Abstract
The HIRA histone chaperone complex is composed of the proteins HIRA, UBN1, and CABIN1 and cooperates with the histone chaperone ASF1a to specifically bind and deposit H3.3/H4 into chromatin. We recently reported that the UBN1 Hpc2-related domain (HRD) specifically binds to H3.3/H4 over H3.1/H4. However, the mechanism for HIRA complex deposition of H3.3/H4 into nucleosomes remains unclear. Here, we characterize a central region of UBN1 (UBN1 middle domain) that is evolutionarily conserved and predicted to have helical secondary structure. We report that the UBN1 middle domain has dimer formation activity and binds to H3/H4 in a manner that does not discriminate between H3.1 and H3.3. We additionally identify a nearby DNA-binding domain in UBN1, located between the UBN1 HRD and middle domain, which binds DNA through electrostatic contacts involving several conserved lysine residues. Together, these observations suggest a mechanism for HIRA-mediated H3.3/H4 deposition whereby UBN1 associates with DNA and dimerizes to mediate formation of an (H3.3/H4)2 heterotetramer prior to chromatin deposition.
Keywords: histone chaperone, histone, chromatin, DNA-binding protein, epigenetics
Introduction
The deposition of histones onto DNA for nucleosome formation and chromatin maintenance is a hallmark of eukaryotic cells and serves to package and protect DNA for the efficient regulation of gene expression. Histone deposition is coordinated by histone chaperone proteins, which serve to bind and protect the basic histone proteins from nonspecific interactions prior to deposition into nucleosomes (1, 2). Additionally, histones have many variant sequences that are incorporated into nucleosomes at specific genomic locations to effect changes on the local chromatin environment, and many distinct histone chaperones have specifically evolved to bind and deposit a single variant histone into chromatin (3, 4). The human HIRA histone chaperone complex specifically binds to and deposits H3.3/H4 into chromatin in a replication-independent manner (5). The complex is composed of the three core subunits, HIRA, UBN1, and CABIN1, with the transient addition of ASF1a, which binds a H3.3/H4 dimer and precludes formation of the (H3.3/H4)2 tetramer (6, 7). ASF1a delivers H3.3/H4 to the complex but is not required for ultimate H3.3/H4 deposition into nucleosomes (5, 8, 9). Much work has been done to elucidate how the HIRA complex assembles (10–13), and we have recently reported that UBN1 specifically binds an H3.3/H4 dimer bound to ASF1a (14), but the mechanism of how the HIRA complex ejects ASF1a and transitions the H3/H4 dimer to an (H3.3/H4)2 tetramer for deposition is not clear. The study of (H3/H4)2 tetramer deposition has been the focus of work for several other histone chaperones system (15–21) because of evidence that the vast majority of H3/H4 deposits as a tetramer onto DNA prior to completion of nucleosome formation by the subsequent addition of two H2A/H2B dimers (22). Several studies have identified that dimerization of histone chaperones may contribute to formation of a (H3/H4)2 tetramer for deposition onto DNA (2, 23–26). We recently reported that the C-terminal region of the HIRA subunit forms a stable trimer in solution, and trimerization is essential for HIRA histone chaperone activity in cells (27), whereas it is still unclear whether this timer contributes to formation an (H3.3/H4)2 tetramer for deposition.
The HIRA complex subunit UBN1 has been reported to bind to the WD40 domain of HIRA through a small domain of about 30 amino acids called the NHRD (11). The NHRD was named after discovery of its HIRA-binding activity because it lies just N-terminal to the UBN1 Hpc2-related domain (HRD),3 which has strong conservation with the Saccharomyces cerevisiae Hir complex subunit Hpc2. After identification of the NHRD as a HIRA-binding domain, we found that the HRD specifically binds to H3.3/H4 and determined a crystal structure of the UBN1/H3.3/H4/Asf1 complex (14). Whereas the crystal structure is informative about the exact nature of the H3.3-specific interactions that are mediated by UBN1, it only contains 20 amino acids from UBN1. Together, our prior studies focus on roughly 50 of the 1134 amino acids comprising the UBN1 polypeptide. We therefore postulate that the rest of the 1134-residue polypeptide may contain additional regions with previously uncharacterized functions in HIRA-associated H3.3/H4 deposition into chromatin (28).
Here, we characterize two UBN1 domains of previously unknown function. Through secondary structure prediction and sequence conservation, we have identified a centrally located region of UBN1, which we call the UBN1 middle domain. Spanning residues ∼300–600, the middle domain is predicted to have helical secondary structure. Through size-exclusion chromatography and analytical ultracentrifugation experiments, we find that the C-terminal region of the UBN1 middle domain, residues 504–584, is responsible for dimer formation. Through pulldown and deletion analysis, we also find that the N-terminal region of the UBN1 middle domain, residues 296–341, is required for H3/H4 binding. Additionally, we identify several vertebrate-specific conserved lysine residues in a very basic loop region of low predicted structure residing between the UBN1 HRD and middle domain. Using fluorescence polarization and size-exclusion chromatography analysis of UBN1/DNA complex formation, we determine that these conserved lysine residues are essential for nonspecific DNA binding by UBN1. Together, our observations of H3/H4 binding and dimer formation by the UBN1 middle domain indicate that UBN1 may be capable of binding two H3.3/H4 molecules to facilitate (H3.3/H4)2 tetramer formation in a pathway to nucleosome deposition that may also involve UBN1-mediated DNA binding by the HIRA complex.
Results
The UBN1 middle domain forms stable monomer and dimer populations
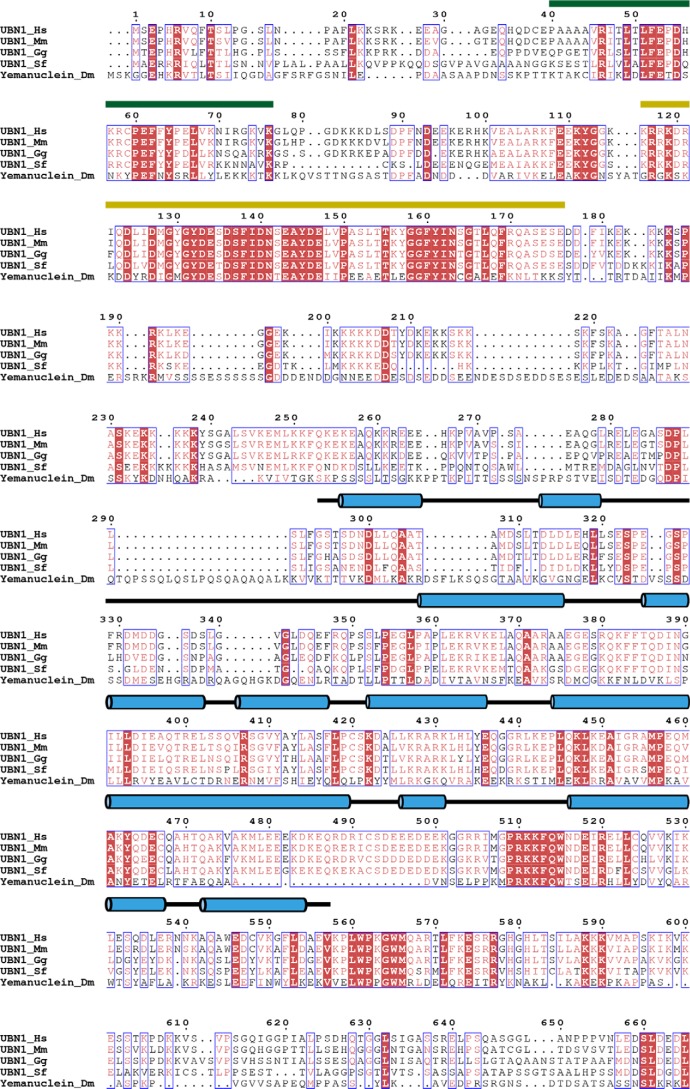
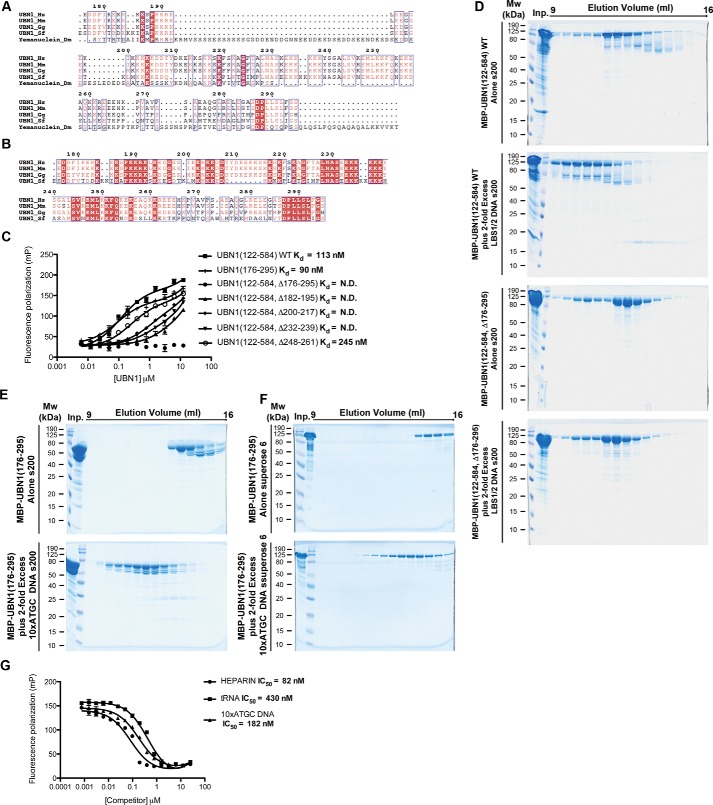
To identify regions of UBN1 that contribute to the biological function of the human HIRA histone chaperone complex, beyond the well-characterized HRD and NHRD domains, we performed a multiple-sequence alignment of metazoan UBN1 homologs to hone in on additional regions of conservation to probe for functional significance (Fig. 1). Hpc2, the UBN1 homolog from S. cerevisiae, only shares conservation in the HRD and NHRD domains; to identify nuanced features of human UBN1 outside of these domains, we omitted UBN1 homologs from yeast from our alignment. Through alignment of Homo sapiens UBN1, Mus musculus UBN1, Gallus gallus UBN1, Scleropages formosus UBN1, and Drosophila melanogaster yemanuclein, it is evident that the NHRD (residues 41–77) and HRD (residues 119–175) are very highly conserved, but there are additional regions of conservation, most notably the region spanning residues ∼300–600 (Fig. 1A).
Figure 1.
Alignment of UBN1 metazoan homologs and secondary structure prediction. The alignment depicts sequence conservation among H. sapiens UBN1, M. musculus UBN1, G. gallus UBN1, S. formosus UBN1, and D. melanogaster yemanuclein. The conserved UBN1 HRD (residues 120–175) (yellow line over sequence) and NHRD (residues 41–71) (green line over sequence) domains have been well-characterized for their functions. UBN1 residues 176–700 are well-conserved among metazoan species, although the biological function of this UBN1 region remains unclear. Predicted helical structure for the human UBN1 middle domain are shown as blue cylinders above sequences. Note that S. formosus represents UBN1 from a fish (Asian bonytongue), as there is not an annotated full-length sequence for UBN1 from Danio rerio.
To complement the multiple sequence alignment, we also performed secondary structure prediction for the human UBN1 protein sequence using the PSIPRED server (29) (Fig. 1). Whereas the majority of human UBN1 has little predicted structure or low-confidence prediction, UBN1 residues ∼300–555 are predicted to form helical secondary structure with high confidence of prediction. This, in combination with the metazoan sequence conservation, identifies this domain of UBN1 as a good target for functional characterization with biochemical and structural studies. As this domain has been uncharacterized for function in human UBN1, we preliminarily name this region the UBN1 middle domain.
