
- 6.Dienstag JL: non-A, Non-B hepatitis. II. Experimental transmission, putative virus agents and markers, and prevention. Gastroenterology 1983, 85:743–768. [PubMed] [Google Scholar]
- 7.Feinstone SM, Mihalik KB, Kamimura T, Alter HJ, London WT, Purcell RH: Inactivation of Hepatitis-B Virus and Non-a, Non-B Hepatitis by Chloroform. Infect. Immun 1983, 41:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley DW, McCaustland KA, Cook EH, Schable CA, Ebert JW, Maynard JE: Posttransfusion non-A, non-B hepatitis in chimpanzees. Physicochemical evidence that the tubule-forming agent is a small, enveloped virus. Gastroenterology 1985, 88:773–779. [PubMed] [Google Scholar]
- ••9. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M: Isolation of a Cdna Clone Derived From a Blood-Borne Non-a, Non-B Viral-Hepatitis Genome. Science 1989, 244:359–362. [DOI] [PubMed] [Google Scholar]
- 10.Pringle CR: Virus taxonomy 1996 - a bulletin from the Xth International Congress of Virology in Jerusalem. 1996:2251–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE, et al. : An Assay for Circulating Antibodies to a Major Etiologic Virus of Human Non-a, Non-B-Hepatitis. Science 1989, 244:362–364. [DOI] [PubMed] [Google Scholar]
- 12.Bruix J, Barrera JM, Calvet X, Ercilla G, Costa J, Sanchez-Tapias JM, Ventura M, Vall M, Bruguera M, Bru C: Prevalence of antibodies to hepatitis C virus in Spanish patients with hepatocellular carcinoma and hepatic cirrhosis. Lancet 1989, 2:1004–1006. [DOI] [PubMed] [Google Scholar]
- ••13. Greenberg HB, Pollard RB, Lutwick LI, Gregory PB, Robinson WS, Merigan TC: Effect of Human Leukocyte Interferon on Hepatitis B Virus-Infection in Patients with Chronic Active Hepatitis. New England Journal of Medicine 1976, 295:517–522. [DOI] [PubMed] [Google Scholar]
- 14.Nagano Y, Kojima Y: *Pouvoir Immunisant Du Virus Vaccinal Inactive Par Des Rayons Ultraviolets. C. R. Seances Soc. Biol. Fil 1954, 148:1700–1702. [PubMed] [Google Scholar]
- 15.Isaacs A, Lindenmann J: Virus interference. I. The interferon. Proc. R. Soc. Lond., B, Biol. Sci 1957, 147:258–267. [PubMed] [Google Scholar]
- 16.HO M: Interferon for the Treatment of Infections. Annu. Rev. Med 1987, 38:51–59. [DOI] [PubMed] [Google Scholar]
- 17.Hoofnagle JH, Mullen KD, Jones DB, Rustgi V, Dibisceglie A, Peters M, Waggoner JG, Park Y, Jones EA: Treatment of Chronic Non-a,Non-B Hepatitis with Recombinant Human Alpha-Interferon - a Preliminary-Report. New England Journal of Medicine 1986, 315:1575–1578. [DOI] [PubMed] [Google Scholar]
- 18.Sharara AI, Hunt CM, Hamilton JD: Hepatitis C. Ann. Intern. Med 1996, 125:658–668. [DOI] [PubMed] [Google Scholar]
- •19. McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK, et al. : Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. New England Journal of Medicine 1998, 339:1485–1492. [DOI] [PubMed] [Google Scholar]
- •20. Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M-H, Albrecht JK: Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. The Lancet 2001, 358:958–965. [DOI] [PubMed] [Google Scholar]
- 21.Heim MH: 25 years of interferon-based treatment of chronic hepatitis C: an epoch coming to an end. Nat Rev Immunol 2013, 13:535–542. [DOI] [PubMed] [Google Scholar]
- 22.Bartenschlager R, Lohmann V: Replication of hepatitis C virus. J. Gen. Virol 2000, 81:1631–1648. [DOI] [PubMed] [Google Scholar]
- 23.Simmonds P: Variability of hepatitis C virus. Hepatology 1995, 21:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •24. Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS: Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol 1993, 74 (Pt 11):2391–2399. [DOI] [PubMed] [Google Scholar]
- •25. Simmonds P, Smith DB, McOmish F, Yap PL, Kolberg J, Urdea MS, Holmes EC: Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol 1994, 75 ( Pt 5):1053–1061. [DOI] [PubMed] [Google Scholar]
- •26. Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM: Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol 1993, 67:1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C, Lindenbach BD, Prágai BM, McCourt DW, Rice CM: Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol 1994, 68:5063–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •28. Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H: Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol 1993, 67:3835–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •29. Tomei L, Failla C, Santolini E, De Francesco R, La Monica N: NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol 1993, 67:4017–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •30. Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K: Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol 1993, 67:4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31. Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM: Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol 1993, 67:2832–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM: A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA 1993, 90:10583–10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirowatari Y, Hijikata M, Tanji Y, Nyunoya H, Mizushima H, Kimura K, Tanaka T, Kato N, Shimotohno K: Two proteinase activities in HCV polypeptide expressed in insect cells using baculovirus vector. Arch Virol 1993, 133:349–356. [DOI] [PubMed] [Google Scholar]
- 34.Suzich JA, Tamura JK, Palmer-Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS: Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol 1993, 67:6152–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DW, Gwack Y, Han JH, Choe J: C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun 1995, 215:160–166. [DOI] [PubMed] [Google Scholar]
