Abstract
Herpes virus entry mediator (HVEM), also called tumor necrosis factor receptor superfamily 14 (TNFRSF14), is highly expressed in various tumor tissues and plays critical roles in tumor biology. However, the role of HVEM in clear cell renal cell carcinoma (ccRCC) is unknown. This study evaluated the clinical importance of HVEM in patients with ccRCC. HVEM expression was assessed in fresh and 140 archived paraffin-embedded ccRCC tissue samples by quantitative RT-PCR, western blot, and immunohistochemical staining. HVEM expression was higher in ccRCC than in paired peritumor tissue. Kaplan-Meier analysis showed that high level of HVEM expression was associated with poor overall survival (OS) and disease-free survival (DFS) in patients with ccRCC (both P < 0.001). Multivariate analysis indicated that HVEM overexpression was independently prognostic of survival in ccRCC patients. Two novel nomogram systems were constructed by integrating HVEM expression and other clinical parameters to predict OS (c-index 0.75) and DFS (c-index 0.74) in these patients, with both having better predictive accuracy than traditional TNM (c-index 0.65 for OS and 0.639 for DFS) and Fuhrman (c-index 0.612 for OS and 0.641 for DFS) systems. In addition, HVEM silencing led to an observable reduction in tumor cells growth in vitro and in vivo. Taken together, these findings indicate that high HVEM expression is a novel and independent adverse predictor of clinical outcomes in patients with ccRCC and that HVEM may be a potential therapeutic target.
Keywords: Clear cell renal cell carcinoma, herpes virus entry mediator, disease-free survival, overall survival, prognostic biomarker
Introduction
Renal cell carcinoma (RCC) is the third most common type of urological cancer in adults [1]. These tumors arise from the proximal tubular epithelium, with RCCs accounting for 90-95% of all renal tumors [2]. The incidence rate of RCC has increased over the last three decades, with approximately 50% of patients eventually developing metastatic disease, despite improvements in diagnosis [3-5]. Clear cell RCC (ccRCC), the most common RCC subtype, accounts for 75-80% of all primary kidney malignancies [6,7]. The evaluation of ccRCC prognosis is primarily based on the TNM staging system and Fuhrman grade [8], although the University of California Integrated Staging System (UCISS) and stage, size, grade, and necrosis (SSIGN) score are also used [9]. However, ccRCC patients with the same clinical stage often have different clinical outcomes, indicating that these systems cannot accurately predict prognosis in patients with ccRCC. The identification of molecular markers may enhance the ability to predict prognosis in patients with ccRCC.
Herpes virus entry mediator (HVEM), also called tumor necrosis factor receptor superfamily 14 (TNFRSF14), is a cellular mediator of herpes simplex virus entry [10,11]. It is widely expressed on several types of cell types, including T cells and B cells, as well as on other hematopoietic cells, such as monocytes, Tregs and NK cells, and non-hematopoietic cells, such as parenchymal cells [12]. HVEM, as a ligand for BTLA and CD160 and a receptor for LIGHT and LTα [13,14], can activate either co-stimulatory or co-inhibitory signaling pathways [15,16]. Previous studies have reported that HVEM pathway involved in various infection, autoimmune, and inflammation related diseases, such as arthritis, hepatitis [12], and colitis [17-19]. Furthermore, it has been shown that HVEM expression is obviously up-regulated in different tumors, including breast cancer, gastric cancer, and hepatocellular carcinoma [20-22]. A recent study reported that HVEM level was elevated in ovarian cancer tissues, and that knock-down of HVEM expression increased the sensitivity of ovarian cancer cells to activated T cells [23]. To date, however, the expression patterns and clinical significance of HVEM in patients with ccRCC have not been evaluated.
This study therefore investigated the expression of HVEM in ccRCC and analyzed the association of HVEM expression with the clinicopathological characteristics, overall survival (OS), and disease-free survival (DFS) in ccRCC patients. Moreover, two novel nomogram systems, formed by integrating HVEM expression with other clinical parameters, were found to predict OS and DFS in patients with ccRCC. In addition, the possible role of HVEM in ccRCC was further investigated in vitro and in vivo.
Materials and methods
Tissue specimens
Thirty paired fresh ccRCC and peritumor tissue samples were collected from patients who underwent partial or radical nephrectomy at the Southwest Hospital, Third Military Medical University (Chongqing, China) during 2016. In addition, tissue specimens from 140 patients with ccRCC who underwent surgery between 2010 and 2011 were examined. Patients were included if they had no history of any other malignancy and had not received any anticancer therapy prior to nephrectomy. Patients were excluded if follow-up was incomplete or they died within 1 month after surgery, or if paraffin-embedded samples contained areas of > 80% tumor necrosis. Clinical tumor stages were classified according to the 2010 TNM system of the American Joint Committee on Cancer (AJCC) [24]. All patients with ccRCC were followed up from the date of diagnosis to the date of death or last follow-up. All procedures complied with the Helsinki declaration. The study protocol was approved by the ethics committee of Southwest Hospital, and all patients provided written informed consent.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissue samples using TRIzol reagent (Takara, Japan), and first-strand cDNA was synthesized using a reverse transcription system (Takara, Japan), according to the manufacturer’s protocol. Levels of mRNA expression were normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in the same samples. The primer sequences for PCR amplification were: HVEM forward, 5’-TCATCGTCATTGTTTGCTCCA-3’; HVEM reverse, 5’-ACCTTGACTACATCACCCCTT-3’; GAPDH forward, 5’-CTCTGCTCCTCCTGTTCGAC-3’ and GAPDH reverse, 5’-GCGCCCAATACGACCAAATC-3’. All samples were assayed in triplicate. Differences in gene expression levels were calculated using 2-ΔΔct method [25].
