Abstract
Ovarian cancer is one of the most lethal malignancies of the female reproductive system. Platinum-resistance is the major obstacle in the successful treatment of ovarian cancer. Previous studies largely failed to identify the key genes associated with platinum-resistance by using candidate genes testing, bioinformatic analysis and GWAS method. The aim of the study was to utilize the whole human Genome-scale CRISPR-Cas9 knockout (GeCKO) library to screen for novel genes involved in cisplatin resistance in ovarian cancer cell lines. The GeCKO library targeted 19052 genes with 122417 unique guide sequences. Six candidate genes had been screened out including one previously validated gene SULF1 and five novel genes ZNF587B, TADA1, SEMA4G, POTEC and USP17L20. After validated by CCK-8 and RT-PCR analysis, two genes (ZNF587B and SULF1) were discovered to be involved in cisplatin resistance. ZNF587B may serve as a new biomarker for predicting cisplatin resistance.
Keywords: CRISPR/Cas 9, ovarian cancer, platinum, SULF1, ZNF587B
Introduction
Ovarian cancer is one of the most common cancers in gynecological malignancy with the highest mortality rate [1-3]. Platinum-based chemotherapy is the recommended first-line treatment for high-grade ovarian cancer (EOC). Despite with a high initial response rate, more than 60% of the patients will relapse within 18 months, while all patients will relapse within 3 years [4]. Thus, understanding the mechanism of platinum-resistance is vital. The former studies had focused on candidate genes testing, bioinformatic analysis and GWAS method to identify key genes, and many genes such as ERCC1, XPD, XRCC1, MDR1 and GSTP1 were extensively studied, as well as their polymorphisms [5-12]. However, no powerful genes have been identified so far. In 2014, Zhang’s research team [13] had constructed the human Genome-scale CRISPR-Cas9 [14-16] knockout (GeCKO) library and applied it to screening for genes whose loss was involved in resistance to vemurafenib resistance. Therefore, the application of GeCKO library provides a robust method to identify novel genes in the setting drug resistance of cancer cells. In this study, we firstly applied this approach coupled with new generation sequencing (NGS) to screening for genes involved in cisplatin resistance in ovarian cancer cells, and identified that loss of ZNF587B or SULF1 contributed to cisplatin resistance in ovarian cancer cell lines.
Materials and methods
Cell lines
The human ovarian carcinoma A2780, SKOV3 cell lines and normal ovarian cell line IOSE80 were obtained from (ATCC, USA). A2780 and IOSE80 cells were cultured in RPMI-1640 medium (HyClone, USA) with 10% fetal bovine serum (Biological Industries, Israel), and SKOV3 cells were cultured in Mccoys’5A medium (Biological Industries, Israel) with 15% fetal bovine serum. Cells were cultivated at 37°C in a humidified atmosphere containing with 5% CO2.
Lentiviral packaging and infection
The Human CRISPR Knockout Pooled Library was acquired from Addgene (http://www.addgene.org/crispr/libraries/geckov2/). GeCKO library plasmids, pVSVg (AddGene, USA) and psPAX2 (AddGene, USA) were added into 100 μL of Opti-MEM in a ratio of 1:0.5:1.5, and then the mixture with lipofectamin 2000 (Invitrogen, USA) were incubated for 20 minutes and added into HEK293T cells. The cell supernatant containing lentiviruses was collected after 48 hours. SKOV3 and A2780 cells were infected at a low multiplicity of infection (MOI=0.3) to ensure that most cells received only 1 viral construct. After transduction, cells were selected with puromycin (1 µg/ml) for 14 days so that only cells transducted with a LentiCRISPR construct could survive [17,18].
Cisplatin treatment and sample preparation
We firstly determined the concentration of cisplatin inhibiting the proliferation of SKOV3 and A2780 cells. The stably transfected cells were exposed to 4 μM of cisplatin for 4 days, and about 20% cells were survived, then they were restored in normal medium without cisplatin for 14 days before genomic DNA extraction and analysis. Genomic DNA was extracted using a Blood & Cell Culture Midi Kit (Qiagen), and PCR was performed according to the manufacturer’s protocol. The forward Primer sequences for amplifying lentiCRISPR sgRNAs in A2780 and SKOV3 were tagged with different barcodes, the primers were as follows, F/A2780 5’-TGACCACTTGGGTAGTTTGCAGTTTT-3’, F/SKOV3 5’-ACAGTGCTTGGGTAGTTTGCAGTTTT-3’, R 5’-CAACTTCTCGGGGACTGTGG-3’.
