Abstract
Background:
Epigenetic clocks have been suggested to capture one feature of the complexity between aging and the epigenome. However, little is known about the epigenetic clock in childhood allergy and asthma.
Objective:
To examine associations of DNA methylation age (DNAmAge) and epigenetic age acceleration with childhood allergy and asthma.
Methods:
We calculated DNAmAge and age acceleration at birth, early and mid-childhood based on IlluminaHumanMethylation450BeadChip in Project Viva. We evaluated epigenetic clock associations with allergy and asthma using covariate-adjusted linear- and logistic-regressions. We attempted to replicate our findings in the ‘Genetics of Asthma in Costa Rica Study’.
Results:
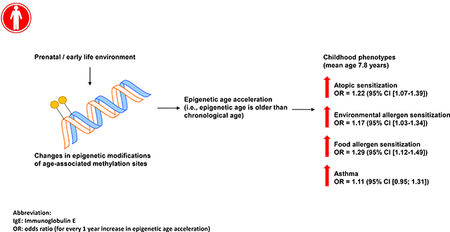
At mid-childhood (mean age=7.8 years) in Project Viva, DNAmAge and age acceleration were cross-sectionally associated with higher total serum IgE, and greater odds of atopic sensitization. Every 1-year increase in intrinsic epigenetic age acceleration was associated with a 1.22 (95% CI [1.07–1.39]), 1.17 (95% CI [1.03–1.34]), and 1.29 (95% CI [1.12–1.49]) higher odds of atopic sensitization, environmental and food allergen sensitization. DNAmAge and extrinsic epigenetic age acceleration were also cross-sectionally associated with current asthma at mid-childhood. DNAmAge and age acceleration at birth and early-childhood were not associated with mid-childhood allergy or asthma. The mid-childhood association between age acceleration and atopic sensitization were replicated in an independent dataset.
Conclusions:
As the epigenetic clock may reflect the immuno-and-developmental components of biological aging, our study suggests pathways through which molecular epigenetic mechanisms of immunity, development and maturation may interact along the age-axis, and associate with childhood allergy and asthma by mid-childhood.
Keywords: epigenetic clock, allergy, asthma, DNA methylation
Capsule summary
Accelerated epigenetic aging may reflect perturbations in epigenetic maintenance. Research should focus on finding determinants of epigenetic aging in childhood and the clinical potential of interventions targeting aging pathways in children with allergy, atopy and asthma.
Graphical Abstract:
Introduction
DNA methylation serves as a molecular marker for biological aging and its manifestations (1). An increasing body of evidence suggests that DNA methylation derived epigenetic clock metrics (which include DNA methylation age (DNAmAge) and epigenetic age acceleration) cross-sectionally associate with adult diseases including physical and cognitive decline (2), and Parkinson’s disease (3); and are predictive of future onsets of lung cancer (4). These epigenetic clock metrics have also predicted cancer, cardiovascular and all-cause mortality independent of chronological age (5–8). More recently, research on the epigenetic clock has been expanded into the early-life spectrum (9–12). Studies have demonstrated that birth weight (9), birth by Caesarean section (9), and other pregnancy-related exposures (maternal smoking, body mass index (BMI), selenium and cholesterol levels) (9) associate with higher DNAmAge in children (age range 7 to 19). Epigenetic age acceleration residuals (the residuals from a linear regression of DNA methylation age on chronological age) are also prospectively associated with faster increase in weight during childhood and adolescence (10), and are predictive of earlier pubertal development in young girls (12). However, epigenetic age and epigenetic age acceleration residuals have not been studied in association with allergy and asthma in children. Further, two other epigenetic clock calculations—intrinsic epigenetic age acceleration (IEAA) and extrinsic epigenetic age acceleration (EEAA)—have also not been linked to childhood allergic diseases. Intrinsic epigenetic age acceleration measures epigenetic aging independent of major blood immune cell counts, while EEAA weights blood cell types that are known to change with age, including naïve cytotoxic T cells (CD45RA+ CCR7+), exhausted cytotoxic T cells (CD28-CD45RA-), and plasma B cells, and measures epigenetic aging in immune-related domains (8, 13, 14).
The development of allergic diseases is likely age-dependent during childhood, beginning in some children with food allergy and atopic dermatitis (infancy to early-childhood), followed by allergic asthma and rhinitis (early to mid-childhood) (15–17). However, chronological age alone does not fully explain disease variability. Epigenetic mechanisms may mark allergic disease susceptibility, and play a role in the onset, progression and manifestations of allergy and asthma (18–22). Studying epigenetic signatures at birth or in early childhood has also been postulated as a molecular approach to distinguish transient wheezing from early asthma symptoms, and help to define age-associated disease endotypes (23). Since the epigenetic clock likely captures a higher order property of the methylome (24) and reflects the immunologic and developmental components of biological aging, DNA methylation age (DNAmAge) may serve as a molecular marker of allergy and asthma in children.
In this study, we sought to evaluate the associations of DNAmAge and epigenetic age acceleration (estimated at birth, early-childhood and mid-childhood) with mid-childhood allergic phenotypes (including total serum IgE, atopic sensitization, environmental allergen sensitization, food allergen sensitization, and asthma) in Project Viva—a longitudinal pre-birth cohort in Boston, Massachusetts. We considered both extrinsic epigenetic age acceleration (EEAA) and intrinsic epigenetic age acceleration (IEAA) to best capture the age-associated metrics reflective and independent of age-related changes in cell count, respectively (3, 8, 13). We hypothesized that older DNAmAge and age acceleration (including IEAA and EEAA) are associated with higher risks of mid-childhood allergy and asthma. We further postulated that maternal lifestyle factors during pregnancy may impact the epigenetic clock trajectory toward allergen sensitization in mid-childhood.