- 36.Behrens SE, Tomei L, De Francesco R: Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 1996, 15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann V, Körner F, Herian U, Bartenschlager R: Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol 1997, 71:8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •38. Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O’Malley ET, et al. : Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 1996, 87:343–355. [DOI] [PubMed] [Google Scholar]
- •39. Love RA, Parge HE, Wickersham JA, Hostomsky Z, Habuka N, Moomaw EW, Adachi T, Hostomska Z: The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 1996, 87:331–342. [DOI] [PubMed] [Google Scholar]
- •40. Lesburg CA Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC: Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol 1999, 6:937–943. [DOI] [PubMed] [Google Scholar]
- 41.Boyer JC, Haenni AL: Infectious transcripts and cDNA clones of RNA viruses. Virology 1994, 198:415–426. [DOI] [PubMed] [Google Scholar]
- 42.Han JH, Shyamala V, Richman KH, Brauer MJ, Irvine B, Urdea MS, Tekamp-Olson P, Kuo G, Choo QL, Houghton M: Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5” untranslated region and poly(A) tails at the 3” end. Proc Natl Acad Sci USA 1991, 88:1711–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43. Kolykhalov AA, Feinstone SM, Rice CM: Identification of a highly conserved sequence element at the 3’ terminus of hepatitis C virus genome RNA. J. Virol 1996, 70:3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •44. Tanaka T, Kato N, Cho MJ, Shimotohno K: A novel sequence found at the 3’ terminus of hepatitis C virus genome. Biochem Biophys Res Commun 1995, 215:744–749. [DOI] [PubMed] [Google Scholar]
- ••45. Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM: Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 1997, 277:570–574. [DOI] [PubMed] [Google Scholar]
- ••46. Yanagi M, Purcell RH, Emerson SU, Bukh J: Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA 1997, 94:8738–8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •47. French R, Ahlquist P: Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic-virus Rna3. J. Virol 1987, 61:1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S: Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol 1993, 67:6439–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liljeström P, Garoff H: A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (N.Y.) 1991, 9:1356–1361. [DOI] [PubMed] [Google Scholar]
- 50.Andino R, Rieckhof GE, Achacoso PL, Baltimore D: Poliovirus RNA synthesis utilizes an RNP complex formed around the 5’-end of viral RNA. EMBO J 1993, 12:3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKnight KL, Lemon SM: Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol 1996, 70:1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khromykh AA, Westaway EG: Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol 1997, 71:1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behrens SE, Grassmann CW, Thiel HJ, Meyers G, Tautz N: Characterization of an autonomous subgenomic pestivirus RNA replicon. J. Virol 1998, 72:2364–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••54. Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R: Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999, 285:110–113. [DOI] [PubMed] [Google Scholar]
- ••55. Blight KJ, Kolykhalov AA, Rice CM: Efficient initiation of HCV RNA replication in cell culture. Science 2000, 290:1972–1974. [DOI] [PubMed] [Google Scholar]
- 56.Frese M, Pietschmann T, Moradpour D, Haller O, Bartenschlager R: Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol 2001, 82:723–733. [DOI] [PubMed] [Google Scholar]
- 57.Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, Strand D, Bartenschlager R: Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol 2002, 76:4008–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lohmann V, Körner F, Dobierzewska A, Bartenschlager R: Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol 2001, 75:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krieger N, Lohmann V, Bartenschlager R: Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol 2001, 75:4614–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •60. Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R: Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol 2003, 77:3007–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •61. Blight KJ, McKeating JA, Rice CM: Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol 2002, 76:13001–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frese M, Barth K, Kaul A, Lohmann V, Schwarzle V, Bartenschlager R: Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol 2003, 84:1253–1259. [DOI] [PubMed] [Google Scholar]
- ••63. Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. : Complete replication of hepatitis C virus in cell culture. Science 2005, 309:623–626. [DOI] [PubMed] [Google Scholar]
- ••64. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich H-G, Mizokami M, et al. : Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 2005, 11:791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••65. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV: Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA 2005, 102:9294–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ: A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proceedings of the National Academy of Sciences 2009, 106:16410–16415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, et al. : Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA 2007, 104:12884–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lv Z, Chu Y, Wang Y: HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV AIDS (Auckl) 2015, 7:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwong AD, Kauffman RS, Hurter P, Mueller P: Discovery and development of telaprevir: an NS3–4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat. Biotechnol 2011, 29:993–1003. [DOI] [PubMed] [Google Scholar]