The amplicons were extracted from 1% agarose gels using an EasyPure Quick Gel Extraction Kit (Transgent, China) and then were sent to company for NGS (Genetalks, China).
Transfection of plasmid DNA and siRNA
pcDNA3.1-ZNF587B-eGFP and SULF1 were constructed by Genechem, China, and the siRNAs of SULF1, TADA1, SEMA4G and ZNF587B were synthesized by RIBOBIO, China. The sequences of siRNAs were listed in Table 1. Transfections were performed on lipofectamine 2000 Reagent.
Table 1.
siRNA sequences of candidate genes for knockdown
| Gene | Sequences |
|---|---|
| ZNF587B | si1 5’-GTTCAAACGTGAACCTTAA-3’ |
| si2 5’-GGAAGCCCAGCCTTAGTTA-3’ | |
| SULF1 | si1 5’-CGAGAAAGATTATGGAACA-3’ |
| si2 5’-GACAAAGAGTGCAGTTGTA-3’ | |
| TADA1 | si1 5’-CCTGAAGAATAGTGTAGTA-3’ |
| si2 5’-GTACGATCTTTTTGAAGCT-3’ | |
| SEMA4G | si1 5’-GGATGCACATTATTGAAGA-3’ |
| si2 5’-GACCCTATATGGAATACCA-3’ |
Real time PCR
Total RNA was extracted using RNAiso Plus reagent (Takara, Japan) and quantified using ND-1000 spectrophotometer (NanoDrop, USA). Then reverse transcription from 1 µg of purified RNA to cDNA synthesis was performed using PrimeScript RT reagent kit (Takara, Japan). The RT-PCR was performed using 2× SYBR Green qPCR Master Mix (Bimake, USA) in Light Cycler 480 machine (Roche, USA). ACTB was used as a control. Primer pairs were listed in Table 2 (Biosune, China). The relative expression was analyzed by 2-ΔΔCt method. All the experiments were replicated triplicate.
Table 2.
Primers of candidate genes for RT-PCR
| Gene | Sequences |
|---|---|
| ZNF587B | F 5’-GCGCCATCAAAAAGTTCACG-3’ |
| R 5’-GCTGGGCTTCCGACTAAAAG-3’ | |
| SULF1 | F 5’-GGGCAGAAGTGGCAATG-3’ |
| R 5’-AACCAGACTCCCTACAA-3’ | |
| TADA1 | F 5’-ATGGCGACCTTTGTGAGC-3’ |
| R 5’-TAGGTTAGCCCAGTATTGTT-3’ | |
| SEMA4G | F 5’-CCGCTACCGATCCTGCTATG-3’ |
| R 5’-CTGTATCAGTGCTGTCCTGC-3’ | |
| POTEC | F 5’-AAGCAGATAGAAGTGGCTGAA-3’ |
| R 5’-AGTTCCAGTCTCCAGAAATTAGC-3’ |
Cytotoxicity assay
After infection, cells were treated with cisplatin (Sigma-Aldrich, USA). Cisplatin was added at different concentrations for 48 h. Cell viability was detected by CCK-8 approach (Takara, Japan) according to the protocol, and IC50 values were calculated using the statistical software GraphPad Prism 7 (GraphPad software, CA). All the experiments were triply replicated.
Statistical analysis
All data were described as mean ± standard deviation (SD). Statistical analysis was performed using SPSS V.22 software (SPSS Inc, USA). Student’s t test or analysis of variance (ANOVA) was used for continuous variables.