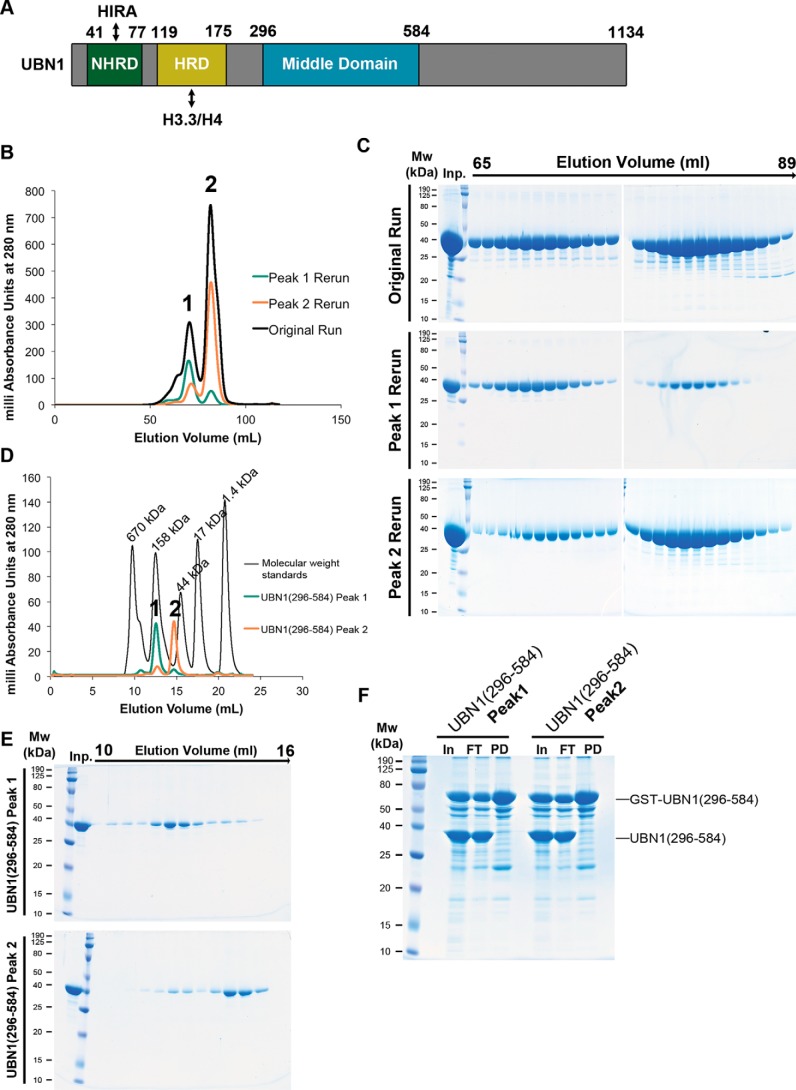
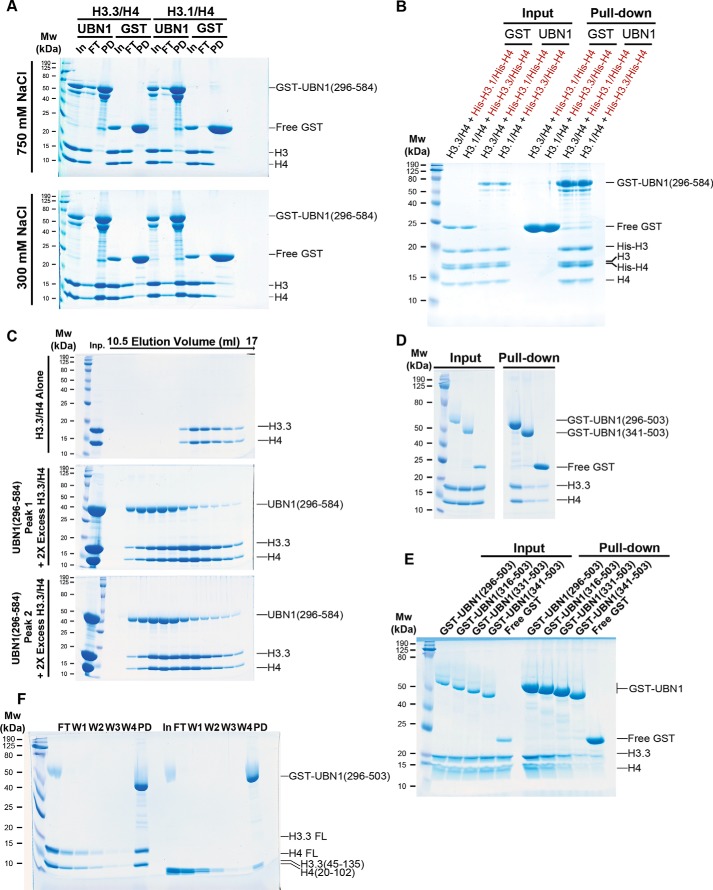
A construct encoding UBN1 middle domain residues 296–584, bearing an N-terminal His6 that is removable with TEV protease, was generated and expressed in Escherichia coli cells (Fig. 2A). This UBN1(296–584) fragment was easily purified and stable in solution. Upon size-exclusion chromatography analysis using a Superdex 200 16/600 column (GE Healthcare), we observed that this fragment of UBN1 forms two monodisperse populations that we call “peak 1” and “peak 2” (Fig. 2B). Upon pooling and rerunning the isolated populations separately on the size-exclusion column, it was observed that these populations are relatively stable and do not readily redistribute back into the same two populations (Fig. 2, B and C). To further analyze these stable UBN1(296–584) peaks, we then injected the isolated populations onto an analytical Superdex 200 10/300 column (GE Healthcare) and compared the elution profiles with a set of molecular weight standards (Bio-Rad) (Fig. 2, D and E). Comparison with the standards indicates that peak 1 has an elution profile similar to that of the 158-kDa standard, and peak 2 elutes just before the 44-kDa standard. To further test whether or not these populations exchange, we performed a pulldown experiment where GST-UBN1(296–584) was incubated with untagged UBN1(296–584) from the peak 1 or peak 2 populations (Fig. 2F). We observed that both peak 1 and peak 2 are unable to form complexes with GST-UBN1(296–584), indicating that these populations have minimal exchange after they are initially formed.
Figure 2.
UBN1(296–584) forms two nonexchanging monodisperse populations. A, domain architecture diagram depicting the characterized UBN1 NHRD, HRD, and middle domains. B, chromatogram of UBN1(296–584) resolved on a HiLoad Superdex 200 16/600 size-exclusion column. Fractions from 65 to 75 ml were pooled and reloaded onto the column for Peak 1 Rerun, whereas fractions from 76 to 89 ml were pooled and reloaded onto the column for Peak 2 Rerun. C, SDS-polyacrylamide gel analysis of 1-ml fractions spanning 65–89 ml for all three runs. D, chromatograms from UBN1(296–584) peak 1 and peak 2 populations resolved on an analytical Superdex 200 10/300 size-exclusion column compared with molecular weight standards. E, SDS-polyacrylamide gel analysis of 0.5-ml fractions spanning 10–16 ml for both runs. F, GST pulldown confirming that both peak 1 and peak 2 are unable to form a complex with GST-UBN1(296–584), further indicating that these are stable and nonexchanging populations.
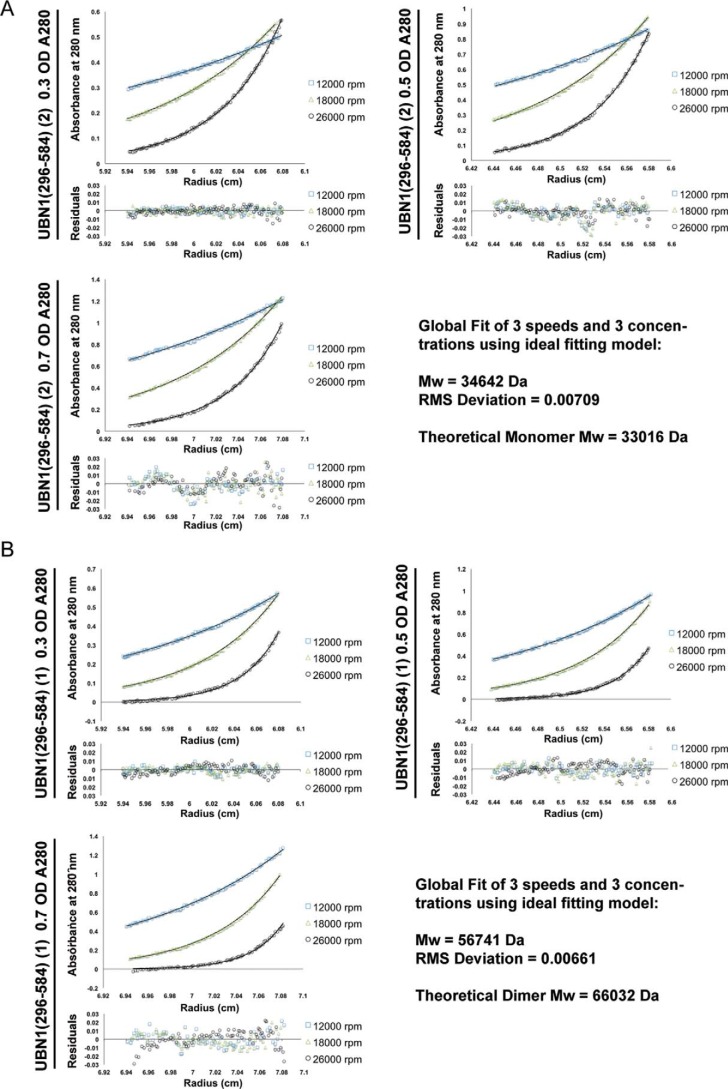
A theoretical monomer of UBN1(296–584) has a molecular mass of 33 kDa. To determine whether these two UBN1 populations represent different oligomerization states, we performed a sedimentation equilibrium analytical ultracentrifugation analysis of the two populations. Both peaks were monitored for the equilibrium distribution of three different protein concentrations (A280 of 0.3, 0.5, and 0.7) at three centrifugation speeds (12,000, 18,000, and 26,000 rpm). The combined equilibrium data were globally fit with an ideal model to determine the average molecular weight of the protein in solution. Analysis of peak 1 determined an experimental molecular mass of 56.7 kDa (Fig. 3A), whereas analysis of peak 2 determined an experimental molecular mass of 34.6 kDa (Fig. 3B). Comparison with the molecular mass of a theoretical UBN1(296–584) monomer (33 kDa) and dimer (66 kDa) indicates that peak 1 contains predominantly a dimer population, whereas peak 2 contains a majority monomer population.
Figure 3.
Analytical ultracentrifugation of UBN1(296–584). A, sedimentation equilibrium of the UBN1(296–584) peak 2 population. Three different protein concentrations (A280 = 0.3, 0.5, and 0.7) were analyzed for equilibrium distribution at three different speeds (12,000, 18,000, and 26,000 rpm). These data were fit to the ideal fitting model using the program HeteroAnalysis. The ideal fit yielded an experimental molecular mass of 34,642 Da, indicating this to be a predominantly monomeric population. B, sedimentation equilibrium of UBN1(296–584) peak 1 population. Three different protein concentrations (A280 = 0.3, 0.5, and 0.7) were analyzed for equilibrium distribution at three different speeds (12,000, 18,000, and 26,000 rpm). These data were fit to the ideal fitting model using the program HeteroAnalysis. The ideal fit yielded an experimental molecular mass of 56,741 Da, indicating that this is a predominantly dimeric population.