Western blot
Total protein was isolated from tissues samples using RIPA buffer (Beyotime, Shanghai, China), according to the manufacturer’s protocol. Protein concentrations were measured using BCA Protein Assay Kits (Beyotime, Shanghai, China). Protein samples (30 μg/lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated overnight at 4°C with mouse anti-human HVEM antibody (1:500; Santa Cruz Biotechnology), rabbit anti-human Bcl-2 antibody (1:1000; Abcam, Cambridge, MA, USA), and rabbit anti-human Bax antibody (1:1000; Abcam, Cambridge, MA, USA). Mouse anti-human GAPDH antibody (1:1000; Abcam, Cambridge, MA, USA) was used as a loading control, followed by incubation with horseradish peroxidase-conjugated goat anti-mouse IgG and goat anti-rabbit IgG secondary antibody (both 1:3000; ZSGB-BIO, Beijing, China), respectively. Immunoblots were visualized using the ECL Western Blot Detection System (Millipore, Billerica, MA, USA).
Immunohistochemical staining
Immunohistochemical staining was performed using routine protocols as described [26]. Briefly, sections (4-μm) were blocked with 5% BSA for 1 h at room temperature and incubated at 4°C overnight with primary antibodies against HVEM (1:100, Santa Cruz Biotechnology), followed by incubation with horseradish peroxidase-conjugated secondary antibody (1:800; Beyotime, Shanghai, China). The results were analyzed using DAB assay kits (ZSGB-BIO, Beijing, China) by two pathologists, who were blinded to the clinicopathological features of the samples. Immunohistochemical scores ranging from 0 to 12 were calculated by multiplying staining intensities (0: negative; 1: weak; 2: moderate; and 3: strong) by the proportion of positively stained tumor cells (0: < 5%; 1: 5%-25%; 2: 26%-50%; 3: 51%-75%; and 4: > 75%). Staining index scores ≥ 6 and ≤ 4 were defined as high and low HVEM expression, respectively.
Knockdown HVEM expression in vitro
786-O and ACHN cells were plated in 6-well plates (1 × 103 cells/cm2) in complete medium without antibiotics. After 70% confluent, cells were transfected with 10 μM siRNA for HVEM (HVEM-siRNA-1 and HVEM-siRNA-2) and scramble control siRNA (Ctrl-siRNA; all from RiboBio, Guangzhou, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, Calif) according to the manufacturer’s protocol. At 6 hours later, the medium was replaced with complete medium for 24 hours. To confirm the efficacy of the siRNA on the expression of HVEM, protein was isolated and assayed by western blot. In order to stably knockdown HVEM expression, lentiviral short hairpin (sh)-HVEM-knockdown constructs (sh-HVEM) and the negative control lentiviruses (vector) were purchased (GeneChem, Shanghai, China). The lentiviral infection was performed according to the manufacturer’s protocol and the knockdown effect of sh-HVEM was determined by western blot.
Cell viability and apoptosis assays
Cell viability was determined using commercially available Cell Counting Kit-8 kits (CCK-8; Dojinodo, Shanghai, China) according to the manufacturer’s protocol. The absorbance at 450 nm was recorded using the microplate reader. Cell apoptosis was measured by using flow cytometry as previously described [27].
In vivo xenograft experiment
The male nude mice (4-5-week-old) used in this study were purchased from the Peking University Animal Center (Beijing, China). A total of 5 × 106 ACHN cells stably transfected with sh-HVEM or negative control (vector) were subcutaneously injected into right oxter in mice. Tumor size was measured and the volume was calculated every four days by tumor length (L) and width (W) as: volume = (L × W2)/2. The mice were sacrificed at 28 days after cells injection. Ki67 levels in tumor tissues were measured by immunohistochemical staining with the anti-Ki67 antibody (1:100, Abcam, Cambridge, MA, USA). All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of the Third Military Medical University.
Statistical analysis
All data were analyzed using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, USA) and SPSS 19.0 (SPSS Inc., IL, Chicago, USA) software. The associations between HVEM expression and clinicopathological characteristics of ccRCC patients were evaluated by Chi-square tests. Survival curves were determined by the Kaplan-Meier method and compared by log-rank tests. Univariate and multivariate Cox proportional hazard models were used to evaluate hazard ratios (HRs) and 95% confidence intervals (CIs). The statistically significant differences between two groups were assessed using the Student’s t-test, the data are represented as means ± standard error of the mean (SEM), and P < 0.05 was considered statistically significant.
Results
HVEM overexpression in ccRCC tissues
HVEM mRNA expression was assessed by qRT-PCR in 30 paired fresh tumor and peritumor tissues. Compared with peritumor tissues, HVEM mRNA expression was significantly higher in tumor tissues (P < 0.001; Figure 1A). Measurements of HVEM protein levels by western blot in tumor and peritumor tissues yielded similar results (Figure 1B). These data indicate that the expression of HVEM is significantly upregulated in fresh ccRCC tissues.
Figure 1.
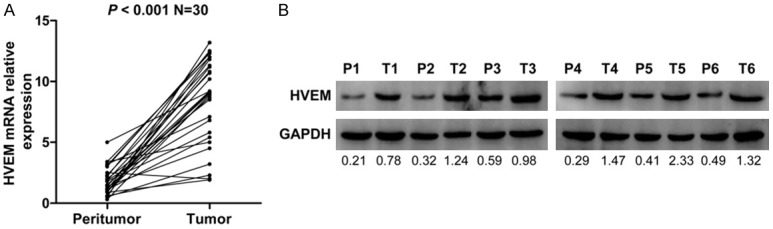
Analysis of the expression of HVEM in fresh ccRCC tissues. A. qRT-PCR measurement of HVEM mRNA expression in 30 paired fresh tumor tissues and peritumor tissues. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as a loading control. P < 0.05 on paired-samples t test was regarded as statistically significant. B. Western blot assay of the expression of HVEM protein in six paired tumor and peritumor tissue samples. GAPDH was used as a loading control. P = Peritumor; T = Tumor.