Methods
Study Population
Our study included mother-child pairs from Project Viva, a prospective longitudinal pre-birth cohort in Boston, Massachusetts. A detailed description of the protocol has been published elsewhere (25). In brief, Project Viva enrolled 2670 pregnant women at their initial obstetrical visit at Atrius Harvard Vanguard Medical Associates in Boston Massachusetts between 19992002 (2128 women had live births). We included women with single gestation, gestational age <22 weeks at enrollment, able to answer questions in English and intended to stay in the study region before delivery. We performed in-person visits during pregnancy, after delivery, during infancy, and at early and mid-childhood.
We collected information on maternal age, smoking habits (never smoker / former smoker / smoked during pregnancy), education status, marital status and history of allergy (asthma, eczema or hay fever) at enrollment. Child date of birth and sex were extracted from hospital records. We calculated gestational age by subtracting the date of the last menstrual period (LMP) from the date of delivery; in situations in which gestational age based on the second trimester ultrasound was greater than 10 days different from that by the LMP, we estimated gestational age based on ultrasound results (26). Mothers reported child race / ethnicity at the early childhood visit. We recorded child age at blood drawn during early and mid-childhood visits. We defined women who never smoked or smoked <100 cigarettes in their lifetime as “never smokers”; women who smoked >100 cigarettes in their lifetime but quit smoking 3 months or more before learning they were pregnant as “former smokers”; women who smoked >100 cigarettes and continued to smoke during the 3 months before learning they were pregnant or reported smoking on the first or second trimester questionnaire as “smokers during pregnancy” (27).
Of the 2128 mother-child pairs enrolled, 485 samples had cord blood DNA methylation measurements, 120 children had peripheral blood DNA methylation measurements at early-childhood, and 460 children had peripheral blood DNA methylation measurements at mid-childhood. Of the 460 children with mid-childhood methylation measurements, we excluded 52 participants, as follows: 51 mother-child pairs with missing information on covariates or allergy measurements at mid-childhood, and 1 participant who had a DNAmAge at mid-childhood that was greater than 20. Thus, 408 mother-child pairs were included in the mid-childhood analysis.
DNA methylation age and age acceleration calculation
Trained personnel collected blood samples at birth (umbilical cord blood), early and mid-childhood (peripheral blood). We extracted DNA from cord blood and child peripheral blood using Qiagen Puregene Kits (Valencia, CA) and performed bisulfite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). Samples were randomized by chips and plates and we generated DNA methylation data using the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA). We removed samples that were technical replicates, samples with low quality, and samples with genotype or sex mismatches. We removed low quality probes (detection p-value >0.05), probes on sex chromosomes, 65 SNP probes, probes within 10 base pairs of a known SNP with minor allele frequency ≥ 1%, non-CpG probes, as well as non-specific and cross-reactive probes. We applied the normal-exponential out-of-band (“noob”) method for background correction and dye bias adjustment (28), and performed sample normalization using Beta Mixture Quantile Dilation (BMIQ) (29) as described previously (30). We controlled for technical variability using Combat—a non-parametric empirical Bayes method (31). We estimated blood cell proportions in peripheral blood from an adult reference panel (Houseman) (32) and calculated blood cell proportions in cord blood using the Bakulski algorithm, which includes nucleated red blood cell counts (33, 34).
We estimated DNA methylation age (DNAmAge) at birth, early and mid-childhood using the Horvath method (1). This method predicted chronological age using 8,000 samples from 82 Illumina DNA methylation array datasets across a broad spectrum of age and tissue samples, and selected 353 methylation sites predictive of chronological age using elastic-net regularized regression (1). This method assumes a logarithmic dependence of epigenetic age on chronologic age until adulthood (age 20), and a linear relationship after age 20 (1). Based on Horvath’s coefficient estimates and intercept values, we estimated DNAmAge at birth, early and mid-childhood separately. Further, we reported two methylation age acceleration metrics using the Horvath and Hannum methods, which we calculated as (1) the residual resulting from a multivariate regression of Horvath DNAmAge on chronological age and blood cell counts (naïve CD8+ T cells, exhausted CD8+ T cells, plasmblasts, CD4+ T cells, natural killer cells, monocytes, and granulocytes) (intrinsic epigenetic age acceleration IEAA) (8), (2) the weighted average of Hannum’s estimate with 3 immune cell types: naïve (CD45RA+CCR7+) cytotoxic T cells, exhausted (CD28-CD45RA-) cytotoxic T cells, and plasmablasts (extrinsic epigenetic age acceleration, EEAA) (8, 35) at birth, early and mid-childhood. Specifically, for EEAA, (a) we first calculated DNA methylation age from Hannum’s estimation (the Hannum epigenetic clock estimation included 656 human samples, aged 19 to 101, and predicts chronological age based on whole blood methylomic information from Illumina’s 450K BeadChip array using elastic-net regression (36)), (b) we up-weight the contributions of age associated blood cell counts (naïve cytotoxic T cells, exhaust cytotoxic T cells and plasmablast) using the Klemera Doubal approach (3) lastly, the residual variation is calculated from the univariate model regressing the age estimation on chronological age (37). We computed the intra-class correlations (ICCs) of IEAA and EEAA at birth, early and mid-childhood. We also used the Knight et al epigenetic clock for gestational age (DNAm-GA) based on 148 methylation sites selected using elastic-net in 6 training datasets and 9 testing datasets (a total of 1434 neonates from 15 independent birth cohorts) for cord blood (38).