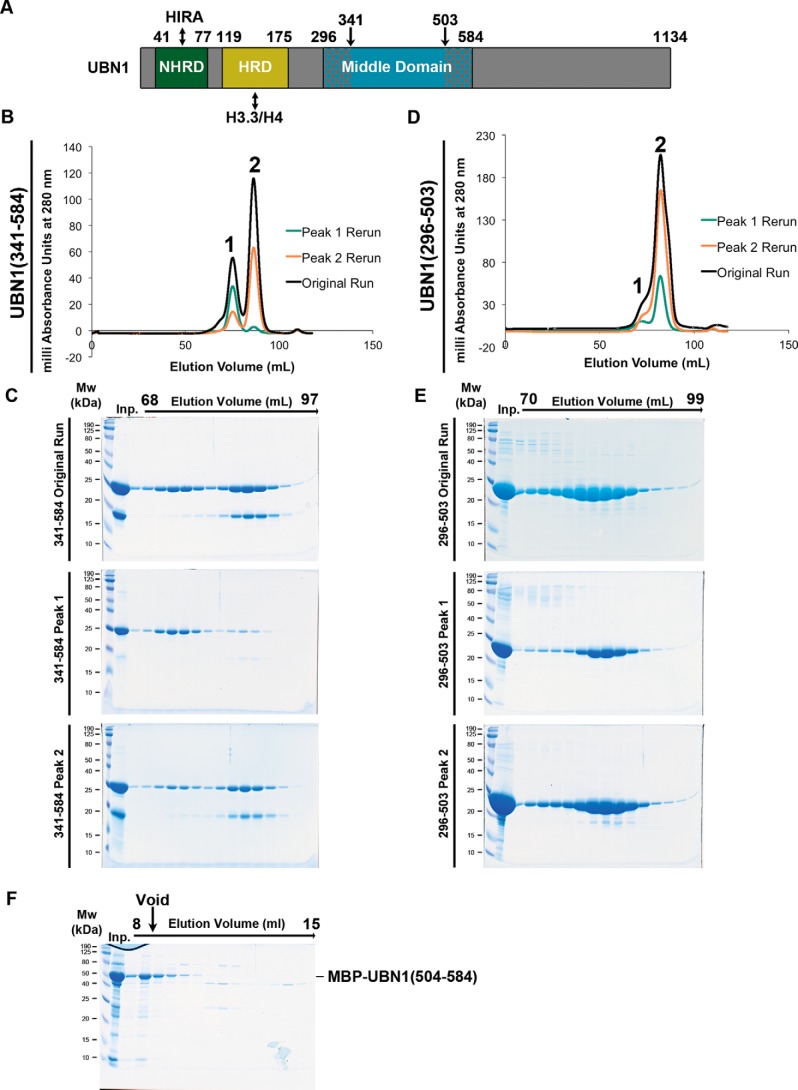
We then expressed and purified UBN1(341–584) in the same manner as UBN1(296–584) to monitor the effect of truncating the predicted N-terminal helices and loop (Fig. 4A). UBN1(341–584) resolves into the same peak 1 and peak 2 populations observed for UBN1(296–584) on a Superdex 200 16/600 column; the only major difference for this fragment was that we observed the co-migration of a smaller protein (between 18 and 12 kDa) that may represent a proteolytic product (Fig. 4, B and C). Indeed, LC-MS analysis on the extracted band revealed it to be UBN1(341–503), which represents a C-terminal degradation.
Figure 4.
Analysis of UBN1(341–584) and UBN1(296–503). A, domain architecture diagram for UBN1 depicting middle domain fragments. B, chromatograph of UBN1(341–584) resolved on a HiLoad Superdex 200 16/600 size-exclusion column. Fractions from 68 to 78 ml were pooled and reloaded onto the column for Peak 1 Rerun, whereas fractions from 80 to 94 ml were pooled and reloaded onto the column for Peak 2 Rerun. C, SDS-polyacrylamide gel analysis of every other 1-ml fraction spanning 68–97 ml for all three runs. D, chromatograph of UBN1(296–503) resolved on a HiLoad Superdex 200 16/600 size-exclusion column. Fractions from 70 to 78 ml were pooled and reloaded onto the column for Peak 1 Rerun, whereas fractions from 80 to 90 ml were pooled and reloaded onto the column for Peak 2 Rerun. E, SDS-PAGE of every other 1-ml fraction spanning 70–99 ml for all three runs. F, MBP-UBN1(504–584) was resolved on a Superose 6 10/300 size-exclusion column. The majority of MBP-UBN1(504–584) eluted in the void volume.
We then prepared UBN1(296–503) in the same manner as UBN1(296–584) and UBN1(341–584) to identify the contribution of UBN1 residues 504–584 to the formation of the monomer and dimer populations, as this region harbors several highly conserved middle domain residues (Fig. 1A). When UBN1(296–503) was resolved on a Superdex 200 16/600 column, only one major peak with a small higher-molecular weight shoulder was observed (Fig. 4, D and E). We did not see the same distinctly separate populations observed for UBN1(296–584) and UBN1(341–584), but we proceeded with analysis of the shoulder as a potentially separate population from the main peak. We pooled and re-injected the shoulder and the main peak as peak 1 and peak 2, respectively. We observed that rerun of both peak 1 and peak 2 resulted in formation of the same distribution as the original run, indicating that the constituents of peaks 1 and 2 are likely in dynamic equilibrium (Fig. 4, D and E). This observation leads to the conclusion that residues 504–584 of UBN1 are required for stable dimer formation. We attempted to purify UBN1(504–584) as an MBP-UBN1(504–584) fusion, but this fragment formed very large soluble aggregates. We were unable to isolate soluble untagged UBN1(504–584) with TEV protease cleavage. When MBP-UBN1(504–584) was resolved on a Superose 6 10/300 column, the majority eluted in the void volume, indicating that this fragment of UBN1 is aggregated and not suitable for biophysical analysis in isolation (Fig. 4F).
The UBN1 middle domain specifically binds H3/H4
Given our observation that the UBN1 middle domain forms stable monomer and dimer populations, together with our previous finding that the UBN-HRD domain specifically binds to H3.3/H4, we decided to test whether the UBN1 middle domain also binds to histones, expecting that it would not, indicative of functional specialization of the HRD and middle domains. We prepared GST-UBN1(296–584) for a pulldown-based histone-binding assay. GST-UBN1(296–584) was mixed with either H3.1/H4 or H3.3/H4, and binding was analyzed in buffer containing either 300 or 750 mm NaCl. Surprisingly, we observed that UBN1(296–584) was able to bind H3.1/H4 and H3.3/H4 with similar affinity, and the interaction was only evident in the 300 mm NaCl buffer (Fig. 5A), indicating that the interaction between H3/H4 and the UBN1 middle domain is electrostatic in nature and not specific for H3.3/H4. To verify our observation that the UBN1 middle domain has no binding preference for H3.3/H4 compared with H3.1/H4, we conducted a competition-binding assay that we previously employed for our analysis of the UBN1 HRD (14). Untagged H3.3/H4 complex was incubated with an equimolar concentration of a His-H3.1/His-H4 complex and subjected to pulldown with GST-UBN1(296–584). Additionally, the pulldown was also conducted with untagged H3.1/H4 in competition with His-H3.3/His-H4. Under both competition conditions, we observed no binding specificity for H3.3 versus H3.1, with comparable amounts of both untagged and His-tagged H3/H4 in the pulldown for both pools (Fig. 5B). We then tested whether the monomer or dimer population of UBN1(296–584) had any influence on this middle domain/histone interaction. We isolated UBN1(296–584) peak 1 or peak 2 and incubated the individual populations with excess H3.3/H4 prior to resolving the mixtures over a Superdex 200 10/300 column; both the monomer and dimer populations were capable of forming a similar complex with H3.3/H4 when resolved on the Superdex 200 column (Fig. 5C).
Figure 5.
UBN1(296–584) binds histones H3/H4 with no specificity for H3.3 versus H3.1. A, GST pulldown conducted with GST-UBN1(296–584) and H3.3/H4 or H3.1/H4 at 300 and 700 mm NaCl. B, GST pulldown conducted with His-tagged and untagged H3.3/H4 and H3.1/H4 complexes in competition. C, size-exclusion histone-binding experiment where UBN1(296–584) peak 1 and peak 2 were mixed with excess H3.3/H4 and run on an analytical Superdex 200 10/300 column to analyze complex formation. D, GST pulldown conducted with H3.3/H4 and GST-UBN1(296–503) or GST-UBN1(341–503). E, GST pulldown conducted with H3.3/H4 and various UBN1 fragments illustrating that residues 296–341 contribute to H3/H4 binding. F, GST pulldown conducted with GST-UBN1(296–584) and FL or ΔN H3.3/H4.
To test the significance of the flanking middle domain regions 296–340 and 504–584 to the H3/H4 interaction, GST-UBN1(296–503) and GST-UBN1(341–503) were incubated with H3.3/H4 and subjected to pulldown. Whereas GST-UBN1(296–503) was able to pull down H3.3/H4, no interaction above background was observed for GST-UBN1(341–503) (Fig. 5D), suggesting that UBN1 residues 504–584 are dispensable for H3/H4 binding, whereas the interaction depends on UBN1 residues 296–340. There is low but significant evolutionary conservation in the region of UBN1 residues 296–340 but no attractive candidate sites to target for individual point mutations (Fig. 1A). There is conservation among vertebrates within this region as the H. sapiens and S. formosus UBN1 share 46.5% sequence identity for residues 296–340, whereas yemanuclein from D. melanogaster shares only 17.4% sequence identity with H. sapiens UBN1 in this region. We employed a deletion strategy in this region and generated several N-terminal deletions to compare for H3/H4 binding: GST-UBN1(296–503), GST-UBN1(316–503), GST-UBN1(331–503), and GST-UBN1(341–503). Each UBN1 fragment was incubated with H3.3/H4 and subjected to GST pulldown (Fig. 5E). We observed that GST-UBN1(296–503) was able to form a stable complex with H3.3/H4, but with each successive N-terminal deletion, we observed less H3.3/H4 being pulled down, with GST-UBN1(341–503) having no interaction above background. This result indicates that many amino acids within UBN1(296–340) contribute to H3/H4 binding.
We then investigated whether the middle domain binds to the folded histone core, lysine-rich N-terminal tails, or perhaps a combination of both. We previously characterized the UBN1-HRD/H3.3/H4 interaction and showed that the HRD binds primarily to the folded histone core with a smaller contribution from the histone tails (14). GST-UBN1(296–503) was incubated with either FL H3.3/H4 or N-terminally truncated H3.3(45–135)/H4(20–102) and subjected to pulldown. The results reveal that GST-UBN1(296–584) is able to pulldown significantly more FL H3.3/H4 compared with the N-terminally truncated complex (Fig. 5F). This suggests that the UBN1 middle domain, in addition to dimerizing, binds H3/H4 with some contribution from the N-terminal H3/H4 tails.
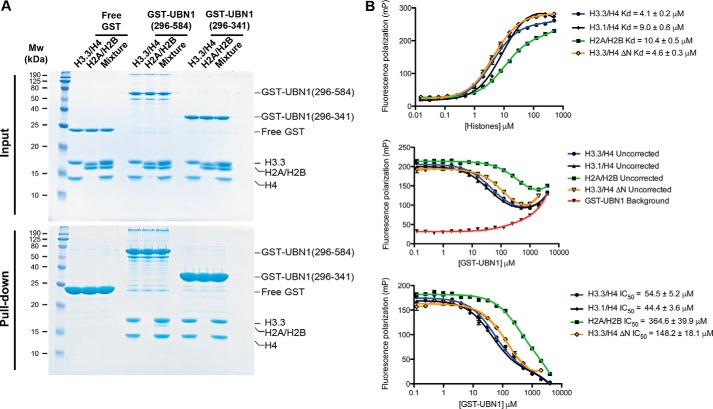
To determine the extent to which the middle domain can discriminate between different types of histone complexes, we conducted a pulldown histone binding assay to compare the middle domain interaction with H3/H4 and H2A/H2B. Both H3/H4 and H2A/H2B have a strong positive charge, but we hypothesized that UBN1 would likely show specificity toward H3/H4 if the interaction is biologically relevant, as the HIRA complex is a H3.3/H4 chaperone. GST-UBN1(296–584) and GST-UBN1(296–341) were incubated with H3.3/H4 or H2A/H2B individually or with a 1:1 pool of H3.3/H4 and H2A/H2B in competition. The pull downs show that both GST-UBN1(296–584) and GST-UBN1(296–341) are able to bind H3.3/H4 but not H2A/H2B when the histones are assayed individually and only bind H3.3/H4 in the competition pulldown (Fig. 6A). These studies demonstrate that the UBN1 middle domain residues 296–341 specifically bind H3/H4 over H2A/H2B.