Immunohistochemical assessment and association between HVEM expression and clinicopathological characteristics
To further evaluate HVEM protein expression in ccRCC tissues, we assayed the expression of HVEM in 140 paraffin-embedded ccRCC tissue samples and 38 peritumor tissue samples by immunohistochemical staining. HVEM expression was mainly localized in the membrane of ccRCC cells (Figure 2C-J). In contrast, no or weak HVEM staining was observed in peritumor tissues (Figure 2A and 2B). Index scores for HVEM staining showed low and high HVEM expression in 41 (29.3%; Figure 2C-F) and 99 (70.7%; Figure 2G-J) tumor specimens, respectively (P < 0.001; Figure 2K). HVEM expression correlated significantly with older age (P = 0.003) and larger tumor size (P < 0.001) (Table 1). No other clinicopathological variables were significantly correlated with HVEM expression.
Figure 2.
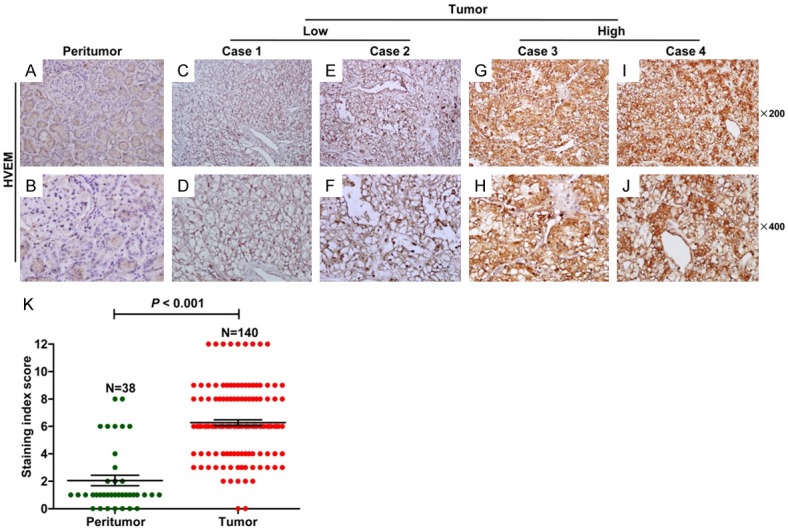
Immunohistochemical assays of HVEM expression in ccRCC tissues. A, B. Expression of HVEM in peritumor tissue. (upper, original magnification × 200; lower, original magnification × 400). C-F. Low HVEM expression in tumor tissues. (upper, original magnification × 200; lower, original magnification × 400). G-J. High HVEM expression in tumor tissues. (upper, original magnification × 200; lower, original magnification × 400). K. Staining index scores of HVEM expression in tumor tissues and peritumor tissues. P < 0.05 by the Mann Whitney test was considered statistically significant.
Table 1.
Correlation between HVEM expression and clinicopathologic characteristics of ccRCC patients
| Characteristic | Cases (n = 140) | HVEM expression | X2 | P ‡ | |
|---|---|---|---|---|---|
|
| |||||
| Low (n = 41) | High (n = 99) | ||||
| Gender | 0.642 | 0.423 | |||
| Male | 85 | 27 (31.8%) | 58 (68.2%) | ||
| Female | 55 | 14 (25.5%) | 41 (74.5%) | ||
| Age (years)† | 8.873 | 0.003* | |||
| ≤ 55 | 91 | 19 (20.9%) | 72 (79.1%) | ||
| > 55 | 49 | 22 (44.9%) | 27 (55.1%) | ||
| Tumor size (cm)† | 14.766 | < 0.001* | |||
| ≤ 4 | 74 | 32 (43.2%) | 42 (56.8%) | ||
| > 4 | 66 | 9 (13.6%) | 57 (86.4%) | ||
| T classification | 1.507 | 0.299 | |||
| T1-T2 | 129 | 36 (27.9%) | 93 (72.1%) | ||
| T3-T4 | 11 | 5 (45.5%) | 6 (54.5%) | ||
| N classification | 0.482 | 0.671 | |||
| N0 | 134 | 40 (29.9%) | 94 (70.1%) | ||
| N1 | 6 | 1 (16.7%) | 5 (83.3%) | ||
| TNM stage | 0.000 | 1.000 | |||
| I-II | 123 | 36 (29.3%) | 87 (70.7%) | ||
| III-IV | 17 | 5 (29.4%) | 12 (70.6%) | ||
| Fuhrman grade | 3.82 | 0.051 | |||
| 1-2 | 96 | 33 (34.4%) | 63 (65.6%) | ||
| 3-4 | 44 | 8 (18.2%) | 36 (81.8%) | ||
| Necrosis | 0.968 | 0.396 | |||
| Absent | 124 | 38 (30.6%) | 86 (69.4%) | ||
| Present | 16 | 3 (18.8%) | 13 (81.2%) | ||
| Vascular invasion | 0.076 | 0.721 | |||
| Absent | 131 | 38 (29.0%) | 93 (71.0%) | ||
| Present | 9 | 3 (33.3%) | 6 (66.7%) | ||
Abbreviations: HVEM = Herpes virus entry mediator.
Split at median;
P-value from Chi-square or Fisher exact test;
Statistically significant (P < 0.05).
Prognostic significance of HVEM in ccRCC patients
The 5-year OS and DFS rates in the 140 patients with ccRCC were 75% and 65%, respectively (Figure 3A). The Kaplan-Meier method was used to determine whether levels of HVEM expression were predictive of clinical outcomes in patients with ccRCC. We found that patients with high HVEM had significantly poorer OS (P < 0.001) and DFS (P < 0.001) than patients with low HVEM expression (Figure 3B). The prognostic value of HVEM was further evaluated by comparing its level of expression in different subgroups of ccRCC patients stratified by clinical TNM stage. We found that high level of HVEM expression strongly correlated with poorer OS (P = 0.003) and DFS (P < 0.001) in patients with TNM stage I+II, or early-stage, ccRCC (Figure 3C). This correlation, however, was not observed in patients with TNM stage III+IV, or advanced-stage, ccRCC (both P = 0.176; Figure 3D), probably due to the small sample size. These findings suggest that HVEM is an important biomarker for ccRCC patients, at least in the early-stage of this disease.
Figure 3.