Allergy phenotypes
Our study includes several allergic phenotypes in mid-childhood, including total serum IgE (on a continuous scale), atopic sensitization, environmental allergen and food allergen sensitization and asthma. We measured total serum IgE concentration in peripheral blood using ImmunoCAP (Phadia, Uppsala, Sweden)—a well-established in vitro sandwich immunoassay to quantitatively measure circulating IgE in serum samples. Atopic sensitization was defined as any serum specific IgE level ≥ 0.35 IU/ml to common indoor allergens (dust mite, cat dander, dog dander, cockroach), mold (Aspergillus, Alternaria tenuis), outdoor allergens (rye grass, ragweed) or food allergens (egg white, milk, wheat, peanut, soybean), or total serum IgE greater than or equal to 100 IU/ml; environmental allergen sensitization was defined as any serum specific IgE level ≥ 0.35 IU/ml to common indoor allergens (dust mite, cat dander, dog dander, cockroach), mold (Aspergillus, Alternaria tenuis) or grass pollen (ryegrass, ragweed) (39); and food allergen sensitization was defined as any serum specific IgE level ≥ 0.35 IU/ml to one of the food allergens listed (egg white, milk, wheat, peanut, soybean) (40). Via questionnaire, we defined current asthma in mid-childhood as ever doctor diagnosis of asthma since birth (reported on the mid-childhood questionnaire), plus any use of asthma medications or wheezing in the past 12 months.
Statistical analysis
We evaluated the association of DNAmAge and age acceleration (IEAA and EEAA) at birth, early-childhood (age range = 2.9 to 5.3 years old) and mid-childhood (age range = 6.7–10.2 years old) with mid-childhood total serum IgE using generalized linear regression, assuming a Gaussian distribution with identity link function. We natural log-transformed total serum IgE to reduce skewness of the residuals and meet model assumptions. For the dichotomized outcomes of atopic sensitization, environmental allergen, food allergen sensitization, and asthma, we performed the analyses using generalized linear regression, assuming a binomial distribution with logit link function and report effect estimates as odds ratios. We adjusted for the following covariates selected a priori: maternal [age at enrollment (continuous), smoking status (never, former or during pregnancy), college graduate (yes / no), atopy history (yes / no)], child [sex (female / male), race/ethnicity (white / black / others), gestational age at birth (continuous)], and estimated cell proportions computed from peripheral blood to capture cellular heterogeneity (percentages of monocytes, CD4T cells, CD8Tcells, B cells, and granulocytes). For the cord blood analysis, we adjusted for cell proportions based on the Bakulski method (which includes nucleated red blood cell counts) (33). As a sensitivity analysis, we further adjusted for child’s age at blood draw when examining the association between DNAmAge and allergic outcomes, and tested for effect modification by child sex, race/ethnicity and maternal smoking status. For the cord blood analysis, we also compared analysis results from the Horvath method with the Knight method for DNAm-GA.
We computed Pearson correlations between chronological age and DNAmAge (the Horvath method) at early-childhood and mid-childhood. However, since chronological age was set to zero in the Horvath estimation for cord blood, a direct comparison between chronological age and DNAmAge at birth was not possible. We computed Pearson correlations between gestational age and the Knight DNAm-GA—an epigenetic clock metric specific for gestation. We also calculated the mean and standard deviation (variability) of chronological age and DNAmAge at early and mid-childhood.
We evaluate the association between chronological age at early-childhood and mid-childhood with mid-childhood allergy and asthma, and adjusted for the following covariates selected a prior: maternal [age at enrollment (continuous), smoking status (never, former or during pregnancy), college graduate (yes / no), atopy history (yes / no)], child [sex (female / male), race/ethnicity (white / black / others), gestational age at birth (continuous)]. For the continuous outcome (total serum IgE), we used covariate-adjusted linear regression; for dichotomized outcomes (atopic sensitization, environmental allergen sensitization and food allergen sensitization, and asthma), we used covariate-adjusted logistic regressions.
Maternal social-economic factors and mid-childhood DNAmAge
As an exploratory analysis, we conducted one-way analysis of variance (ANOVA) to examine whether DNAmAge differed by maternal sociodemographic factors, including maternal smoking status, marital status during pregnancy, and maternal education level.
All statistical analyses were performed in R version 3.5.0 (https://www.r-project.org/), except for intra-class correlation calculations, which were performed in SAS 9.4.
Replication
We replicated our findings from Project Viva in a subset of subjects from the ‘Genetics of Asthma in Costa Rica Study’ (GACRS) (41). In brief, from February 2001 to December 2006, screening questionnaires were sent to 13,125 parents whose children were aged 6 to 14 years old and were enrolled in 113 schools in Costa Rica. Children were eligible if they had physician-diagnosed asthma and at least two episodes of respiratory symptoms or asthma attacks in the prior year, and a high probability of having six or more great-grandparents born in the Central Valley of Costa Rica; 592 subjects subsequently underwent genome-wide genotyping as described and the methylation set is a subset of these individuals (41, 42). Our nested study represented 159 subjects with genome-wide DNA methylation measured using Illumina’s 450K platform that passed quality control. We applied “noob” for background correction and dye bias adjustment (28), and performed sample normalization using BMIQ (29). Further, we estimated blood cell proportions in peripheral blood using Houseman’s method (32). Analogous to the epigenetic clock estimations in Project Viva, we calculated DNAmAge, IEAA and EEAA at mid-childhood (mean=9.1; range=(6.0–13.0)) using the Horvath’s epigenetic clock calculator (1). We measured total serum IgE using the UniCAP 250 system (Pharmacia & Upjohn, Kalamazoo, Mich). Atopic sensitization was based on skin test (positive skin test to environmental allergens [D. pteroyssinus, D. farina, Cat, A. tenuis, mixed grasses, mixed trees, P. Americana, dogs, B. Germania, dust mite, and cockroach] or total serum IgE greater than or equal to 100 IU/ml) or serum test (positive serum IgE test to environmental allergens [D. pteroyssinus, cockroach, and Ascaris] and total serum IgE greater than or equal to 100 IU/ml). Given that the cohort was ascertained on the basis of parent report of physician’s diagnosis of asthma, for the analysis of the association between DNAmAge and age acceleration with asthma, we used a strict definition of asthma based on physician, diagnosis, wheeze in the last year and airway responsiveness to methacholine (measured as a 20% decrement in FEV1 (forced expiratory volume in one second) after the administration of ≤16.8 mg/ml of methacholine) or bronchodilator responsiveness. We examined the association of DNAmAge and age acceleration (IEAA and EEAA) at mid-childhood with mid-childhood total serum IgE using generalized linear regression, assuming a Gaussian distribution with an identity link function. We natural log-transformed total serum IgE to reduce skewness of the residuals and meet model assumptions. For binary outcomes (asthma, atopic sensitization), we performed the analyses using generalized linear regression, assuming a binomial distribution with a logit link function and report effect estimates as odds ratios. The study was approved by the Partners Human Research Committee at Brigham and Women’s Hospital (Boston, MA; protocol No. 2000-P-001130/55) and the Hospital Nacional de Niños (San José, Costa Rica).
