Abstract
Transcription factors are key determinants of gene expression that recognize and bind to short DNA sequence motifs, thereby regulating many biological processes including differentiation, development, and metabolism. Transcription factors are increasingly recognized for their roles in cancer progression. Here, we describe a subfamily of zinc finger transcription factors named zinc finger and SCAN domain containing (ZSCAN) transcription factors. In this review, we summarize the identified members of the ZSCAN family of transcription factors and their roles in cancer progression. Due to the complex regulation mechanisms, ZSCAN transcription factors may show promotive or prohibitive efforts in angiogenesis, cell apoptosis, cell differentiation, cell migration and invasion, cell proliferation, stem cell properties, and chemotherapy sensitivity. The upstream regulation mechanisms of their varied expression levels may include gene mutation, DNA methylation, alternative splicing, and miRNA regulation. What’s more, to clarify their diverse functions, we summarize the modulation mechanisms of their activity in downstream genes transcription, including protein-protein interactions mediated by their SCAN box, recruitment of co-regulating molecules and post-translational modifications. A better understanding of the widespread regulatory mode of these transcription factors will provide further insight into the mechanism of transcriptional regulation and suggest novel therapeutic strategies against tumor progression.
Keywords: ZSCAN transcription factor, transcriptional regulation, SCAN domain, cancer progression
Introduction
Transcription factors play a crucial role in controlling gene expression from DNA to mRNA by recognizing specific DNA sequences. Transcription factors are classified into families according to their conserved DNA-binding domains. New technologies such as large-scale chromatin immunoprecipitation (ChIP)-seq and DNase protection assays have revealed that promoters overlap with transcription factors. Transcription factors can turn genes on or off in specific environments and at specific times as part of their accurate transcriptional regulation [1]. They can also function as promoters and/or suppressors of downstream gene expression, and therefore possess a broad range of properties [2].
Zinc finger (ZNF) transcription factors, which form the largest transcription factor family, are characterized by finger-like DNA binding domains that require one or more zinc ions to stabilize the structure; they play an important role in many biological processes [3]. ZNF family transcription factors are divided into several classes according to the manner in which the zinc ions bind to the cysteine or histidine residues of the finger-like domain motif, such as C2H2 and Cys6 among others [4]. ZNF transcription factors contain N-terminal domains that interact with other proteins to regulate expression, subcellular localization, and transcriptional activity [5], including the Kruppel-associated box domain (KRAB) [6], the poxvirus and zinc finger domain (POZ) [7], the insect zinc finger associated domain (ZAD) [8], and the SCAN domain [9]. In this review, we will focus on zinc finger and SCAN domain-containing (ZSCAN) transcription factors, which comprise the smallest and most recently defined subfamilies [9]. The members of the mouse SCAN family, a highly conserved protein family, were previously described [5], whereas the human ZSCAN transcription factors are not well-organized. Providing additional insight into their roles and regulatory function is important as an increasing number of studies have reported the relationship between the ZSCAN transcription factors and cancer progression.
In this review, we phylogenetically classified the ZSCAN family and summarized the role of all the well-studied members in cancer. In addition, we described the potential underlying mechanisms from several aspects, namely, upstream regulation of these transcription factors, modulation of the transcriptional activity through protein interactions, and regulation of downstream genes.
The structure and members of the ZSCAN transcription factor family
ZSCAN transcription factors contain two main domains, the zinc finger domain and the SCAN box (Figure 1).
Figure 1.
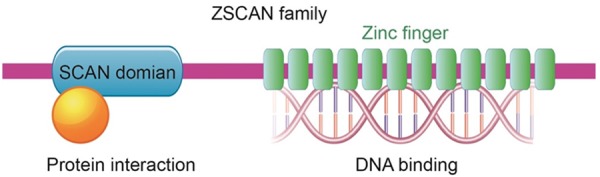
Model of the structures of the ZSCAN family. The ZSCAN family of transcription factors shares a similar DNA binding domain consisting of three or more zinc fingers (green). At the N-terminus, the SCAN domain (blue) functions as an interaction domain.