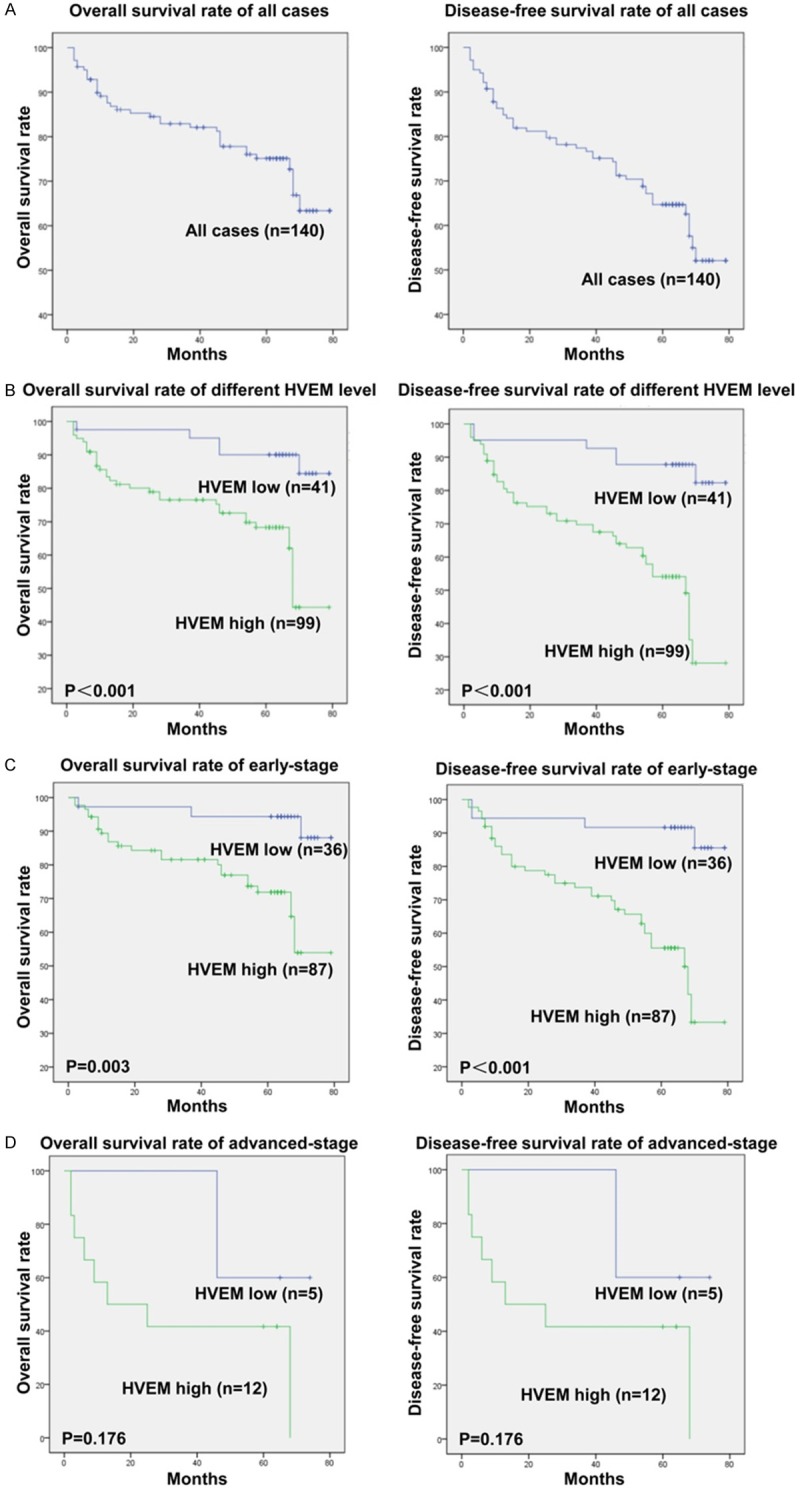
Kaplan-Meier survival analysis of the effects of HVEM expression on OS and DFS in ccRCC patients. A. Five-year OS and DFS rates in all patients. B. OS and DFS in patients with high and low tumor HVEM expression. C, D. OS and DFS in patients with early-stage (TNM stage I+II) and advanced-stage (TNM stage III+IV) ccRCC and with high and low tumor HVEM expression. P < 0.05 by the log rank test was regarded as statistically significant.
Univariate and multivariate Cox regression analyses were also performed to evaluate the correlation between HVEM expression and postoperative survival (Table 2). Univariate Cox regression analysis showed that tumor size, N classification, TNM stage, Fuhrman grade, and high HVEM expression were independent predictors of OS (HR, 4.396, 95% CI, 1.643-11.766, P = 0.003) and of DFS (HR, 5.525; 95% CI, 2.290-13.329; P < 0.001) (Table 2). Multivariate analysis showed that HVEM expression was an independent predictor of OS (HR, 3.249; 95% CI, 1.078-9.793; P = 0.036) and DFS (HR, 4.748; 95% CI, 1.795-12.562; P = 0.002). Other predictors of survival included tumor size (P = 0.035 for OS), N classification (P = 0.011 for DFS), TNM staging (P = 0.002 for OS and P = 0.017 for DFS) and Fuhrman grade (P = 0.028 for OS and P < 0.001 for DFS) (Table 2).
Table 2.