All statistical analyses were performed in R version 3.5.0 (https://www.r-project.org/), and SAS 9.4.
Results
Study population
Four-hundred and eight children had complete information on mid-childhood DNA methylation measurements and mid-childhood atopic phenotype measurements (Table 1). Ten percent of mothers smoked during pregnancy, 20% were former smokers. Forty percent of mothers reported a history of atopy (including asthma, eczema, and hay fever). Among children, mean gestational age at delivery was 39.6 (± 1.6) weeks, 49% were female, and 63% were white. Mean ± SD age at blood draw at mid-childhood was 7.8 ± 0.7 years. Total serum IgE at mid-childhood showed a right-skewed distribution, with a mean of 152.2 kU/L (± 287.0). Fifty-five percent of children were classified as atopic, 43% were environmental allergen sensitized and 26% were food allergen sensitized. Fifteen percent of children had asthma at mid-childhood.
Table 1.
Characteristics of children and their mothers in Project Viva.
| Participants characteristics | N=408 |
|---|---|
| Maternal | |
| Age (years), mean (SD) | 32.2 (5.6) |
| Smoking status, N (%) | |
| Smoking during pregnancy | 41 (10%) |
| Former smoker | 83 (20%) |
| Never smoker | 284 (70%) |
| Education, N (%) | |
| College or graduate | 270 (66%) |
| Not college or graduate | 138 (34%) |
| Atopy | |
| Yes | 162 (40%) |
| No | 246 (60%) |
| Child (at mid-childhood) | |
| Gestational age (weeks), mean (SD) | 39.6 (1.6) |
| Age at blood drawn (years), mean (SD) | 7.8 (0.7) |
| Sex, N (%) | |
| Female | 201 (49%) |
| Male | 207 (51%) |
| Race, N (%) | |
| White | 255 (63%) |
| Black | 77 (19%) |
| Other | 76 (18%) |
| Total serum IgE (kU/L), mean (SD) | 152.2 (287.0) |
| Atopic sensitization, N (%) | |
| Yes | 224 (55%) |
| No | 184 (45%) |
| Environmental allergen positive, N (%) | |
| Yes | 175 (43%) |
| No | 233 (57%) |
| Food allergen positive, N (%) | |
| Yes | 108 (26%) |
| No | 300 (74%) |
| Current asthma, N (%)† | |
| Yes | 62 (15%) |
| No | 300 (74%) |
number of missing = 46
Association between DNA methylation age and age acceleration at birth, early-childhood and mid-childhood with mid-childhood allergy and asthma
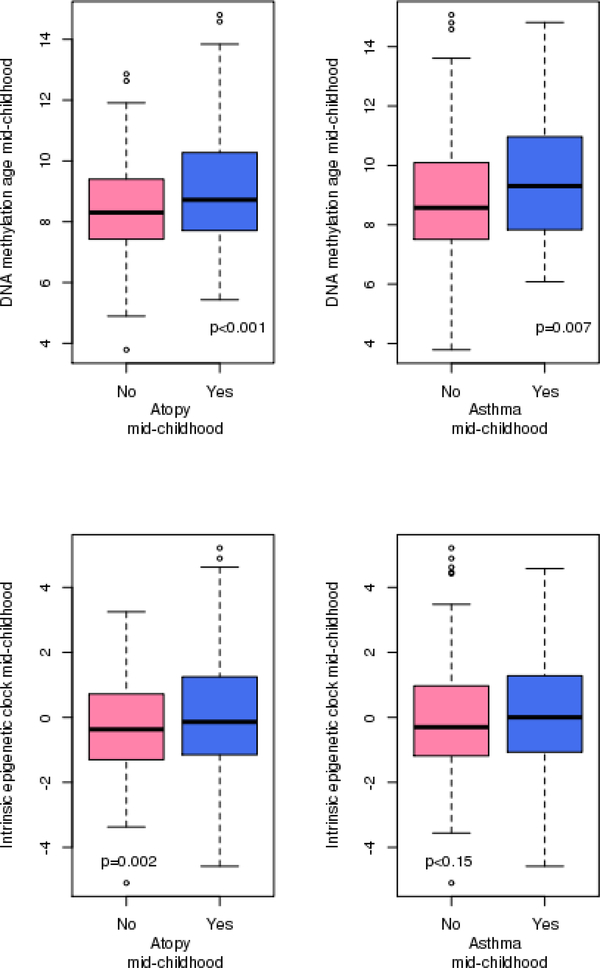
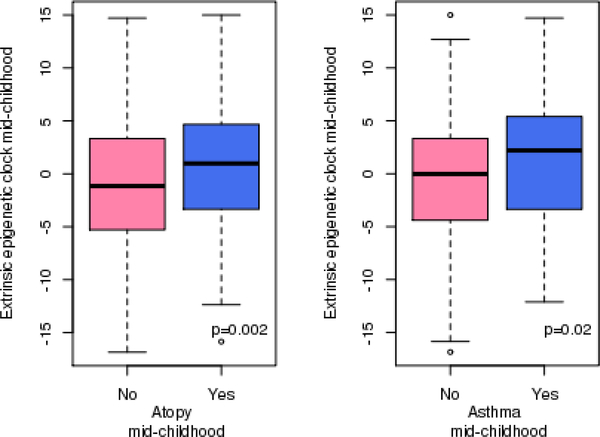
DNAmAge at mid-childhood was cross-sectionally associated with allergy and asthma in mid-childhood (Table 2; Figure 1; Supplementary Figure 1; Supplementary Figure 2). DNAmAge had a mean of 8.8 years old (SD=1.8) at mid-childhood (Supplementary Table 2). A one-year increase in DNAmAge was associated with a 1.21 (95% CI [1.07; 1.38%], p-value=0.003), 1.20 (95% CI [1.06; 1.37], p-value=0.004), 1.21 (95% CI [1.06; 1.39], p-value=0.006) and 1.16 (95% CI [1.00; 1.34], p-value=0.044) higher of odds of atopic sensitization, environmental allergen and food allergen, and asthma, respectively (Table 2). Additionally, adjusting for chronological age did not alter the results (Supplementary Table 1). In addition to DNAmAge, epigenetic age accelerations (IEAA and EEAA) in mid-childhood were also cross-sectionally associated with the allergic phenotypes at mid-childhood (Table 2). For example, for every 1-year increase in IEAA, we observed a 1.22 (95% CI [(1.07; 1.39), p-value=0.004], 1.17 (95% CI [(1.03; 1.34), p-value=0.018], and 1.29 (95 CI [1.12; 1.49), p-value<0.0001) higher odds of atopic sensitization, environmental allergen and food allergen, respectively (Table 2). We observed relatively high correlations between DNAmAge and IEAA in mid-childhood (Supplementary Table 2). The extrinsic epigenetic age acceleration at mid-childhood—which is thought to capture immune development and response—was additionally associated with asthma at mid-childhood (OR: 1.07; 95% CI [(1.02; 1.12), p-value=0.008)]). We did not observe effect modification by gender, race/ethnicity or maternal smoking status with a p-value threshold of 0.05.