Figure 6.
UBN1(296–584) binds histones H3/H4 specifically over H2A/H2B. A, GST pulldown conducted using GST-UBN1(296–584) or GST-UBN1(296–341) with H3.3/H4 or H2A/H2B alone or in combination to analyze binding specificity. B, fluorescence polarization binding assay monitoring UBN1(296–341) affinity for H3.3/H4, H3.3(45–135)/H4(20–102), and H2A/H2B by direct binding and competition binding.
To quantitatively analyze the UBN1 middle domain interaction with H3/H4, we employed a fluorescence polarization histone-binding assay similar to one we used in our previous study of the interaction between the UBN1 HRD and H3.3/H4 (14). UBN1(296–341) has no cysteine residues; we added a cysteine to the C terminus of this UBN1 fragment and employed the reactivity of the free thiol to label it with fluorescein. The fluorescein-labeled UBN1(296–341) was incubated with H3.3/H4, H3.1/H4, H3.3(45–135)/H4(20–102), or H2A/H2B in a series of concentrations to generate binding curves. From these curves we observed that H3.3/H4, H3.1/H4, H3.3(45–135)/H4(20–102), and H2A/H2B all have relatively similar Kd values of 4.1, 9.0, 4.6, and 10.4 μm, respectively (Fig. 6B, top). These data are contradictory to our pulldown results, where we observed that the UBN1 middle domain binds comparably to H3.3/H4 and H3.1/H4 while exhibiting significantly less affinity for H3.3(45–135)/H4(20–102) and H2A/H2B. We have observed a similar phenomenon previously in our fluorescence polarization studies of the UBN1 HRD, where direct binding to both H3.3/H4 and H3.1/H4 had comparable Kd values, but isothermal calorimetry and pulldown studies showed that the UBN1 HRD bound with high specificity to H3.3/H4 (14). We determined that the fluorescein molecule itself had enhanced affinity for the histones likely due to their high positive charge. To get around this nonspecific interaction between the histones and fluorescein, we were able to determine IC50 values by a fluorescence polarization competition-binding assay that more accurately reflected our isothermal calorimetry and pulldown results (14). We used a similar strategy here and generated competition binding curves using GST-UBN1(296–341) to compete the histones away from the fluorescein-labeled UBN1(296–341). In this experiment, we observed that GST-UBN1(296–341) had some background interaction with the fluorescein-labeled UBN1(296–341) (Fig. 6B, middle). We subtracted that background from the competition curves to generate our final IC50 curves (Fig. 6B, bottom). These experiments reveal that GST-UBN1(296–341) binds to H3.3/H4 and H3.1/H4 with comparable IC50 values of 54.5 and 44.4 μm, respectively (Fig. 6B, bottom), whereas the affinity for H3.3(45–135)/H4(20–102) is roughly 3-fold weaker with an IC50 value of 148.2 μm, and affinity for H2A/H2B is ∼7-fold weaker with an IC50 value of 364.6 μm (Fig. 6B, bottom). These experiments more qualitatively demonstrate that the UBN1 middle domain preferentially binds H3/H4 over H2A/H2B with binding contributions from the H3/H4 N-terminal tails.
UBN1 residues 176–295 bind to DNA without sequence specificity
The UBN1 region of 176–295, located between the HRD and middle domain, has little predicted secondary structure and is extremely lysine-rich. Similar to the H3/H4-binding region of the middle domain, there is not significant conservation in this region with the UBN1 homolog yemanuclein from D. melanogaster, but if yemanuclein is excluded from the multiple sequence alignment, it becomes clear that the basic loop is well-conserved among vertebrate UBN1 homologs (Fig. 7, A and B). This observation in combination with the report that UBN1 has been shown to have stronger non-sequence-specific interaction with DNA compared with other members of the HIRA complex (28) leads us to hypothesize that this region of UBN1 may be involved in non-sequence-specific binding to DNA. To address this hypothesis, we generated MBP-UBN1(122–584), harboring the HRD, basic loop, and middle domain. Within MBP-UBN1(122–584), we made several deletions, including the entire 175–296 loop as well as smaller stretches of conserved lysine residues. Additionally, we generated MBP-UBN1(176–195) to address whether the loop alone would be sufficient for DNA binding. We employed a fluorescence polarization–based DNA binding assay to test interaction with DNA. A FAM-labeled 46-bp double-stranded LBS1/2 DNA, originally generated to study binding with the Kaposi's sarcoma–associated herpesvirus DNA-binding protein LANA (30), was repurposed for our study as a non-sequence-specific DNA binding FP probe. We incubated the probe with increasing MBP-UBN1(122–584) to monitor the change in fluorescence polarization as the UBN1/DNA complex formed, and from the resulting FP curve, we were able to determine the Kd of the UBN1/DNA interaction to be 113 nm (Fig. 7C). Using the same FP assay, we were able to show that MBP-UBN1(176–295) alone is able to bind DNA with a highly comparable Kd of 90 nm, whereas MBP-UBN1(122–584, Δ176–295) did not have any detectable binding to DNA (Fig. 7C). Additionally, deletion of conserved lysine patches 182–195, 200–217, and 232–239 resulted in incomplete binding curves that could not be fit to determine Kd values, whereas deletion of 248–261 resulted in reduced binding with a Kd of 245 nm (Fig. 7C). These data combined indicate that UBN1 region 176–295 is necessary and sufficient for DNA binding, and the lysine-rich UBN1 region of 176–295 is essential for non-sequence-specific DNA binding.
Figure 7.
UBN1 residues 176–295 bind to DNA with nanomolar affinity. A, alignment of UBN1 residues 176–295 with metazoan homologs. B, alignment of UBN1(176–295) with vertebrate homologs. Lysine residues are clearly conserved among vertebrates. C, fluorescence polarization binding experiment conducted with MBP-UBN1 fragments and a double-stranded LBS1/2 FP probe. Deletion analysis of UBN1 shows that conserved lysine residues within the 176–295 region contribute to DNA-binding affinity. D, MBP-UBN1(122–584) resolved on a Superdex 200 10/300 column alone or with 2-fold excess LBS1/2 dsDNA (top) and MBP-UBN1(122–584, Δ176–295) resolved on a Superdex 200 10/300 column alone or with 2-fold excess LBS1/2 dsDNA (bottom). E, MBP-UBN1(176–295) resolved on a Superdex 200 column 10/300 alone (top) or with 2-fold excess 10xATGC dsDNA (bottom). F, MBP-UBN1(176–295) resolved on a Superpose 6 10/300 column alone (top) or with 2-fold excess 10xATGC dsDNA (bottom). G, a fluorescence polarization competition experiment was conducted by preforming a complex of UBN1 with FAM-tagged LBS1/2 DNA. This complex was then titrated with heparin, tRNA, or double-stranded 10xATGC DNA to determine the IC50 for the competitors.
To further confirm the veracity of our findings from the FP DNA-binding assay, we analyzed MBP-UBN1(122–584) WT and Δ176–295 mutant for binding with LBS1/2 dsDNA without a FAM label on size-exclusion chromatography. MBP-UBN1(122–584) WT was incubated with a 2-fold molar excess of LBS1/2 dsDNA, and the complex was resolved on a Superdex 200 10/300 column. In this experiment, we observed a significant shift in the elution volume of the MBP-UBN1(122–584) upon incubation with DNA, indicating the formation of a higher-molecular weight complex with DNA (Fig. 7D). In contrast, the elution profile of MBP-UBN1(122–584, Δ176–295) was not altered after DNA incubation, indicating a lack of binding with DNA (Fig. 7D).
Whereas binding of the “nonrelevant” LBS1/2 DNA strongly suggests UBN1 binding of DNA to be non-sequence-specific, to build better confidence, we proceeded to test UBN1 binding to DNA with random sequence. Specifically, we prepared a dsDNA 40 bp long, composed of 10 repeats of ATGC, which we named 10×ATGC. Excess 10×ATGC was incubated with MBP-UBN1(176–295), and the mixture was resolved on both the Superdex 200 and the Superose 6 10/300 size-exclusion column. Similar to what we have observed with LBS1/2 and MBP-UBN1(122–584), we have observed earlier elution of MBP-UBN1(176–295) after incubation with 10xATGC, which suggests a complex formation between the UBN1 protein and the DNA (Fig. 7, E and F). The binding of UBN1 to this random DNA fragment further suggests the non-sequence-specific nature of UBN1 binding to DNA.
To quantitatively compare the 10xATGC and LBS1/2 interactions as well as probe the DNA specificity of the interaction, a competition FP experiment was performed to compare the ability of 10xATGC DNA, heparin, and tRNA to compete UBN1 from the FAM-LBS1/2 DNA. Heparin and tRNA represent non-DNA molecules with strong negative charge. 10xATGC DNA was able to compete UBN1(122–584) away from FAM-LBS1/2 with an IC50 of 182 nm, whereas heparin had an IC50 of 80 nm and tRNA had an IC50 of 430 nm (Fig. 7G). Whereas there is some variability among the IC50 values for the three competitors, all interactions are on the same order of magnitude, indicating that UBN1 residues 176–295 have a propensity to nonspecifically bind negatively charged species. Although the interaction is based on nonspecific charge, these results, together with the known biological activity of UBN1 to participate in histone deposition into chromatin is most consistent with a biologically significant role of UBN1 in DNA binding.
