Abstract
The filovirus family includes some of the deadliest viruses known, including Ebola virus and Marburg virus. These viruses cause periodic outbreaks of severe disease that can be spread from person to person, making the filoviruses important public health threats. There remains a need for approved drugs that target all or most members of this virus family. Small molecule inhibitors that target conserved functions hold promise as pan-filovirus therapeutics. To date, compounds that effectively target virus entry, genome replication, gene expression and virus egress have been described. The most advanced inhibitors are nucleoside analogs that target viral RNA synthesis reactions.
The filovirus family.
Filoviruses, zoonotic pathogens associated with severe disease in humans, are filamentous, enveloped viruses with non-segmented, negative-sense RNA genomes [1]. Included in filovirus family is the genus Ebolavirus, which has six species. Among these, Zaire ebolavirus (EBOV), Sudan virus (SUDV) and Bundibugyo virus (BDBV) have caused substantial outbreak with significant morbidity and mortality in humans. Marburgvirus is another genus with members that have caused that includes Marburg virus (MARV) and Ravn virus (RAVV). Cuevavirus, which contains Lloviu virus (LLOV) and proposed genus Dianlovirus, which contains a single member, Měnglà virus (MLAV), have not to date been associated with human disease [2–4]. LLOV and MLAV, both identified in bats, have not been isolated or cultured and their significance with regard to human health is unknown.
The largest filovirus outbreak on record was caused by EBOV in West Africa between 2013–2016. This resulted in more than 28,000 infections, more than 11,000 deaths and the export of infected cases to the United States and Europe [5]. In pregnant women the fatality rate is estimated to be 70%, and survivors are known to exhibit persistent infection with virus residing in immune privileged sites, including the eye and testes [6–10]. The only treatments available were supportive care and experimental therapies, hampering patient treatment and leaving healthcare workers at severe risk. The West Africa epidemic reinforced the threat posed by the filovirus family and demonstrated that in addition to being a bio-terrorism threat, emergences from natural sources can have a profound public health impact.
Because of their extreme virulence, fully-replication competent filoviruses are studied in Biosafety Level 4 (BSL4) containment. This limits the number of investigators who have direct access to virus and makes screening of antiviral against live virus challenging, even for those investigators with BSL4 access. Therefore, much effort has been devoted to targeting specific steps in the virus replication cycle, such as viral entry, viral RNA synthesis and virus assembly and release, that can be reconstituted in transfection-based studies that do not require live virus. This approach can facilitate the discovery of small molecules that target specific viral functions that must then be tested for efficacy against live virus in cell culture and animal models.
Summary of relevant filovirus biology.
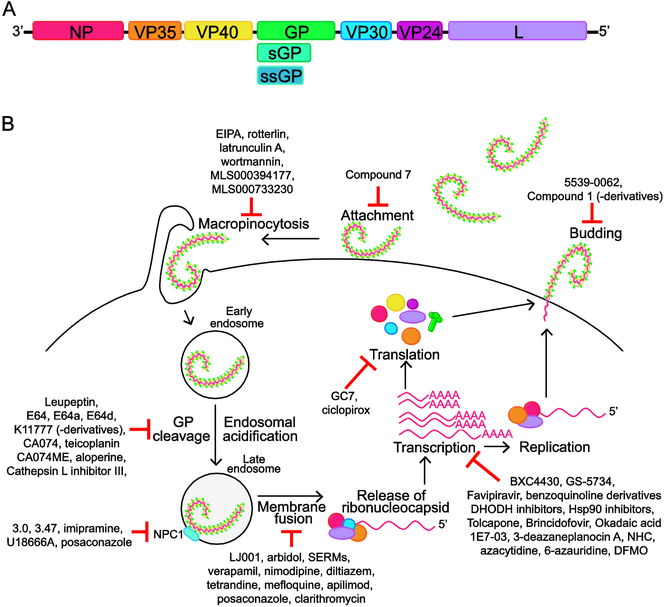
The filovirus genome is approximately 19 kilobases in length and encodes up to nine translation products from seven separate transcriptional units [1,11]. These genes encode the viral nucleoprotein (NP), viral protein of 35 kDa (VP35), VP40, a type I transmembrane glycoprotein (GP), VP30, VP24, and the large protein (L), which is the viral polymerase. Members of the Ebolavirus genus, and also likely LLOV, produce secreted forms of the GP protein [1,12,13] (Fig 1A).
Figure 1. Filovirus genome, replication cycle and small molecules inhibitors.
A. Schematic of filovirus genome. The negative-sense RNA genome has seven transcriptional units that encode for the nucleoprotein, NP; viral protein 35, VP35; VP40; glycoprotein/soluble glycoprotein, GP/sGP (sGP is not encoded by Marburg virus); VP30; VP24; Large protein, L (viral polymerase). Note that sGP and ssGP is produced by members of the Ebolavirus genus, and predicted to be produced by LLOV. Genome schematic is not to scale. B. Schematic of the steps in the filovirus lifecycle. GP mediates attachment of the filovirus to the surface of the cell. The virus is then taken up by macropinocytosis. Following acidification of the endosome, cathepsins B and L cleave GP, a requirement for its interaction with the host protein NPC1 that facilitates fusion of viral and endosomal membranes. Endosomal calcium channels, known as two-pore channels (TPCs), play a role in trafficking the virus particle to the site of membrane fusion. Following fusion, the ribonucleocapsid is released into the cytoplasm where 5’-capped, 3’polyadenylated mRNAs are transcribed for each viral gene and a copy of the full-length genomic RNA is produced, which acts as a template for synthesis of new negative-sense viral genomes. Transcription requires NP, VP35, VP30 and L, while replication does not need VP30. Viral proteins are translated from the viral mRNAs and new viral particles are formed at the cell surface. VP40 drives viral budding and is assisted by GP. Viral ribonucleoproteins containing genomic RNA, NP, VP35, VP30 and VP24 are incorporated into the budding particles. The steps in the filovirus lifecycle are potential targets for therapeutic intervention; small molecules that target these processes are noted on the schematic. Greater detail on the filovirus lifecycle and these small molecules can be found in the review.
Viral entry is mediated by GP which acts as an attachment factor and mediates fusion of viral and host cell membranes within an endosomal compartment [14] (Fig 1B). The viral genome is released into the cytoplasm as a ribonucleoprotein complex. This serves as the template for the RNA synthesis reactions that replicate the viral genomic RNA and transcribe the mRNAs that lead to viral gene expression. Replication requires NP, which associates with the viral genomic and antigenomic RNAs throughout the course of infection; VP35, a non-enzymatic cofactor for the viral RNA-dependent RNA polymerase that also serves as a potent suppressor of innate antiviral signaling pathways and L, which possesses all the enzymatic activities required for viral transcription and genome replication, including RNA-dependent RNA polymerase (RdRp) activity, guanyltransferase and methyltransferase activities [15,16]. Viral transcription (mRNA synthesis) involves the production of distinct 5’-capped, 3’polyadenylated mRNAs from each of the viral genes and requires, in addition to NP, VP35 and L, the VP30 protein [16] (Fig 1B). Co-transfection of these four viral proteins with a model viral genomic RNA can recapitulate the filovirus RNA synthesis machinery in cell-based “minigenome” assays in biosafety level 2 (BSL2), enabling the study of filovirus RNA synthesis [17–20]. In addition to the required viral proteins, host factors modulate viral RNA synthesis through interaction with viral factors, however, a complete understanding as to how host factors contribute to viral RNA synthesis remains elusive [21–23].
Other viral functions include filovirus assembly and release [24]. The VP40 matrix protein drives the membrane budding events that lead to release of new virus particles. GP is incorporated into the membrane of viral particles and enhances budding. Viral ribonucleoproteins (RNPs) that contain genomic RNA, NP, VP35, VP30 and VP24 are recruited into the budding particles. In addition to playing roles in replication and assembly, several filovirus proteins counteract host innate antiviral defenses [25]. The filovirus VP35 proteins block interferon (IFN)-α/β production and the VP24 proteins of Ebolavirus and Cuevavirus genus members and the VP40 proteins of the Marburgvirus genus block IFN-induced antiviral signaling [15,26–34]. Marburgvirus VP24 proteins also modulate host antioxidant response pathways through interaction with the host protein Keap1 [35–37]. While these viral functions can be studied independently by transfection based assays, inclusion of the viral genes for VP40, GP and VP24 into the model viral genomic RNA results in an advanced system that produces replication and transcription-competent virus-like particles (trVLPs), allowing for BSL2 study of most viral lifecycle steps, including entry and budding, in addition to RNA synthesis [38].