Table 2.
Cross-sectional associations between DNA methylation age, intrinsic epigenetic age acceleration (IEAA) and extrinsic epigenetic age acceleration (EEAA) with total serum IgE, atopic sensitization, environmental allergen sensitization, food allergen sensitization and asthma measured at mid-childhood.
| “Epigenetic clock” metrics at mid-childhood |
||||||
|---|---|---|---|---|---|---|
| DNA methylation age | IEAA (intrinsic) | EEAA (extrinsic) | ||||
| Mid-childhood outcomes | Estimate (95% CI) | P value | Estimate (95% CI) | P value | Estimate (95% CI) | P value |
| Total serum IgE (log-scale) | 0.11 (0.02; 0.20) | 0.017 | 0.13 (0.03; 0.22) | 0.010 | 0.024 (−0.002; 0.05) | 0.076 |
| Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | |
| Atopic sensitization | 1.21 (1.07; 1.38) | 0.003 | 1.22 (1.07; 1.39) | 0.004 | 1.06 (1.02; 1.10) | 0.002 |
| Environmental allergen sensitization | 1.20 (1.06; 1.37) | 0.004 | 1.17 (1.03; 1.34) | 0.018 | 1.07 (1.03; 1.11) | <0.001 |
| Food allergen sensitization | 1.21 (1.06; 1.39) | 0.006 | 1.29 (1.12; 1.49) | <0.001 | 1.04 (1.00; 1.08) | 0.055 |
| Current asthma | 1.16 (1.00; 1.34) | 0.044 | 1.11 (0.95; 1.31) | 0.193 | 1.07 (1.02; 1.12) | 0.008 |
For DNAmAge, model adjusted for maternal [age at enrollment (continuous), smoking status (never, former, or during pregnancy), education (college / graduate level or not college / graduate level), and atopy history (asthma, eczema or hay fever)], and for child [sex (female or male), race / ethnicity (white, black or others), gestational age (continuous)], and cell type proxies estimated for peripheral blood (percentages of monocytes, granulocytes, CD8T cells, CD4T cells, and B cells).
For IEAA and EEAA, models adjusted for maternal [age at enrollment (continuous), smoking status (never, former, or during pregnancy), education (college / graduate level or not college / graduate level), and atopy history (asthma, eczema or hay fever)], child [sex (female or male), race / ethnicity (white, black or others), gestational age (continuous)].
Figure 1.
Box and whisker plots of DNA methylation age and age acceleration at mid-childhood by atopic status.
We did not observe statistically significant associations of DNAmAge or age acceleration (IEAA and EEAA) at birth or in early-childhood with allergic phenotypes in mid-childhood (Supplementary Table 3 and 4). The Knight DNA methylation age for gestational age (DNAmGA), which is specific for epigenetic age at birth, was also not associated allergic phenotypes in mid-childhood (Supplementary Table 3).
To examine the between and within person variability of epigenetic age acceleration, we computed the ICCs for IEAA and EEAA. For IEAA, the overall ICC across all three time points was 0.08. We observed modest ICC between birth and early-childhood (ICC=0.13); relatively high ICC of IEAA between early and mid-childhood (ICC=0.44); and low ICC between birth and mid-childhood (ICC=0.03). For EEAA, the overall ICC across the three time points was 0.18. Intra-class correlations between birth and early-childhood, early-childhood and mid-childhood, and birth and mid-childhood were 0.21, 0.28 and 0.16, respectively.
Chronological age versus DNAmAge, and association between chronological age at earlychildhood and mid-childhood with mid-childhood allergy and asthma
DNA methylation age for gestational age (DNAm-GA; the Knight method) was positively correlated with actual gestational age (ρ=0.58, p-value<0.0001) (Supplementary Figure 3). DNA methylation age (DNAmAge; the Horvath method) in early childhood and mid-childhood was also positively correlated with chronological age in early childhood and mid-childhood (ρ=0.55, p-value<0.0001; ρ=0.47, p-value<0.0001, respectively) (Supplementary Figure 4 and 5). We observed higher mean level and wider range (standard deviation) [mean±SD] of DNAmAge at early [3.8±0.9 years] and mid-childhood [9.0±1.9 years] compared to chronological age at early [3.4±0.5 years] and mid-childhood [7.9±0.8 years]. A q-q plot of the association between chronological age and the 353 methylation sites contributing to DNAmAge showed no epigenomic inflation (lamba=0.864) (Supplementary Figure 6).