The UBN1 HRD and middle domain bind H3/H4 in a mutually exclusive manner
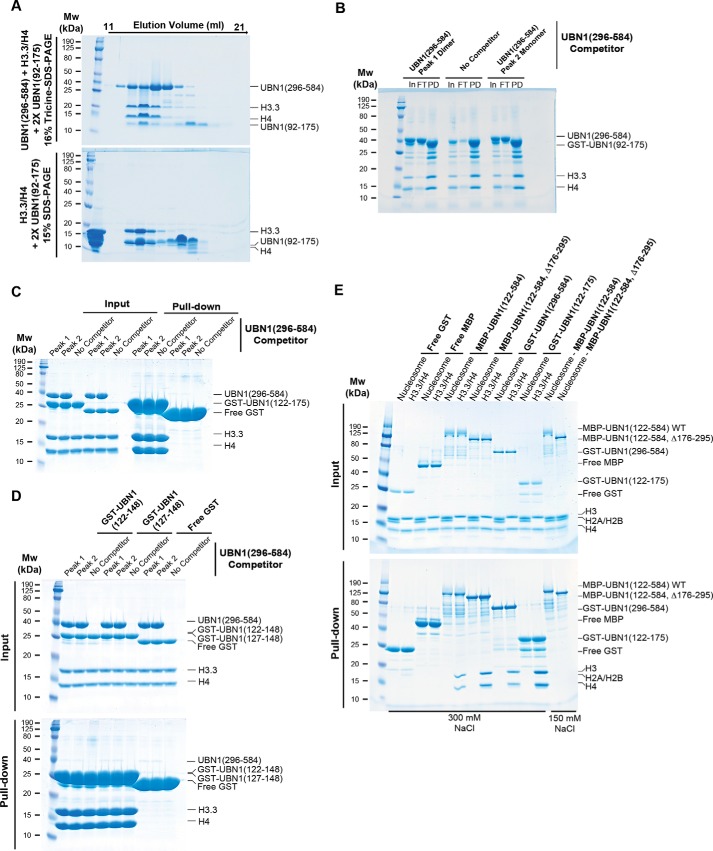
As both the UBN1 HRD and middle domain have histone-binding activity, we investigated whether they represent partial interactions of a larger UBN1/H3.3/H4 complex. First, we mixed UBN1(296–584), UBN1(92–175), and H3.3/H4 in a 2:2:1 ratio and resolved the mixture over a Superdex 200 10/300 size-exclusion column. We observed that UBN1(92–175) appears to form complex with H3.3/H4, but UBN1(296–584) does not appear to be incorporated in the complex (Fig. 8A, top). To further verify this result, we prepared just the 2:1 ratio of UBN1 (92–175) and H3.3/H4 and resolved the mixture over the same Superdex 200 10/300 column. We observed that the UBN1(92–175)/H3.3/H4 complex elutes in the same fractions, showing that there is no shift upon the addition of UBN1(296–584) (Fig. 8A, compare top and bottom). Following this result, we analyzed whether either the UBN1(296–584) monomer or dimer populations could alter the mutually exclusive nature of the UBN1 HRD and middle domain histone interactions. A pulldown histone-binding assay was performed through incubating either UBN1(296–584) monomer or dimer population with GST-UBN1(92–175) and H3.3/H4 prior to GST pulldown. Neither the UBN1(296–584) monomer nor dimer was able to be incorporated into the GST-UBN1(92–175)/H3.3/H4 complex, and the interaction level for GST-UBN1(92–175) with H3.3/H4 was not altered by the addition of a UBN1(296–584) monomer or dimer competitor (Fig. 8B). Further, we performed similar competition pulldown experiments, incubating UBN1(296–584) peak 1 dimer or peak 2 monomer with GST-UBN1(122–175), GST-UBN1(122–148), and GST-UBN1(127–148) (Fig. 8, C and D). In all three of these competition pulldown experiments, we observed that neither the UBN1(296–584) peak 1 dimer or peak 2 monomer was able to be incorporated into the UBN1 HRD complex with H3.3/H4. These data suggest that the UBN1 middle domain associates with a similar region of H3.3/H4 as the HRD. Our previous binding studies with the HRD have demonstrated that whereas the HRD binds specifically to the globular core of H3.3/H4, deletion of the N-terminal tails of H3.3/H4 does result in a roughly 2-fold reduction of HRD affinity for H3.3/H4, suggesting that the HRD does have some interaction with the histone tails that was not observed in our crystal structure of the UBN1 HRD bound to H3.3/H4/Asf1 (14). From these results and our observation that the middle domain binds with higher affinity to FL H3/H4 than to H3/H4 with the N-terminal tails removed, we hypothesize that the UBN1 middle domain and HRD may compete for similar binding sites on the H3/H4 core and N-terminal tails. These observations combined led us to conclude that the UBN1-HRD and middle domain interact with H3.3/H4 in a mutually exclusive manner.
Figure 8.
The UBN1 and HRD show mutually exclusive interaction with H3/H4 and do not bind to nucleosomes. A, size-exclusion chromatography experiment performed by adding 2-fold excess UBN1(296–584) and UBN1(92–175) to H3.3/H4 and resolving the mixture on a Superdex 200 10/300 column (top). Control was run by adding 2-fold UBN1(92–175) to H3.3/H4 without UBN1(296–584) (bottom). B, GST pulldown conducted by mixing GST-UBN1(92–175), H3.3/H4, and either UBN1(296–584) peak 1 or peak 2. C, GST pulldown conducted by mixing GST-UBN1(122–175), H3.3/H4, and either UBN1(296–584) peak 1 or peak 2. D, GST pulldown conducted by mixing GST-UBN1(122–148) or GST-UBN1(127–148) with H3.3/H4, and either UBN1(296–584) peak 1 or peak 2. E, pulldown experiment comparing the ability of MBP-UBN1(122–584, WT), MBP-UBN1(122–584, Δ176–295), GST-UBN1(296–584), and GST-UBN1(122–175) to bind free H3.3/H4 or nucleosomes. As UBN1 residues 176–295 have been shown to contribute to DNA binding, the pulldown with MBP-UBN1(122–584, WT) and MBP-UBN1(122–584, Δ176–295) was attempted at 150 mm NaCl as well as 300 mm NaCl.
UBN1 binds free H3/H4 but does not bind nucleosomes or tetrasomes
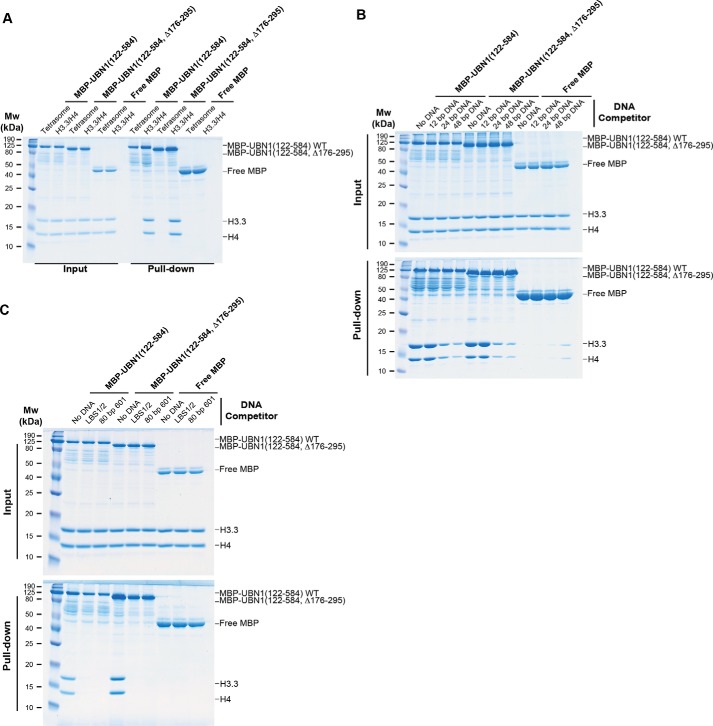
Our identification and characterization of the additional histone binding and DNA binding regions in UBN1 prompted us to investigate whether these newly characterized domains in UBN1 could be involved in nucleosome binding. Nucleosomes were prepared with histones H3.1, H4, H2A, and H2B and a 147-bp dsDNA fragment bearing the 601-nucleosome positioning sequence (31). Although UBN1-HRD binding is selective for H3.3/H4 (14), the UBN1 middle domain binds H3.1/H4 and H3.3/H4 with similar affinity, and the H3.3-specific contact surface of the UBN1-HRD is not accessible in the nucleosome core particle. We compared binding of nucleosomes versus H3.3/H4 alone with several different UBN1 fragments in a pulldown-based binding assay. MBP-UBN1(122–584, WT), MBP-UBN1(122–584, Δ176–295), GST-UBN1(296–584), and GST-UBN1(122–175) were incubated with nucleosomes or H3.3/H4 and subjected to pulldown. We observed that whereas all four UBN1 fragments were capable of binding free H3.3/H4, none showed any detectable binding with nucleosomes (Fig. 8E). We also performed the nucleosome pulldown for MBP-UBN1(122–584, WT) and MBP-UBN1(122–584, Δ176–295) at 150 mm NaCl in addition to 300 mm, and we still observed no nucleosome binding at the lower salt concentration (Fig. 8E). Based on these results, we conclude that UBN1 likely does not participate in nucleosome binding. Further, we assembled H3.3/H4 tetrasomes with an 80-bp 601-nucleosome positioning sequence. We employed a pulldown assay to compare association of free H3.3/H4 or H3.3/H4 tetrasomes with either MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP. We observed that both MBP-UBN1(122–584) and MBP-UBN1(122–584, Δ176–295) were able to bind the free H3.3/H4 but were not able to bind to the H3.3/H4 tetrasomes (Fig. 9A). This result further indicates that UBN1 is unable to bind to H3.3/H4 after it has been deposited onto DNA.
Figure 9.
UBN1 does not bind to tetrasomes and competes for H3. 3/H4 binding with DNA fragments of 24 bp or larger. A, MBP pulldown conducted by mixing free H3.3/H4 or H3.3/H4 tetrasomes with either MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP. B, MBP pulldown conducted by mixing MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP with H3.3/H4 alone or in competition with excess DNA competitor of 12, 24, or 48 bp. C, MBP pulldown conducted by mixing MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP with H3.3/H4 alone or in competition with excess LBS1/2 or 80-bp 601-nucleosome positioning DNA.
DNA fragments of 24 bp or longer are sufficient to compete H3.3/H4 away from UBN1
To further analyze the mechanism of H3.3/H4 deposition by UBN1, we conducted a series of competition pulldown experiments with dsDNA fragments of varied length composed of repeats of ATGC measuring 12, 24, or 48 bp. MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP was incubated with H3.3/H4 alone or in competition with 3-fold molar excess DNA and subjected to pulldown. We observed that both MBP-UBN1(122–584) and MBP-UBN1(122–584, Δ176–295) were able to associate well with free H3.3/H4 or H3.3/H4 in competition with the 12-bp DNA fragment, but the 24- and 48-bp fragments were able to compete a significant portion of the H3.3/H4 away from UBN1 (Fig. 9B). This result indicates that stretches of DNA measuring 24 bp or more are sufficient to bind H3.3/H4 and prevent association with UBN1. We also tested the LBS1/2 and 80-bp 601-nucleosome positioning DNA sequences by the same competition pulldown assay and observed that both of these fragments are able to prevent UBN1 association with H3.3/H4 (Fig. 9C). These results suggest a mechanism of deposition where UBN1 disassociates from H3.3/H4 immediately upon transfer to DNA.
The UBN1 HRD, middle domain, and basic loop are required for senescent-associated heterochromatin focus (SAHF) formation
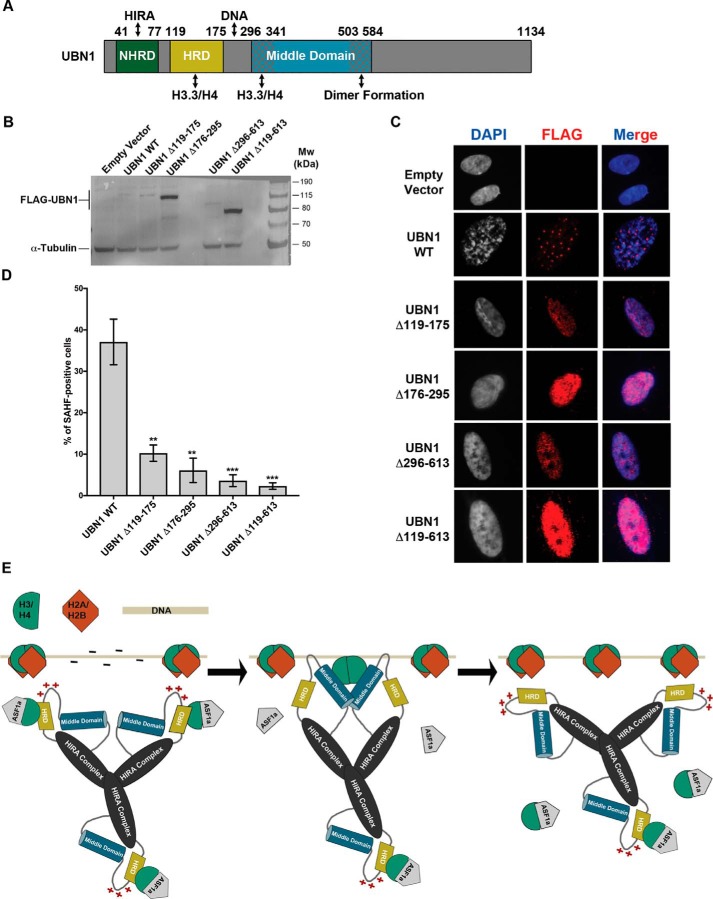
Senescent human cells often accumulate distinct DAPI-dense nuclear foci known as SAHFs (32). Ectopic expression of either HIRA, UBN1, or H3.3 can induce the formation of SAHF in a manner that depends on integrity of the HIRA histone chaperone complex (12, 13, 33–35). Conversely, disruption of the chaperone complex or its subnuclear localization by RNAi or a dominant negative mutant blocks SAHF formation (35, 36). Whereas the HIRA complex does not obviously co-localize with SAHF, in senescent cells, the chaperone complex is recruited to PML bodies, subnuclear regions implicated in diverse nuclear functions, including SAHF formation (12, 13, 34–37). Having shown the H3.3/H4-binding activity of both the UBN1 HRD and middle domain and the DNA-binding activity of basic loop, we next asked about the role of these domains inside the cell. We prepared the deletion constructs of HRD (Δ119–175), basic loop (Δ176–295), middle domain (Δ296–613), and a combination of all three (Δ119–613) and investigated whether these domains are involved in formation of SAHF in senescent human cells (Fig. 10A). UBN1 WT and the different domain deletions were produced in cells through ectopic expression by retroviral infection; protein expression was monitored by Western blotting (Fig. 9B).