The zinc finger domain
The zinc finger domain in the ZSCAN transcription factor contains a C2H2 motif. The C-terminus of the ZSCAN transcription factor contains three or more fingers consisting of two conserved cysteine residues and two histidine residues coordinated with a zinc ion [10]. The C2H2 ZNF transcription factors are the main type of ZNF transcription factors. According to the Inter Pro database (updated on February 2018), there are approximately 2,443 genes in the human genome encoding proteins with a C2H2 motif (http://www.ebi.ac.uk/interpro/entry/IPR036236/proteins-matched?species =9606). This zinc finger domain functions as a DNA binding domain and helps the ZSCAN transcription factor target specific cis-acting elements.
The SCAN domain
Based on the four members initially identified (SRE-ZBP, CT n-51, AW-1, and Number 18 cDNA), another domain was named the SCAN box [9]. It was described more than 20 years ago. In the human genome, approximately 244 protein products containing the SCAN domain have been identified, among which approximately 50 ZSCAN transcription factors were described (http://www.ebi.ac.uk/interpro/entry/IPR003309/proteins-matched?species=9606). The highly conserved SCAN domain consists of 84 amino acids rich in leucine residues; therefore, this domain is also known as the leucine rich region (LeR) [11]. The SCAN domain has an amphipathic secondary structure and is involved in protein-protein interactions, in particular for self-association and to mediate oligomerization [12-14]. Increasing research on this domain revealed that it can interact with an isolated SCAN domain, such as SCAN domain protein 1 (SDP1), or with other family members with the SCAN domain [15]. Furthermore, this interaction is selective, indicating that not all the family members can form oligomers. These varied interactions between transcription factors lead to diverse transcriptional activities.
The human ZSCAN transcription factor family
There are 54 identified members of the human ZSCAN transcription factor family, and because they are all zinc finger proteins, they are referred to as ZNF proteins. In addition, specific proteins may have several special names according to their domains. Examination of the NCBI gene database revealed that family members have unified names ranging from ZSCAN1 to ZSCAN54 (Table 1). In this family, 24 members containing both the KRAB domain and the SCAN domain are named ZKSCAN proteins. In addition, certain proteins have unique names such as myeloid zinc finger 1 (MZF1) and paternally expressed 3 (PEG3).
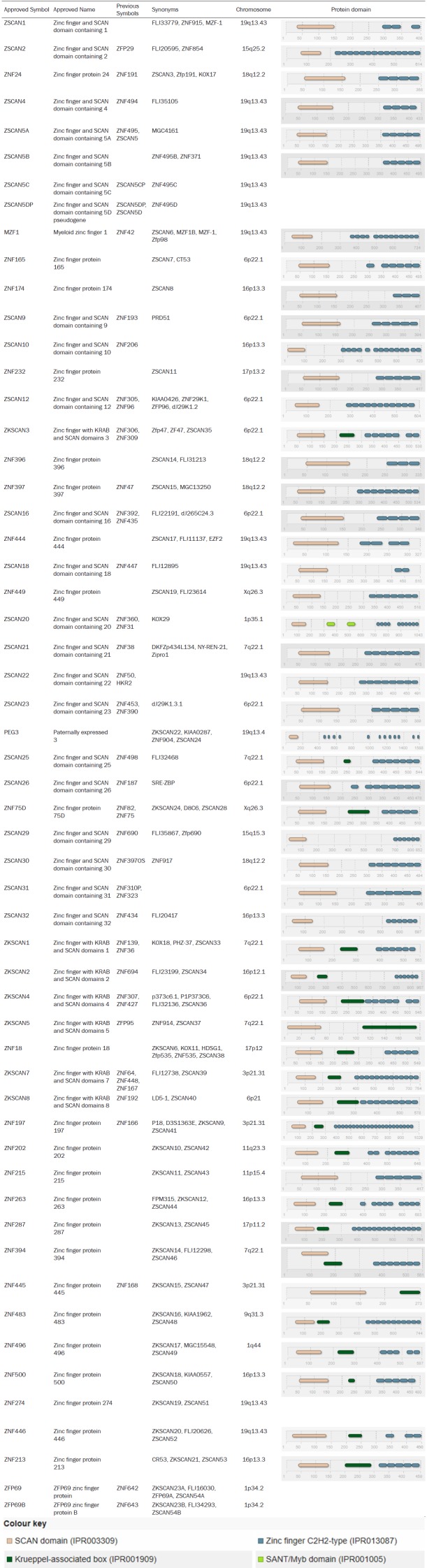
Table 1.