After adjusting for covariates, chronological age at early-childhood visits was not prospectively associated with allergy or asthma in mid-childhood (Supplementary Table 5). A cross-sectional analysis of chronological age and allergic phenotypes—both measured at mid-childhood—also yielded null results (Supplementary Table 5).
Maternal social-economic factors and mid-childhood DNAmAge
Maternal education level and marital status during pregnancy were significantly associated with DNAmAge (p-value=0.03 and p-value<0.0001, respectively). Children whose mother had lower education level (less than college) or were not married had higher DNAmAge in mid-childhood (Supplementary Figure 7). Active maternal smoking during pregnancy was not associated with higher DNAmAge in mid-childhood (Supplementary Figure 7).
Replication
We replicated our mid-childhood findings from Project Viva in an independent cohort—the ‘Genetics of Asthma in Costa Rica’ cohort (N=159). Participants’ characteristics are shown in Supplementary Table 6. The age distribution in the Costa Rica study (mean ± SD = 9.1 ± 1.8 years) denoted that the children were slightly older than the children in Project Viva (mean ± SD = 7.8 ± 0.7 years). Increases in DNAmAge and age acceleration at mid-childhood were cross-sectionally associated with higher odds of atopic sensitization as measured by the skin test (p≤0.01 for both IEAA and EEAA, Supplementary Table 7). Increases in IEAA was also positively associated with higher odds of asthma (Supplementary Table 7). We observed same signs of direction and comparable effect estimates in Project Viva and the Costa Rica Study for the relationship between EEAA, IEAA and total serum IgE and atopic sensitization based on serum specific IgE tests, although these associations did not reach a statistical significance threshold (Supplementary Table 7).
Discussion
Our study demonstrates that (i) at mid-childhood, DNA methylation age in peripheral blood was cross-sectionally associated with higher total serum IgE, as well as higher odds of atopic sensitization, environmental allergen and food allergen, and asthma; (ii) mid-childhood epigenetic age acceleration metrics (including EEAA and IEAA) were associated with atopic phenotypes; (iii) extrinsic epigenetic age acceleration, which weights blood cell types and measures epigenetic aging in immune-related domains, was additionally associated with asthma; (iii) maternal sociodemographic factors (lower education level, being unmarried) were associated with older DNA methylation age at mid-childhood.
Previous epigenome-wide association studies have linked blood, nasal and bronchial mucosal tissue methylomes with childhood allergy and asthma (18–20, 22, 43–46). However, most of these studies were conducted at a single time point, and may not capture postnatal age-related biological processes, which often coincide with early immune development. Indeed, there is increasing evidence showing age-related intra-individual change in DNA methylation profiles over time, often capturing epigenetic pathways regulating immunity (47, 48). Hence, our study of examining DNA methylation age and childhood allergy adds an important piece of evidence to the literature and furthers our understanding of the role of epigenetic regulation in immune system development and allergy.
The epigenetic clock has been studied extensively in age-related biological processes (weight change during childhood and adolescence (10) pubertal development in childhood (12)), aging-associated diseases (cognitive decline (2), Parkinson’s disease (3), lung cancer (4)), as well as cancer, cardiovascular and all-cause mortality (5–8). Although research on psychosocial outcomes have been performed in childhood cohorts (49), little research has been done on chronic diseases of childhood. Here, we demonstrated for the first time that at mid-childhood, DNA methylation age and age acceleration associated with allergic phenotypes. The age dependent allergic progression in children is well documented in the clinical literature (15), but may be difficult to observe in birth cohorts, as participants in birth cohorts usually come to research visits based on their developmental stages within a narrow age window. Therefore, in our study, we did not observe associations between chronological age and childhood allergy at the three research visit windows (at birth, early-childhood and mid-childhood) in Project VIVA. On the other hand, DNAmAge integrates additional sources of variabilities from environmental and life-style factors, and has wider variations at birth, early-childhood, and mid-childhood.
An Ingenuity Pathway analysis from the original epigenetic clock manuscript by Horvath suggested the 353 methylation sites used to compute the epigenetic clock showed significant enrichment for immune cell trafficking, hematological system development and function, organismal development, embryonic development and tissue development (1). In alignment with the pathway findings, our study showed DNAmAge and age acceleration were associated with mid-childhood allergy. Intrinsic epigenetic age acceleration is suggested to capture cell/intrinsic aging independent of shifts in cell proportion. A shift of Th1/Th2 immuno-dominance towards Th2 responses has been postulated as one primary mechanism of allergy. Further, the extrinsic epigenetic age acceleration, which weights blood cell types that are known to change with age, (13), was additionally associated with asthma. Due to its weighting of age-related blood cell types, EEAA is considered to capture immune-related biological processes and would be a highly relevant biomarker for immune-related phenotypes, such as allergy and asthma. On the other hand, previous literature has shown that anti-inflammatory treatment (such as oral or inhaled corticosteroids) does not work for every single child with allergy or asthma (50), suggesting that other biological processes besides immune/inflammatory pathways may contribute to the development and progression of allergy and asthma. Intrinsic epigenetic age acceleration may help to capture the non-immune part of the story.
Our study demonstrated that maternal sociodemographic factors (i.e., lower education level; not married during pregnancy) were correlated with higher DNA methylation age and age acceleration in children and should be considered in future studies of epigenetic age acceleration and chronic diseases in children. A previous study found differential epigenome-wide DNA methylation patterns in children raised since birth in institutional care compared with children raised by their biological parents, with most of the significant methylation sites involved in the control of immune responses and cell signaling (51). Maternal depression, anxiety, and maternal antidepressant use also have been associated with changes in DNA methylation in sites related to oxidative stress, mRNA translocation from the nucleus, and cell division (52, 53). Among 292 African American youths transitioning into adulthood (i.e., spanning ages 17 to 22), lower socioeconomic status (measured by household income below the federal poverty line, receipt of Temporary Assistance for Needy Families, caregiver report of income as insufficient to meet all needs, and primary caregiver without high school education or current employment) was associated with higher epigenetic age acceleration (49). The impact of maternal factors on asthma and atopy may be partially accounted for by epigenetic effects.