Results
GeCKO library screening for genes associated with cisplatin resistance
To identify genes referred to cisplatin resistance in ovarian cancer, GeCKO library lentiviruses were used to infect A2780 and SKOV3 cell lines. After 14 days of puromycin selection, infected cells were plated into separate dishes for drug treatment, then cells were treated with cisplatin for 4 days. As the result, 80% of the cells died, and DNA of the survived cells was extracted for PCR and NGS. After NGS analysis, a list of gene expression was obtained for each sample. The genes were ranked according to the number of sgRNA and the NGS reads (Figure 1). As shown in the scatter diagram of sgRNA number and corresponding sequencing reads of genes, the detected genes were well-distributed in every sgRNA (Figure 2A). We replicated the experiments four times. Among them, we failed to extract the DNA from one sample. Thus, 7 gene lists were obtained finally. We calculated the intersection of 7 genes lists online (http://bioinformatics.psb.ugent.be/webtools/Venn/) and identified 6 candidate genes (ZNF587B, SULF1, TADA1, SEMA4G, POTEC, and USP17L20) (Figure 2B). Among the 6 candidate genes, USP17L20 was enriched in almost every screen regardless of the studied phenotype, so it was a false positive gene from screen background rate and wasn’t included in our following experiments. Searching the NCBI gene database, we found that POTEC was only expressed in testis, and our experiments confirmed that it was undetectable in ovarian cancer cells. Therefore, POTEC was also not included in subsequent research.
Figure 1.
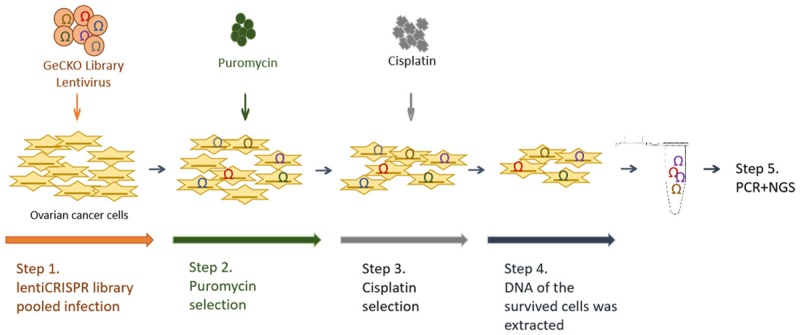
Schematic of cisplatin-resistant ovarian cancer cells construction for high-throughput sequencing analysis.
Figure 2.
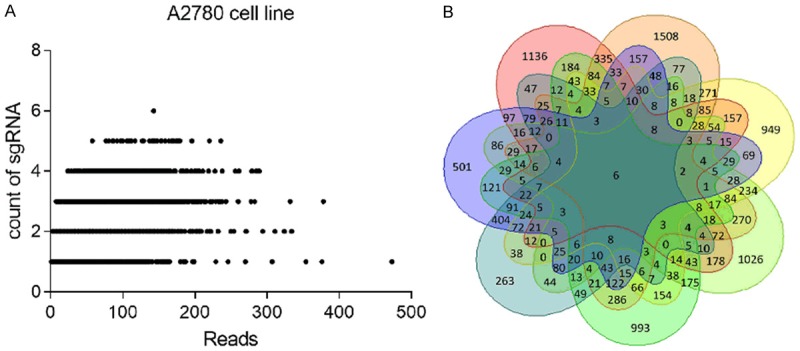
GeCKO screen in ovarian cancer cells reveals genes whose loss confers cisplatin resistance. A. Scatter diagram of sgRNA and corresponding sequencing reads of genes; B. The intersection of 7 gene lists was calculated, and presented as a Venn diagram.