Figure 10.
UBN1 induced SAHF formation in IMR90 cells and a proposed model for H3.3/H4 deposition by the HIRA complex. A, domain architecture diagram depicting the characterized UBN1 domains and proposed functions. B, immunoblot of IMR90 cells stably expressing full-length UBN1 (WT) and deletion mutants or empty pBabe vector with FLAG. α-Tubulin was used as loading control. C, immunofluorescence of IMR90 cells stably expressing full-length UBN1 (WT) and deletion mutants or empty pBabe vector with FLAG. D, quantification of SAHF; mean ± S.D. (error bars) (n = 3 biological replicates). p values are as indicated; two-tailed unpaired Student's t test. **, p < 0.01; ***, p < 0.001. E, proposed model for H3.3/H4 deposition coordinated by the HIRA complex based on our observations and previously reported data. The three-step model first shows an H3.3/H4 dimer bound by ASF1 and the UBN1-HRD. Next, UBN1 binds DNA, and the middle domain forms a dimer and associates with H3.3/H4 to displace ASF1a and the UBN1-HRD to allow for (H3.3/H4)2 tetramer formation. Finally, the HIRA complex disassociates from the (H3.3/H4)2 tetramer and DNA to allow additional chaperones to deposit H2A/H2B and coordinate final nucleosome formation.
Expression of UBN1 WT markedly induced SAHF formation over a 10-day period in about 40% of cells (in line with previous observations (13)), whereas the absence of the tested domains heavily perturbed SAHF formation (Fig. 10, C and D)). In addition, as shown previously (13), ectopically expressed UBN1 WT was recruited in the PML nuclear bodies in the cells (25%), but the mutants were unable to localize to PML bodies and were expressed throughout the nucleoplasm in a fine speckled pattern (Fig. 10C). Hence, the HRD, lysine-rich loop, and middle domain are required for SAHF formation and localization to the PML bodies. Because localization of the HIRA chaperone complex to PML bodies is thought to be a prerequisite for SAHF formation (36), these results do not allow us to pinpoint a role for the HRD, lysine-rich loop, and/or middle domain directly in SAHF formation. As another caveat, two of the UBN1 mutants were expressed at a substantially higher level than WT, perhaps suggesting their misfolding in cells. Despite these caveats, these results underscore the importance of these domains for normal function of the UBN1 protein in cells. Together, these in vitro and in vivo data suggest that UBN1, via direct interaction with H3.3 through the HRD and middle domain, participates in the incorporation of H3.3 into chromatin, and the binding of UBN1 to chromatin may also be mediated by the basic loop.
Discussion
UBN1 is an essential member of the HIRA complex, but only the very small HRD and NHRD domains have been studied in any depth. In our attempt to characterize regions of UBN1 outside of the well-studied HRD and NHRD domains, we have identified the middle domain as a structured region of UBN1 that has dimerization activity, with residues 504–584 necessary for dimer formation. We have also demonstrated that residues 296–241 of UBN1 middle domain are sufficient for binding histones H3/H4 in a manner that is mutually exclusive with the HRD. Although this mutually exclusive interaction is somewhat confounding, the observations may be explained by a hand-off mechanism. Whereas we observe preferential interaction of H3.3/H4 with the HRD over the middle domain, there may be some sort of biological switch, such as a post-translational modification or allosteric process, that allows for hand-off from one domain to the other, although further investigation is necessary to confirm this hypothesis. Additionally, we identified that the loop separating the HRD and middle domain harbors many conserved lysine residues that have nonspecific DNA-binding activity. We have shown that although UBN1 is capable of binding H3/H4 and DNA in isolation, it does not associate with assembled nucleosomes. Finally, we have confirmed the functional relevance of the UBN1 middle domain and basic loop with a SAHF formation assay in senescent cells. Several studies have identified dimer-formation in other histone chaperones that bind to an (H3.3/H4)2 tetramer for nucleosome deposition (17, 19, 21, 23, 25, 26), and it was previously demonstrated that the majority of H3.3/H4 is deposited into nucleosomes in the tetramer form (22). We recently reported that a C-terminal domain in the HIRA subunit forms a stable trimer and is required for H3.3/H4 deposition in cells (27). This HIRA trimerization domain may cooperate with the UBN1 middle domain dimer to deposit (H3.3/H4)2 tetramers onto DNA, although further experiments are necessary to fully elucidate the mechanism of H3.3/H4 deposition by the HIRA complex. Human UBN1 has been previously shown to exhibit DNA-binding activity (28), as have several other histone chaperones, such as FACT (38) and CAF-1 (15, 39). From these observations in combination with ours, we propose a model for HIRA complex function in which UBN1 binds and stabilizes a (H3.3/H4)2 tetramer prior to H3.3/H4 deposition, UBN1 releases from H3.3/H4 following deposition onto DNA, and final nucleosome assembly occurs with the aid of additional histone chaperones and cellular factors (Fig. 10E). Because we have observed no difference between UBN1(122–584) WT and Δ176–295 in our tetrasome/nucleosome and DNA competition binding studies, it is possible that the DNA-binding domain of UBN1 may function to target the HIRA complex to chromatin or even stretches of nucleosome-free DNA, as the HIRA complex has been previously reported to target nucleosome-free stretches of DNA for deposition of H3.3/H4 in a gap-filling mechanism (28, 40).
Experiments with the full-length HIRA complex and different HIRA subcomplexes are needed to rigorously confirm our model, as we have only observed a lack of nucleosome and tetrasome binding for UBN1 in isolation. It has recently been reported that the WD40 domain of HIRA is essential for nucleosome assembly (41), although this observation could be due to the function of HIRA as a molecular scaffold for the HIRA complex, as the UBN1-NHRD binds to the HIRA-WD40 domain (11). Additionally, our observation of mutually exclusive H3.3/H4 binding by the UBN1-HRD and middle domain is somewhat confounding. Further investigation will be required to determine whether there is a potential hand-off or other allosteric mechanism that could be driven by a yet to be determined factor. Of particular interest to our studies is that the CAF-1 complex has recently been reported to form an H3/H4 tetramer only upon binding to an H3/H4 dimer together with DNA, which allows a CAF-1 dimer drive (H3/H4)2 tetramer formation prior to deposition (16). Perhaps UBN1 could operate through a similarly triggered mechanism. Although there is still much that remains unknown about how the HIRA complex and other histone chaperones mediate histone deposition, our data suggest that UBN1 may function similarly to other H3/H4 histone chaperones through a mechanism that employs both dimerization and DNA-binding activities to efficiently deposit (H3.3/H4)2 onto DNA for nucleosome formation.
Experimental procedures
Multiple-sequence alignments
Alignments for Figs. 1A and 6A were generated using Clustal Omega (42) and formatted for better visualization using ESPript version 3.0 (43).
Protein secondary structure prediction
The predicted secondary structure of UBN1 depicted in Fig. 1B was generated using the PSIPRED secondary structure prediction server (29).
Generation of expression plasmids
The plasmids encoding His-UBN1(296–584), His-UBN1(341–584), and His-UBN1(296–503) were generated by PCR amplification from an FL UBN1 construct (13) and ligation into the BamHI/XhoI sites of a custom-engineered pCDFduet-1 (Novagen) E. coli expression vector carrying an N-terminal His6 tag that is removable by TEV protease cleavage. The plasmids encoding GST-UBN1(296–584), GST-UBN1(296–503), GST-UBN1(316–503), GST-UBN1(330–503), GST-UBN1(341–503), GST-UBN1(296–341), GST-UBN1(296–341 with C-terminal cystine), GST-UBN1(92–175), GST-UBN1(122–148), GST-UBN1(127–148), and GST-UBN1(122–175) were generated by PCR amplification and ligation into the BamHI/XhoI sites of a custom engineered pCDFduet-1 (Novagen) E. coli expression vector carrying an N-terminal GST tag that is removable by TEV protease cleavage. The plasmid encoding His-MBP-UBN1(122–584) was generated by PCR amplification and ligation into the BamHI/XhoI sites of a pDB.His.MBP E. coli expression vector carrying an N-terminal His-MBP tag that is removable by TEV protease cleavage (44). The plasmids encoding His-MBP-UBN1(122–584, Δ176–295), His-MBP-UBN1(122–584, Δ182–195), His-MBP-UBN1(122–584, Δ200–217), His-MBP-UBN1(122–584, Δ232–239), and His-MBP-UBN1(122–584, Δ248–261) were generated by site-directed mutagenesis (45). The plasmids encoding FL H3.1, H3.3, and H4 were generated as described previously (14). The plasmid encoding H2B was a gift from Don Cleveland (University of California San Diego, La Jolla, CA). The plasmids encoding H3.3(45–135) and H4(20–102) were generated by PCR amplification of the H3.3 and H4 sequences and ligation into the BamHI/XhoI sites of custom engineered pETduet-1 (Novagen) E. coli expression vector carrying an N-terminal His tag that is removable by TEV protease cleavage. The retrovirus construct encoding FL FLAG-UBN1 WT was generated by PCR amplification and ligation into the BamHI/SalI sites of the pBABE-puro plasmid (Addgene). pBABE FLAG-UBN1 deletions (Δ119–175, Δ176–295, Δ296–613, and Δ119–613) were generated through site-directed mutagenesis.