Status of Promising Anti-Filovirus Approaches.
The typical progression of anti-filovirus therapeutics is demonstration of efficacy in cell culture, then in mice, followed by guinea pigs and then non-human primates (NHPs), the “gold standard” for efficacy in animals [39]. Historically, NHPs have proven much more difficult to protect than rodents. The focus of this review is small molecule inhibitors of EBOV and other filoviruses, which are summarized in Figure 1 and Table 1. It is nonetheless important to recognize that monoclonal antibodies and nucleic acid-based therapeutics have also successfully protected NHPs from lethal filovirus challenge. A notable achievement was the finding that the three monoclonal antibody cocktail ZMAPP could protect NHPs from lethal challenge even after the onset of clinical symptoms [40]. Antibody-based approaches are sufficiently promising that three such treatments are being investigated in a clinical trial in the 2018–2019 EBOV outbreak in the Democratic Republic of Congo. These are ZMAPP (Mapp Biopharmaceutical, Inc.), Mab 114 (National Institute of Allergy and Infectious Diseases) and the three monoclonal antibody cocktail REGN-EB3 (Regeneron).
Table 1.
Status of small molecule drugs developed for filovirus infection.
| Targeted step of virus lifecycle | Small Molecule | MOAa | Efficacy in vitro | Efficacy in vivo | Clinical Trialb |
|---|---|---|---|---|---|
| RNA synthesis | BCX4430[43] | Inhibition of viral RdRp - chain terminator | IC50 3.4–11.8 μMc,d,e,f IC90 10.3–25.4 μMc,d,e,f |
Protection in mice, guinea pigs. NHPs: 15 mg/kg, twice daily via i.m.g confers protection against MARV when administered up to 48hr post-infection. | Phase I (NCT02319772, completed; NCT03800173, recruiting) |
| GS-5734[42] | Inhibition of viral RdRp - chain terminator | IC50 0.06–0.14 μMd,h,i IC90 0.18–0.41 μMd,h,i |
Confers complete protection in NHPs against EBOV when administered 72hr post-infection at a 10 mg/kg loading dose followed by 3 or 10 mg/kg daily dose i.v.j | Phase II (NCT02818582, recruiting) | |
| Favipiravir[41,48,49] | Either inhibition of viral RdRp and/or causes lethal mutagenesis | IC50 67 μMd,k
IC90 110 μMd,k |
Complete protection of IFNAR knockout mice, immunocompetent C57BL/6 mice. NHPs: Three p.o.l dosing regimens (400 mg/kg loading dose at day −3 followed by 200 mg/kg daily, 250 mg/kg loading dose at day 0 followed by 150 mg/kg twice daily, 125 mg/kg loading dose at day 0 followed by 75 mg/kg twice daily) protected 1 of 18 EBOV infected NHPs, despite plasma levels above EC50 values. I.V. dosing of a 250 mg/kg loading dose followed by 150 mg/kg twice daily protected 5 of 6 MARV infected NHPs. | Phase III (influenza, NCT02008344, completed) Phase II (EBOV, NCT02329054, completed; NCT02662855, completed) | |
| 3-deazaneplanocin A[52,53] | Inhibition of SAH hydrolase | IC50 2 μMd,k | Complete protection of BALB/c mice against MA-EBOVaa when treated once with 1 mg/kg up to 48hr post-infection. | N/A | |
| (β-d-N4-hydroxycytidine (NHC)[60] | Unknown | IC50 3 μMd,k | N/A | N/A | |
| Azacytidine[17] | Unknown | IC50 4 μMm,u
86% inhibition at 50 μMd,k |
N/A | N/A | |
| 6-azauridine[18] | Unknown | IC50 5 μMn,p and 14 μMm,p
98% inhibition at 80 μMc,k 99% inhibition at 80 μMd,k |
N/A | N/A | |
| Benzoquinoline compounds[61] | Unknown | IC50 0.5–5.6 μMm,u
>2 log reduction of viral titer at 1 μMd,k |
N/A | N/A | |
| HSP90 inhibitors (such as geldanamycin, 17AAG and radicicol)[23] | Destabilization of EBOVL | Geldanamycin: IC50 1.6 μMd,k
17AAG: IC50 5.3 μMd,k Radicicol: IC50 1.7 μMd,k |
N/A | N/A | |
| Inhibitors of polyamine biosynthesis (such as DFMO)[63,64] | Defect in EBOV RdRp dependent mRNA accumulation | DFMO: 90% inhibition at 500 μMm,p | N/A | N/A | |
| Inhibitors of hypusination (such as GC7 and ciclopirox)[63,64] | Ineffective translation of EBOV mRNA | GC7: 85% inhibition at 10 μMm,p
Ciclopirox: >2 log reduction of viral titer at 30 μMc,d,o |
N/A | N/A | |
| DHODH inhibitors (such as GSK983 and Brequinar)[65] | Inhibition of de novo pyrimidine synthesis | GSK983: IC50 0.007 μMm,u and <0.02 μMd,f
Brequinar: IC50 0.15 μMm,u and 0.1 μMd,f |
N/A | N/A | |
| Inhibitors of VP30 dephosphorylation (such as Okadaic acid (OA) and 1E7–03)[71,72] | Inhibition of viral transcription as phosphorylated VP30 does not participate in transcription | OA: IC50 0.13μM OAm,p
>90% inhibition at 0.08 μMd,p,* 1E7–03: 200-fold reduction of viral titerat 10 μMd,k |
N/A | N/A | |
| Tolcapone[77] | Inhibition of NP:NPBP interaction, necessary for polymerase activity | IC50 2 μMq
At 10 μM, >100 fold decrease at MOI 0.01 and >5 fold reduction at MOI 2d,k |
N/A | N/A | |
| Virus Entry | Compound 7[93] | Direct interaction with GP | IC50 10 μMd,kand 12 μMc,k | N/A | N/A |
| LJ001 [94,95] | Inhibition of virus fusion - type II photosensitizer that modifies unsatu rated phospholipids, negatively impacting viral membrane | 0.5 μM < IC50 < 1 μMc,d,k | At 20 μM, protects 80% of mice infected with MA-EBOV. | N/A | |
| Arbidol[97] | Unknown | IC50 2.7 μMd,o | N/A | N/A | |
| EIPA[79] | Inhibition of macropinocytosis prevents cellular uptake of virus | ~50–75% inhibition at 100 μMr,f,k,*
~70% inhibition at 200 μMd,k,* |
N/A | N/A | |
| Latrunculin A[79] | Inhibition of macropinocytosis prevents cellular uptake of virus | ~30–65% inhibition at 0.5 μMr,f,k,*
~40% inhibition at 0.5 μMd,k,* |
N/A | N/A | |
| Wortmannin[79] | Inhibition of macropinocytosis prevents cellular uptake of virus | ~50% inhibition at 0.1 μMr,f,k,* | N/A | N/A | |
| Rottlerin[82] | Inhibition of macropinocytosis prevents cellular uptake of virus | >50% inhibition at 2.5 μMr,k | N/A | N/A | |
| Leupeptin[105] | Inhibition of cysteine-serine protease, inhibits GP proteolysis | >95% inhibition at 10 μMk,s,* | N/A | N/A | |
| E64, E64a, E64d[84] | Inhibition of cysteine protease, inhibits GP proteolysis | E64d: >90% inhibition at 50 |jMk,s,* | N/A | N/A | |
| K11777 and derivates[101] | Inhibition of cysteine protease, inhibits GP proteolysis | K11777: 0.87–5.91 nMu,s,t,w
Derivatives: 0.1–2.69 nMu,s |
N/A | N/A | |
| CA074[83], CA074Me[84] | Cathepsin B inhibitor | CA074: ~90% inhibition at 10 μMs,k,* >10-fold decrease at 80 μMd,k,* CA074Me: >80% inhibition at 0.5 μMs,k,* |
N/A | N/A | |
| Cathepsin L inhibitor lll[108] | Cathepsin L inhibitor | IC50 7 μMs,u | N/A | N/A | |
| Aloperine and derivatives[107] | Cathepsin B inhibitor | Derivative 2e: IC50 4.8 μMu,s and 7.1 μMu,t | Aloperine derivative 2e administered half by i.v. (50 μg) and half by i.p.v (50 μg) on day of challenge with pHIV-EBOVGP-Fluc or pHIV-MARVGP-Fluc reduced bioluminescence by 58% 4 days post-infection and 45% 5 days post-infection, respectively. | N/A | |
| 3.0 and 3.47[91,114] | Interacts with NPC1, inhibiting binding of cleaved GP | 3.0: 1 μM < IC50 < 10 μMk,s