Effects of candidate genes on cellular response to cisplatin
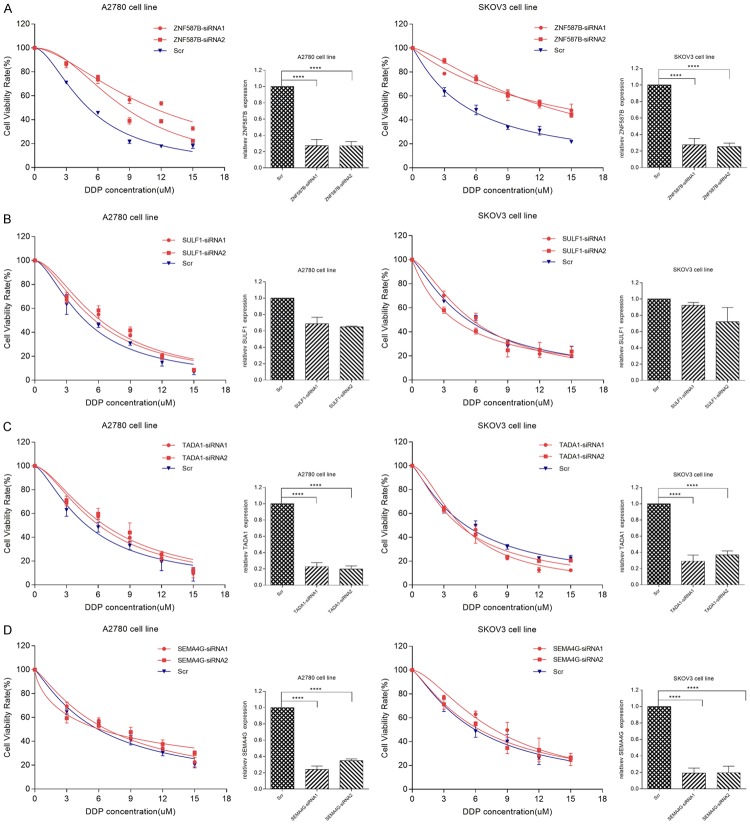
To investigate the effect of 4 candidate genes on cisplatin resistance in EOC cells, siRNAs were applied to knockdown their expression in A2780 and SKOV3 cells. After transfection for 48 hours, the gene expression was examined by RT-PCR. Our results showed that siRNAs of genes except SULF1 effectively decreased their expression (Figure 3, P < 0.0001). After knockdown, cells were subjected to CCK-8 assay in the absence or presence of different concentrations of cisplatin. As shown in Figures 3 and 4, the IC50 values were 9.87 μM and 13.83 μM respectively in A2780 and SKOV3 cells in ZNF687B-siRNA groups, and 5.07 μM and 5.16 μM in Scr groups, respectively. The results showed that down-regulation of ZNF587B significantly increased cisplatin-resistance of ovarian cancer cells (P < 0.01). There was no correlation between TADA1 or SEMA4G expression and cisplatin resistance. On the other hand, the Ct values of SULF1 in our study were nearly 29 in A2780 and SKOV3 parental cell lines which suggested that the expression of SULF1 was very low, and it might be the reason that siRNAs for SULF1 were unable to knockdown its expression. Some previous pertinent studies reported that SULF1 was dysregulated in many cancers, such as ovarian cancer, hepatocellular cancer and breast cancer [19-22], and was undetected in A2780 and SKOV3, but it could be detected in normal ovarian cell lines (IOSE80) [23]. So we firstly tested the SULF1 expression of IOSE80, A2780 and SKOV3 cell lines, and found that the expression of SULF1 in IOSE80 cells was 32 and 17 times higher than that of A2780 and SKOV3 cells respectively. As we expected, A2780 and SKOV3 were more resistant to cisplatin than IOSE80 (IC50, 4.68 μM and 6.29 μM vs 1.30 μM, P < 0.0001, Figure 5A). After knockdown of the SULF1 expression of IOSE80, we found that the SULF1 expression was significantly decreased by siRNA, and cells with reduced SULF1 expression were more resistant to cisplatin than the Scr group (IC50, 3.72 μM vs 1.91 μM, P < 0.01, Figure 5B). This demonstrated that loss of SULF1 could increase cisplatin resistance.
Figure 3.
Cells were transiently transfected with ZNF587B-siRNAs (A), SULF1-siRNAs (B), TADA1-siRNAs (C), SEMA4G-siRNAs (D), or scrambled control (Scr) siRNAs. Knockdown effeciency of gene expression was measured by RT-PCR. Dose response curves were depicted and half maximal inhibitory concentration (IC50) was calculated by Graphpad 7.0 software from 3 independent experiments. All the results were reproducible in three independent experiments. ****P < 0.0001.
Figure 4.