ZSCAN family members and protein structures
Involvement of ZSCAN transcription factors in cancer
Although these family members share a similar structure (Figure 1), ZSCAN transcription factors show great diversity of biological functions, especially in cancer progression. Recently, an increasing number of studies detected aberrant expression of ZSCAN transcription factors in different kinds of cancer. Notably, in different cancer types or even in the same cancer type, ZSCAN transcription factors can lead to opposite outcomes. Next, we discuss the well-researched members and their impacts on cancer. To summarize, ZSCAN transcription factors may take part in angiogenesis, cell apoptosis, cell differentiation, cell migration and invasion, cell proliferation, stem cell properties, and chemotherapy sensitivity (Figure 2). We list all the members and their functions in Table 2 (sorted by biological process).
Figure 2.
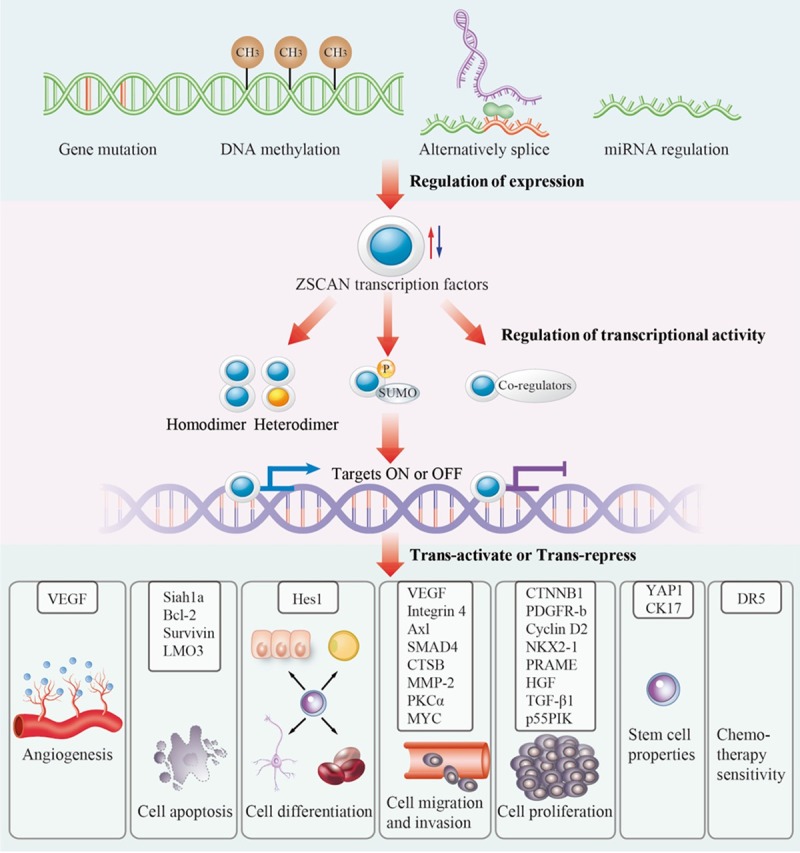
Roles of ZSCAN transcription factors in cancer and the potential mechanism. Regulation of the expression of ZSCAN transcription factors: gene mutation, DNA methylation, alternative splicing, or microRNA regulation may up- or down-regulate the expression of ZSCAN transcription factors. Regulation of transcriptional activity: according to the context of the environment, ZSCAN transcription factors function as transcriptional activators or repressors: i) they may interaction through the SCAN domain to form a homodimer or heterodimer; ii) post-translational modifications, such as phosphorylation and SUMOylation, may affect transcription, resulting in different outcomes; and iii) they may recruit co-regulators including SETDB1, NSD1, H2A.Z, and HDAC1 to form transcriptional complexes. Transactivation or transrepression of a wide variety of downstream genes: by targeting different genes, ZSCAN transcription factors are involved in i) angiogenesis; ii) cell apoptosis; iii) cell differentiation; iv) cell migration and invasion; v) cell proliferation; vi) stem cell properties; and vii) chemotherapy sensitivity.