Our study has several strengths. (1) We computed a range of epigenetic clock estimators, with a focus on the intrinsic epigenetic clock from Horvath’s estimation (1, 3), the extrinsic epigenetic age acceleration from Hannum’s estimation (3), and DNA methylation age for gestational age using the method of Knight (38). Our results were consistent across the different epigenetic age estimators. (2) We examined a comprehensive range of atopic and asthma phenotypes at mid-childhood. We showed repeatedly that the epigenetic clock metrics were associated with Th2-cell driven phenotypes. This is particularly evident for extrinsic epigenetic age acceleration, which measures epigenetic aging in immune-related domains. (3) Our mid-childhood associations between atopy and age acceleration in Project Viva replicated in an independent children cohort—the ‘Genetics of Asthma in Costa Rica’ cohort, support the robustness of our findings.
Our study has several limitations. (1) At mid-childhood, peripheral blood DNA methylation and allergic outcomes are measured at the same time, and it is hard to disentangle the temporal relationships between the two. However, although not statistically significant (possible due to the small sample size), DNA methylation age and age acceleration at early-childhood were also associated with higher odds of allergy and asthma in-mid-childhood, suggesting that the observed associations were unlikely due to reverse causation. As for future direction, better powered longitudinal studies are needed to fully address the issue of reverse causation. (2) Current asthma at mid-childhood was defined via parent-reported questionnaire, and measurement errors are inevitable. Better phenotyping of asthma is warranted in future studies to reduce measurement errors. (3) We did not find chronological age associated with childhood allergy at the three clinical visits (at birth, early-childhood and mid-childhood), possibly related to the narrow chronological age windows we studied. However, this did not impact our ability to identify epigenetic age associations, likely due to the wider range of DNA methylation age range. (4) Our null findings at early childhood could also be due to a lack of power. (5) Importantly, our replication cohort was ascertained based on physician-diagnosed asthma, potentially limiting the generalizability of our findings.
Conclusion
Our study showed that DNA methylation age and epigenetic age acceleration in mid-childhood were cross-sectionally associated with higher levels of total serum IgE, as well as higher odds of atopic sensitization, environmental allergen and food allergen sensitizations, and asthma in children. As the epigenetic clock is thought to reflect the immuno- and developmental components of biological aging, our study shed light on the potential molecular mechanisms through which immunity, development and maturation interact along the age axis, and associates with childhood allergy or asthma. Taken together, accelerated epigenetic aging in allergic and asthmatic children may reflect perturbations in epigenetic maintenance, and raises the speculation as to whether anti-aging therapies may eventually find a role in mitigating the allergic march.
Supplementary Material
Key message.
In this study, we showed that epigenetic age acceleration at mid-childhood is cross-sectionally associated with higher levels of total serum IgE, as well as higher odds of asthma and atopy in Project Viva.
Accelerated epigenetic aging in allergic and asthmatic children may reflect perturbations in epigenetic maintenance, and raises the speculation as to whether anti-aging therapies may eventually find a role in mitigating the allergic march.
Acknowledgments
FUNDING
The Project Viva study is supported by grants from the National Institutes of Health (NIH R01 HL 111108, P01 HL 132825, R01 NR013945, R01 HD 034568, UG3OD023286, K23 ES022242, R01 AI102960).
Abbreviations
- DNAmAge
DNA methylation age
- EEAA
extrinsic age acceleration
- FEV1
forced expiratory volume in one second
- GACRS
‘Genetics of Asthma in Costa Rica Study’
- IEAA
intrinsic epigenetic age acceleration
Footnotes
Declaration: The authors have declared that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Horvath S DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44(4):1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany NY). 2015;7(12):1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine ME, Hosgood HD, Chen B, Absher D, Assimes T, Horvath S. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY). 2015;7(9):690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15(1):149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8(9):1844–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpkin AJ, Hemani G, Suderman M, Gaunt TR, Lyttleton O, McArdle WL, et al. Prenatal and early life influences on epigenetic age in children: a study of mother-offspring pairs from two cohort studies. Hum Mol Genet. 2016;25(1):191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simpkin AJ, Howe LD, Tilling K, Gaunt TR, Lyttleton O, McArdle WL, et al. The epigenetic clock and physical development during childhood and adolescence: longitudinal analysis from a UK birth cohort. Int J Epidemiol. 2017;46(2):549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpkin AJ, Cooper R, Howe LD, Relton CL, Davey Smith G, Teschendorff A, et al. Are objective measures of physical capability related to accelerated epigenetic age? Findings from a British birth cohort. BMJ Open. 2017;7(10):e016708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binder AM, Corvalan C, Mericq V, Pereira A, Santos JL, Horvath S, et al. Faster ticking rate of the epigenetic clock is associated with faster pubertal development in girls. Epigenetics. 2017:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu AT, Xue L, Salfati EL, Chen BH, Ferrucci L, Levy D, et al. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9(1):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnetson RS, Rogers M. Childhood atopic eczema. BMJ. 2002;324(7350):1376–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bantz SK, Zhu Z, Zheng T. The Atopic March: Progression from Atopic Dermatitis to Allergic Rhinitis and Asthma. J Clin Cell Immunol. 2014;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng T, Yu J, Oh MH, Zhu Z. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. Allergy Asthma Immunol Res. 2011;3(2):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everson TM, Lyons G, Zhang H, Soto-Ramirez N, Lockett GA, Patil VK, et al. DNA methylation loci associated with atopy and high serum IgE: a genome-wide application of recursive Random Forest feature selection. Genome Med. 2015;7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martino D, Joo JE, Sexton-Oates A, Dang T, Allen K, Saffery R, et al. Epigenome-wide association study reveals longitudinally stable DNA methylation differences in CD4+ T cells from children with IgE-mediated food allergy. Epigenetics. 