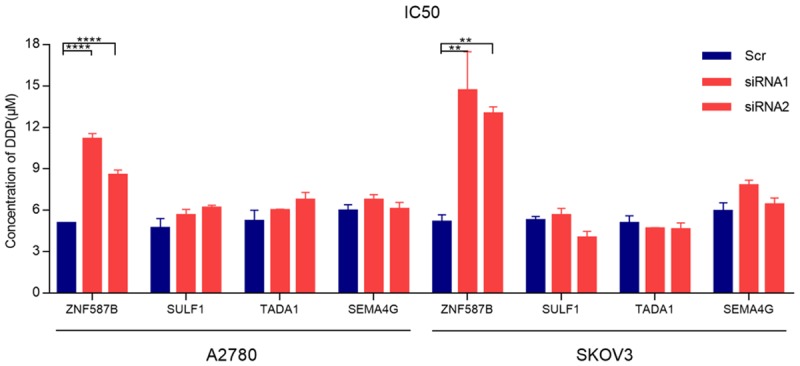
Histogram presented the differences of IC50 between four candidate genes-siRNA groups and scrambled control (Scr) siRNA groups in A2780 or SKOV3 cell lines. All the results were reproducible in three independent experiments. **P < 0.01, ****P < 0.0001.
Figure 5.
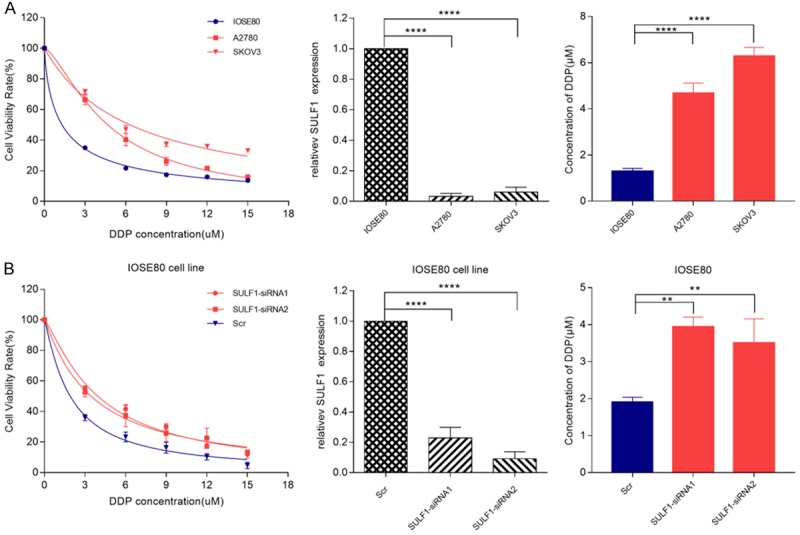
Correlation between SULF1 expression and response in ovarian cell line. A. The IC50 values and SULF1 expression of IOSE80, A2780 and SKOV3 cell lines; B. The IC50 values and SULF1 expression of SULF1-siRNA group and Scr group in IOSE80 cells. All the results were reproducible in three independent experiments. **P < 0.01, ****P < 0.0001.
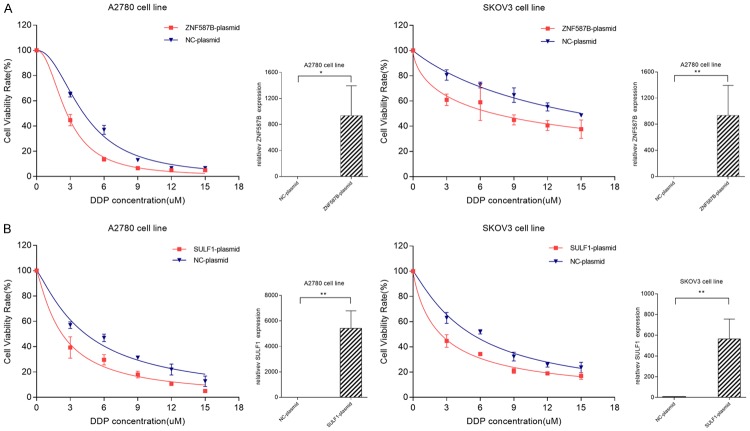
According to the above results, we confirmed that knockdown of SULF1 and ZNF587B in cells increased cisplatin resistance. In order to observe whether the over-expression of the two genes would increase cisplatin sensitivity, we constructed pcDNA3.1-ZNF587B and pcDNA3.1-SULF1 plasmids, and transfected them into A2780 and SKOV3 cell lines. The transfection efficiency of SULF1 was validated by RT-PCR (Figure 6). After transfection for 72 hours, cells were also subjected to cytotoxicity assay, a significant association was found between over-expression group and control group. The IC50 values of ZNF587B over-expression groups were 1.35 and 2.86 times higher than that of control groups. Similarly, the IC50 values of the SULF1 over-expression groups were 1.43 and 3.00 times higher than that of the control groups (Figure 6, P < 0.05).
