Abstract
Oleate hydratases (OhyAs) belong to a large family of bacterial proteins catalyzing the hydration or isomerization of double bonds in unsaturated fatty acids. A Staphylococcus aureus gene (Sa0102) is predicted to encode an OhyA. Here, we recombinantly expressed and purified SaOhyA and found that it forms a homodimer that requires FAD for activity. SaOhyA hydrates only unsaturated fatty acids containing cis-9 double bonds, but not fatty acids with trans-9 double bonds or cis double bonds at other positions. SaOhyA products were not detected in S. aureus phospholipids and were released into the growth medium. S. aureus does not synthesize unsaturated fatty acids, and the SaOhyA substrates are derived from infection sites. Palmitoleate (16:1(9Z)) is a major mammalian skin–produced antimicrobial fatty acid that protects against S. aureus infection, and we observed that it is an SaOhyA substrate and that its hydroxylated derivative is not antimicrobial. Treatment of S. aureus with 24 μm 16:1(9Z) immediately arrested growth, followed by growth resumption after a lag period of 2 h. The ΔohyA mutant strain did not recover from the 16:1(9Z) challenge, and increasing SaOhyA expression using a plasmid system prevented the initial growth arrest. Challenging S. aureus with sapienic acid (16:1(6Z)), an antimicrobial fatty acid produced only by human skin, arrested growth without recovery in WT, ΔohyA, and SaOhyA-overexpressing strains. We conclude that SaOhyA protects S. aureus from palmitoleic acid, the antimicrobial unsaturated fatty acid produced by most mammals, and that sapienic acid, uniquely produced by humans, counters the OhyA-dependent bacterial defense mechanism.
Keywords: Staphylococcus aureus (S. aureus), fatty acid metabolism, bacterial metabolism, host defense, host-pathogen interaction, innate immunity, antimicrobial fatty acid, FAD, hydroxy fatty acid, oleate hydratase, sebum
Introduction
Bacterial oleate hydratase (OhyA)2 activity (EC 4.2.1.53) was first detected in 1962 (1), and the product was characterized as 10(R)-hydroxy-18:0 using soluble enzyme preparations from Pseudomonas sp. (2) (Fig. 1). This organism is now called Elizabethkingia meningoseptica, and E. meningoseptica OhyA is a well-characterized member of a large family of related bacterial genes predicted to encode unsaturated fatty acid hydratases (3, 4). OhyA has utility in the synthesis of hydroxy fatty acids of commercial interest by bioconversion (5, 6). OhyA genes are widely distributed in bacteria, and recently a Hydratase Database was established (https://hyed.biocatnet.de/)3 to accelerate the characterization of OhyAs as biocatalysts. This bioinformatic analysis sorts sequences of known and putative hydratases into 11 subfamilies (3). Members of the OhyA family belong to the subset of FAD-dependent enzymes that are not oxidoreductases (4) (Fig. 1). FAD oxidation/reduction does not have an active role in substrate conversion, and it is thought that FAD binding promotes organization of the active site or stabilizes the transition state (4, 7, 8). Although the bacterial OhyA proteins are highly related and all use an FAD cofactor (4), there are clear differences in the products formed by the individual enzymes. Some OhyA enzymes are selective for 9Z double bonds, whereas the Streptococcus pyogenes OhyA catalyzes hydration of both (9Z) and (12Z) double bonds (9). E. meningoseptica OhyA catalyzes the reversible hydration of the cis double bond of oleate (18:1(9Z))4 to 10(R)-hydroxy-18:0 (h18:0), which may also be converted to 18:1(10E) or 18:1(9Z) by a reverse dehydration (7, 8). The OhyA family member from Lactobacillus plantarum also functions as an isomerase that forms 18:2(9Z,11E) (conjugated linoleic acid) in a four-enzyme pathway for polyunsaturated fatty acid saturation (10).
Figure 1.
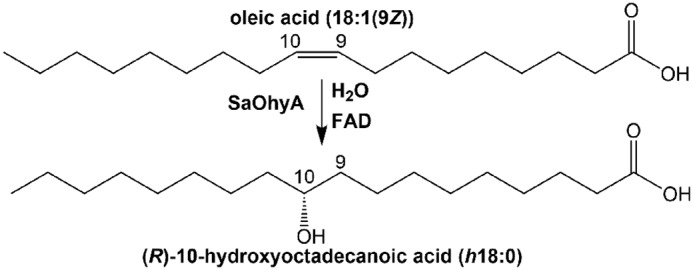
The oleate hydratase reaction. OhyAs are a group of bacterial enzymes that hydrate and/or isomerize the double bonds present in unsaturated fatty acids. SaOhyA catalyzes the addition of water to cis-9 double bonds, but not to trans double bonds or cis double bonds at other positions.
Staphylococcus aureus oleate hydratase (SaOhyA) is a member of the HF#2 hydratase subfamily that contains 1188 sequences including Lactobacillus and Streptococcus species (3) (Fig. 1). The substrate specificity and biological function of SaOhyA are unknown. However, S. aureus does not synthesize unsaturated fatty acids, and therefore the system is present to metabolize environmental fatty acids that would be encountered during infection. S. aureus is the leading cause of skin and soft tissue infections (11), and unsaturated 16-carbon fatty acids are a potent innate immune defense deployed by the skin to prevent infection. In mice (and other mammals), 16:1(9Z) is the most potent antimicrobial fatty acid produced by the skin (12). Antimicrobial fatty acids permeabilize the cells, leading to the leakage of low-molecular-weight solutes and proteins <20 kDa into the medium (12). However, human sebum is unique in that it is the only documented place where sapienic acid (16:1(6Z)) is produced in the animal kingdom where it is deployed instead of 16:1(9Z) as a major skin antimicrobial fatty acid (13, 14). This is because fatty acid desaturase 2 (FADS2), the same enzyme that is involved in the formation of polyunsaturated fatty acids, is highly expressed in human skin, leading to the desaturation of palmitate at carbon-6 (15). Humans (16) and mice (17) deficient in the production of these 16-carbon monounsaturated fatty acids are more susceptible to S. aureus skin infections. The goal of this project is to biochemically characterize SaOhyA and determine whether it functions as a countermeasure used by S. aureus to combat host antimicrobial fatty acids.
Results
Identification and purification of SaOhyA
S. aureus has a single ohyA gene homolog in its genome (Sa0102, SaOhyA). An N-terminal His-tagged version of SaOhyA was cloned, expressed in Escherichia coli, and purified to determine its biochemical properties. SaOhyA was found in the cytosol of the E. coli expression system and was purified using affinity chromatography on Ni2+–nitrilotriacetic acid–agarose. The protein was further purified by gel filtration chromatography on a Sepharose S-200 column to yield a protein of >95% purity based on gel electrophoresis (Fig. 2A). The elution position in gel filtration chromatography indicated that SaOhyA was a homodimer (Fig. 2A), a configuration that is characteristic of bacterial oleate hydratases (4, 5, 7, 8).
Figure 2.