Univariate and multivariate Cox regression analysis of factors prognostic of overall survival (OS) and disease free survival (DFS) in ccRCC patients
| Characteristic | OS | DFS | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Univariate | Multivariate | Univariate | Multivariate | |||||
|
|
|
|
|
|||||
| HR (95% CI) | P ‡ | HR (95% CI) | P ‡ | HR (95% CI) | P ‡ | HR (95% CI) | P ‡ | |
| Gender (Male vs Female) | 0.814 (0.406-1.633) | 0.563 | 1.105 (0.634-1.927) | 0.725 | ||||
| Age, years (> 55 vs ≤ 55) | 1.032 (0.524-2.033) | 0.926 | 1.299 (0.748-2.257) | 0.354 | ||||
| Tumor size, cm (> 4 vs ≤ 4) | 1.116 (1.458-5.986) | 0.049* | 2.524 (1.066-5.977) | 0.035* | 2.892 (1.616-5.179) | < 0.001* | 1.814 (0.911-3.613) | 0.090 |
| T classification (T3+T4 vs T1+T2) | 2.307 (0.96-5.547) | 0.062 | 1.532 (0.654-3.590) | 0.326 | ||||
| N classification (N1 vs N0) | 5.007 (1.733-14.464) | 0.003* | 2.998 (0.933-9.632) | 0.065 | 4.656 (1.818-11.922) | 0.001* | 3.936 (1.375-11.263) | 0.011* |
| TNM stage (III+IV vs I+II) | 3.452 (1.662-7.173) | 0.001* | 8.579 (2.256-32.623) | 0.002* | 2.172 (1.088-4.335) | 0.028* | 4.147 (1.289-13.342) | 0.017* |
| Fuhrman grade (3+4 vs 1+2) | 2.956 (1.526-5.728) | 0.001* | 2.339 (1.098-4.981) | 0.028* | 3.496 (2.017-6.061) | < 0.001* | 3.287 (1.760-6.139) | < 0.001* |
| Necrosis (Present vs Absent) | 1.208 (0.469-3.107) | 0.696 | 1.165 (0.525-2.584) | 0.708 | ||||
| Vascular invasion (Present vs Absent) | 0.394 (0.054-2.877) | 0.358 | 0.266 (0.037-1.929) | 0.190 | ||||
| HVEM expression (High vs low) | 4.396 (1.643-11.766) | 0.003* | 3.249 (1.078-9.793) | 0.036* | 5.525 (2.290-13.329) | < 0.001* | 4.748 (1.795-12.562) | 0.002* |
Abbreviations: HVEM = Herpes virus entry mediator; OS = overall survival; DFS = disease-free survival; CI = confidence interval; HR = hazard ratio.
P-value from the Cox proportional hazards model;
Statistically significant (P < 0.05).
Construction of novel prognostic nomogram systems for OS and DFS and evaluation of their accuracy
Two novel nomogram systems for predicting 3-year and 5-year OS and DFS in patients with ccRCC were constructed by integrating the independent parameters identified in multivariate analysis (Figure 4A and 4B). These nomogram systems showed that HVEM expression was a negative indicator of OS and DFS. To evaluate the prognostic accuracy of these nomogram systems, we compared the concordance index (c-index) of the novel system with that of TNM stage and the Fuhrman grade (Table 3). The integrated nomogram system had a higher c-index (0.75) than the TNM system (0.65) and Fuhrman system (0.612) for OS, as well as having a higher c-index (0.74) than the TNM system (0.639) and Fuhrman system (0.641) for DFS (Table 3). These findings demonstrate that the novel nomogram systems are more accurate than traditional TNM stage and the Fuhrman grade for predicting OS and DFS in patients with ccRCC.
Figure 4.
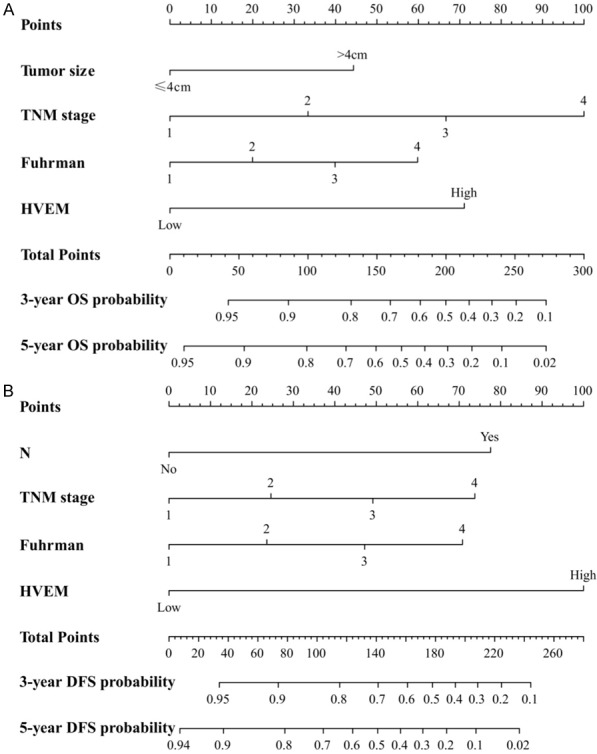
Nomogram systems for predicting 3-year and 5-year OS and DFS in patients with ccRCC and evaluation of their accuracy. A. Nomogram predictive of OS. B. Nomogram predictive of DFS.
Table 3.
Comparison of the nomogram model with TNM and Fuhrman systems by Concordance index (C-index)
| C-index | C-index 95% CI | P ‡ | |||
|---|---|---|---|---|---|
| OS | Nomogram | 0.750 | 0.672 | 0.828 | < 0.001* |
| TNM | 0.650 | 0.563 | 0.737 | < 0.001* | |
| Fuhrman | 0.612 | 0.522 | 0.702 | 0.015* | |
| DFS | Nomogram | 0.740 | 0.671 | 0.809 | < 0.001* |
| TNM | 0.639 | 0.569 | 0.709 | < 0.001* | |
| Fuhrman | 0.641 | 0.568 | 0.714 | < 0.001* | |
Abbreviations: OS = overall survival; DFS = disease-free survival; C-index = Concordance index; CI = confidence interval.
P-value from the z test;
Statistically significant (P < 0.05).
The correlation of HVEM expression with ccRCC cells growth
In order to investigate the role of HVEM in promoting ccRCC, the expression of HVEM in tumor cell lines 786-O and ACHN cells were knockdown by siRNA in vitro (Figure 5A). It was found that the knockdown of HVEM expression in 786-O and ACHN cells markedly led to a significant reduction in cells viability and an increase in cells apoptosis, accompanied with a significant reduction in the anti-apoptosis protein Bcl-2 expression and an increase in the apoptosis-promoting protein Bax expression (Figure 5B-D).