>99% inhibition at 20 μMd,k 3.47: 0.01 μM < IC50 < 0.1 μMk,s |
3.47: Mice treated i.p. with 1, 5 or 25 mg/kg daily, showed no protection from MA-EBOV infection. | N/A | |
| lmipramine[114] | Unknown, mimics NPC1 deficiency | ~50% inhibition at 10 μMc,d,x | Mice treated i.p. with 20 mg/kg daily or every other day showed no significant protection from MA-EBOV infection, although lower viral replication at day 3 and 5 was detected. | N/A | |
| U18666A[90,114] | Unknown, mimics NPC1 deficiency | ~99% inhibition at 10 μMs,k | Mice treated i.p with 2 mg/kg daily or every other day showed no significant protection from MA-EBOV infection, although lower viral replication at day 3 was detected. | N/A | |
| Toremifene[115] | Unknown, off-target effect | IC50 0.973–6.17 μMc,d,e,k | Mice treated i.p. with 60 mg/kg on day 0 and 1 and every other day after showed 50% protection from MA-EBOV infection. | N/A | |
| Clomiphene[115] | Unknown, off-target effect | IC50 3.83–11.1 μMc,d,e,k | Mice treated i.p. with 60 mg/kg on day 0 and 1 and every other day after showed 90% protection from MA-EBOV infection. | N/A | |
| Mefloquine[116] | Unknown | IC50 2.73 μMf,r | N/A | N/A | |
| Posaconazole[116] | Unknown | IC50 7.69 μMf,r | N/A | N/A | |
| Clarithromycin[116] | Unknown | IC50 4.53 μMf,r | N/A | N/A | |
| Verapamil[121] | Prevents EBOV escape from endosome | >80% inhibition at 60 μg/mls,t,y | N/A | N/A | |
| Nimodipine[120] | Prevents EBOV escape from endosome | IC50 ~25 μMd,f,* | N/A | N/A | |
| Diltiazem[120] | Prevents EBOV escape from endosome | IC50 ~25 μMd,f,* | N/A | N/A | |
| Tetrandrine[120] | Prevents EBOV escape from endosome | IC50 55 μMd,f | Mice treated i.p. with 30 mg/kg on day 0 and every second day showed 75% protection from MA-EBOV infection. Treatment of 90 mg/kg starting 1 day post-infection protected 50% of mice. | N/A | |
| Apilimod[123] | Prevents trafficking of incoming virus to NPC1/membrane fusion sites | IC50 0.01–0.14 μMc,d,k,i | N/A | N/A | |
| Genistein[128] | Unknown | >70% inhibition at 100 μMd,u | N/A | N/A | |
| Tyrphostin AG1478[128] | Unknown | 96–99% inhibition at 100|jMd,u | N/A | N/A | |
| Virus Egress | 5539–0062[132] | Inhibits EBOV VP40:tsg101 interaction | >90% inhibition at 100 μMd,r | N/A | N/A |
| Compound 1 and derivatives[133] | Inhibits VP40:Nedd4 interaction | Compound 1: 5-fold decrease at 20 μMu,z Compound 4: at 1 μM, 100-foldu,z and 10-foldu,r decrease |
N/A | N/A |
Mechanism of Action
Phase and NCT identifier of clinical trial and status information from ClinicalTrials.gov
MARV
EBOV
SUDV
Hela cells
Intramuscular route
HFF-1, HMVEC-TERT cells
Primary macrophages and HUH7 cells
Intravenous
Vero/Vero E6 cells
Oral route
EBOV minigenome assay
MARV minigenome assay
HepG2 cells
BSR T7/5 cells
Fluorescent polarization assay for the interaction of NP and NPBP
EBOV VLP
EBOV GP pseudotyped virions
MARV GP pseudotyped virions
293T/293FT cells
Intraperitoneal route
SUDV, TAFV, RESTV and BDBV GP pseudotyped virions
HUVEC cells
EAhy cells
MARV VLP
Mouse-adapted EBOV
estimated from figure
Targeting filovirus RNA synthesis.
Targeting viral RNA synthesis reactions shows substantial promise, including the targeting of viral RNA polymerase function by nucleoside analogues and small molecules that affect viral protein expression, stability and post-translational modifications necessary for viral replication.
Nucleoside analogues BXC4430, an adenosine analogue, GS-5734 (Remdesivir, Gilead, USA), a monophosphoramidate prodrug of an adenosine analogue and favipiravir (T-705, Toyama Chemical, Japan), a synthetic guanidine nucleoside analogue, exhibit anti-filovirus activity, likely through inhibition of viral polymerase activity [41–43]. BXC4430 was the first small molecule demonstrated to protect non-human primates from lethal filovirus challenge, even when administered up to two days post-infection [43]. This compound has progressed to Phase I clinical trials (URL:ClinicalTrials.gov Identifier:NCT02319772 and NCT03800173).
GS-5734 exhibits antiviral activity against a number of RNA viruses and has been shown to inhibit respiratory syncytial virus (RSV) and hepatitis C virus (HCV) polymerases, with greater selectivity for the viral polymerase than the cellular [42,44,45]. GS-5734 demonstrated protection of rhesus macaques from EBOV challenge across several dosing regimens, including complete protection from lethal disease of animals that received GS-5734 beginning three days post infection [42]. Interestingly, in macaques, administration led to distribution in tissues such as testes, epididymis, eyes and brain where EBOV may reside after recovery from illness. GS-5734 was provided to some human patients infected during the West Africa epidemic, and has been used in the 2018–2019 outbreak in DRC under an emergency use protocol for experimental medical interventions [7,46] (WHO; URL:https://www.who.int/ebola/drc-2018/treatments-approved-for-compassionate-use/en/). It is also currently in clinical trials, including a study to evaluate treatment of male survivors of Ebola virus disease (EVD) with persistent EBOV in their semen (URL:ClinicalTrials.gov Identifier:NCT02818582).
Favipiravir has broad spectrum activity against a number of RNA viruses and has been studied in Phase 3 clinical trials in Japan and the United States, with approval in Japan for treatment of influenza virus infection (URL:ClinicalTrials.gov Identifier:NCT02008344; Toyama Chemical Co Ltd.; URL:https://www.toyama-chemical.co.jp/eng/news/news140324e.html). It likely acts as a “pseudo purine”, inhibiting influenza virus RdRp activity with selectivity towards viral over cellular polymerases [47]. Favipiravir demonstrated protection of type I interferon receptor (IFNAR) knockout mice and immunocompetent C57BL/6 mice from challenge with EBOV and mouse-adapted EBOV (MA-EBOV), respectively [41,48,49]. When tested in macaques, favipiravir administered orally once or twice daily resulted in only one survivor out of eighteen EBOV infected animals, although delayed time to death and reduced viral levels were documented [41]. In contrast, intravenous administration of favipiravir led to five of six MARV infected animals surviving, with reduced viral loads and symptoms of MARV disease in survivors and delayed time to death for the animal that succumbed. Two clinical trials examined favipiravir as a treatment for EVD during the West Africa epidemic [50,51]. Due to the design of the trials, definitive conclusions could not be made. However, favipiravir treatment correlated with decreased viremia in subjects with low initial viral load, improved survival rates and reduced symptoms. Together, these studies suggest that continued examination of favipiravir as an anti-EBOV therapy is warranted.