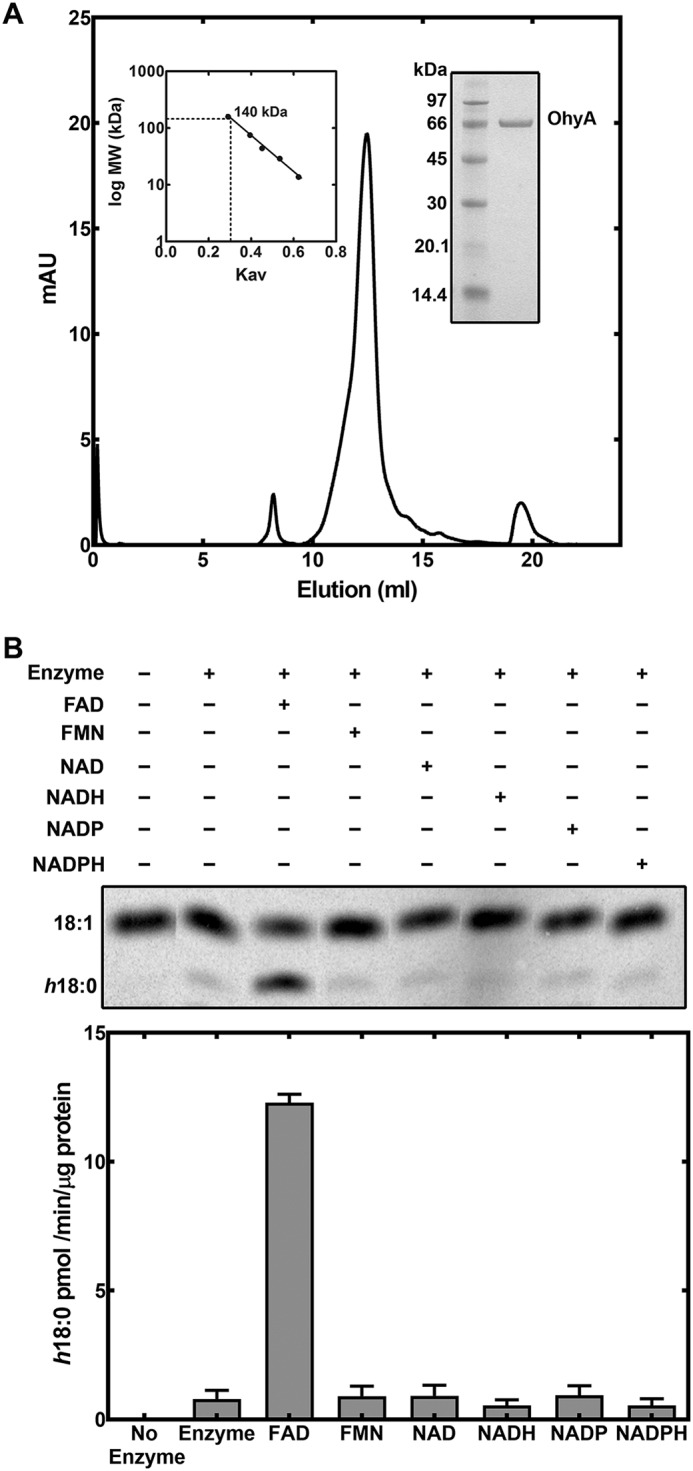
Purification and cofactor requirement for SaOhyA. A, His-tagged SaOhyA was purified by Ni2+-affinity chromatography followed by gel filtration chromatography on Sephadex S-200. Elution volume of SaOhyA compared with a standard curve (left inset) indicated that SaOhyA is a 140-kDa dimer. SaOhyA was a 68-kDa monomer and was >95% pure based on gel electrophoresis (right inset). B, the cofactor requirement for SaOhyA. An example showing the separation of [14C]18:1 from [14C]h18:0 by TLC. The concentrations of the cofactors were 50 μm, and the quantitation of the reaction rates from three experiments is shown in the bar graph. There was no effect of NAD(H) or NADP(H) when added in the presence of 50 μm FAD (not shown). The data presented are the results from triplicate experiments. Error bars, S.E.
The activity of SaOhyA was analyzed in biochemical assays using [1-14C]oleate as the substrate. SaOhyA required FAD for activity and catalyzed the hydroxylation of 18:1(9Z) to h18:0 (Fig. 2B). FMN did not substitute for FAD, and the presence of oxidized or reduced nicotinamide adenine dinucleotides did not influence enzyme activity either alone (Fig. 2B) or when added with FAD (not shown). The SaOhyA protein preparations were not visibly colored, but the activity of purified SaOhyA in the absence of FAD was ∼6% of the maximum rate with FAD present (Fig. 2B). We determined whether the FAD-independent SaOhyA activity (Fig. 2B) was due to the presence of FAD in the protein preparation or FAD-independent enzyme activity by measuring the amount of FAD in the SaOhyA preparation by LC-MS/MS. The amount of FAD present was determined following extraction of the purified protein and quantification of the FAD amount using a standard curve as described under “Experimental procedures.” This analysis showed that the FAD level was sufficient to occupy 3.5% of the SaOhyA active sites. Thus, the residual activity in the absence of added FAD was attributed to a small amount of FAD that copurified with SaOhyA. The apparent Km for FAD was 2.1 ± 0.2 μm (Fig. 3A), consistent with the significant loss of the FAD cofactor during the purification of SaOhyA. The apparent Km for oleate was 5.9 ± 0.6 μm (Fig. 3B). The kinetics with respect to 18:1(9Z) were highly cooperative with a Hill number of 2.2 ± 0.3.
Figure 3.
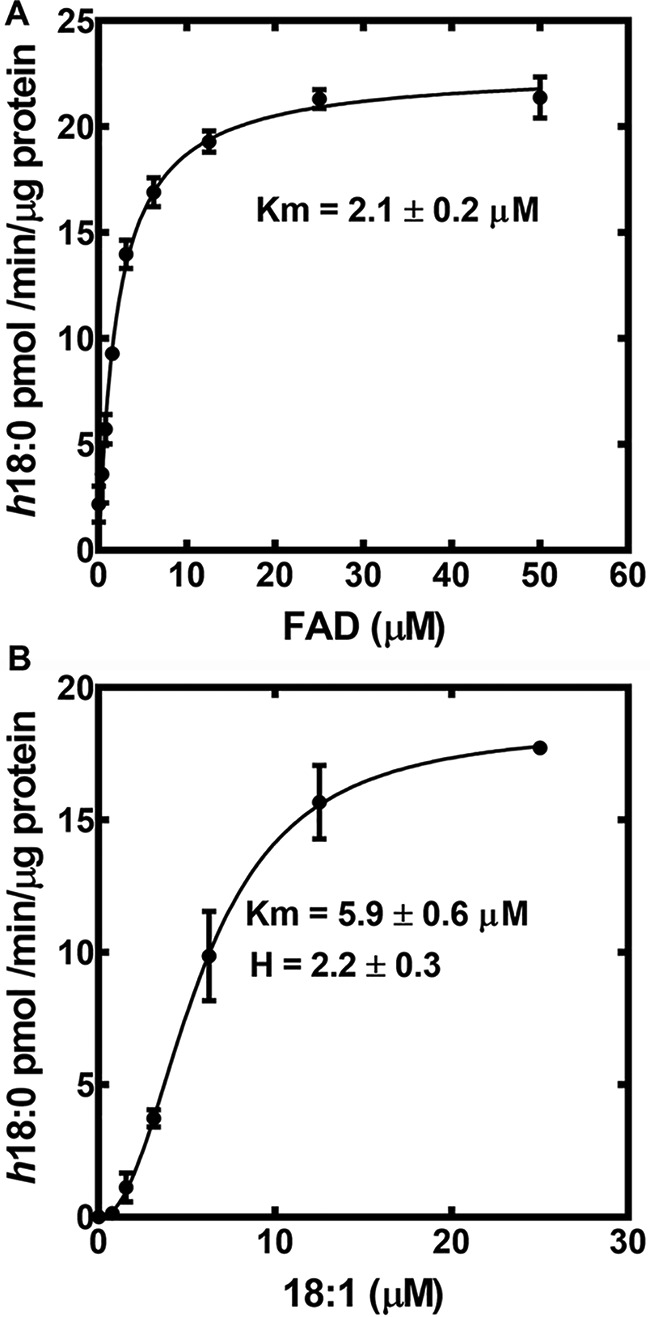
Kinetic analysis of SaOhyA. A, the apparent Km for FAD calculated using 20 μm 18:1(9Z) substrate. The line represents the fit of the data points to the Michaelis–Menten equation. B, the apparent 18:1(9Z) Km determined at 50 μM FAD substrate. The line represents the fit of the data points to the Hill equation. The dependence of the reaction on 18:1(9Z) was highly cooperative with a Hill number of 2.2 ± 0.3. Error bars, S.E.
SaOhyA acts on cis-9 double bonds
An LC-MS system was developed to measure hydroxy-fatty acid formation from unsaturated fatty acids and identify the location of the hydroxyl groups in the product. Hydroxy-fatty acids clearly separated from the parent unsaturated fatty acids in the chromatographic system (Fig. 4A). The reaction product from the in vitro reaction was identified as h18:0 based on its characteristic mass spectrum (Fig. 4B). The parent mass of the SaOhyA product formed from 18:1(9Z) had a m/z = 299.1 consistent with the addition of water to 18:1(9Z), and the fragmentation pattern was characteristic for h18:0 primarily based on the m/z = 185.1 fragment that arises from cleavage adjacent to the hydroxyl as diagramed in Fig. 4B (inset). Similarly, the product of SaOhyA action on 18:2(9Z,12Z) was identified as 10-hydroxy-18:1 (h18:1, m/z = 297.1) based on its mass spectrum (Fig. 4C), and the presence of the m/z = 185.1 that is derived from cleavage adjacent to the hydroxyl group at carbon 10 (Fig. 4C, inset). These mass spectra exactly match the published mass spectra of h18:0 and h18:1 (8, 10).
Figure 4.