Figure 5.
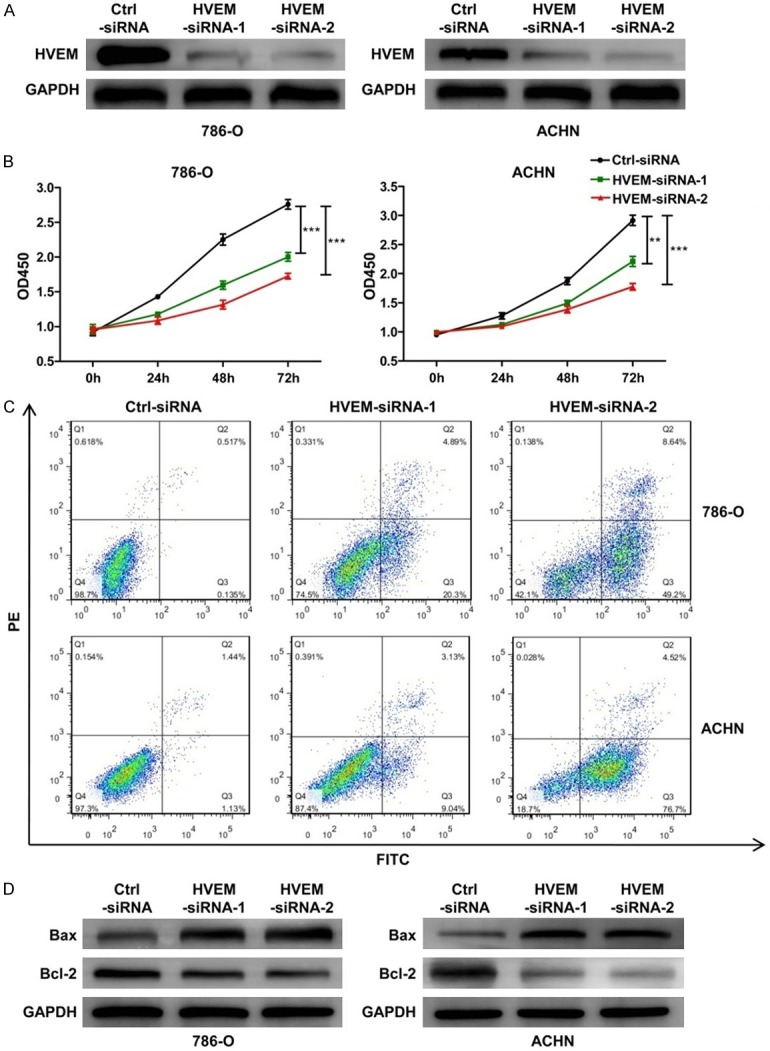
The biological role of HVEM in ccRCC cells. (A) The levels of HVEM protein in 786-O and ACHN cells were detected by western blot analysis. Representative western blot and quantitative data are presented. (B) 786-O and ACHN cells viability and (C) apoptosis in different groups were measured by CCK-8 kit and flowcytometry analysis, respectively. (D) The expression of Bcl-2 and Bax protein were detected by western blot analysis. Representative western blot and quantitative data are presented. Results are representative of three replicate experiments. All values are represented as mean ± SEM. **P < 0.01 and ***P < 0.001.
To further explore the effects of HVEM on tumor formation ability in vivo, the HVEM expression in ACHN cells was stable knockdown with shRNA and the knockdown efficiency was confirmed by western blot (Figure 6A). It was showed that HVEM knockdown led to a significant reduction in tumors size and weight (Figure 6B-D), and Ki67 levels (Figure 6E). Collectively, these results indicate that HVEM promotes tumor formation via enhancing ccRCC cells proliferation.
Figure 6.
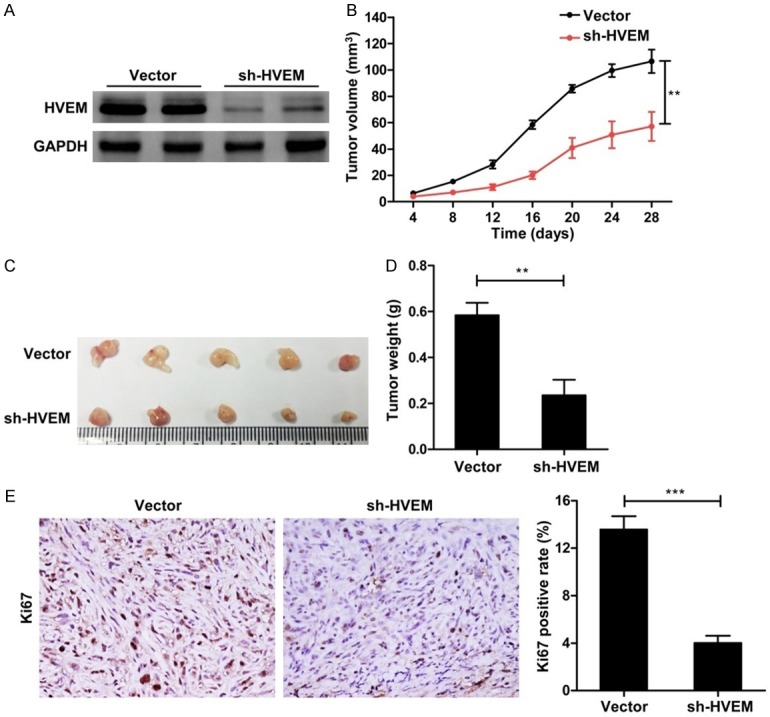
The correlation of HVEM knockdown with ccRCC cells growth in vivo. A. Stable knockdown of HVEM expression in ACHN cells was demonstrated by western blot. Representative western blots were presented. B. The growth curve of the tumor within 28 days. C and D. Representative graph of tumor size and tumor weight. E. Ki67 levels in tumor tissues were measured with immunohistochemical staining. Original magnification × 400. All values are represented as mean ± SEM, n = 5/group. **P < 0.01 and ***P < 0.001.