Nucleoside analogues also exhibit antiviral activity by mechanisms other than direct inhibition of viral polymerase activity. Carbocyclic nucleosides, such as 3-deazaneplanocin A, inhibit replication of a number of negative sense RNA viruses including filoviruses [52–57]. The mechanism of action is thought to be inhibition of cellular S-adenosylhomocysteine (SAH) hydrolase (SAHase), which breaks down SAH produced from S-adenosylmethionine (SAM), a molecule required for macromolecular methylation reactions. Inhibition of SAHase raises intracellular SAH levels, blocking cellular methylation reactions via a feedback inhibition mechanism. This causes diminished methylation of viral mRNAs 5’ cap moieties and impairs viral protein synthesis [58]. 3-deazaneplanocin A has been demonstrated to exhibit anti-EBOV activity in vivo, as it can protect mice from EBOV challenge [52].
Several other nucleoside analogues with anti-EBOV activity have been identified through antiviral screens using EBOV or minigenome assays confirmed in cell culture with EBOV. These include the cytidine analogues β-d-N4-hydroxycytidine (NHC) and azacytidine and the uridine analogue 6-azauridine [17,18,59,60]. Further study is required to demonstrate mechanism of action and efficacy in animal models for these compounds.
A number of non-nucleoside compounds have also been demonstrated to inhibit filovirus replication in cell culture. Benzoquinoline compounds were identified by minigenome assay as inhibitors of EBOV RNA synthesis, with activity shown against a variety of RNA virus families [61]. While their mechanism of action is unknown, the broad-spectrum activity suggests a host target. Hsp90 inhibitors demonstrate inhibition of EBOV in cell culture, likely through the destabilization of EBOV L [23,62]. It has also been demonstrated that inhibition of polyamine biosynthesis, such as by 2-difluoromethylornithine (DFMO), and hypusination, by N1-guanyl-1,7-diamineheptane (GC7) and ciclopirox, reduces EBOV replication [63,64].
Inhibitors of the host enzyme dihydroorotate dehydrogenase (DHODH), which has a role in de novo pyrimidine biosynthesis, have broad-spectrum antiviral activity that includes inhibition of EBOV in cell culture [65,66]. The anti-EBOV activity of DHODH inhibitors such as GSK983 and brequinar is related to depletion of pyrimidine pools [65]. Interestingly, a genetic screen also identified de novo pyrimidine biosynthesis as critical for EBOV replication [67]. Although DHODH inhibitors have been used clinically for other applications, in vivo studies have not demonstrated convincing antiviral activity, possibly because uracil is available systemically in vivo to feed the salvage pathway, overcoming the block to the de novo pyrimidine synthesis pathway [68–70].
Filovirus replication can also be targeted through the VP30 protein, which is required for EBOV and MARV growth and plays roles in viral mRNA synthesis. This function is regulated by VP30 phosphorylation, with dephosphorylated VP30 promoting viral mRNA synthesis and phosphorylated VP30 promoting viral genome RNA replication. Compounds that prevent VP30 dephosphorylation, such as Okadaic acid (OA) and 1E7–03, impair virus growth in cell culture [71–73].
Another strategy being pursued is the targeting of protein-protein interactions involved in filovirus RNA synthesis. This has been enabled by the increasing numbers of virus-virus and virus-host protein-protein interactions that have been identified and characterized by structural, biophysical and molecular biology methods [1,74,75]. In one example, a fluorescence polarization assay was developed around the interaction of NP and the NP binding peptide (NPBP) derived from VP35, an interaction critical for viral RNA synthesis [75]. Screening for inhibitors of the interaction identified Tolcapone, an FDA-approved drug that is used in the treatment of Parkinson’s disease [76,77]. Tolcapone was demonstrated to impair EBOV replication in cell culture. As the NP:NPBP interaction site is well-conserved among filoviruses, the NP:NPBP interaction has potential as a pan-filovirus target [77].
Entry Inhibitors.
There has been substantial effort devoted to developing small molecule inhibitors of EBOV entry. The entry process itself has been studied in depth and is relatively unique [78]. A number of cell surface molecules, including lectins and phosphatidyl serine (PS)-binding proteins have been described to mediate virus attachment to the cell surface, via interactions with GP or PS on the virus surface [78]. Uptake is by macropinocytosis or a macropinocytosis-like process [79–82]. Within endosomes, GP undergoes cleavage by proteases cathepsin B and cathepsin L, although EBOV replication in both cell culture and mouse models may not absolutely require that both these proteases be active [83–89]. Cleaved GP then interacts with host protein Niemann–Pick C1 (NPC1) within the endosomal lumen [90–92]. This interaction is necessary for fusion of viral and endosomal membrane and release of virus particles. Each of these steps, as well as cellular functions associated with these steps, are potential targets for therapeutic intervention.
Compounds directly targeting GP or the viral membrane.
A benzodiazepine derivative called compound 7 was identified in a screen for entry inhibitors through the use of GP pseudotyped lentiviruses [93]. Compound 7 demonstrated selectivity towards inhibition of EBOV and MARV in cell culture over other RNA and DNA viruses and was shown to directly bind GP [93]. In contrast, LJ001, a rhodanine derivative identified in a screen for inhibitors of Nipah virus entry, was demonstrated to have broad-spectrum activity against enveloped but not non-enveloped viruses. It was shown to bind to lipid membranes, acting as a type II photosensitizer modifying unsaturated phospholipids, negatively affecting enveloped virus entry without significant host cell cytotoxicity[94]. LJ001 was also shown to inhibit EBOV replication in cell culture and to protect 80 percent of mice challenged with MA-EBOV [95].
Arbidol is a small molecule used clinically in Russia and China to prevent and treat influenza virus infections [96]. It has broad-spectrum antiviral activity in cell culture, including anti-EBOV activity that is suggested to be through inhibition of EBOV entry [97]. This may be through the capacity of arbidol to bind lipid membranes, but as it has also been demonstrated to directly bind influenza A virus hemagglutinin protein preventing fusion, the possible interaction with filovirus GPs may warrant exploration [98,99].
Inhibitors of macropinocytosis.
Compounds that inhibit macropinocytosis, such as ethylisopropylamiloride (EIPA), an inhibitor of the Na+/H+ exchanger that specifically inhibits macropinocytosis, PKC inhibitor rottlerin, actin polymerization inhibitor latrunculin A, and PI3-kinase inhibitor wortmannin all inhibit EBOV entry [79,82]. Recent screening with MARV GP pseudotyped VSV and retrovirus particles identified 17 compounds able to inhibit MARV and EBOV in cell culture, two of which were novel macropinocytosis inhibitors [100].
Inhibitors of cathepsins.
The proteolysis of EBOV GP by cathepsins B and L, and the critical role of this activity in EBOV entry, was demonstrated in part through the use of pharmacological inhibitors of proteases. These inhibitors include the cysteine-serine protease inhibitor leupeptin, cysteine protease inhibitors E64, E64a, E64d, K11777 and K11777-derivatives, cathepsin B inhibitors CA074 and CA074Me and cathepsin L inhibitor III [83,84,101–105]. Several more studies have been undertaken to identify inhibitors of cathepsin B and L cleavage of GP and inhibition of filovirus entry [106–108]. These include the natural product aloperine and its derivatives which target cathepsin B and the glycopeptide antibiotic teicoplanin that is suggested to inhibit cathepsin L. Finally, it should be noted that cathepsins B and L are activated by low pH and inhibitors of endosomal acidification also inhibit filovirus entry [109,110].
Inhibitors that target NPC1 interaction with GP or mimic the phenotype of NPC1 deficiency.
NPC1 was initially identified as an essential receptor for EBOV entry by both a genetic and a chemical screen [90,91]. NPC1 is an endosomal and lysosomal cholesterol transporter and mutations in NPC1 are associated with Niemann–Pick disease, a neurovisceral atypical lysosomal lipid storage disorder where cholesterol and sphingolipids accumulate in lysosomes [111]. A benzylpiperazine adamantane diamide compound, called 3.0, and an analogue, 3.47, identified in the chemical screen caused cholesterol accumulation in cells and were demonstrated to interact with NPC1, inhibiting binding of cleaved GP [91]. Although the cholesterol transport function of NPC1 is not required for EBOV entry, compounds that mimic NPC1 deficiency in cells, including imipramine and U18666A, the latter of which also binds NPC1, have been shown to block EBOV entry [90,112,113]. However, while imipramine and U18666A treatment of mice infected with MA-EBOV led to lower viral replication, no significant protection was demonstrated [114].