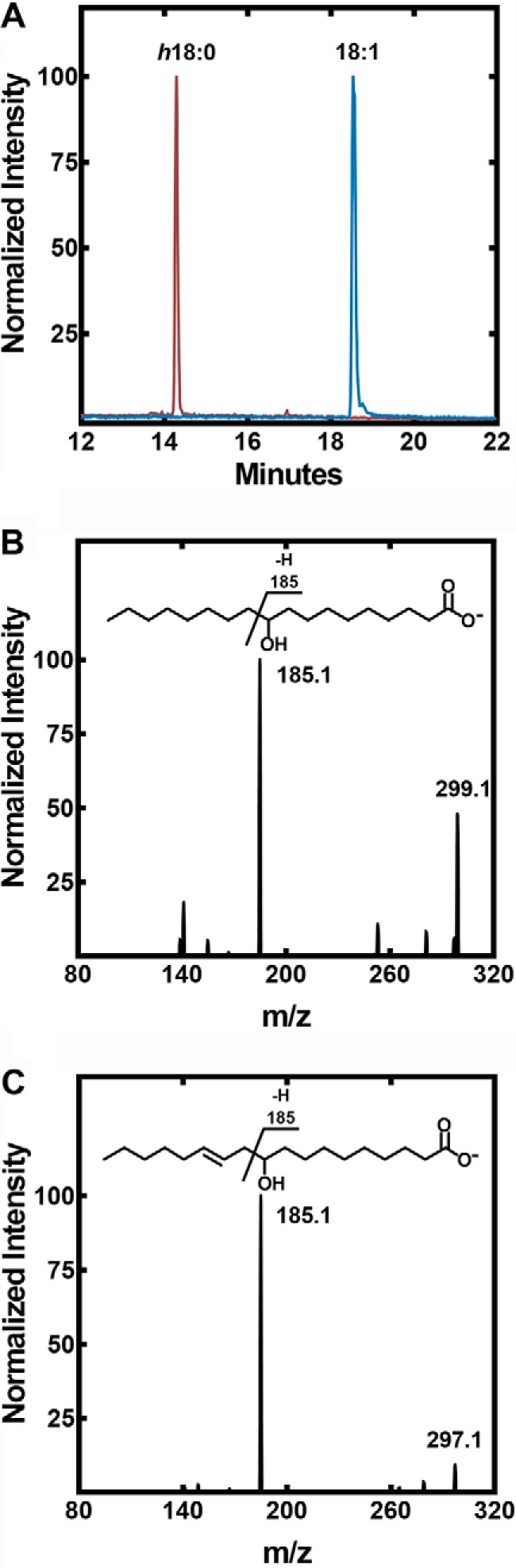
Formation and structure of SaOhyA products. A, an example illustrating the LC-MS method for separating and detecting hydroxy-fatty acids in SaOhyA assays (Table 1). The experiments shown are samples from an assay containing 18:1(9Z) in the presence (vermilion trace) or absence (blue trace) of SaOhyA (2.5 μg). There was almost complete conversion of 18:1(9Z) to h18:0 in this experiment. B, mass spectrum of h18:0 product peak from an SaOhyA assay containing 18:1(9Z). The molecular ion (m/z = 299.1) corresponds to the addition of water, and the m/z = 185.1 fragment was diagnostic for hydroxylation at carbon-10 as indicated in the fragmentation diagram (inset). C, mass spectrum of h18:1 from an SaOhyA assay containing 18:2(9Z,12Z). The same m/z = 185.1 peak was diagnostic for hydroxylation at carbon-10 in this fatty acid as indicated in the fragmentation diagram (inset). Chromatographic conditions and MS parameters are detailed under “Experimental procedures.”
The LC-MS SaOhyA assay was used to examine the substrate specificity of the enzyme toward a panel of unlabeled mammalian unsaturated fatty acids (Table 1). In these experiments, the SaOhyA (2.5 μg) was used to yield ∼80–90% conversion of 18:1 to h18:0, so that even poor substrates for the reaction would be detected. If a hydroxy-fatty acid was produced by SaOhyA, then the mass spectrum was obtained to identify the location of the hydroxyl group along the acyl chain. The only unsaturated fatty acids that were substrates were those containing a cis-9 double bond (Table 1). 16:1(9Z) was an excellent substrate, but product formation was not detected with either 18:1(11Z) or 16:1(6Z) substrates. SaOhyA did not hydroxylate 16:1(9E) or 18:1(9E) substrates, showing that trans double bonds were not substrates. Both α-linolenic (18:3(9Z,12Z,15Z)) and γ-linolenic (18:3(6Z,9Z,12Z)) acids were SaOhyA substrates and hydrated the cis-9 double bond in these fatty acids (Table 1). Arachidonic and docosahexaenoic acids are two prevalent mammalian unsaturated fatty acids that lack a 9Z double bond, and they were not SaOhyA substrates (Table 1). We did not determine the stereochemistry of the hydroxyl group, but members of the oleate hydratase family exclusively produce the R-hydroxy isomer (18). Thus, SaOhyA was specific for the formation of 10-hydroxy fatty acids only from unsaturated fatty acids with cis-9 double bonds.
Table 1.
Unsaturated fatty acids as substrates of SaOhyA
SaOhyA assays were performed as described under “Experimental procedures” using LC–MS to detect hydroxy-fatty acid formation and identify the location of the hydroxyl group (see Fig. 4).
| Fatty acid | Product |
|---|---|
| 16:1(6Z) | Not detected |
| 16:1(9Z) | 10-Hydroxy-16:0 (h16:0) |
| 16:1(9E) | Not detected |
| 18:1(9Z) | 10-Hydroxy-18:0 (h18:0) |
| 18:1(9E) | Not detected |
| 18:1(11Z) | Not detected |
| 18:2(9Z,12Z) | 10-Hydroxy-18:1 (12Z) (h18:1) |
| 18:3(9Z,12Z,15Z) | 10-Hydroxy-18:2 (12Z,15Z) |
| 18:3(6Z,9Z,12Z) | 10-Hydroxy-18:2 (6Z,12Z) |
| 20:4(5Z,8Z,11Z,14Z) | Not detected |
| 22:6(4Z,7Z,10Z,13Z,16Z,19Z) | Not detected |
Metabolism of antimicrobial fatty acids
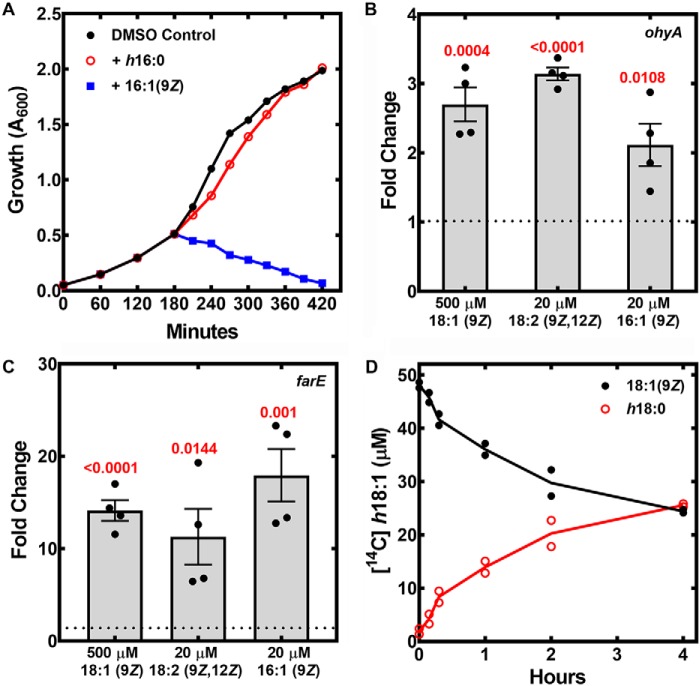
The first step was to determine whether h16:0, the SaOhyA product derived from the antimicrobial 16:1(9Z), possessed antimicrobial activity. The SaOhyA assay was scaled, and h16:0 was purified and quantified as described under “Experimental procedures.” Strain AH1263 was grown to mid-log phase and treated with 30 μm of either 16:1(9Z) or h16:0, and the growth was monitored (Fig. 5A). Treatment with 16:1(9Z) triggered the abrupt cessation of cell growth, whereas h16:0 was without effect. These data illustrate that the hydroxylated products of SaOhyA had attenuated antimicrobial activity, suggesting that SaOhyA may function as a countermeasure against growth inhibition by unsaturated antimicrobial fatty acids.
Figure 5.
Growth, gene expression, and metabolism of unsaturated fatty acids by S. aureus. A, strain AH1263 was grown to mid-log phase, treated with 30 μm 16:1(9Z), h16:0, or vehicle (DMSO), and the growth of the cultures was monitored. B, qRT-PCR measurement of ohyA mRNA levels in strain AH1263 exposed to the indicated concentrations of fatty acids. The data are representative of four independent cultures. C, qRT-PCR measurement of farE mRNA in strain AH1263 exposed to the indicated concentrations of fatty acids. The calibrator was glyA, and the p values calculated using the Student's t test (GraphPad) are shown in red. The data are graphed as means ± S.E. D, the conversion of [14C]18:1(9Z) to [14C]h18:0 by strain JLB2 (ΔohyA)/pPJ490 and its release into the culture media. We used a ΔfakA strain to prevent incorporation of fatty acids into phospholipid and introduced the SaOhyA expression plasmid pPJ490 to amplify product formation. Culture supernatants were sampled at the indicated times and extracted, and [14C]18:1(9Z) was separated from [14C]h18:0 by TLC and quantitated using a Typhoon PhosphorImager. The results from two experiments are plotted.