Discussion
To the best of our knowledge, this study is the first to show an association between high HVEM expression and poor prognosis of ccRCC patients. We found that HVEM expression was significantly higher in ccRCC than in peritumor tissue samples and that a high level of HVEM protein correlated with ccRCC tumor stages. Moreover, univariate and multivariate Cox regression analyses showed that HVEM expression was an independent prognostic factor in patients with ccRCC. These findings led to the construction of two novel nomogram systems, which integrated HVEM expression with other pathologic factors predictive of OS and DFS. Comparisons of their c-indexes showed that these nomograms were more accurate than TNM stage and the Fuhrman grade for predicting OS and DFS. Moreover, we found that HVEM promoted ccRCC cells growth both in vitro and in vivo, suggesting that HVEM aggravates the pathogenesis of ccRCC.
HVEM is an immunoregulatory factor expressed on T cells, B cells, monocytes, and immature DCs [28]. The level of HVEM expression was found to correlate negatively with the infiltration of CD4+ and CD8+ cells, and with interferon-γ (IFN-γ) level in hepatocellular carcinoma [20]. In ovarian cancer tissues, HVEM expression was found to be elevated, and HVEM blockade was found to enhance tumor-reactive T-cell activation as well as the secretion of TNF-α and IFN-γ [23], and to inhibit tumor growth [29,30]. Results of the 2013 TCGA cohort showed that HVEM mRNA expression was upregulated in ccRCC samples and that the levels of HVEM were closely associated with the expression of the tumor-promoting factors IL-11RA, IL-32, and IL-17RC in ccRCC tissues (Figure S1). Indeed, we found that HVEM mRNA and protein levels were markedly increased in ccRCC tissues. These findings indicate that aberrant HVEM expression in ccRCC tissues interferes with tumor progression and that HVEM expression may be clinically significant in patients with ccRCC.
Immunotherapy targeting co-inhibitory molecules, which inhibit T-cell proliferation and cytokine production, is one of the most promising strategies to improve the survival of patients with malignant tumors [31,32]. Tumor cell expression of HVEM has been found to correlate with local T cell suppression in primary tumors [20]. The BTLA/HVEM network is thought to inhibit T-cell proliferation and cytokine production [33], which are critical for the process of tumorigenesis, suggesting that HVEM may be an interesting target in tumor immunotherapy [28,34], including in ccRCC. Indeed, we found that HVEM silencing led to an observable increase in cells apoptosis in ccRCC cells and a remarkable reduction in ccRCC cells growth both in vitro and in vivo.
This study had several limitations, including its retrospective design and the relatively small number of patients, especially those with advanced-stage ccRCC. Large, prospective, and multicenter studies are needed to determine the clinical significance of HVEM in ccRCC.
In conclusion, we found that HVEM expression was markedly increased in ccRCC tissues. High level of HVEM was independently prognostic of OS and DFS and may represent a novel and useful prognostic marker for ccRCC patients. Prognostic nomogram systems integrating HVEM expression and other pathologic features were found to be more predictive of OS and DFS than traditional TNM stage and the Fuhrman grade in patients with ccRCC. HVEM is expected to become a potential marker for prognosis and one possible therapeutic target for the patients with ccRCC.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NO. 81670684).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Zhang D, Xia X, Wang X, Zhang P, Lu W, Yu Y, Deng S, Yang H, Zhu H, Xu N, Liang S. PGRMC1 is a novel potential tumor biomarker of human renal cell carcinoma based on quantitative proteomic and integrative biological assessments. PLoS One. 2017;12:e0170453. doi: 10.1371/journal.pone.0170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedke J, Gauler T, Grunwald V, Hegele A, Herrmann E, Hinz S, Janssen J, Schmitz S, Schostak M, Tesch H, Zastrow S, Miller K. Systemic therapy in metastatic renal cell carcinoma. World J Urol. 2017;35:179–188. doi: 10.1007/s00345-016-1868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss RH, Lin PY. Kidney cancer: identification of novel targets for therapy. Kidney Int. 2006;69:224–232. doi: 10.1038/sj.ki.5000065. [DOI] [PubMed] [Google Scholar]
- 4.Lindblad P. Epidemiology of renal cell carcinoma. Scand J Surg. 2004;93:88–96. doi: 10.1177/145749690409300202. [DOI] [PubMed] [Google Scholar]
- 5.Parker AS, Eckel-Passow JE, Serie D, Hilton T, Parasramka M, Joseph RW, Wu KJ, Cheville JC, Leibovich BC. Higher expression of topoisomerase II alpha is an independent marker of increased risk of cancer-specific death in patients with clear cell renal cell carcinoma. Eur Urol. 2014;66:929–935. doi: 10.1016/j.eururo.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Liu Y, Xu L, An H, Chang Y, Yang Y, Zhang W, Xu J. P2X7 receptor predicts postoperative cancer-specific survival of patients with clear-cell renal cell carcinoma. Cancer Sci. 2015;106:1224–1231. doi: 10.1111/cas.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun M, Shariat SF, Cheng C, Ficarra V, Murai M, Oudard S, Pantuck AJ, Zigeuner R, Karakiewicz PI. Prognostic factors and predictive models in renal cell carcinoma: a contemporary review. Eur Urol. 2011;60:644–661. doi: 10.1016/j.eururo.2011.06.041. [DOI] [PubMed] [Google Scholar]
- 9.Frank I, Blute ML, Cheville JC, Lohse CM, Weaver AL, Zincke H. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol. 2002;168:2395–2400. doi: 10.1016/S0022-5347(05)64153-5. [DOI] [PubMed] [Google Scholar]