Figure 6.
Effects of candidate genes over-expression on cellular response to cisplatin. Cells transiently transfected with ZNF587B-plasmid (A), SULF1-plasmid (B), or negative control-plasmids (NC-plasmid) were subjected to treatment with cisplatin. *P < 0.05, **P < 0.01.
Discussion
In this study, we used the GeCKO library in ovarian cancer cell lines to screen for genes involving in cisplatin resistance, and found that SULF1 and ZNF587B were associated with cisplatin resistance.
Lots of studies had demonstrated that SULF1 was associated with platinum resistance. Down-regulation of SULF1 in ovarian cancer cells led to an attenuation of cisplatin-induced cytotoxicity. Moreover, EOC patients with higher levels of SULF1 showed a better response rate to chemotherapy [24]. Some researchers used heparin-polyethyleneimine (HPEI) nanogels to deliver SULF1 combined with cisplatin in ovarian cancer nude mice, the combination of SULF1/HPEI complexes with cisplatin exhibited the enhanced antitumor activity than DDP alone [25]. The specific mechanism of SULF1 regulating cisplatin sensitivity is unclear. However, SULF1 is an extracellular sulfatase that selectively removes 6-O-sulfate groups from heparan sulfate (HS) chains of heparan sulfate proteoglycans (HSPGs). HS chains act as co-receptors for heparin-binding growth factors such as fibroblast growth factor and vascular endothelial growth factor, so SULF1 might regulate cellular signaling by modifying the sulfation state of HS chains to affect platinum sensitivity.
ZNF587B is a C2H2-type zinc finger protein (ZFP). ZFPs are a group of transcription factors with zinc finger domains which regulate gene expression at the transcriptional and translation levels by binding specifically to DNA, RNA, DNA-RNA, and itself or other ZFPs [26-29]. Our results demonstrated that knockdown of ZNF587B significantly decreased the sensitivity of cisplatin in ovarian cancer cells, and the over-expression of ZNF587B promoted the cytotoxicity of cisplatin. These results showed that ZNF587B might play a key role in platinum resistance. However, the specific mechanism is unclear. HKR1, a C2H2-type ZFP, was proven over-expressed in lung cancers than in normal lung tissues, and the mRNA level of HKR1 in lung cancers cell lines was increased after exposure to cisplatin [30]. ZNF93, another C2H2-type ZFP, was overexpressed in a cisplatin-resistant ovarian cancer cell lines [31]. In 2018, a C2H2 protein GLI2 was found to promote platinum-based combination chemotherapy in colorectal cancer [32]. These studies indicated that role of ZFP in chemotherapeutic resistance was gradually being discovered. In order to analyze the functional domains of ZNF587B, we retrieved in SMART database (http://smart.embl-heidelberg.de/smart/set_mode.cgi?GENOMIC=1), and found that ZNF587B contained a Krüppel-associated box (or KRAB domain) which is believed to be one of the most extensive and powerful transcriptional inhibition domains found in mammals up to now. We speculated that the KRAB domain might also be related to the resistant mechanism [33].