2014;9(7):998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong X, Hao K, Ladd-Acosta C, Hansen KD, Tsai HJ, Liu X, et al. Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children. Nat Commun. 2015;6:6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang L, Willis-Owen SAG, Laprise C, Wong KCC, Davies GA, Hudson TJ, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520(7549):670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W, Wang T, Pino-Yanes M, Forno E, Liang L, Yan Q, et al. An epigenome-wide association study of total serum IgE in Hispanic children. J Allergy Clin Immunol. 2017;140(2):571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeVries A, Vercelli D. Early predictors of asthma and allergy in children: the role of epigenetics. Curr Opin Allergy Clin Immunol. 2015;15(5):435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018. [DOI] [PubMed] [Google Scholar]
- 25.Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, De Meo D, et al. Cohort profile: project viva. Int J Epidemiol. 2015;44(1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oken E, Kleinman KP, Rich-Edwards J, Gillman MW. A nearly continuous measure of birth weight for gestational age using a United States national reference. BMC Pediatr. 2003;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oken E, Huh SY, Taveras EM, Rich-Edwards JW, Gillman MW. Associations of maternal prenatal smoking with child adiposity and blood pressure. Obes Res. 2005;13(11):2021–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Triche TJ Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41(7):e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardenas A, Rifas-Shiman SL, Godderis L, Duca RC, Navas-Acien A, Litonjua AA, et al. Prenatal Exposure to Mercury: Associations with Global DNA Methylation and Hydroxymethylation in Cord Blood and in Childhood. Environ Health Perspect. 2017;125(8):087022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. [DOI] [PubMed] [Google Scholar]
- 32.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, S LM, et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardenas A, Allard C, Doyon M, Houseman EA, Bakulski KM, Perron P, et al. Validation of a DNA methylation reference panel for the estimation of nucleated cells types in cord blood. Epigenetics. 2016;11(11):773–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klemera P, Doubal S. A new approach to the concept and computation of biological age. Mech Ageing Dev. 2006;127(3):240–8. [DOI] [PubMed] [Google Scholar]
- 36.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY). 2017;9(2):419–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knight AK, Craig JM, Theda C, Baekvad-Hansen M, Bybjerg-Grauholm J, Hansen CS, et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol. 2016;17(1):206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bunyavanich S, Rifas-Shiman SL, Platts-Mills TA, Workman L, Sordillo JE, Camargo CA Jr., et al. Prenatal, perinatal, and childhood vitamin D exposure and their association with childhood allergic rhinitis and allergic sensitization. J Allergy Clin Immunol. 2016;137(4):106370 e2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bunyavanich S, Rifas-Shiman SL, Platts-Mills TA, Workman L, Sordillo JE, Camargo CA Jr., et al. Peanut, milk, and wheat intake during pregnancy is associated with reduced allergy and asthma in children. J Allergy Clin Immunol. 2014;133(5):1373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brehm JM, Ramratnam SK, Tse SM, Croteau-Chonka DC, Pino-Yanes M, Rosas-Salazar C, et al. Stress and Bronchodilator Response in Children with Asthma. Am J Respir Crit Care Med. 2015;192(1):47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunninghake GM, Soto-Quiros ME, Avila L, Ly NP, Liang C, Sylvia JS, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol. 2007;119(3):654–61. [DOI] [PubMed] [Google Scholar]
- 43.Baccarelli A, Rusconi F, Bollati V, Catelan D, Accetta G, Hou L, et al. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics. 2012;4(1):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang IV, Pedersen BS, Liu AH, O’Connor GT, Pillai D, Kattan M, et al. The nasal methylome and childhood atopic asthma. J Allergy Clin Immunol. 2017;139(5):1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim YJ, Park SW, Kim TH, Park JS, Cheong HS, Shin HD, et al. Genome-wide methylation profiling of the bronchial mucosa of asthmatics: relationship to atopy. BMC Med Genet. 2013;14:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng C, Cardenas A, Rifas-Shiman SL, Hivert MF, Gold DR, Platts-Mills TA, et al. Epigenome-wide association study of total serum immunoglobulin E in children: a life course approach. Clin Epigenetics. 2018;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martino DJ, Tulic MK, Gordon L, Hodder M, Richman TR, Metcalfe J, et al. Evidence for age-related and individual-specific changes in DNA methylation profile of mononuclear cells during early immune development in humans. Epigenetics. 2011;6(9):1085–94. [DOI] [PubMed] [Google Scholar]
- 48.Urdinguio RG, Torro MI, Bayon GF, Alvarez-Pitti J, Fernandez AF, Redon P, et al. Longitudinal study of DNA methylation during the first 5 years of life. J Transl Med. 2016;14(1):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller GE, Yu T, Chen E, Brody GH. Self-control forecasts better psychosocial outcomes but faster epigenetic aging in low-SES youth. Proc Natl Acad Sci U S A. 2015;112(33):10325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Aalderen WM. Childhood asthma: diagnosis and treatment. Scientifica (Cairo). 2012;2012:674204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naumova OY, Lee M, Koposov R, Szyf M, Dozier M, Grigorenko EL. Differential patterns of whole-genome DNA methylation in institutionalized children and children raised by their biological parents. Dev Psychopathol. 2012;24(1):143–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Non AL, Binder AM, Kubzansky LD, Michels KB. Genome-wide DNA methylation in neonates exposed to maternal depression, anxiety, or SSRI medication during pregnancy. Epigenetics. 2014;9(7):964–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurnot C, Martin-Subero I, Mah SM, Weikum W, Goodman SJ, Brain U, et al. Prenatal antidepressant exposure associated with CYP2E1 DNA methylation change in neonates. Epigenetics. 2015;10(5):361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.