Table 2.
ZSCAN family target genes and biological progress
| Member | Target | Cancer type | Biological process | Tumor suppresser or promoter | Reference |
|---|---|---|---|---|---|
| ZNF24 | VEGF | Breast cancer | Angiogenesis | Inhibitor | [61,62] |
| ZKSCAN3 | VEGF | Colorectal cancer | Angiogenesis | Promoter | [53] |
| PEG3 | Siah1a | Fibroblast cell | Cell apoptosis | Inhibitor | [48] |
| ZKSCAN1 | Bcl-2 and survivin | Gastric cancer | Cell apoptosis | Promoter | [67,69] |
| MZF1 | LMO3 | Glioma | Cell apoptosis | Promoter | [19] |
| ZNF206 | Neuroblastoma | Cell differentiation | Promoter | [63] | |
| ZKSCAN4 | Hes1 | Basal cell carcinoma | Cell differentiation | Promoter | [64] |
| ZNF24 | VEGF | Gastric cancer | Cell migration and invasion | Inhibitor | [44] |
| ZKSCAN3 | Colorectal cancer | Cell migration and invasion | Promoter | [73] | |
| ZKSCAN3 | Integrin 4 | Colorectal cancer | Cell migration and invasion | Promoter | [74] |
| ZKSCAN3 | Prostate cancer | Cell migration and invasion | Promoter | [76] | |
| MZF1 | Axl | Cervical cancer | Cell migration and invasion | Promoter | [21] |
| MZF1 | Axl | Colorectal cancer | Cell migration and invasion | Promoter | [22] |
| MZF1 | SMAD4 | Gastric cancer | Cell migration and invasion | Inhibitor | [41] |
| MZF1 | CTSB | Breast cancer | Cell migration and invasion | Promoter | [27-29] |
| MZF1 | N-cadherin | Esophageal cancer | Cell migration and invasion | Promoter | [33] |
| MZF1 | MMP-2 | Cervical cancer | Cell migration and invasion | Inhibitor | [40] |
| ZNF24 | PDGFR-b | Breast cancer | Cell proliferation | Inhibitor | [63] |
| ZNF24 | CTNNB1 (β-catenin) | Hepatocellular carcinoma | Cell proliferation | Promoter | [65] |
| ZKSCAN3 | Colorectal cancer | Cell proliferation | Promoter | [74] | |
| ZKSCAN3 | Cyclin D2 | Multiple myeloma | Cell proliferation | Promoter | [75] |
| ZKSCAN3 | Breast cancer | Cell proliferation cell migration and invasion | Promoter | [79] | |
| ZKSCAN1 | Hepatocellular carcinoma | Cell proliferation cell migration and invasion | Inhibitor | [71] | |
| PEG3 | β-catenin | Glioma | Cell proliferation | Inhibitor | [53] |
| PEG3 | Ovarian cancer | Cell proliferation | Inhibitor | [56] | |
| ZNF496 | Breast cancer | Cell proliferation | Inhibitor | [102] | |
| MZF1 | NKX2-1 | Cervical cancer | Cell proliferation | Promoter | [20] |
| MZF1 | PRAME | Melanoma | Cell proliferation | Promoter | [34] |
| MZF1 | HGF | Multiple myeloma | Cell proliferation | Promoter | [35] |
| MZF1 | TGF-β1 | Breast cancer | Cell proliferation | Promoter | [26] |
| MZF1 | p55PIK | Colorectal cancer | Cell proliferation | Promoter | [36] |
| MZF1 | PKCα | Hepatocellular carcinoma | Cell proliferation cell migration and invasion | Promoter | [23] |
| MZF1 | PKCα | Bladder transitional cell carcinoma | Cell proliferation cell migration and invasion | Promoter | [24] |
| MZF1 | PKCα | Breast cancer | Cell proliferation cell migration and invasion | Promoter | [25] |
| MZF1 | MYC | Lung adenocarcinomas | Cell proliferation cell migration and invasion | Promoter | [31,32] |
| MZF1 | Lung squamous cell carcinoma | Cell proliferation cell migration and invasion | Promoter | [37] | |
| MZF1 | NFKB | Gastric cancer | Cell proliferation cell migration and invasion | Inhibitor | [42] |
| MZF1 | DR5 | Colorectal cancer | Chemotherapy sensitivity | Inhibitor | [44] |
| MZF1 | YAP1 | Osteosarcomas | Stem cell properties | Promoter | [38] |
| MZF1 | CK17 | Cervical cancer | Stem cell properties | Promoter | [39] |
MZF1
MZF1, also known as ZNF42, ZFP98, or ZSCAN6, was first reported in myeloid differentiation and leukemia [16]. Since then, it has attracted increasing attention for its involvement in several mammalian cancers [15,17,18]. Data from the TCGA database show significant alterations of MZF1 gene copy numbers in several tumors such as breast, lung, bladder, glioma, and colon cancer. However, whether MZF1 acts as a tumor promoter or suppressor remains unclear because of its specific expression in different tissues and the complexity of its function. We will then discuss the roles of MZF1 in different biological processes during tumorigenesis from two aspects: the promoters or the inhibitors of tumors.