Our study utilize human GeCKO library coupled with new generation sequencing (NGS) to screen genes for cisplatin resistance in ovarian cancer, we identified ZNF587B as a new potential mechanism for cisplatin sensitivity. We hope that ZNF587B could be served as a new biomarker to predict platinum-resistance. Further studies are needed to perform in vivo and clinical studies to explore the specific mechanism.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81402968), the Hunan Natural Science Foundation (2017JJ3503), Graduate students independently explore innovative projects of central south university, China (2017zzts862, 2018zzts905).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Yao Y, Sun L, Zhou J, Miao M, Luo S, Deng G, Li J, Wang J, Tang J. Snail driving alternative splicing of CD44 by ESRP1 enhances invasion and migration in epithelial ovarian cancer. Cell Physiol Biochem. 2017;43:2489–2504. doi: 10.1159/000484458. [DOI] [PubMed] [Google Scholar]
- 3.Xie X, Yang M, Ding Y, Yu L, Chen J. Formyl peptide receptor 2 expression predicts poor prognosis and promotes invasion and metastasis in epithelial ovarian cancer. Oncol Rep. 2017;38:3297–3308. doi: 10.3892/or.2017.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes D. The problem with platinum. Nature. 2015;527:S218–219. doi: 10.1038/527S218a. [DOI] [PubMed] [Google Scholar]
- 5.Mo D, Fang H, Niu K, Liu J, Wu M, Li S, Zhu T, Aleskandarany MA, Arora A, Lobo DN, Madhusudan S, Balajee AS, Chi Z, Zhao Y. Human helicase RECQL4 drives cisplatin resistance in gastric cancer by activating an AKT-YB1-MDR1 signaling pathway. Cancer Res. 2016;76:3057–3066. doi: 10.1158/0008-5472.CAN-15-2361. [DOI] [PubMed] [Google Scholar]
- 6.Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC. ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer. 2005;4:18. doi: 10.1186/1476-4598-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasello M, Michelacci F, Scionti I, Hattinger CM, Zuntini M, Caccuri AM, Scotlandi K, Picci P, Serra M. Overcoming glutathione s-transferase P1-related cisplatin resistance in osteosarcoma. Cancer Res. 2008;68:6661–6668. doi: 10.1158/0008-5472.CAN-07-5840. [DOI] [PubMed] [Google Scholar]
- 8.Li T, Peng J, Zeng F, Zhang K, Liu J, Li X, Ouyang Q, Wang G, Wang L, Liu Z, Liu Y. Association between polymorphisms in CTR1, CTR2, ATP7A, and ATP7B and platinum resistance in epithelial ovarian cancer. Int J Clin Pharmacol Ther. 2017;55:774–780. doi: 10.5414/CP202907. [DOI] [PubMed] [Google Scholar]
- 9.Yin JY, Li X, Zhou HH, Liu ZQ. Pharmacogenomics of platinum-based chemotherapy sensitivity in NSCLC: toward precision medicine. Pharmacogenomics. 2016;17:1365–1378. doi: 10.2217/pgs-2016-0074. [DOI] [PubMed] [Google Scholar]
- 10.Yin JY, Huang Q, Zhao YC, Zhou HH, Liu ZQ. Meta-analysis on pharmacogenetics of platinum-based chemotherapy in non small cell lung cancer (NSCLC) patients. PLoS One. 2012;7:e38150. doi: 10.1371/journal.pone.0038150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YB, Mei Y, Tian ZW, Long J, Luo CH, Zhou HH. Downregulation of RIF1 enhances sensitivity to platinum-based chemotherapy in epithelial ovarian cancer (EOC) by regulating nucleotide excision repair (NER) pathway. Cell Physiol Biochem. 2018;46:1971–1984. doi: 10.1159/000489418. [DOI] [PubMed] [Google Scholar]
- 12.Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey PJ, Kassahn KS, Newell F, Quinn MC, Kazakoff S, Quek K, Wilhelm-Benartzi C, Curry E, Leong HS Australian Ovarian Cancer Study Group. Hamilton A, Mileshkin L, Au-Yeung G, Kennedy C, Hung J, Chiew YE, Harnett P, Friedlander M, Quinn M, Pyman J, Cordner S, O’Brien P, Leditschke J, Young G, Strachan K, Waring P, Azar W, Mitchell C, Traficante N, Hendley J, Thorne H, Shackleton M, Miller DK, Arnau GM, Tothill RW, Holloway TP, Semple T, Harliwong I, Nourse C, Nourbakhsh E, Manning S, Idrisoglu S, Bruxner TJ, Christ AN, Poudel B, Holmes O, Anderson M, Leonard C, Lonie A, Hall N, Wood S, Taylor DF, Xu Q, Fink JL, Waddell N, Drapkin R, Stronach E, Gabra H, Brown R, Jewell A, Nagaraj SH, Markham E, Wilson PJ, Ellul J, McNally O, Doyle MA, Vedururu R, Stewart C, Lengyel E, Pearson JV, Waddell N, deFazio A, Grimmond SM, Bowtell DD. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 13.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard AP, Hood L. Sequence to array: probing the genome’s secrets. Nat Biotechnol. 1996;14:1649. doi: 10.1038/nbt1296-1649. [DOI] [PubMed] [Google Scholar]