Compounds inhibiting late steps in the entry process.
Selective estrogen receptor modulators (SERMs), including clomiphene and toremifene, were identified during screening of FDA-approved drugs as inhibitors of EBOV entry in cell culture [115–117]. When tested in mice, clomiphene protected 90% of animals and toremifene protected 50% of animals from death [115]. Mechanistically, the compounds function in cells lacking estrogen receptor, suggesting an off-target effect and were shown in one study to inhibit a late step in EBOV entry, such that fusion does not occur, although in another, toremifene was found to bind and destabilize GP [115,118].
Repurposing of FDA drugs is complicated by an inability to achieve in vivo levels sufficient for anti-EBOV activity, therefore combinations of inhibitors have been assessed for efficacy [119]. Two 3-drug combinations, toremifene-mefloquine-posaconazole and toremifene-clarithromycinposaconazole, were identified as being active at clinically achievable concentrations. Mechanistic studies suggested all inhibit NAADP-AM stimulated lysosomal calcium release, while posaconazole inhibits NPC1 function and posaconazole, toremifene and mefloquine inhibit acid sphingomyelinase activity.
EBOV entry requires endosomal calcium channels known as two-pore channels (TPCs) [120]. Inhibition of TPCs by genetic or pharmacological approaches, including the FDA-approved drugs verapamil, nimodipine and diltiazem, as well as the natural product tetrandrine, prevent EBOV escape from endosomes, thereby aborting infection [110,120–122]. Tetrandrine, the most potent of these, was demonstrated to protect mice from lethal challenge with mouse-adapted EBOV [120].
Other genes potentially important for EBOV entry were described in the genetic screen that identified NPC1, including phosphatidylinositol-3-phosphate 5-kinase (PIKfyve) [90]. Based on this, the small molecule apilimod, which inhibits PIKfyve, was examined and demonstrated potent inhibition of EBOV and MARV entry in human macrophages [123]. The mechanism of inhibition appears to be impaired trafficking of incoming virus to sites where NPC1 resides and membrane fusion takes place [123]. Apilimod has been well-tolerated in phase I and II clinical trials as an interleukin-12/23 inhibitor for the treatment of Crohn’s disease, psoriasis, and rheumatoid arthritis [124–127]. Given these activities, it will be of interest to determine whether the antiviral activity of apilimod outweighs its immune suppressing activities in the context of filovirus infection in vivo.
Other entry inhibitors.
A combination of the kinase inhibitors genistein and tyrphostin AG1478 was demonstrated to inhibit infection by EBOV and MARV GP-pseudotyped VSV and to inhibit EBOV growth, although the mechanism remains to be elucidated [128].
Inhibitors of EBOV egress.
The VP40 protein of filoviruses serves as the major matrix protein that is responsible for the budding of new virus particles from the cell surface. Budding is facilitated by the interaction of proline-rich “late domain” motifs on VP40 with components of the host cell vacuolar protein sorting (vps) pathway, such as Tsg101 and Nedd4 [129–131]. An in silico screen using the NMR structure of the PTAP peptide-binding pocket in human tsg101 identified a compound, 5539–0062, that was demonstrated to inhibit the interaction between tsg101 and EBOV VP40 and to prevent budding of VP40 from cells in transfection studies [132]. High concentrations of compound were needed; however, the inhibitory concentrations were not cytotoxic [132]. A second in silico screen using the structure of a WW domain of Nedd4 in complex with a PPxY motif identified compound 1 that inhibits MARV VP40 budding [133]. Structure/activity studies led to the generation of compounds with enhanced potency and inhibition of both MARV and EBOV VP40 interaction with Nedd4 and budding [133,134]. This inhibition extended to other viruses where matrix proteins rely on late domain-Nedd4 interactions for budding, including VSV and rabies virus [133].
Conclusions.
The development of therapeutic antibodies and small molecule inhibitors that protect non-human primates from EBOV challenge is a major milestone as is advancement of therapeutics into clinical trials. Given that none is yet approved for human use and the facts that propensities of these approached to elicit resistance, continued efforts at drug development for filoviruses remains a necessity. Small molecule approaches targeting conserved viral functions would seem to offer the greatest possibility for pan-filovirus efficacy. Therefore, small molecule drug development should be a priority.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Messaoudi I, Amarasinghe GK, Basler CF: Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat Rev Microbiol 2015, 13:663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rougeron V, Feldmann H, Grard G, Becker S, Leroy EM: Ebola and Marburg haemorrhagic fever. J Clin Virol 2015, 64:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand FX, Briese T, et al. : Taxonomy of the order Mononegavirales: update 2016. Arch Virol 2016, 161:2351–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XL, Tan CW, Anderson DE, Jiang RD, Li B, Zhang W, Zhu Y, Lim XF, Zhou P, Liu XL, et al. : Characterization of a filovirus (Mengla virus) from Rousettus bats in China. Nat Microbiol 2019. [DOI] [PubMed] [Google Scholar]
- 5.Spengler JR, Ervin ED, Towner JS, Rollin PE, Nichol ST: Perspectives on West Africa Ebola Virus Disease Outbreak, 2013–2016. Emerg Infect Dis 2016, 22:956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayden FG, Friede M, Bausch DG: Experimental Therapies for Ebola Virus Disease: What Have We Learned? J Infect Dis 2017, 215:167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs M, Rodger A, Bell DJ, Bhagani S, Cropley I, Filipe A, Gifford RJ, Hopkins S, Hughes J, Jabeen F, et al. : Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 2016, 388:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uyeki TM, Erickson BR, Brown S, McElroy AK, Cannon D, Gibbons A, Sealy T, Kainulainen MH, Schuh AJ, Kraft CS, et al. : Ebola Virus Persistence in Semen of Male Survivors. Clin Infect Dis 2016, 62:1552–1555. [DOI] [PubMed] [Google Scholar]
- 9.Yeh S, Varkey JB, Crozier I: Persistent Ebola Virus in the Eye. N Engl J Med 2015, 373:1982–1983. [DOI] [PubMed] [Google Scholar]
- 10.Zeng X, Blancett CD, Koistinen KA, Schellhase CW, Bearss JJ, Radoshitzky SR, Honnold SP, Chance TB, Warren TK, Froude JW, et al. : Identification and pathological characterization of persistent asymptomatic Ebola virus infection in rhesus monkeys. Nat Microbiol 2017, 2:17113. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann H, Sanchez A, Geisbert TW: Filoviridae: Marburg and Ebola Viruses. Edited by Knipe DM, Howley P. Philadelphia: :: Wolters Kluwer; 2015. [Google Scholar]
- 12.Negredo A, Palacios G, Vazquez-Moron S, Gonzalez F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martinez M, et al. : Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog 2011, 7:e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehedi M, Falzarano D, Seebach J, Hu X, Carpenter MS, Schnittler HJ, Feldmann H: A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J Virol 2011, 85:5406–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey RA, Shtanko O, Anantpadma M, Sakurai Y, Chandran K, Maury W: Mechanisms of Filovirus Entry. Curr Top Microbiol Immunol 2017, 411:323–352. [DOI] [PubMed] [Google Scholar]
- 15.Cardenas WB, Loo YM, Gale M Jr., Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF: Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 2006, 80:5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhlberger E: Filovirus replication and transcription. Future Virol 2007, 2:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards MR, Pietzsch C, Vausselin T, Shaw ML, Bukreyev A, Basler CF: High-Throughput Minigenome System for Identifying Small-Molecule Inhibitors of Ebola Virus Replication. ACS Infect Dis 2015, 1:380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uebelhoer LS, Albarino CG, McMullan LK, Chakrabarti AK, Vincent JP, Nichol ST, Towner JS: High-throughput, luciferase-based reverse genetics systems for identifying inhibitors of Marburg and Ebola viruses. Antiviral Res 2014, 106:86–94. [DOI] [PubMed] [Google Scholar]