Tumor promoter
Cell apoptosis
MZF1 is associated with poor prognosis in gliomas, where MZF1 directly binds to the promoter of LIM domain only protein 3 (LMO3), which induces apoptosis by upregulating the expression of the caspase signal pathway [19].
Cell migration and invasion
In cervical cancer, the induction of forkhead box M1 (FOXM1) by the MZF1/NK2 homeobox 1 (NKX2-1) axis is involved in cell invasion [20]. It is consistent with another aspect of cervical cancer, in which MZF1 binds directly to the AXL receptor tyrosine kinase (AXL) promoter, which promotes cervical cell invasion, and the expression of MZF1 is closely correlated with the clinical stage in cervical cancer [21]. In colon cancer, MZF1 regulates the expression of AXL, leading to enhanced migratory, invasive, and metastatic potentials, as demonstrated in vivo and in vitro [22]. A research team from Taiwan suggested that MZF1 cooperates with Elk-1 and significantly upregulates protein kinase C alpha (PKCα), promoting cell migration and invasion in hepatocellular, breast, and bladder transitional cell carcinoma [23-25]. In breast cancer, MZF1 is a key transcription factor mediating the expression of transforming growth factor beta 1 (TGF-β) and it is involved in the transformation of mesenchymal stem cells to cancer associated fibroblasts [26]. Similarly, another group found that in ErbB2-driven invasion of breast cancer, MZF1 plays a decisive role by activating the downstream gene cathepsin B (CTSB) [27-29]. In non-small cell lung cancer, MZF1 is an important prognostic marker in early stage patients [30]. Microarray analysis of the c-Myc regulatory gene network in lung adenocarcinoma identified MZF1 as a putative indirect target of c-Myc action, and its involvement in the Myc tumorigenic phenotype was demonstrated [31]. The involvement of the MZF1/c-Myc axis in migration, invasion, and soft agar growth of lung adenocarcinoma cells was also confirmed in another study [32]. Research in esophageal cancer cells found that MZF1 may upregulates N-cadherin during epithelial-mesenchymal transition [33].
Cell proliferation
The cooperation of MZF1 and Elk-1 may also promote cell proliferation in hepatocellular, breast, and bladder transitional cell carcinomas [24-26]. In melanoma cells, MZF1 binds to the hypomethylated promoter of the tumor antigen preferentially expressed antigen of melanoma (PRAME) and transactivates the expression of PRAME, increasing cell proliferation [34]. Hepatocyte growth factors (HGFs), whose expressions are elevated in multiple myeloma cells, are potential targets of MZF1. Mechanistic studies have shown that DNA mutations in the promoter alleles of HGF provide new functional binding sites for MZF1 [35]. Another group found that MZF1 transcriptionally activates p55PIK and increases the proliferation of colorectal cancer cells [36]. In lung squamous cell carcinoma, MZF1 acts as an oncogene to promote cell proliferation and invasion [37].