In our previous work with antimicrobial fatty acids, we analyzed the global gene expression response to 18:1(9Z) (12) (accession number GSE36231 in the NCBI Gene Expression Omnibus database). The ohyA gene was increased 2.5-fold in the published array experiment, and we validated the result obtained with the microarray using qRT-PCR (Fig. 5B). The levels of ohyA mRNA were elevated slightly by three fatty acids (18:1(9Z), 16:1(6Z), and 18:2(9Z,12Z)). Although we detected induction of ohyA expression in the presence of these unsaturated fatty acids, the up-regulation of ohyA transcription was not robust compared with the highly regulated farE gene that is controlled by the FarR transcriptional regulator (12, 19) (Fig. 5C). In the published microarray experiment (12), there were numerous genes that were elevated by 2–3-fold, including many genes encoding ribosomal proteins. These results suggest that the unsaturated fatty acid effect on ohyA expression may be tied to the oleate-dependent increase in growth rate and cellular yield (12) rather than a specific transcriptional response to an extracellular SaOhyA substrate.
Our laboratory has published the molecular species composition of S. aureus membrane phospholipids derived from cultures grown with 18:1(9Z) or 16:1(9Z) supplements and has not observed a phosphatidylglycerol molecular species with a mass consistent with the incorporation of hydroxy-fatty acids (12, 20, 21). Thus, metabolic labeling experiments were performed to determine whether SaOhyA products are released into the growth medium. Strain JLB2 (ΔfakA)/pPJ490 was used in this experiment to prevent the incorporation of 18:1(9Z) into phospholipid by deleting fakA (the kinase component of fatty acid kinase) and to elevate cellular SaOhyA activity to more clearly determine the fate of hydroxy-fatty acids. Strain JLB2 (ΔfakA)/pPJ490 was grown to mid-log phase and labeled with [14C]18:1(9Z), and at the indicated times following fatty acid addition, samples were removed and cells were separated from media by centrifugation. The label remained constant in the cell culture supernatant at all time points, and the small amount of radioactivity associated with the cell pellet was removed from the cells by resuspension and washing (not shown). TLC analysis of the label in the supernatant showed a steady increase in [14C]h18:0 with a corresponding decrease in [14C]18:1(9Z) (Fig. 5D). These data show that [14C]18:1(9Z) was taken up by the cells and converted to [14C]h18:0, and the [14C]h18:0 was then released into the medium. Similarly, h18:1(12Z), the product of SaOhyA action on 18:2(9Z,12Z), was found in the culture supernatants and was not incorporated into membrane phospholipids (not shown).
SaOhyA confers resistance to antimicrobial fatty acids
The role of ohyA in resistance to antimicrobial fatty acids was evaluated using a ΔohyA knockout strain PDJ68. The inactivation of the ohyA gene was achieved by the deletion of the ohyA coding sequence as diagramed in Fig. 6A, and the ΔohyA gene deletion was verified by PCR analysis (Fig. 6A). An expression vector was also prepared containing the ohyA gene driven by the sarA promoter (pPJ490) to complement the gene deletion as described under “Experimental procedures.” The ability of the WT, knockout, and complemented strains to hydroxylate 16:1(9Z) was determined by LC-MS (Fig. 6B). Strain AH1263/pPJ480 (WT) formed h16:0 from 16:1, whereas h16:0 formation was not detected in the supernatant of strain PDJ68 (ΔohyA)/pPJ480. The complemented mutant strain PDJ68 (ΔohyA)/pPJ490 produced increased amounts of h16:0, showing that ohyA expression was the only gene required for S. aureus to produce hydroxylated fatty acids.
Figure 6.
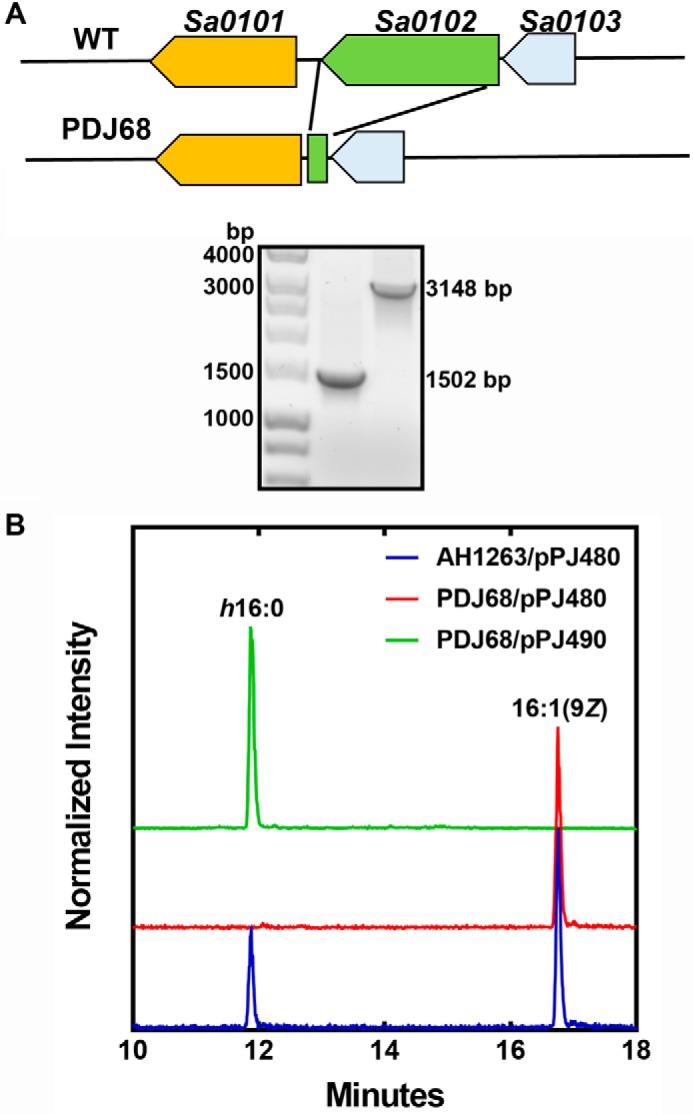
Validation of the ohyA gene deletion in S. aureus strain AH1263. A, top panel, a diagram of the genomic region surrounding ohyA (locus tag Sa0102) and the genomic structure following excision of the ohyA coding sequence. Bottom panel, PCR verification of the ohyA deletion in strain PDJ68 (1502 bp). B, analysis of the formation of h16:0 following a challenge with 20 μm 16:1(9Z). Strains AH1263/pPJ480 (blue trace), PDJ68 (ΔohyA)/pPJ480 (red trace), and PDJ68/pPJ490 (green trace) were grown to A600 of 0.5 and exposed to 16:1(9Z) for 1 h. The cell supernatants were harvested, and the formation of h16:0 was detected by LC-MS as described under “Experimental procedures.”