Protein expression and purification
Individual histone proteins H2A, H2B, H3, and H4 were expressed to the inclusion bodies in BL21-Gold(DE3) cells (Agilent). The individual histone proteins were purified and refolded to form H3/H4 and H2A/H2B as described previously (14, 31). These proteins were additionally used to form tetrasomes and nucleosomes with the addition of 601-nucleosome positioning sequence dsDNA according to previously reported protocols (31, 46). His-UBN1(296–584), His-UBN1(341–584), and His-UBN1(296–503) were generated in BL21-Gold(DE3) cells (Agilent) induced with 0.8 mm isopropyl 1-thio-β-d-galactopyranoside and expressed overnight at 18 °C. Cells were resuspended in buffer containing 20 mm Tris, pH 8.0, 500 mm NaCl, 5 mm BME, and 10 mm imidazole and lysed with sonication. Lysate was clarified with centrifugation, and the supernatant was subjected to nickel affinity chromatography to isolate the His-tagged protein. Proteins were eluted with 200 mm imidazole and dialyzed overnight with the addition of TEV protease to remove the His tag. Dialysis buffer contained 20 mm Tris, pH 7.5, 50 mm NaCl, and 5 mm BME. Proteins were then subjected to ion-exchange chromatography using a HiTrap SP column (GE Healthcare). Following ion-exchange chromatography, proteins were concentrated using a spin concentrator (Millipore) and loaded either onto either a Superdex 200 10/300 or 16/600 column (GE Healthcare) in buffer with 20 mm Tris, pH 7.5, 300 mm NaCl, and 1 mm TCEP for further purification and analysis by gel filtration. GST-UBN1(296–584), GST-UBN1(296–503), GST-UBN1(316–503), GST-UBN1(330–503), GST-UBN1(341–503), GST-UBN1(92–175), GST-UBN1(296–341), GST-UBN1(296–341 with C-terminal cystine), GST-UBN1(122–148), GST-UBN1(127–148), and GST-UBN1(122–175) were generated in BL21-Gold(DE3) cells (Agilent) induced with 0.8 mm isopropyl 1-thio-β-d-galactopyranoside and expressed overnight at 18 °C. Cells were resuspended in 1× PBS supplemented with 5 mm BME and lysed by sonication. Lysate was clarified by centrifugation, and supernatant was subjected to GST affinity chromatography. Protein was eluted with 20 mm reduced GSH and dialyzed into a buffer containing 20 mm Tris, pH 7.5, 300 mm NaCl, and 5 mm BME for storage. UBN1(92–175) was generated by adding TEV protease to GST-UBN1(92–175) bound to GST affinity resin followed by incubation at 4 °C overnight. Cleaved protein was eluted with a buffer containing 20 mm Tris, pH 7.5, 300 mm NaCl, and 5 mm BME. Eluted protein was concentrated using a spin concentrator (Millipore) and loaded onto a Superdex 75 10/300 (GE Healthcare) for further purification by gel filtration in buffer with 20 mm Tris, pH 7.5, 300 mm NaCl, and 1 mm TCEP. His-MBP-UBN1(122–584, Δ176–295), His-MBP-UBN1(122–584, Δ182–195), His-MBP-UBN1(122–584, Δ200–217), His-MBP-UBN1(122–584, Δ232–239), and His-MBP-UBN1(122–584, Δ248–261) were expressed and purified by the same method as His-UBN1(296–584), His-UBN1 (341–584), and His-UBN1(296–503) with omission of the TEV cleavage step.
Size-exclusion analysis of UBN1 peak 1 and peak 2 distribution
UBN1(296–584) was purified through the ion-exchange step, and protein eluted from the ion-exchange column was pooled and loaded onto a Superdex 200 16/600 column (GE Healthcare) in buffer containing 20 mm Tris, pH 7.5, 300 mm NaCl, and 1 mm TCEP. Protein eluting in the range of 65–75 ml was pooled as peak 1, and protein eluting in the range of 76–89 ml was pooled as peak 2. Both peak 1 and peak 2 were reloaded back onto the column for further analysis of distribution. UBN1(341–584) was analyzed by the same method as UBN1(296–584) with the exception that peak 1 was defined as 68–78 ml and peak 2 was defined as 80–94 ml. UBN1(296–503) was analyzed by the same method as UBN1(296–584) with the exception that peak 1 was defined as 70–78 ml and peak 2 was defined as 80–90 ml. Analysis of UBN1(296–584) peak 1 and peak 2 on the analytical Superdex 200 10/300 column was conducted by loading protein isolated from the first run on the Superdex 200 16/600 column onto the analytical column for comparison with molecular weight standards (Bio-Rad).
Analytical ultracentrifugation–sedimentation equilibrium
UBN1(296–584) peak 1 was prepared for sedimentation equilibrium analysis by conducting an initial gel filtration run on a Superdex 200 16/600 column. Protein eluting in the region of 65–75 ml was pooled and reloaded onto the same Superdex 200 16/600 column; the protein eluting in the same region for the second run was then pooled and analyzed for concentration. Samples were prepared at A280 nm of 0.3, 0.5, and 0.7, loaded into a 6-channel ultracentrifugation cell with buffer blanks, and analyzed for equilibrium distribution analysis at 12,000, 18,000, and 26,000 rpm using an Optima XL-I analytical ultracentrifuge (Beckman). Equilibrium distribution data were fit with the program HeteroAnalysis using the ideal fitting model (47). UBN1(296–584) peak 2 was prepared for sedimentation equilibrium analysis by conducting an initial gel filtration run on a Superdex 200 16/600 column. Protein eluting in the region of 76–89 ml was pooled and reloaded onto the Superdex 200 16/600 column. The protein eluting in the same region for the second run was then pooled and analyzed for concentration. Protein was analyzed for equilibrium distribution similarly to peak 1.
LC-MS analysis of protein samples
LC-MS digestion and peptide analysis were performed by the proteomics and metabolomics facility at the Wistar Institute (Philadelphia, PA). We provided the facility with an SDS-polyacrylamide gel stained with Coomassie Brilliant Blue G-250 and indicated the protein bands for analysis.
GST/MBP pulldown binding assay
Pulldown experiments were conducted by incubating 2 μm tagged bait protein with 4 μm untagged prey protein for 30 min at 4 °C. Proteins were then subjected to pulldown by incubation with GSH agarose (Fisher Scientific) or amylose (New England Biolabs) resin for 30 min. Bound resin was washed with 120 column volumes of buffer before elution of bound proteins by boiling resin in SDS loading buffer. Pulldowns were analyzed by visualization using SDS-PAGE stained with Coomassie Brilliant Blue G-250. All pulldowns were conducted in buffer with 20 mm Tris, pH 7.5, 300 mm NaCl, and 1 mm TCEP with the exception of those in Fig. 5A (conducted in both this buffer and one with 750 mm NaCl) and Fig. 7C (conducted in both this buffer and one with 150 mm NaCl). The pulldown conducted in Fig. 7B was conducted by mixing 2 μm GST-UBN1(92–175), 2 μm H3.3/H4, and 4 μm UBN1(296–584) peak 1 or peak 2. Pulldowns conducted in Fig. 7 (C and D) were conducted by mixing 10 μm GST-UBN1(122–175), GST-UBN1(122–148), or GST-UBN1(127–148) with 10 μm H3.3/H4 and 10 μm UBN1(296–584) peak 1 or peak 2. The pulldown conducted in Fig. 9A was carried out by mixing 1 μm MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP with 1 μm free H3.3/H4 or 1 μm H3.3/H4 tetrasome. Pulldowns conducted in Fig. 9 (B and C) were carried out by mixing 5 μm MBP-UBN1(122–584), MBP-UBN1(122–584, Δ176–295), or free MBP with 5 μm H3.3/H4 and 15 μm competitor DNA.
Formation of protein/protein complexes using size-exclusion chromatography
To form complexes with UBN1(296–584) peak 1 or peak 2 and H3.3/H4 for Fig. 5B, 200 μm UBN1 (296–584) was incubated with 400 μm H3.3/H4 and resolved on a Superdex 200 10/300 column (GE Healthcare) in a buffer with 20 mm Tris, pH 7.5, 300 mm NaCl, and 1 mm TCEP. H3.3/H4 was run alone at a concentration of 100 μm. For analysis of UBN1(92–175) and UBN1(296–584) H3.3/H4 binding cooperation for Fig. 7A, 200 μm UBN1(296–584), 200 μm UBN1(92–175), and 100 μm H3.3/H4 were incubated in a buffer containing 20 mm Tris, pH 7.5, 300 mm NaCl, and 1 mm TCEP and resolved on a Superdex 200 10/300 column (GE Healthcare). Analysis of UBN1(92–175) and H3.3/H4 alone was conducted by incubating 200 μm UBN1(92–175) with 100 μm H3.3/H4 and resolving the mixture on the column in the same 20 mm Tris, pH 7.5, 300 mm NaCl, 1 mm TCEP buffer.
DNA probes
The FAM-labeled LBS1/2 (LANA-binding sites 1 and 2) dsDNA used for the fluorescence polarization DNA-binding assay was assembled from synthesized DNA fragments (IDT). This FAM-labeled probe contains a DNA sequence recognized by the KSHV protein LANA and was originally used to study this interaction (30). It was repurposed for our study, as UBN1 has nonspecific DNA-binding activity. Unlabeled LBS1/2, 10xATGC, 12-bp ATGC, 24-bp ATGC, 48-bp ATGC, and 80-bp 601-nucleosome positioning dsDNA fragments were assembled from synthesized sense and antisense fragments. Sense fragment sequences were as follows: LBS1/2, AGCGGCCCCATGCCCGGGCGGGAGGCGCCGCAGGCCCCGGCGGCGT; 10xATGC, ATGCATGCATGCATGCATGCATGCATGCATGCATGCATGCATGCATGC; 12-bp ATGC, ATGCATGCATGC; 24-bp ATGC, ATGCATGCATGCATGCATGCATGC; 48-bp ATGC, ATGCATGCATGCATGCATGCATGCATGCATGCATGCATGCATGCATGC; 80-bp 601, GTCGTAGACAGCTCTAGATCCGCTTAATCGAACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTA.
Sense and antisense fragments were dissolved in a buffer with 10 mm Tris, pH 7.5, 10 mm NaCl, and 1 mm EDTA, mixed in a 1:1, ratio and boiled followed by slow cooling to room temperature to anneal and form dsDNA.
Fluorescence polarization binding assays
To quantitatively analyze the UBN1/DNA interaction, a fluorescence polarization-based binding assay was performed. Protein and DNA used for this assay were prepared in buffer containing 20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm TCEP, and 2 mg/ml BSA. The change in fluorescence polarization was monitored as increasing concentration of UBN1 protein was titrated into FAM-LBS1/2 DNA (10 nm) to determine the Kd of the UBN1/DNA interaction. A competition-binding assay was also performed to determine the IC50 for the interaction between UBN1 and heparin (Fisher), tRNA, and 10xATGC DNA. To monitor the ability of each competitor to compete UBN1 from LBS1/2 DNA, increasing concentration of competitor was titrated into solution containing 10 nm FAM-LBS1/2 and 300 nm MBP-UBN1(122–584). The tRNA used for this study was a pooled tRNA purified from yeast (Invitrogen).
To quantitatively analyze the UBN1/histone interaction, we site-specifically labeled a fragment of UBN1 (296–341 with a C-terminal cysteine) with fluorescein. Labeling of the free cysteine was conducted with 25-fold excess fluorescein-5-maleimide (Thermo Fisher Scientific) and UBN1(296–341 with C-terminal cysteine) incubated overnight in buffer with 20 mm HEPES, pH 7.5, 300 mm NaCl. After conjugation was complete, the reaction mixture was concentrated and run on a Superdex 75 10/300 GL size-exclusion column (GE Healthcare) in buffer containing 20 mm HEPES, pH 7.5, 300 mm NaCl, and 1 mm TCEP. UBN1(296–341) and histone complexes used for this assay were prepared in buffer containing 20 mm HEPES, pH 7.5, 300 mm NaCl, 1 mm TCEP, and 2 mg/ml BSA. The change in fluorescence polarization was monitored as increasing concentration histones were titrated into fluorescein-labeled UBN1(296–341) (10 nm) to determine the Kd of the UBN1/histone interaction. To obtain a suitable fluorescence polarization range in the competition binding assay, the concentration of H3/H4, H3.3(45–135)/H4(20–102), or H2A/H2B was held at or slightly above the relative dissociation constant for fluorescein-labeled UBN1(296–341). Increasing concentration of GST-UBN1(296–341) competitor was titrated into solutions containing constant concentrations of fluorescein-labeled UBN1(296–641) (10 nm) and either H3.3/H4 (15 μm), H3.1/H4 (15 μm), H3.3(45–135)/H4(20–102) (25 μm), or H2A/H2B (75 μm) to estimate IC50 values by competition binding. All data were collected using a PerkinElmer EnVision Xcite Multilabel plate reader, and curves were fit using a one site–total binding model with GraphPad Prism (version 5.0a).