- 17.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinhart Z, Pavlovic Z, Chandrashekhar M, Hart T, Wang X, Zhang X, Robitaille M, Brown KR, Jaksani S, Overmeer R, Boj SF, Adams J, Pan J, Clevers H, Sidhu S, Moffat J, Angers S. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat Med. 2017;23:60–68. doi: 10.1038/nm.4219. [DOI] [PubMed] [Google Scholar]
- 19.Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, Shridhar V. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 20.Lai JP, Thompson JR, Sandhu DS, Roberts LR. Heparin-degrading sulfatases in hepatocellular carcinoma: roles in pathogenesis and therapy targets. Future Oncol. 2008;4:803–814. doi: 10.2217/14796694.4.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aghajanian C. Clinical update: novel targets in gynecologic malignancies. Semin Oncol. 2004;31:22–26. doi: 10.1053/j.seminoncol.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Khurana A, Beleford D, He X, Chien J, Shridhar V. Role of heparan sulfatases in ovarian and breast cancer. Am J Cancer Res. 2013;3:34–45. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu P, Khurana A, Rattan R, He X, Kalloger S, Dowdy S, Gilks B, Shridhar V. Regulation of HSulf-1 expression by variant hepatic nuclear factor 1 in ovarian cancer. Cancer Res. 2009;69:4843–4850. doi: 10.1158/0008-5472.CAN-08-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staub J, Chien J, Pan Y, Qian X, Narita K, Aletti G, Scheerer M, Roberts LR, Molina J, Shridhar V. Epigenetic silencing of HSulf-1 in ovarian cancer: implications in chemoresistance. Oncogene. 2007;26:4969–4978. doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Gou M, Yi T, Qi X, Xie C, Zhou S, Deng H, Wei Y, Zhao X. The enhanced antitumor effects of biodegradable cationic heparin-polyethyleneimine nanogels delivering HSulf-1 gene combined with cisplatin on ovarian cancer. Int J Oncol. 2012;41:1504–1512. doi: 10.3892/ijo.2012.1558. [DOI] [PubMed] [Google Scholar]
- 26.Konieczny P, Stepniak-Konieczna E, Sobczak K. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014;42:10873–10887. doi: 10.1093/nar/gku767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Font J, Mackay JP. Beyond DNA: zinc finger domains as RNA-binding modules. Methods Mol Biol. 2010;649:479–491. doi: 10.1007/978-1-60761-753-2_29. [DOI] [PubMed] [Google Scholar]
- 28.Matthews JM, Sunde M. Zinc fingers--folds for many occasions. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 30.Oguri T, Katoh O, Takahashi T, Isobe T, Kuramoto K, Hirata S, Yamakido M, Watanabe H. The Krüppel-type zinc finger family gene, HKR1, is induced in lung cancer by exposure to platinum drugs. Gene. 1998;222:61–7. doi: 10.1016/s0378-1119(98)00464-8. [DOI] [PubMed] [Google Scholar]
- 31.Cordes N, Duan Z, Choy E, Harmon D, Yang C, Ryu K, Schwab J, Mankin H, Hornicek FJ. ZNF93 increases resistance to ET-743 (Trabectedin; Yondelis) and PM00104 (Zalypsis) in human cancer cell lines. PLoS One. 2009;4:e6967. doi: 10.1371/journal.pone.0006967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang YA, Chen YF, Bao Y, Mahara S, Yatim S, Oguz G, Lee PL, Feng M, Cai Y, Tan EY, Fong SS, Yang ZH, Lan P, Wu XJ, Yu Q. Hypoxic tumor microenvironment activates GLI2 via HIF-1alpha and TGF-beta2 to promote chemoresistance in colorectal cancer. Proc Natl Acad Sci U S A. 2018;115:E5990–E5999. doi: 10.1073/pnas.1801348115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]