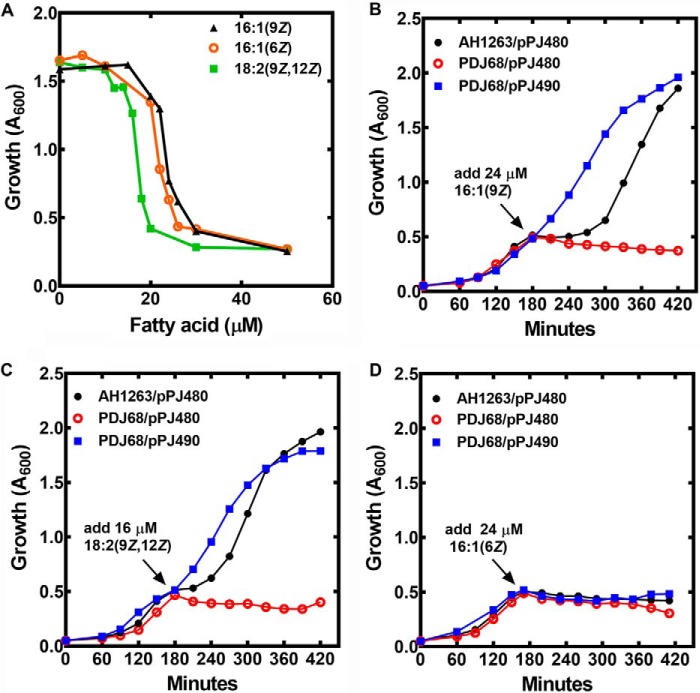
The major antimicrobial fatty acids produced by the innate immune system to protect skin from infection are 16:1(9Z) and 16:1(6Z) in mice and humans, respectively (16, 17). These two fatty acids differ by the location of their double bond, but they do not have a different minimum inhibitory concentrations for S. aureus using a standard microbroth dilution assay method (12). Although 18:2(9Z,12Z) does not have an established physiological role in innate skin defense, it was included in our study because it is a potent antimicrobial fatty acid (12, 19) and an SaOhyA substrate (Table 1). Antimicrobial fatty acids act to permeabilize the S. aureus membrane, resulting in the release of intracellular metabolites (like ATP) and low-molecular-weight proteins (like ACP) into the medium (12). Growth arrest triggered by the three fatty acids occurred over a very narrow concentration range in liquid culture (Fig. 7A). Linoleate (18:2(9Z,12Z)) was slightly more potent than the two equivalently potent 16:1 isomers. At concentrations below the transition concentration, cell growth was not affected, and above the transition point, the cells did not recover. At a 16:1(9Z) concentration within the transition range (24 μm), the growth of strain AH1263 (WT) was immediately arrested, but after 2 h of continued incubation, the cells recovered and resumed growth at the same rate as 16:1(9Z) concentrations below the transition point (not shown). This observation led us to test the role of OhyA in the growth response of S. aureus to antimicrobial fatty acids.
Figure 7.
Effect of SaOhyA on antimicrobial activity. A, concentration-dependent growth inhibition of strain AH1263/pPJ480 when challenged with either 16:1(9Z), 18:2(9Z,12Z), or 16:1(6Z). Strain AH1263/pPJ480 was grown to an A600 of 0.3, and the indicated concentrations of fatty acids were added. The A600 values of the cultures were then determined after 2 h as described under “Experimental procedures.” B–D, three strains were used in the following experiments. Strain AH1263/pPJ480 (empty expression vector) was the WT control, strain PDJ68 (ΔohyA)/pPJ480 (empty vector) was the ohyA knockout, and PDJ68/pPJ490 was the ohyA knockout with the complementing ohyA expression vector. Each of the three strains were grown to mid-log phase (A600 = 0.5) and then challenged with the indicated antimicrobial fatty acids. B, strains were challenged with 24 μm 16:1(9Z). C, strains were challenged with 16 μm 18:2(9Z,12Z). D, strains were challenged with 24 μm 16:1(6Z). The data shown are examples of the growth curves in one experiment, and the results were verified in a second experiment.
A series of plasmid-bearing strains harboring either the empty control plasmid (pPJ480) or pPJ490, the expression vector containing the ohyA gene under the control of the sarA promoter (Table S1) was used. Like strain AH1263, the growth of strain AH1263/pPJ480 immediately arrested when challenged with 24 μm 16:1(9Z), and growth resumed at the normal rate after a lag of 2 h (Fig. 7B). Strain PDJ68 (ΔohyA)/pPJ480 lacked OhyA and did not recover from 16:1(9Z) growth arrest triggered by 24 μm 16:1(9Z) (Fig. 7B). Introduction of the ohyA gene into the knockout strain (PDJ68/pPJ490) eliminated the lag phase, rendering the cells refractory to 16:1(9Z) growth inhibition by 24 μm 16:1(9Z). This result indicates that the combination of the sarA promoter and a multicopy plasmid increased ohyA expression to higher levels than present in the WT strain. Challenging strain AH1263/pPJ480 with 16 μm 18:2(9Z,12Z) also resulted in transient growth arrest (Fig. 7C). Elevated SaOhyA expression rendered cells refractory to growth arrest by 16 μm 18:2(9Z,12Z), whereas the ΔohyA knockout strain PDJ68 was unable to recover from the 18:2(9Z,12Z) challenge (Fig. 7C). However, when the three strains were treated with 24 μm 16:1(6Z), growth arrest was immediate, and none of the strains recovered from 16:1(6Z) (Fig. 7D). These results showed that SaOhyA protected cells from the action of 16:1(9Z) and 18:2(9Z,12Z), but not 16:1(6Z), consistent with the substrate specificity of SaOhyA (Table 1), and the fact that hydroxy-fatty acids were not antimicrobial (Fig. 5A).
Discussion
This work establishes SaOhyA as an active countermeasure against 16:1(9Z) through its conversion to h16:0 and efflux from the cell (Fig. 8). An incorporation pathway into membrane phospholipids is one mechanism S. aureus uses to detoxify antimicrobial fatty acids. At concentrations of 16:1(9Z) that do not inhibit cell growth (<20 μm), free fatty acids were not detected in the cells, and there was no evidence for h16:0 in the membrane phospholipids (12). 16:1(9Z) is incorporated into phospholipid by PlsY, following its activation by fatty acid kinase, transfer to ACP by PlsX, and elongation to 18:1(11Z) and 20:1(13Z) by FASII (12). However, 16:1(9Z) itself is a poor substrate for phospholipid biosynthesis, and the capacity of this pathway to dispose of the antimicrobial fatty acids will be overwhelmed as the concentration of extracellular fatty acids rises. SaOhyA is a FAD-dependent hydratase that acts as a primary line of defense by hydrating unsaturated fatty acids with cis-double bonds located at carbon-9. Hydration of the double bond eliminates antimicrobial activity, and the hydroxy products are not used for phospholipid synthesis and are released into the medium. Palmitoleic acid (16:1(9Z)) is the principle antimicrobial fatty acid that protects against S. aureus skin infections in mammals. Human skin has deployed a counter-countermeasure to SaOhyA by uniquely producing the equally effective 16:1(6Z) as a substitute for 16:1(9Z) (13, 14). 16:1(6Z) is not an SaOhyA substrate, making human skin lipids a more effective deterrent against S. aureus infection than the skin lipids of other mammals (13, 14). SaOhyA is not the only mechanism used by S. aureus to avoid toxic fatty acids. Antimicrobial fatty acids may also be inactivated by their incorporation into phospholipid following their activation of fatty acid kinase (Fig. 8). Efflux pumps like FarE (19) and potentially Tet38 (22) are additional active countermeasures to prevent the cellular accumulation and toxicity of antimicrobial fatty acids. Thus, the SaOhyA defense against skin antimicrobial fatty acids is one mechanism in a layered defense deployed by S. aureus against antimicrobial fatty acids deployed by the innate immune system.
Figure 8.
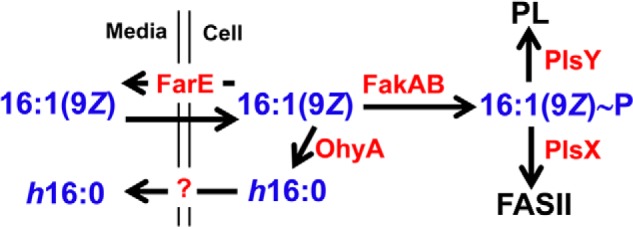
Model illustrating the active defense mechanisms used by S. aureus to detoxify antimicrobial fatty acids. Antimicrobial fatty acids (16:1(9Z)) are produced by the skin to block the growth of S. aureus and are thought to enter the cell by flipping across the membrane bilayer. S. aureus deploys three active countermeasures to combat 16:1(9Z); 1) SaOhyA hydroxylates and inactivates 16:1(9Z) to h16:0, which then exits the cell into the medium by an unknown mechanism; 2) FarE is a fatty acid efflux pump that protects against antimicrobial fatty acid by returning them to the medium (19); and 3) 16:1(9Z) can also be inactivated by incorporation into phospholipid by the PlsX and PlsY enzymes. The data show that most of the 16:1(9Z) incorporated into phospholipid (PL) is first converted to acyl-ACP by PlsX and elongated by FASII before it is utilized for phospholipid synthesis by PlsY (12). Human skin produces 16:1(6Z), which is not an SaOhyA substrate, to circumvent the OhyA-dependent resistance mechanism.
An expanding area of research focuses on how commensal bacteria signal the immune system to tolerate their presence, and OhyA products appear to have an important role in this process (23). Linoleate (18:2(9Z,12Z)) is an abundant host fatty acid that is hydrated by SaOhyA, although this fatty acid does not have a defined role in innate skin defense. It was realized decades ago that 18:2(9Z,12Z) metabolites, like 18:2(9Z,11E) (conjugated 18:2) and h18:1, are produced by commensal lactic acid bacteria that inhabit the gut microbiome (10, 24). More recently, it has become clear that commensal bacteria use signaling molecules like h18:1 to educate the immune system to create a tolerant environment for the bacterium (23, 25). Commensal bacteria produce PPARγ-activating ligands that dampen the innate immune response against bacteria (26–28), and hydroxy fatty acids potently activate PPARγ (29, 30). The anti-inflammatory action of h18:1 blocks tumor necrosis factor–induced intestinal barrier impairment (31, 32) and activates GPR40 (32). The discovery that SaOhyA produces the same immunomodulatory metabolite as commensal bacteria illustrates how this pathogen has coopted a biochemical signalling pathway in symbionts to evade immune surveillance. Thus, SaOhyA joins the growing list of countermeasures deployed by S. aureus not only to resist attack by the innate immune system but also to alter host response to infection (33).