- 19.Nelson EV, Pacheco JR, Hume AJ, Cressey TN, Deflube LR, Ruedas JB, Connor JH, Ebihara H, Muhlberger E: An RNA polymerase II-driven Ebola virus minigenome system as an advanced tool for antiviral drug screening. Antiviral Res 2017, 146:21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S: Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol 1999, 73:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luthra P, Jordan DS, Leung DW, Amarasinghe GK, Basler CF: Ebola virus VP35 interaction with dynein LC8 regulates viral RNA synthesis. J Virol 2015,89:5148–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, et al. : Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 2013,14:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith DR, McCarthy S, Chrovian A, Olinger G, Stossel A, Geisbert TW, Hensley LE, Connor JH: Inhibition of heat-shock protein 90 reduces Ebola virus replication. Antiviral Res 2010, 87:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolesnikova L, Nanbo A, Becker S, Kawaoka Y: Inside the Cell: Assembly of Filoviruses. Curr Top Microbiol Immunol 2017,411:353–380. [DOI] [PubMed] [Google Scholar]
- 25.Olejnik J, Hume AJ, Leung DW, Amarasinghe GK, Basler CF, Muhlberger E: Filovirus Strategies to Escape Antiviral Responses. Curr Top Microbiol Immunol 2017,411:293–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basler CF, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk HD, Garcia-Sastre A, Palese P: The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A 2000,97:12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung DW, Prins KC, Borek DM, Farahbakhsh M, Tufariello JM, Ramanan P, Nix JC, Helgeson LA, Otwinowski Z, Honzatko RB, et al. : Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol 2010,17:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateo M, Reid SP, Leung LW, Basler CF, Volchkov VE: Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J Virol 2010,84:1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reid SP, Leung LW, Hartman AL, Martinez O, Shaw ML, Carbonnelle C, Volchkov VE, Nichol ST, Basler CF: Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J Virol 2006,80:5156–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF: Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J Virol 2007,81:13469–13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W, Edwards MR, Borek DM, Feagins AR, Mittal A, Alinger JB, Berry KN, Yen B, Hamilton J, Brett TJ, et al. : Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 2014,16:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valmas C, Basler CF: Marburg virus VP40 antagonizes interferon signaling in a species-specific manner. J Virol 2011,85:4309–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valmas C, Grosch MN, Schumann M, Olejnik J, Martinez O, Best SM, Krahling V, Basler CF, Muhlberger E: Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog 2010,6:e1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feagins AR, Basler CF: Lloviu virus VP24 and VP35 proteins function as innate immune antagonists in human and bat cells. Virology 2015,485:145–152. [DOI] [PubMed] [Google Scholar]
- 35.Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF: The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep 2014,6:1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson B, Li J, Adhikari J, Edwards MR, Zhang H, Schwarz T, Leung DW, Basler CF, Gross ML, Amarasinghe GK: Dimerization Controls Marburg Virus VP24-dependent Modulation of Host Antioxidative Stress Responses. J Mol Biol 2016, 428:3483–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page A, Volchkova VA, Reid SP, Mateo M, Bagnaud-Baule A, Nemirov K, Shurtleff AC, Lawrence P, Reynard O, Ottmann M, et al. : Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep 2014, 6:1026–1036. [DOI] [PubMed] [Google Scholar]
- 38.Hoenen T, Watt A, Mora A, Feldmann H: Modeling the lifecycle of Ebola virus under biosafety level 2 conditions with virus-like particles containing tetracistronic minigenomes. J Vis Exp 2014:52381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bente D, Gren J, Strong JE, Feldmann H: Disease modeling for Ebola and Marburg viruses. Dis Model Mech 2009, 2:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. : Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514:47–53.This study demonstrated that an anti-glycoprotein monoclonal antibody cocktail was effective even after onset of disease symptoms in non-human primates.
- 41.Bixler SL, Bocan TM, Wells J, Wetzel KS, Van Tongeren SA, Dong L, Garza NL, Donnelly G, Cazares LH, Nuss J, et al. : Efficacy of favipiravir (T-705) in nonhuman primates infected with Ebola virus or Marburg virus. Antiviral Res 2018, 151:97–104. [DOI] [PubMed] [Google Scholar]
- *42.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, et al. : Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531:381–385.GS-5734 has advanced to clinical trials in humans. This study paved the way for the advancement of GS-5734 by demonstraing efficacy in non-human primates.
- *43.Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, et al. : Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014, 508:402–405.This study privded the first proof that small molecule inhibitors could prove effective to treat filovirus disease in non-human primates.
- 44.Cho A, Saunders OL, Butler T, Zhang L, Xu J, Vela JE, Feng JY, Ray AS, Kim CU: Synthesis and antiviral activity of a series of 1’-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett 2012, 22:2705–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang L, Neville S, Carra E, Lew W, Ross B, et al. : Discovery and Synthesis of a Phosphoramidate Prodrug of a Pyrrolo[2,1-f][triazin-4-amino] Adenine C-Nucleoside (GS-5734) for the Treatment of Ebola and Emerging Viruses. J Med Chem 2017, 60:1648–1661. [DOI] [PubMed] [Google Scholar]
- 46.Dornemann J, Burzio C, Ronsse A, Sprecher A, De Clerck H, Van Herp M, Kolie MC, Yosifiva V, Caluwaerts S, McElroy AK, et al. : First Newborn Baby to Receive Experimental Therapies Survives Ebola Virus Disease. J Infect Dis 2017, 215:171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL: Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res 2013, 100:446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oestereich L, Ludtke A, Wurr S, Rieger T, Munoz-Fontela C, Gunther S: Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res 2014, 105:17–21. [DOI] [PubMed] [Google Scholar]
- 49.Smither SJ, Eastaugh LS, Steward JA, Nelson M, Lenk RP, Lever MS: Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antiviral Res 2014, 104:153–155. [DOI] [PubMed] [Google Scholar]
- 50.Sissoko D, Laouenan C, Folkesson E, M’Lebing AB, Beavogui AH, Baize S, Camara AM, Maes P, Shepherd S, Danel C, et al. : Experimental Treatment with Favipiravir for Ebola Virus Disease (the JIKI Trial): A Historically Controlled, Single-Arm Proof-of-Concept Trial in Guinea. PLoS Med 2016, 13:e1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai CQ, Mu JS, Kargbo D, Song YB, Niu WK, Nie WM, Kanu A, Liu WW, Wang YP, Dafae F, et al. : Clinical and Virological Characteristics of Ebola Virus Disease Patients Treated With Favipiravir (T-705)-Sierra Leone, 2014. Clin Infect Dis 2016, 63:1288–1294. [DOI] [PubMed] [Google Scholar]
- 52.Bray M, Driscoll J, Huggins JW: Treatment of lethal Ebola virus infection in mice with a single dose of an S-adenosyl-L-homocysteine hydrolase inhibitor. Antiviral Res 2000, 45:135–147. [DOI] [PubMed] [Google Scholar]
- 53.Huggins J, Zhang ZX, Bray M: Antiviral drug therapy of filovirus infections: S-adenosylhomocysteine hydrolase inhibitors inhibit Ebola virus in vitro and in a lethal mouse model. J Infect Dis 1999, 179 Suppl 1:S240–247. [DOI] [PubMed] [Google Scholar]