- •70. Steinkühler C, Biasiol G, Brunetti M, Urbani A, Koch U, Cortese R, Pessi A, De Francesco R: Product inhibition of the hepatitis C virus NS3 protease. Biochemistry 1998, 37:8899–8905. [DOI] [PubMed] [Google Scholar]
- •71. Llinàs-Brunet M, Bailey M, Fazal G, Goulet S, Halmos T, Laplante S, Maurice R Poirier M, Poupart MA, Thibeault D, et al. : Peptide-based inhibitors of the hepatitis C virus serine protease. Bioorg. Med. Chem. Lett 1998, 8:1713–1718. [DOI] [PubMed] [Google Scholar]
- 72.Randolph JT, DeGoey DA: Peptidomimetic inhibitors of HIV protease. Curr Top Med Chem 2004, 4:1079–1095. [DOI] [PubMed] [Google Scholar]
- ••73. Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P, Bös M, Cameron DR, Cartier M, Cordingley MG, et al. : An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 2003, 426:186–189. [DOI] [PubMed] [Google Scholar]
- 74.Ascione A: Boceprevir in chronic hepatitis C infection: a perspective review. Ther Adv Chronic Dis 2012, 3:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilby KJ, Partovi N, Ford J-AE, Greanya E, Yoshida EM: Review of boceprevir and telaprevir for the treatment of chronic hepatitis C. Can. J. Gastroenterol 2012, 26:205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Leuw P, Stephan C: Protease inhibitor therapy for hepatitis C virus-infection. Expert Opin Pharmacother 2018, 19:577–587. [DOI] [PubMed] [Google Scholar]
- 77.Heim MH: Innate immunity and HCV. J Hepatol 2013, 58:564–574. [DOI] [PubMed] [Google Scholar]
- 78.Eltahla AA, Luciani F, White PA, Lloyd AR, Bull RA: Inhibitors of the Hepatitis C Virus Polymerase; Mode of Action and Resistance. Viruses 2015, 7:5206–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sofia MJ: Enter Sofosbuvir: The Path to Curing HCV. Cell 2016, 167:25–29. [DOI] [PubMed] [Google Scholar]
- 80.Borgia G, Maraolo AE, Nappa S, Gentile I, Buonomo AR: NS5B polymerase inhibitors in phase II clinical trials for HCV infection. Expert Opin Investig Drugs 2018, 27:243–250. [DOI] [PubMed] [Google Scholar]
- ••81. Lemm JA, O’Boyle D, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St Laurent DR, Serrano-Wu MH, et al. : Identification of hepatitis C virus NS5A inhibitors. J. Virol 2010, 84:482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••82. Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun J-H, O’Boyle DR, et al. : Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 2010, 465:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ross-Thriepland D, Harris M: Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J. Gen. Virol 2015, 96:727–738. [DOI] [PubMed] [Google Scholar]
- 84.Pawlotsky J-M: New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology 2014, 146:1176–1192. [DOI] [PubMed] [Google Scholar]
- 85.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ: Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA 1991, 88:2451–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •86. Lohmann V: Hepatitis C virus cell culture models: an encomium on basic research paving the road to therapy development. Med Microbiol Immunol 2018, doi: 10.1007/s00430-018-0566-x. [DOI] [PubMed] [Google Scholar]
- ••87. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P: Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- •88. Janssen HLA, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. : Treatment of HCV infection by targeting microRNA. New England Journal of Medicine 2013, 368:1685–1694. [DOI] [PubMed] [Google Scholar]
- 89.Xu Y, Zhong J: Innate immunity against hepatitis C virus. Curr. Opin. Immunol 2016, 42:98–104. [DOI] [PubMed] [Google Scholar]
- 90.Rehermann B, Thimme R: Insights From Antiviral Therapy into Immune Responses to HBV and HCV Infection. Gastroenterology 2018, doi: 10.1053/j.gastro.2018.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Brien TR, Prokunina-Olsson L, Donnelly RP: IFN-λ4: the paradoxical new member of the interferon lambda family. J. Interferon Cytokine Res 2014, 34:829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •92. Harak C, Meyrath M, Romero-Brey I, Schenk C, Gondeau C, Schult P, Esser-Nobis K, Saeed M, Neddermann P, Schnitzler P, et al. : Tuning a cellular lipid kinase activity adapts hepatitis C virus to replication in cell culture. Nat Microbiol 2016, 2:16247. [DOI] [PubMed] [Google Scholar]
- 93.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R: Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol 2018, 16:125–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scheel TKH, Rice CM: Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 2013, 19:837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knight A: The beginning of the end for chimpanzee experiments? Philos Ethics Humanit Med 2008, 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miner JJ, Diamond MS: Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe 2017, 21:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abbink P, Stephenson KE, Barouch DH: Zika virus vaccines. Nat. Rev. Microbiol 2018, 16:594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]