Experimental procedures
Materials
FAD, FMN, NAD, NADH, NADP, NADPH, and 18:1(9Z) were purchased from Sigma–Aldrich. Antibiotics, high density nickel resin, and DTT were purchased from GoldBio (St. Louis, MO). [1-14C]Oleic acid (59 mCi/mmol, 0.1 mCi/ml) was purchased from PerkinElmer Life Sciences. 16:1(9Z), 16:1(6Z), 16:1(9E), 18:1(9E), 18:1(6Z), 18:1(11Z), and 18:2(9Z, 12Z) were purchased from Matreya (State College, PA). Bacteria media supplies were purchased from BD Medical Technologies (Franklin Lakes, NJ). All chemicals and solvents were reagent grade or better. Bacterial strains, plasmids, and primers used in this study are listed in Table S1.
Molecular biology
Oleate hydratase knockout, in which the first 1646 bases of the coding sequence are deleted, was generated in AH1263 by allelic replacement (Fig. 6A). Briefly, 900 bp from either side of ohyA was amplified by PCR using primers OhyA-Up-F, OhyA-Up-R, OhyA-Dn-F, and OhyA-Dn-R. Plasmid vector pJB38 was digested with SacI and SalI and gel-purified. The PCR products were moved into pJB38 by Gibson Assembly (New England Biolabs) yielding pPJ523. Plasmid pPJ523 was transformed into AH1263 by electroporation, and the knockout was generated as previously described (34, 35). The ΔohyA knockout strain PDJ68 was confirmed by PCR using primers Ohy-F3 and OhyA-R6. The DNA sequence of locus tag Sa0102 was synthesized with the appropriate restriction sites (Invitrogen) and cloned into the NdeI and XhoI sites of pET28a to construct pPJ520 for the purification of SaOhyA with an N-terminal His tag from E. coli. The SaOhyA expression vector for strain AH1263 (36) was constructed by first modifying plasmid pCS119 (37) using QuikChange Lightning multisite-directed mutagenesis kit (Agilent Technologies) using specific primers pCS119-a2398t-F and pCS119-g3141c-F to delete the NcoI and HindIII sites in the body of the plasmid by introducing silent mutations. The resulting plasmid was digested with EcoRI and HindIII and gel-purified. A DNA sequence was synthesized containing the SarAP1 promoter, a ribosome-binding site, and multiple cloning sites (5′-GGAAACAGCTATGACATGATTACGAATTTGATATTTTTGACTAAACCAAATGCTAACCCAGAAATACAATCACTGTGTCTAATGAATAATTTGTTTTATAAACACTTTTTTGTTTACTTCTCATTTTTAATTAGTTATAATTAACGCTAGAAAGGAGGTGGATCCATGGCCGCGGGAATTCGAGCTCCTGCAGCGTCGACAAGCTTGCGGCCGCACTCGAGCACCACCACCACCACCACTGAGATAGCTTTTAAAAAGCAAATATGAGCCAAA) and was moved into the digested plasmid via Gibson assembly to obtain plasmid pPJ520. The His-tagged ohyA gene was subcloned from pPJ520 to pPJ480 by NcoI and XhoI to yield pPJ490. All plasmids were confirmed by sequencing.
Cloning, expression, and purification of SaOhyA
The expression of N-terminal His-tagged SaOhyA was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside in strain BL21(DE3) harboring pPJ520 for 3 h and purified using Ni2+-affinity chromatography. The supernatant was loaded on to a Ni2+–nitrilotriacetic acid column and was washed with 20 column volumes of each 10, 20, and 40 mm imidazole in 20 mm Tris-HCl, pH 7.5, 500 mm NaCl. The protein was eluted with 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 250 mm imidazole and dialyzed overnight against 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, and 100 mm EDTA at 4 °C. EDTA was sequentially removed by dialyzing against 20 mm Tris-HCl, pH 7.5, 400 mm NaCl, and 50 mm EDTA; then with 20 mm Tris, pH 7.5, 300 mm NaCl, and 25 mm EDTA; and finally with 20 mm Tris, pH 7.5, and 200 mm NaCl. Size-exclusion chromatography was performed by loading affinity-purified SaOhyA onto a Sephadex S-200 column that was eluted with 20 mm Tris-HCl, pH 7.5, and 200 mm NaCl. Protein standards used for estimating the size of SaOhyA were thyroglobulin (669 kDa), IgG (150 kDa), BSA (68.6 kDa), and myoglobin (17 kDa).
SaOhyA assay
The SaOhyA assay contained 50 mm potassium phosphate buffer, pH 6.0, 10 mm NaCl, 10 mm DTT, 50 μm FAD, 0.2 mg/ml BSA, 20 μm [14C]18:1(9Z), and 0.05 mg/ml of SaOhyA in a final volume of 20 μl. The reactions were incubated at 37 °C for 15 min and were spotted onto silica gel H TLC plates developed with choloroform:methanol (90/10, v/v). The distributions of radioactivity on the dried plates and the extent of product formation were quantified using a Typhoon PhosphorImager. The cofactor FAD was replaced with FMN, NAD, NADH, NADP, and NADPH at final concentrations of 50 μm to determine the cofactor specificity. To determine the FAD Km, FAD was varied from 0 to 50 μm, and to determine the fatty acid Km, [14C]18:1(9Z) was varied from 0 to 25 μm.
Mass spectrometry of OhyA products
The substrate specificity of SaOhyA was determined using LC-MS to measure the formation of hydroxy products. SaOhyA reactions contained 2.5 μg of SaOhyA, 50 mm potassium phosphate buffer, pH 6.0, 10 mm NaCl, 10 mm DTT, 50 μm FAD, 0.2 mg/ml BSA, and 400 μm fatty acid in a final volume of 20 μl. The reactions were incubated at 37 °C for 10 min and stopped by the addition of 80 μl of acetonitrile followed by centrifugation at 4000 × g for 10 min. The supernatant containing the fatty acid substrate and hydroxy-fatty acid product were analyzed with a Shimadzu Prominence UFLC attached to a QTrap 4500 equipped with a Turbo V ion source (Sciex). Samples were injected onto an XSelect® HSS C18, 2.5 μm, 3.0 × 150-mm column (Waters) at 45 °C with a flow rate of 0.4 ml/min. Solvent A was water, and solvent B was acetonitrile. The HPLC program was as follows: starting solvent mixture of 60% B, 0–1 min isocratic with 60% B; 1–16 min linear gradient to 100% B; 16–21 min isocratic with 100% B; 21–23 min linear gradient to 0% B; and 23–28 min isocratic with 0% B. The Sciex QTrap 4500 was operated in the negative mode, and the ion source parameters were: ion spray voltage, −4500 V; curtain gas, 30 p.s.i.; temperature, 320 °C; collision gas, medium; ion source gas 1, 20 p.s.i.; ion source gas 2, 35 p.s.i; and declustering potential, −35 V. The system was controlled by Analyst® software (Sciex).
The Sciex QTrap 4500 mass spectrometer was operated in the negative mode using the product scan to determine the position of the hydroxyl group in the SaOhyA reactions. The source parameters were: ion spray voltage, −4500 V; curtain gas, 15 p.s.i.; temperature, 250 °C; collision gas, high; ion source gas 1, 15 p.s.i.; ion source gas 2, 20 p.s.i.; declustering potential, −25 V; and collision energy, −35 V. The system was controlled by Analyst® software (Sciex).
The phenotype of the ΔohyA strains was confirmed by analyzing the formation of h16:0 from 16:1(9Z) in the medium. Strains AH1263/pPJ480, PDJ68/pPJ480, and PDJ68/pPJ490 were grown to an A600 value of 0.5 in Tryptone broth containing 1% DMSO. 16:1(9Z) in DMSO was added to a final concentration of 20 μm, and the cultures were grown for 1 h at 37 °C with shaking. The cells were separated from media by centrifugation, and the medium was extracted by adding methanol to a final concentration of 80%. Extracts were centrifuged to pellet debris, and the supernatant was analyzed by LC-MS as described above to detect the presence of the h16:0.