- 54.Ye W, Schneller SW: The enantiomers of the 1’,6’-isomer of neplanocin A: synthesis and antiviral properties. Bioorg Med Chem 2014, 22:5315–5319. [DOI] [PubMed] [Google Scholar]
- 55.De Clercq E: Antiviral and antimetabolic activities of neplanocins. Antimicrob Agents Chemother 1985, 28:84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C, Chen Q, Schneller SW: Enantiomeric 3-deaza-1’,6’-isoneplanocin and its 3-bromo analogue: Synthesis by the Ullmann reaction and their antiviral properties. Bioorg Med Chem Lett 2016, 26:928–930. [DOI] [PubMed] [Google Scholar]
- 57.Liu C, Chen Q, Cardinale S, Bowlin TL, Schneller SW: 6’-Fluoro-3-deazaneplanocin: Synthesis and antiviral properties, including Ebola. Bioorg Med Chem Lett 2018, 28:3674–3675. [DOI] [PubMed] [Google Scholar]
- 58.De Clercq E: Antivirals and antiviral strategies. Nat Rev Microbiol 2004, 2:704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welch SR, Guerrero LW, Chakrabarti AK, McMullan LK, Flint M, Bluemling GR, Painter GR, Nichol ST, Spiropoulou CF, Albarino CG: Lassa and Ebola virus inhibitors identified using minigenome and recombinant virus reporter systems. Antiviral Res 2016, 136:9–18. [DOI] [PubMed] [Google Scholar]
- 60.Reynard O, Nguyen XN, Alazard-Dany N, Barateau V, Cimarelli A, Volchkov VE: Identification of a New Ribonucleoside Inhibitor of Ebola Virus Replication. Viruses 2015, 7:6233–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luthra P, Liang J, Pietzsch CA, Khadka S, Edwards MR, Wei S, De S, Posner B, Bukreyev A, Ready JM, et al. : A high throughput screen identifies benzoquinoline compounds as inhibitors of Ebola virus replication. Antiviral Res 2018, 150:193–201. [DOI] [PubMed] [Google Scholar]
- 62.Connor JH, McKenzie MO, Parks GD, Lyles DS: Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 2007, 362:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olsen ME, Filone CM, Rozelle D, Mire CE, Agans KN, Hensley L, Connor JH: Polyamines and Hypusination Are Required for Ebolavirus Gene Expression and Replication. MBio 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olsen ME, Cressey TN, Muhlberger E, Connor JH: Differential Mechanisms for the Involvement of Polyamines and Hypusinated eIF5A in Ebola Virus Gene Expression. J Virol 2018, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luthra P, Naidoo J, Pietzsch CA, De S, Khadka S, Anantpadma M, Williams CG, Edwards MR, Davey RA, Bukreyev A, et al. : Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antiviral Res 2018, 158:288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harvey R, Brown K, Zhang Q, Gartland M, Walton L, Talarico C, Lawrence W, Selleseth D, Coffield N, Leary J, et al. : GSK983: a novel compound with broad-spectrum antiviral activity. Antiviral Res 2009, 82:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin S, Chiramel AI, Schmidt ML, Chen YC, Whitt N, Watt A, Dunham EC, Shifflett K, Traeger S, Leske A, et al. : A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med 2018, 10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grandin C, Hourani ML, Janin YL, Dauzonne D, Munier-Lehmann H, Paturet A, Taborik F, Vabret A, Contamin H, Tangy F, et al. : Respiratory syncytial virus infection in macaques is not suppressed by intranasal sprays of pyrimidine biosynthesis inhibitors. Antiviral Res 2016, 125:58–62. [DOI] [PubMed] [Google Scholar]
- 69.Wang QY, Bushell S, Qing M, Xu HY, Bonavia A, Nunes S, Zhou J, Poh MK, Florez de Sessions P, Niyomrattanakit P, et al. : Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J Virol 2011, 85:6548–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang C, Chu M: Leflunomide: A promising drug with good antitumor potential. Biochem Biophys Res Commun 2018, 496:726–730. [DOI] [PubMed] [Google Scholar]
- *71.Modrof J, Muhlberger E, Klenk HD, Becker S: Phosphorylation of VP30 impairs ebola virus transcription. J Biol Chem 2002, 277:33099–33104.Demonstrated that the phosphorylation state of VP30 is a potential target for therapeutic intervention.
- 72.Ilinykh PA, Tigabu B, Ivanov A, Ammosova T, Obukhov Y, Garron T, Kumari N, Kovalskyy D, Platonov MO, Naumchik VS, et al. : Role of protein phosphatase 1 in dephosphorylation of Ebola virus VP30 protein and its targeting for the inhibition of viral transcription. J Biol Chem 2014, 289:22723–22738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tigabu B, Ramanathan P, Ivanov A, Lin X, Ilinykh PA, Parry CS, Freiberg AN, Nekhai S, Bukreyev A: Phosphorylated Vp30 of Marburg Virus Is a Repressor of Transcription. J Virol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *74.Batra J, Hultquist JF, Liu D, Shtanko O, Von Dollen J, Satkamp L, Jang GM, Luthra P, Schwarz TM, Small GI, et al. : Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell 2018, 175:1917–1930 e1913.Identified novel virus-host interactions including an interaction between viral protein VP30 and host protein RBBP6 that is critical for viral RNA synthesis. Targeting the interaction with peptides demosntrated that the interaction could be a therapeutic target.
- 75.Leung DW, Borek D, Luthra P, Binning JM, Anantpadma M, Liu G, Harvey IB, Su Z, Endlich-Frazier A, Pan J, et al. : An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions. Cell Rep 2015, 11:376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Politi C, Ciccacci C, Novelli G, Borgiani P: Genetics and Treatment Response in Parkinson’s Disease: An Update on Pharmacogenetic Studies. Neuromolecular Med 2018, 20:1–17. [DOI] [PubMed] [Google Scholar]
- *77.Liu G, Nash PJ, Johnson B, Pietzsch C, Ilagan MX, Bukreyev A, Basler CF, Bowlin TL, Moir DT, Leung DW, et al. : A Sensitive in Vitro High-Throughput Screen To Identify Pan-filoviral Replication Inhibitors Targeting the VP35-NP Interface. ACS Infect Dis 2017, 3:190–198.Demonstrated that a virus protein-protein interface can be effectively targeted with a small molecule drug.
- 78.Moller-Tank S, Maury W: Ebola virus entry: a curious and complex series of events. PLoS Pathog 2015, 11:e1004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H, Schnittler HJ: Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis 2011, 204 Suppl 3:S957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y: Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 2010, 6:e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA: Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog 2010, 6:e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mulherkar N, Raaben M, de la Torre JC, Whelan SP, Chandran K: The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology 2011, 419:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM: Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005, 308:1643–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J: Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 2006, 80:4174–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM: The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J Virol 2009, 83:2883–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bornholdt ZA, Ndungo E, Fusco ML, Bale S, Flyak AI, Crowe JE Jr., Chandran K, Saphire EO: Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies. MBio 2016, 7:e02154–02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marzi A, Reinheckel T, Feldmann H: Cathepsin B & L are not required for ebola virus replication. PLoS Negl Trop Dis 2012, 6:e1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez O, Johnson J, Manicassamy B, Rong L, Olinger GG, Hensley LE, Basler CF: Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cell Microbiol 2010, 12:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K: A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J Virol 2010, 84:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *90.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. : Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011, 477:340–343.Demonstrated that NPC1 is essential for Ebola virus entry.
- *91.Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, et al. : Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011, 477:344–348.Demonstrated that NPC1 is essential for Ebola virus entry and that NPC1-dependent enty can be targeted by small molecules.