- 10.Shui JW, Kronenberg M. HVEM is a TNF receptor with multiple regulatory roles in the mucosal immune system. Immune Netw. 2014;14:67–72. doi: 10.4110/in.2014.14.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 12.Steinberg MW, Cheung TC, Ware CF. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol Rev. 2011;244:169–187. doi: 10.1111/j.1600-065X.2011.01064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedy JR, Gavrieli M, Potter KG, Hurchla MA, Lindsley RC, Hildner K, Scheu S, Pfeffer K, Ware CF, Murphy TL, Murphy KM. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6:90–98. doi: 10.1038/ni1144. [DOI] [PubMed] [Google Scholar]
- 14.Cai G, Anumanthan A, Brown JA, Greenfield EA, Zhu B, Freeman GJ. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol. 2008;9:176–185. doi: 10.1038/ni1554. [DOI] [PubMed] [Google Scholar]
- 15.Cai G, Freeman GJ. The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunol Rev. 2009;229:244–258. doi: 10.1111/j.1600-065X.2009.00783.x. [DOI] [PubMed] [Google Scholar]
- 16.Murphy TL, Murphy KM. Slow down and survive: enigmatic immunoregulation by BTLA and HVEM. Annu Rev Immunol. 2010;28:389–411. doi: 10.1146/annurev-immunol-030409-101202. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Anders RA, Wang Y, Turner JR, Abraham C, Pfeffer K, Fu YX. The critical role of LIGHT in promoting intestinal inflammation and Crohn’s disease. J Immunol. 2005;174:8173–8182. doi: 10.4049/jimmunol.174.12.8173. [DOI] [PubMed] [Google Scholar]
- 18.Shang Y, Guo G, Cui Q, Li J, Ruan Z, Chen Y. The expression and anatomical distribution of BTLA and its ligand HVEM in rheumatoid synovium. Inflammation. 2012;35:1102–1112. doi: 10.1007/s10753-011-9417-2. [DOI] [PubMed] [Google Scholar]
- 19.Xu H, Cao D, Guo G, Ruan Z, Wu Y, Chen Y. The intrahepatic expression and distribution of BTLA and its ligand HVEM in patients with HBV-related acute-on-chronic liver failure. Diagn Pathol. 2012;7:142. doi: 10.1186/1746-1596-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hokuto D, Sho M, Yamato I, Yasuda S, Obara S, Nomi T, Nakajima Y. Clinical impact of herpesvirus entry mediator expression in human hepatocellular carcinoma. Eur J Cancer. 2015;51:157–165. doi: 10.1016/j.ejca.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Heo SK, Ju SA, Kim GY, Park SM, Back SH, Park NH, Min YJ, An WG, Nguyen TT, Kim SM, Kim BS. The presence of high level soluble herpes virus entry mediator in sera of gastric cancer patients. Exp Mol Med. 2012;44:149–158. doi: 10.3858/emm.2012.44.2.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang JYS, Chan KW, Ni YB, Hlaing T, Hu J, Chan SK, Cheung SY, Tse GM. Expression and clinical significance of herpes virus entry mediator (HVEM) in breast cancer. Ann Surg Oncol. 2017;24:4042–4050. doi: 10.1245/s10434-017-5924-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T, Ye L, Han L, He Q, Zhu J. Knockdown of HVEM, a lymphocyte regulator gene, in ovarian cancer cells increases sensitivity to activated T cells. Oncol Res. 2016;24:189–196. doi: 10.3727/096504016X14641336229602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SP, Alt AL, Weight CJ, Costello BA, Cheville JC, Lohse C, Allmer C, Leibovich BC. Independent validation of the 2010 American joint committee on cancer TNM classification for renal cell carcinoma: results from a large, single institution cohort. J Urol. 2011;185:2035–2039. doi: 10.1016/j.juro.2011.02.059. [DOI] [PubMed] [Google Scholar]
- 25.Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005;5:209–219. doi: 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- 26.Pan JS, Huang L, Belousova T, Lu L, Yang Y, Reddel R, Chang A, Ju H, DiMattia G, Tong Q, Sheikh-Hamad D. Stanniocalcin-1 inhibits renal ischemia/reperfusion injury via an AMP-activated protein kinase-dependent pathway. J Am Soc Nephrol. 2015;26:364–378. doi: 10.1681/ASN.2013070703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Wu H, Li W, Yin L, Guo S, Xu X, Ouyang Y, Zhao Z, Liu S, Tian Y, Tian Z, Ju J, Ni B, Wang H. Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-kappaB signaling. Oncogene. 2016;35:5501–5514. doi: 10.1038/onc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasero C, Speiser DE, Derre L, Olive D. The HVEM network: new directions in targeting novel costimulatory/co-inhibitory molecules for cancer therapy. Curr Opin Pharmacol. 2012;12:478–485. doi: 10.1016/j.coph.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Migita K, Sho M, Shimada K, Yasuda S, Yamato I, Takayama T, Matsumoto S, Wakatsuki K, Hotta K, Tanaka T, Ito M, Konishi N, Nakajima Y. Significant involvement of herpesvirus entry mediator in human esophageal squamous cell carcinoma. Cancer. 2014;120:808–817. doi: 10.1002/cncr.28491. [DOI] [PubMed] [Google Scholar]
- 30.Lasaro MO, Sazanovich M, Giles-Davis W, Mrass P, Bunte RM, Sewell DA, Hussain SF, Fu YX, Weninger W, Paterson Y, Ertl HC. Active immunotherapy combined with blockade of a coinhibitory pathway achieves regression of large tumor masses in cancer-prone mice. Mol Ther. 2011;19:1727–1736. doi: 10.1038/mt.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoos A. Development of immuno-oncology drugs-from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15:235–247. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 32.Su S, Wu W, He C, Liu Q, Song E. Breaking the vicious cycle between breast cancer cells and tumor-associated macrophages. Oncoimmunology. 2014;3:e953418. doi: 10.4161/21624011.2014.953418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croft M. The evolving crosstalk between co-stimulatory and co-inhibitory receptors: HVEM-BTLA. Trends Immunol. 2005;26:292–294. doi: 10.1016/j.it.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 34.del Rio ML, Lucas CL, Buhler L, Rayat G, Rodriguez-Barbosa JI. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J Leukoc Biol. 2010;87:223–235. doi: 10.1189/jlb.0809590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