FAD measurement by LC/MS/MS
To duplicate samples of 100 μl of 5 mg/ml SaOhyA, acetonitrile was added to a final concentration of 80% and incubated on ice for 5 min. Samples were centrifuged at 3500 × g for 5 min, and the supernatant was transferred to a glass vial. An FAD standard curve was created by having known amounts of FAD (0.01–50 pmol) in the protein buffer and extracted similarly as described above. FAD was analyzed using a Shimadzu Prominence UFLC attached to a QTrap 4500 equipped with a Turbo V ion source (Sciex). The samples were injected onto an XSelect® HSS C18, 2.5 μm, 3.0 × 150-mm column at 40 °C using a flow rate of 0.3 ml/min. Solvent A was 100 mm ammonium formate, pH 5.0, 2% acetonitrile, and 0.1% tributylamine, and solvent B was 95% acetonitrile, 50 mm ammonium formate, pH 6.3, and 0.1% tributylamine. The HPLC program was as follows: starting solvent mixture of 0% B, 0–2 min isocratic with 0% B; 2–12 min linear gradient to 5% B; 12–17 min linear gradient to 90% B; 17–25 min isocratic with 90% B; 25–27 min linear gradient to 0% B; and 27–30 min isocratic with 0% B. The Sciex QTrap 4500 was operated in the negative mode, and the ion source parameters were: ion spray voltage, −4500 V; curtain gas, 40 p.s.i.; temperature, 500 °C; collision gas, medium; ion source gas 1, 50 p.s.i.; and ion source gas 2, 50 p.s.i.. The multiple reaction monitoring transition for FAD was 784/437 m/z with a declustering potential of −20 V and a collision energy of −41 V. The system was controlled by Analyst® software (Sciex) and analyzed with MultiQuantTM 3.0.2 software (Sciex).
Growth strains in the presence of fatty acids
Selected WT, knockout, and plasmid-bearing strains were inoculated in Tryptone broth containing 1% DMSO at A600 0.05 and grown to an A600 of 0.5. Fatty acids in DMSO were added, and the A600 was monitored every 30 min. For the experiments with the plasmids pPJ480 and pPJ490, the plasmids were electroporated into AH1263 or PDJ68 and selected on 10 μg/ml chloramphenicol.
The concentration of the antimicrobial fatty acids that inhibited growth was determined by growing strain AH1263/pPJ480 in Tryptone broth containing 1% DMSO with 10 μg/ml chloramphenicol to an A600 of 0.3. Then, 16:1(9Z) or 16:1 (6Z) were added at 10, 15, 20, 22, 24, 26, 30, or 50 μm, 18:2 (9Z, 12Z) was added at 5, 10, 12, 14, 16, 18, 20, 30, or 50 μm, and the A600 was measured after growing for 2 h at 37 °C with shaking. DMSO was added at 0.1% as zero fatty acid control.
Quantitative real-time PCR
Strain AH1263 was grown to an A600 of 0.6 in 1% Tryptone broth and treated with 500 μm 18:1(9Z), 20 μm 16:1(9Z), and 20 μm 18:2(9Z,12Z) for 20 min. RNA was isolated with an Ambion RNAqueous purification kit (Ambion, Austin, TX) according to the manufacturer's specifications. Purified RNA was then mixed with a 0.5 volume of LiCl precipitation solution (Ambion) and left at −20 °C for 30 min. Turbo DNase (Ambion) was added to the precipitated RNA to a final concentration of 1 unit of DNase per 5 μg of RNA. This mixture was incubated at 37 °C for 30 min. Integrity of the RNA was assessed before its use in qRT-PCR by agarose gel electrophoresis using the Agilent Technologies 2100 Bioanlyzer. Primers used are listed in Table S1. Each 20-μl RT reaction contained 500 ng of RNA, 12.5 ng/μl of random hexamers (Invitrogen), 0.5 mm dNTPs (Sigma), 40 units of RNaseout (Invitrogen), and 10 units/μl of Superscript II reverse transcriptase (Invitrogen). After the RT reaction, 10 ng of the cDNA product was added to a RT-PCR with SYBR Green PCR master mixes (Applied Biosystems), 150 nm of each forward and reverse primer. Specific products were detected on an ABI Prism 7700 sequence detection system (Applied Biosystems) using the following conditions: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and then a final dissociation curve. The samples were processed in 96-well plates, three RT-PCRs were performed on each cDNA sample. cDNA was prepared from four separate cultures for each biological condition with no template, and reactions without reverse transcriptase were run as negative controls. Real-time values were evaluated using the threshold cycle (CT) method, with genes being normalized to a glyA calibrator.
Efflux of hydroxy-fatty acids
Strain JLB2 (37) (ΔfakA)/pPJ490 was grown to an A600 of 0.6 in LB containing 10 mg/ml BSA, and [14C]18:1(9Z) was added to a final concentration of 50 μm. Aliquots (100 μl) were removed at the indicated times and centrifuged to collect the cell pellet and media. An equal volume of methanol was added to the media, and the samples were centrifuged to remove debris. The cells were washed twice with buffered saline and 10 mg/ml fatty acid free BSA, extracted with 200 μl of 50% methanol, and centrifuged to pellet cell debris. Equal volumes (10 μl) were spotted on silica gel H thin-layer plates developed with choloroform:methanol (90/10, v/v). The distributions of radioactivity on the dried plates and extent of product formation were quantified using a Typhoon PhosphorImager.
Purification of hydroxy-fatty acids
h16:0 was purified from an SaOhyA reaction using a 2795 Alliance HT (Waters) equipped with the 2424 evaporative light scattering detector and fraction collector III. The sample was injected onto an XSelect® HSS C18, 2.5-μm, 3.0 × 150-mm column (Waters) at 45 °C using a flow rate of 0.25 ml/min. Solvent A was water, and solvent B was acetonitrile. The HPLC program was as follows: starting solvent mixture of 60% B, 0–1 min isocratic gradient with 60% B, 1–26 min linear gradient to 100% B, 26–31 min isocratic gradient with 100% B, 31–35 min linear gradient to 60% B, and 35–40 min isocratic gradient with 60% B. Detector parameters were: detector gain, 20; gas pressure, 50 p.s.i.; nebulizer mode, heating; power level, 50%; and drift tube temperature, 65 °C. Fractions containing h16:0 were dried under nitrogen, and to remove any residual water, h16:0 was suspended in 100% ethanol and dried under nitrogen. The mass of the h16:0 was determined from a standard curve constructed with 10-hydroxy-18:0 (AA Blocks, LLC, San Diego, CA) using the chromatography and detection system described above.
Author contributions
C. S., M. W. F., J. L. B., S. G. W., and C. O. R. formal analysis; C. S. and C. O. R. supervision; C. S., M. W. F., J. L. B., S. G. W., and C. O. R. investigation; C. S., M. W. F., and S. G. W. methodology; C. S., M. W. F., and C. O. R. writing-original draft; C. S. and C. O. R. project administration; C. S., M. W. F., J. L. B., S. G. W., and C. O. R. writing-review and editing; C. O. R. funding acquisition.
Supplementary Material
Acknowledgments
We thank Roubing Zhou for OhyA assays, Karen Miller for protein purification, and Pam Jackson for strain construction.
This work was supported by National Institutes of Health Grant GM034496, Cancer Center Support Grant CA21765, and the American Lebanese Syrian Associated Charities. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
Fatty acids are designated by number of carbons:number of double bonds(double bond location/configuration).
- OhyA
- oleate hydratase
- SaOhyA
- S. aureus oleate hydratase
- ACP
- acyl carrier protein
- h18:0
- 10-hydroxyoctadecanoic acid
- h18:1
- 10-hydroxy-12(Z)-octadecenoic acid
- qRT-PCR
- quantitative real-time PCR.