- 92.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, et al. : Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 2012, 31:1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basu A, Li B, Mills DM, Panchal RG, Cardinale SC, Butler MM, Peet NP, Majgier-Baranowska H, Williams JD, Patel I, et al. : Identification of a small-molecule entry inhibitor for filoviruses. J Virol 2011, 85:3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vigant F, Lee J, Hollmann A, Tanner LB, Akyol Ataman Z, Yun T, Shui G, Aguilar HC, Zhang D, Meriwether D, et al. : A mechanistic paradigm for broad-spectrum antivirals that target virus-cell fusion. PLoS Pathog 2013, 9:e1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, et al. : A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A 2010, 107:3157–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boriskin YS, Leneva IA, Pecheur EI, Polyak SJ: Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr Med Chem 2008, 15:997–1005. [DOI] [PubMed] [Google Scholar]
- 97.Pecheur EI, Borisevich V, Halfmann P, Morrey JD, Smee DF, Prichard M, Mire CE, Kawaoka Y, Geisbert TW, Polyak SJ: The Synthetic Antiviral Drug Arbidol Inhibits Globally Prevalent Pathogenic Viruses. J Virol 2016, 90:3086–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Teissier E, Zandomeneghi G, Loquet A, Lavillette D, Lavergne JP, Montserret R, Cosset FL, Bockmann A, Meier BH, Penin F, et al. : Mechanism of inhibition of enveloped virus membrane fusion by the antiviral drug arbidol. PLoS One 2011, 6:e15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kadam RU, Wilson IA: Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc Natl Acad Sci U S A 2017, 114:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anantpadma M, Kouznetsova J, Wang H, Huang R, Kolokoltsov A, Guha R, Lindstrom AR, Shtanko O, Simeonov A, Maloney DJ, et al. : Large-Scale Screening and Identification of Novel Ebola Virus and Marburg Virus Entry Inhibitors. Antimicrob Agents Chemother 2016, 60:4471–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R Jr., Nunneley JW, Barnard D, Pohlmann S, McKerrow JH, Renslo AR, et al. : Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 2015, 116:76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gnirss K, Kuhl A, Karsten C, Glowacka I, Bertram S, Kaup F, Hofmann H, Pohlmann S: Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology 2012, 424:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barrientos LG, Rollin PE: Release of cellular proteases into the acidic extracellular milieu exacerbates Ebola virus-induced cell damage. Virology 2007, 358:1–9. [DOI] [PubMed] [Google Scholar]
- 104.Shah PP, Wang T, Kaletsky RL, Myers MC, Purvis JE, Jing H, Huryn DM, Greenbaum DC, Smith AB 3rd, Bates P, et al. : A small-molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and ebola pseudotype virus infection into human embryonic kidney 293T cells. Mol Pharmacol 2010, 78:319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaletsky RL, Simmons G, Bates P: Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol 2007, 81:13378–13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.van der Linden WA, Schulze CJ, Herbert AS, Krause TB, Wirchnianski AA, Dye JM, Chandran K, Bogyo M: Cysteine Cathepsin Inhibitors as Anti-Ebola Agents. ACS Infect Dis 2016, 2:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang X, Liu Q, Zhang N, Li QQ, Liu ZD, Li YH, Gao LM, Wang YC, Deng HB, Song DQ: Discovery and evolution of aloperine derivatives as novel anti-filovirus agents through targeting entry stage. Eur J Med Chem 2018, 149:45–55. [DOI] [PubMed] [Google Scholar]
- 108.Elshabrawy HA, Fan J, Haddad CS, Ratia K, Broder CC, Caffrey M, Prabhakar BS: Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay. J Virol 2014, 88:4353–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Long J, Wright E, Molesti E, Temperton N, Barclay W: Antiviral therapies against Ebola and other emerging viral diseases using existing medicines that block virus entry. F1000Res 2015, 4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Madrid PB, Chopra S, Manger ID, Gilfillan L, Keepers TR, Shurtleff AC, Green CE, Iyer LV, Dilks HH, Davey RA, et al. : A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One 2013, 8:e60579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vanier MT: Niemann-Pick disease type C. Orphanet J Rare Dis 2010, 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu F, Liang Q, Abi-Mosleh L, Das A, De Brabander JK, Goldstein JL, Brown MS: Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shoemaker CJ, Schornberg KL, Delos SE, Scully C, Pajouhesh H, Olinger GG, Johansen LM, White JM: Multiple cationic amphiphiles induce a Niemann-Pick C phenotype and inhibit Ebola virus entry and infection. PLoS One 2013, 8:e56265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Herbert AS, Davidson C, Kuehne AI, Bakken R, Braigen SZ, Gunn KE, Whelan SP, Brummelkamp TR, Twenhafel NA, Chandran K, et al. : Niemann-pick C1 is essential for ebolavirus replication and pathogenesis in vivo. MBio 2015, 6:e00565–00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johansen LM, Brannan JM, Delos SE, Shoemaker CJ, Stossel A, Lear C, Hoffstrom BG, Dewald LE, Schornberg KL, Scully C, et al. : FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci Transl Med 2013, 5:190ra179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kouznetsova J, Sun W, Martinez-Romero C, Tawa G, Shinn P, Chen CZ, Schimmer A, Sanderson P, McKew JC, Zheng W, et al. : Identification of 53 compounds that block Ebola virus-like particle entry via a repurposing screen of approved drugs. Emerg Microbes Infect 2014, 3:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johansen LM, DeWald LE, Shoemaker CJ, Hoffstrom BG, Lear-Rooney CM, Stossel A, Nelson E, Delos SE, Simmons JA, Grenier JM, et al. : A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med 2015, 7:290ra289. [DOI] [PubMed] [Google Scholar]
- 118.Zhao Y, Ren J, Harlos K, Jones DM, Zeltina A, Bowden TA, Padilla-Parra S, Fry EE, Stuart DI: Toremifene interacts with and destabilizes the Ebola virus glycoprotein. Nature 2016, 535:169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun W, He S, Martinez-Romero C, Kouznetsova J, Tawa G, Xu M, Shinn P, Fisher E, Long Y, Motabar O, et al. : Synergistic drug combination effectively blocks Ebola virus infection. Antiviral Res 2017, 137:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *120.Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, Klugbauer N, Grimm C, Wahl-Schott C, Biel M, Davey RA: Ebola virus. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science 2015, 347:995–998.Demonstrated that two-pore channels are important for virus trafficking during the entry process and that these can be targeted with small molecules.
- 121.Gehring G, Rohrmann K, Atenchong N, Mittler E, Becker S, Dahlmann F, Pohlmann S, Vondran FW, David S, Manns MP, et al. : The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J Antimicrob Chemother 2014, 69:2123–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salata C, Baritussio A, Munegato D, Calistri A, Ha HR, Bigler L, Fabris F, Parolin C, Palu G, Mirazimi A: Amiodarone and metabolite MDEA inhibit Ebola virus infection by interfering with the viral entry process. Pathog Dis 2015, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nelson EA, Dyall J, Hoenen T, Barnes AB, Zhou H, Liang JY, Michelotti J, Dewey WH, DeWald LE, Bennett RS, et al. : The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Negl Trop Dis 2017, 11:e0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sands BE, Jacobson EW, Sylwestrowicz T, Younes Z, Dryden G, Fedorak R, Greenbloom S: Randomized, double-blind, placebo-controlled trial of the oral interleukin-12/23 inhibitor apilimod mesylate for treatment of active Crohn’s disease. Inflamm Bowel Dis 2010, 16:1209–1218. [DOI] [PubMed] [Google Scholar]
- 125.Burakoff R, Barish CF, Riff D, Pruitt R, Chey WY, Farraye FA, Shafran I, Katz S, Krone CL, Vander Vliet M, et al. : A phase 1/2A trial of STA 5326, an oral interleukin-12/23 inhibitor, in patients with active moderate to severe Crohn’s disease. Inflamm Bowel Dis 2006, 12:558–565. [DOI] [PubMed] [Google Scholar]
- 126.Krausz S, Boumans MJ, Gerlag DM, Lufkin J, van Kuijk AW, Bakker A, de Boer M, Lodde BM, Reedquist KA, Jacobson EW, et al. : Brief report: a phase IIa, randomized, double-blind, placebo-controlled trial of apilimod mesylate, an interleukin-12/interleukin-23 inhibitor, in patients with rheumatoid arthritis. Arthritis Rheum 2012, 64:1750–1755. [DOI] [PubMed] [Google Scholar]
- 127.Wada Y, Cardinale I, Khatcherian A, Chu J, Kantor AB, Gottlieb AB, Tatsuta N, Jacobson E, Barsoum J, Krueger JG: Apilimod inhibits the production of IL-12 and IL-23 and reduces dendritic cell infiltration in psoriasis. PLoS One 2012, 7:e35069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kolokoltsov AA, Adhikary S, Garver J, Johnson L, Davey RA, Vela EM: Inhibition of Lassa virus and Ebola virus infection in host cells treated with the kinase inhibitors genistein and tyrphostin. Arch Virol 2012, 157:121–127. [DOI] [PubMed] [Google Scholar]
- 129.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN: Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol 2003, 77:1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J: Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol 2007, 81:4895–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Urata S, Yasuda J: Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J Gen Virol 2010, 91:228–234. [DOI] [PubMed] [Google Scholar]
- 132.Liu Y, Lee MS, Olson MA, Harty RN: Bimolecular Complementation to Visualize Filovirus VP40-Host Complexes in Live Mammalian Cells: Toward the Identification of Budding Inhibitors. Adv Virol 2011, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, et al. : Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol 2014, 88:7294–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Loughran HM, Han Z, Wrobel JE, Decker SE, Ruthel G, Freedman BD, Harty RN, Reitz AB: Quinoxaline-based inhibitors of Ebola and Marburg VP40 egress. Bioorg Med Chem Lett 2016, 26:3429–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]