UBN1 ectopic expression SAHF formation assay
IMR90 fibroblasts were procured from Coriell Institute and grown in an environment of 5% CO2 and 3.5% oxygen using Dulbecco's modified Eagle's medium with 10% fetal bovine serum. To produce the retroviruses, HEK293-T cells were co-transfected with retroviral vector pBabe-FLAG-UBN1 full-length and deletion mutants, VSV-G envelope plasmid, and M57 (MLV gag-pol expression plasmid). Medium was changed after 6 h. Supernatants containing viral particles were collected 24–48 h after transfection and filtered through a 0.45-μm membrane. IMR-90 cells were infected with these supernatants using Polybrene (8 μg/ml). After 10 days of infection, cells were fixed in neutral buffered formalin for immunofluorescence and harvested in EBC500 lysis buffer for Western blotting. Monoclonal anti-FLAG antibody was purchased from Sigma (catalog no. F3165).
Author contributions
M. D. R., N. D., J. F., J. H., M. G., and Y. T. carried out experiments reported in the manuscript; M. D. R. and N. D. prepared the first draft of the manuscript figures and text; R. M. designed and supervised experiments by M. D. R., J. F., J. H., and Y. T.; P. D. A. designed and supervised experiments by N. D.; M. G. provided essential reagents and methodologies; B. E. B. provided essential methodologies and edited the manuscript. All authors read and approved the submitted manuscript.
Acknowledgments
The plasmid encoding H2B was a gift from Don Cleveland (University of California San Diego, La Jolla, CA), and the plasmid encoding H4 was a gift from Karolin Luger (University of Colorado, Boulder, CO). We acknowledge the Proteomics and Metabolomics Facility at the Wistar Institute for preparing gel/LC-MS/MS data and the Molecular Screening and Protein Expression Facility at the Wistar Institute for use of the PerkinElmer EnVision Xcite Multilabel plate reader.
This work was supported by National Institutes of Health Grants AG031862 (to R. M.) and GM082989 (to B. E. B.) and American Heart Association Predoctoral Fellowship 12PRE12030157 (to M. D. R.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HRD
- Hpc2-related domain
- SAHF
- senescent-associated heterochromatin focus
- TEV
- tobacco etch virus
- BME
- β-mercaptoethanol
- TCEP
- tris(2-carboxyethyl)phosphine
- MBP
- maltose-binding protein
- FAM
- fluorescein amidite
- DAPI
- 4′,6-diamidino-2-phenylindole
- PML
- promyelocytic leukemia protein.
References
- 1. Laskey R. A., Honda B. M., Mills A. D., and Finch J. T. (1978) Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275, 416–420 10.1038/275416a0 [DOI] [PubMed] [Google Scholar]
- 2. Burgess R. J., and Zhang Z. (2013) Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 10.1038/nsmb.2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mattiroli F., D'Arcy S., and Luger K. (2015) The right place at the right time: chaperoning core histone variants. EMBO Rep. 16, 1454–1466 10.15252/embr.201540840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maze I., Noh K. M., Soshnev A. A., and Allis C. D. (2014) Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 15, 259–271 10.1038/nrg3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tagami H., Ray-Gallet D., Almouzni G., and Nakatani Y. (2004) Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 10.1016/S0092-8674(03)01064-X [DOI] [PubMed] [Google Scholar]
- 6. English C. M., Adkins M. W., Carson J. J., Churchill M. E., and Tyler J. K. (2006) Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 10.1016/j.cell.2006.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. English C. M., Maluf N. K., Tripet B., Churchill M. E., and Tyler J. K. (2005) ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry 44, 13673–13682 10.1021/bi051333h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonnefoy E., Orsi G. A., Couble P., and Loppin B. (2007) The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 3, 1991–2006 10.1371/journal.pgen.0030182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ray-Gallet D., Quivy J. P., Silljé H. W., Nigg E. A., and Almouzni G. (2007) The histone chaperone Asf1 is dispensable for direct de novo histone deposition in Xenopus egg extracts. Chromosoma 116, 487–496 10.1007/s00412-007-0112-x [DOI] [PubMed] [Google Scholar]
- 10. Tang Y., Poustovoitov M. V., Zhao K., Garfinkel M., Canutescu A., Dunbrack R., Adams P. D., and Marmorstein R. (2006) Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13, 921–929 10.1038/nsmb1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang Y., Puri A., Ricketts M. D., Rai T. S., Hoffmann J., Hoi E., Adams P. D., Schultz D. C., and Marmorstein R. (2012) Identification of an ubinuclein 1 region required for stability and function of the human HIRA/UBN1/CABIN1/ASF1a histone H3.3 chaperone complex. Biochemistry 51, 2366–2377 10.1021/bi300050b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rai T. S., Puri A., McBryan T., Hoffman J., Tang Y., Pchelintsev N. A., van Tuyn J., Marmorstein R., Schultz D. C., and Adams P. D. (2011) Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol. Cell. Biol. 31, 4107–4118 10.1128/MCB.05546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banumathy G., Somaiah N., Zhang R., Tang Y., Hoffmann J., Andrake M., Ceulemans H., Schultz D., Marmorstein R., and Adams P. D. (2009) Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell. Biol. 29, 758–770 10.1128/MCB.01047-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ricketts M. D., Frederick B., Hoff H., Tang Y., Schultz D. C., Singh Rai T., Grazia Vizioli M., Adams P. D., and Marmorstein R. (2015) Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6, 7711 10.1038/ncomms8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sauer P. V., Timm J., Liu D., Sitbon D., Boeri-Erba E., Velours C., Mücke N., Langowski J., Ochsenbein F., Almouzni G., and Panne D. (2017) Insights into the molecular architecture and histone H3-H4 deposition mechanism of yeast Chromatin assembly factor 1. eLife 6, e23474 10.7554/eLife.23474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mattiroli F., Gu Y., Yadav T., Balsbaugh J. L., Harris M. R., Findlay E. S., Liu Y., Radebaugh C. A., Stargell L. A., Ahn N. G., Whitehouse I., and Luger K. (2017) DNA-mediated association of two histone-bound complexes of yeast Chromatin Assembly Factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. eLife 6, e22799 10.7554/eLife.22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang H., Strømme C. B., Saredi G., Hödl M., Strandsby A., González-Aguilera C., Chen S., Groth A., and Patel D. J. (2015) A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 22, 618–626 10.1038/nsmb.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen S., Rufiange A., Huang H., Rajashankar K. R., Nourani A., and Patel D. J. (2015) Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2. Genes Dev. 29, 1326–1340 10.1101/gad.261115.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zasadzińska E., Barnhart-Dailey M. C., Kuich P. H., and Foltz D. R. (2013) Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 32, 2113–2124 10.1038/emboj.2013.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., Wang M., Yang N., and Xu R. M. (2015) Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell 6, 693–697 10.1007/s13238-015-0190-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richet N., Liu D., Legrand P., Velours C., Corpet A., Gaubert A., Bakail M., Moal-Raisin G., Guerois R., Compper C., Besle A., Guichard B., Almouzni G., and Ochsenbein F. (2015) Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 43, 1905–1917 10.1093/nar/gkv021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu M., Long C., Chen X., Huang C., Chen S., and Zhu B. (2010) Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 10.1126/science.1178994 [DOI] [PubMed] [Google Scholar]
- 23. Deleted in proof.
- 24. Quivy J. P., Grandi P., and Almouzni G. (2001) Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 20, 2015–2027 10.1093/emboj/20.8.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su D., Hu Q., Li Q., Thompson J. R., Cui G., Fazly A., Davies B. A., Botuyan M. V., Zhang Z., and Mer G. (2012) Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106. Nature 483, 104–107 10.1038/nature10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fazly A., Li Q., Hu Q., Mer G., Horazdovsky B., and Zhang Z. (2012) Histone chaperone Rtt106 promotes nucleosome formation using (H3-H4)2 tetramers. J. Biol. Chem. 287, 10753–10760 10.1074/jbc.M112.347450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ray-Gallet D., Ricketts M. D., Sato Y., Gupta K., Boyarchuk E., Senda T., Marmorstein R., and Almouzni G. (2018) Functional activity of the H3.3 histone chaperone complex HIRA requires trimerization of the HIRA subunit. Nat. Commun. 9, 3103 10.1038/s41467-018-05581-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D. C., Pchelintsev N. A., Adams P. D., Jansen L. E., and Almouzni G. (2011) Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 10.1016/j.molcel.2011.12.006 [DOI] [PubMed] [Google Scholar]
- 29. Buchan D. W., Minneci F., Nugent T. C., Bryson K., and Jones D. T. (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Domsic J. F., Chen H. S., Lu F., Marmorstein R., and Lieberman P. M. (2013) Molecular basis for oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog. 9, e1003672 10.1371/journal.ppat.1003672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luger K., Rechsteiner T. J., and Richmond T. J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 10.1016/S0076-6879(99)04003-3 [DOI] [PubMed] [Google Scholar]
- 32. Narita M., Nũnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., and Lowe S. W. (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 10.1016/S0092-8674(03)00401-X [DOI] [PubMed] [Google Scholar]
- 33. Duarte L. F., Young A. R., Wang Z., Wu H. A., Panda T., Kou Y., Kapoor A., Hasson D., Mills N. R., Ma'ayan A., Narita M., and Bernstein E. (2014) Histone H3.3 and its proteolytically processed form drive a cellular senescence programme. Nat. Commun. 5, 5210 10.1038/ncomms6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang R., Chen W., and Adams P. D. (2007) Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2343–2358 10.1128/MCB.02019-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang R., Poustovoitov M. V., Ye X., Santos H. A., Chen W., Daganzo S. M., Erzberger J. P., Serebriiskii I. G., Canutescu A. A., Dunbrack R. L., Pehrson J. R., Berger J. M., Kaufman P. D., and Adams P. D. (2005) Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 10.1016/j.devcel.2004.10.019 [DOI] [PubMed] [Google Scholar]
- 36. Ye X., Zerlanko B., Zhang R., Somaiah N., Lipinski M., Salomoni P., and Adams P. D. (2007) Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2452–2465 10.1128/MCB.01592-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams P. D. (2007) Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene 397, 84–93 10.1016/j.gene.2007.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Safina A., Cheney P., Pal M., Brodsky L., Ivanov A., Kirsanov K., Lesovaya E., Naberezhnov D., Nesher E., Koman I., Wang D., Wang J., Yakubovskaya M., Winkler D., and Gurova K. (2017) FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res. 45, 1925–1945 10.1093/nar/gkw1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang K., Gao Y., Li J., Burgess R., Han J., Liang H., Zhang Z., and Liu Y. (2016) A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks. Nucleic Acids Res. 44, 5083–5094 10.1093/nar/gkw106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schneiderman J. I., Orsi G. A., Hughes K. T., Loppin B., and Ahmad K. (2012) Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc. Natl. Acad. Sci. U.S.A. 109, 19721–19726 10.1073/pnas.1206629109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu R., Iwabuchi M., and Ohsumi K. (2017) The WD40 domain of HIRA is essential for RI-nucleosome assembly in Xenopus egg extracts. Cell Struct. Funct. 42, 37–48 10.1247/csf.17001 [DOI] [PubMed] [Google Scholar]
- 42. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gouet P., Robert X., and Courcelle E. (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 10.1093/nar/gkg556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seiler C. Y., Park J. G., Sharma A., Hunter P., Surapaneni P., Sedillo C., Field J., Algar R., Price A., Steel J., Throop A., Fiacco M., and LaBaer J. (2014) DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42, D1253–D1260 10.1093/nar/gkt1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weiner M. P., and Costa G. L. (1994) Rapid PCR site-directed mutagenesis. PCR Methods Appl. 4, S131–S136 10.1101/gr.4.3.S131 [DOI] [PubMed] [Google Scholar]
- 46. Donham D. C. 2nd, Scorgie J. K., and Churchill M. E. (2011) The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4-DNA complexes. Nucleic Acids Res. 39, 5449–5458 10.1093/nar/gkr097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cole J. L. (2004) Analysis of heterogeneous interactions. Methods Enzymol. 384, 212–232 10.1016/S0076-6879(04)84013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]