References
- 1. Wallen L. L., Benedict R. G., and Jackson R. W. (1962) The microbiological production of 10-hydroxystearic acid from oleic acid. Arch. Biochem. Biophys. 99, 249–253 10.1016/0003-9861(62)90006-1 [DOI] [PubMed] [Google Scholar]
- 2. Mortimer C. E., and Niehaus W. G. Jr. (1974) Enzymatic interconversion of oleic acid, 10-hydroxyoctadecanoic acid, and trans-Δ10-octadecenoic acid: reaction pathway and stereospecificity. J. Biol. Chem. 249, 2833–2842 [PubMed] [Google Scholar]
- 3. Schmid J., Steiner L., Fademrecht S., Pleiss J., Otte K. B., and Hauer B. (2016) Biocatalytic study of novel oleate hydratases. J. Mol. Catal. B Enzym. 133, S243–S249 10.1016/j.molcatb.2017.01.010 [DOI] [Google Scholar]
- 4. Demming R. M., Fischer M. P., Schmid J., and Hauer B. (2018) (De)hydratases-recent developments and future perspectives. Curr. Opin. Chem. Biol. 43, 43–50 10.1016/j.cbpa.2017.10.030 [DOI] [PubMed] [Google Scholar]
- 5. Zorn K., Oroz-Guinea I., Brundiek H., and Bornscheuer U. T. (2016) Engineering and application of enzymes for lipid modification, an update. Prog. Lipid Res. 63, 153–164 10.1016/j.plipres.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 6. Kishino S., Park S. B., Takeuchi M., Yokozeki K., Shimizu S., and Ogawa J. (2011) Novel multi-component enzyme machinery in lactic acid bacteria catalyzing C=C double bond migration useful for conjugated fatty acid synthesis. Biochem. Biophys. Res. Commun. 416, 188–193 10.1016/j.bbrc.2011.11.022 [DOI] [PubMed] [Google Scholar]
- 7. Volkov A., Khoshnevis S., Neumann P., Herrfurth C., Wohlwend D., Ficner R., and Feussner I. (2013) Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus. Acta Crystallogr. D Biol. Crystallogr. 69, 648–657 10.1107/S0907444913000991 [DOI] [PubMed] [Google Scholar]
- 8. Engleder M., Pavkov-Keller T., Emmerstorfer A., Hromic A., Schrempf S., Steinkellner G., Wriessnegger T., Leitner E., Strohmeier G. A., Kaluzna I., Mink D., Schürmann M., Wallner S., Macheroux P., Gruber K., et al. (2015) Structure-based mechanism of oleate hydratase from Elizabethkingia meningoseptica. Chembiochem. 16, 1730–1734 10.1002/cbic.201500269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Volkov A., Liavonchanka A., Kamneva O., Fiedler T., Goebel C., Kreikemeyer B., and Feussner I. (2010) Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J. Biol. Chem. 285, 10353–10361 10.1074/jbc.M109.081851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kishino S., Takeuchi M., Park S. B., Hirata A., Kitamura N., Kunisawa J., Kiyono H., Iwamoto R., Isobe Y., Arita M., Arai H., Ueda K., Shima J., Takahashi S., Yokozeki K., et al. (2013) Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. U.S.A. 110, 17808–17813 10.1073/pnas.1312937110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller L. S., and Cho J. S. (2011) Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 11, 505–518 10.1038/nri3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parsons J. B., Yao J., Frank M. W., Jackson P., and Rock C. O. (2012) Membrane disruption by antimicrobial fatty acids releases low molecular weight proteins from Staphylococcus aureus. J. Bacteriol. 194, 5294–5304 10.1128/JB.00743-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Drake D. R., Brogden K. A., Dawson D. V., and Wertz P. W. (2008) Skin lipids: antimicrobial lipids at the skin surface. J. Lipid Res. 49, 4–11 10.1194/jlr.R700016-JLR200 [DOI] [PubMed] [Google Scholar]
- 14. Wille J. J., and Kydonieus A. (2003) Palmitoleic acid isomer (C16:1Δ6) in human skin sebum is effective against gram-positive bacteria. Skin Pharmacol. Appl. Skin Physiol. 16, 176–187 10.1159/000069757 [DOI] [PubMed] [Google Scholar]
- 15. Ge L., Gordon J. S., Hsuan C., Stenn K., and Prouty S. M. (2003) Identification of the Δ6 desaturase of human sebaceous glands: expression and enzyme activity. J. Invest. Dermatol. 120, 707–714 10.1046/j.1523-1747.2003.12123.x [DOI] [PubMed] [Google Scholar]
- 16. Takigawa H., Nakagawa H., Kuzukawa M., Mori H., and Imokawa G. (2005) Deficient production of hexadecenoic acid in the skin is associated in part with the vulnerability of atopic dermatitis patients to colonization by Staphylococcus aureus. Dermatology 211, 240–248 10.1159/000087018 [DOI] [PubMed] [Google Scholar]
- 17. Georgel P., Crozat K., Lauth X., Makrantonaki E., Seltmann H., Sovath S., Hoebe K., Du X., Rutschmann S., Jiang Z., Bigby T., Nizet V., Zouboulis C. C., and Beutler B. (2005) A Toll-like receptor 2-responsive lipid effector pathway protects mammals against skin infections with Gram-positive bacteria. Infect. Immun. 73, 4512–4521 10.1128/IAI.73.8.4512-4521.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bevers L. E., Pinkse M. W., Verhaert P. D., and Hagen W. R. (2009) Oleate hydratase catalyzes the hydration of a nonactivated carbon-carbon bond. J. Bacteriol. 191, 5010–5012 10.1128/JB.00306-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alnaseri H., Arsic B., Schneider J. E., Kaiser J. C., Scinocca Z. C., Heinrichs D. E., and McGavin M. J. (2015) Inducible expression of a resistance-nodulation-division-type efflux pump in Staphylococcus aureus provides resistance to linoleic and arachidonic acids. J. Bacteriol. 197, 1893–1905 10.1128/JB.02607-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parsons J. B., Frank M. W., Subramanian C., Saenkham P., and Rock C. O. (2011) Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U.S.A. 108, 15378–15383 10.1073/pnas.1109208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parsons J. B., Frank M. W., Jackson P., Subramanian C., and Rock C. O. (2014) Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol. Microbiol. 92, 234–245 10.1111/mmi.12556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Truong-Bolduc Q. C., Villet R. A., Estabrooks Z. A., and Hooper D. C. (2014) Native efflux pumps contribute resistance to antimicrobials of skin and the ability of Staphylococcus aureus to colonize skin. J. Infect. Dis. 209, 1485–1493 10.1093/infdis/jit660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brestoff J. R., and Artis D. (2013) Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 14, 676–684 10.1038/ni.2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim K. R., and Oh D. K. (2013) Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 31, 1473–1485 10.1016/j.biotechadv.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 25. Zhang L. S., and Davies S. S. (2016) Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med. 8, 46 10.1186/s13073-016-0296-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kidani Y., and Bensinger S. J. (2012) Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol. Rev. 249, 72–83 10.1111/j.1600-065X.2012.01153.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong C., Kidani Y., A-Gonzalez N., Phung T., Ito A., Rong X., Ericson K., Mikkola H., Beaven S. W., Miller L. S., Shao W. H., Cohen P. L., Castrillo A., Tontonoz P., and Bensinger S. J. (2012) Coordinate regulation of neutrophil homeostasis by liver X receptors in mice. J. Clin. Invest. 122, 337–347 10.1172/JCI58393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kelly D., Campbell J. I., King T. P., Grant G., Jansson E. A., Coutts A. G., Pettersson S., and Conway S. (2004) Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 5, 104–112 10.1038/ni1018 [DOI] [PubMed] [Google Scholar]
- 29. Itoh T., Fairall L., Amin K., Inaba Y., Szanto A., Balint B. L., Nagy L., Yamamoto K., and Schwabe J. W. (2008) Structural basis for the activation of PPARγ by oxidized fatty acids. Nat. Struct. Mol. Biol. 15, 924–931 10.1038/nsmb.1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marion-Letellier R., Savoye G., and Ghosh S. (2016) Fatty acids, eicosanoids and PPARγ. Eur. J. Pharmacol. 785, 44–49 10.1016/j.ejphar.2015.11.004 [DOI] [PubMed] [Google Scholar]
- 31. Bergamo P., Luongo D., Miyamoto J., Cocca E., Kishino S., Ogawa J., Tanabe S., and Rossi M. (2014) Immunomodulatory activity of a gut microbial metabolite of dietary linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, associated with improved antioxidant/detoxifying defences. J. Funct. Foods 11, 192–202 10.1016/j.jff.2014.10.007 [DOI] [Google Scholar]
- 32. Miyamoto J., Mizukure T., Park S. B., Kishino S., Kimura I., Hirano K., Bergamo P., Rossi M., Suzuki T., Arita M., Ogawa J., and Tanabe S. (2015) A gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, ameliorates intestinal epithelial barrier impairment partially via GPR40–MEK–ERK pathway. J. Biol. Chem. 290, 2902–2918 10.1074/jbc.M114.610733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thammavongsa V., Kim H. K., Missiakas D., and Schneewind O. (2015) Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543 10.1038/nrmicro3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bae T., and Schneewind O. (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 35. Bose J. L., Fey P. D., and Bayles K. W. (2013) Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 79, 2218–2224 10.1128/AEM.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parsons J. B., Broussard T. C., Bose J. L., Rosch J. W., Jackson P., Subramanian C., and Rock C. O. (2014) Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111, 10532–10537 10.1073/pnas.1408797111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ericson M. E., Subramanian C., Frank M. W., and Rock C. O. (2017) Role of fatty acid kinase in cellular lipid homeostasis and SaeRS-dependent virulence factor expression in Staphylococcus aureus. mBio 8, e00988–00917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., and Bayles K. W. (2013) A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.