Stem cell properties
MZF1 is amplified in osteosarcoma and may promote stem cell properties by inducing yes associated protein 1 (YAP1) expression, whereas silencing of MZF1 promotes cell differentiation [38]. In cervical cancer, MZF1 is a key transcriptional activator of CK17, which promotes the stemness of cervical cancer cells [39]. In vivo and in vitro experiments demonstrated that the MZF1/NKX2-1/FOXM1 axis mentioned above is also responsible for cell stemness in human papilloma virus 16/18 (HPV16/18)-infected cervical cancer [20].
Tumor suppressor
Although its oncogenic function has been well-studied, many studies describe an opposite role for MZF1 in cancer. In human cervical cancer, MZF1 may suppress matrix metalloproteinase-2 (MMP2) and repress cell migration and invasion [40]. Recently, a study in gastric cancer cells showed that overexpression of MZF1 upregulates the tumor suppressor SMAD family member 4 (SMAD4), which directly suppresses Wnt/β-catenin signaling and inhibits cell migration [41]. This finding indicates that MZF1 can act as a negative regulator of tumorigenesis. Another study of gastric cancer found that MZF1 suppressed cell proliferation and migration through targeting NFKB associated with MT2A [42]. Immunohistochemical analysis of 274 patients with oral squamous cell carcinoma using tissue microarrays concluded that low expression of MZF1 is associated with advanced clinical stage and lower overall survival rates [43]. Different roles of MZF1 were even found in the same cancer. Although MZF1 acts as an oncogene in colon cancer, upregulation of MZF1 by sulindac sulfide induces the expression of the receptor for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), sensitizing cancer cells to TRAIL-based therapy [44].
PEG3
PEG3 is an imprinted gene with tumor suppressor activity, and it interacts with many well-known proteins. Recently, a transcriptional amplification system aimed to improve PEG3 was developed to target Androgen Receptor+ and Androgen Receptor-prostate cancer cells [45]. In a study of cervical cancer, 213 women from Tanzania with cervical intraepithelial neoplasia I/II/III or invasive cervical cancer (ICC) were analyzed to identify DNA methylation markers associated with cancer persistence or progression. The study showed that a 5% increase in PEG3 DNA methylation was associated with a 1.6-fold increase of ICC risk [46]. Downregulation of PEG3 is found in glioma cell lines and glioma tissues, and its expression correlates with glioma subtype or grade [47]. PEG3 can also affect other cancers with the following biological behaviors.
Cell apoptosis
PEG3 is a potential regulator of p53/c-Myc mediated apoptosis through cooperating with Siah1 to regulate Tax translocation in fibroblast cell lines [48]. Other studies found that PEG3 is upregulated in response to DNA damage in a p53-dependent manner, leading to decreased neuronal survival [49].
Angiogenesis
PEG3 is also involved in endothelial cell autophagy, which is related to angiogenesis and tumorigenesis, by regulating transcription factor EB (TFEB) induction and nuclear translocation [50]. Another study investigating its role in endothelial cell autophagy showed that PEG3 increases Beclin 1 promoter activity and expression, which concurrently inhibits endothelial cell migration and invasion [51].
Cell proliferation
PEG3 regulates the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signal transduction pathway related with cell growth by interacting with tumor necrosis factor receptor-associated factor 2 (TRAF2) [52]. Because of its high expression in glioma [47,53], it may lead to increased Wnt/β-catenin signaling and promote proliferation [53]. In vitro experiments showed that transfection of PEG3 inhibited colony forming ability in glioma cell lines [54]. In addition, low expression of PEG3 was reported in ovarian and endometrial cancers by global gene expression analysis [55]. Pyrosequencing demonstrated that hypermethylation of the CpG island may account for its silence, and re-expression of PEG3 inhibited the growth and proliferation of ovarian cancer cells [56]. A different study confirmed that PEG3 functions as a tumor suppressor in ovarian cancer because of hypermethylation of the PEG3-CpG island [57].
Stem cell properties
A recent study showed that PEG3 is related to mesoangioblast stem cell differentiation and migratory potential by acting as a transcription factor repressing target genes [58].
ZNF protein 24 (ZNF24)
ZNF24, also known as ZNF191 or KOX17, is located on chromosome 18q12.1, which is frequently deleted in colorectal cancer [59,60]. Microarray analysis after overexpression or knockdown of ZNF24 showed that ZNF24 may modulate numerous genes associated with tumor growth, including inhibitory and positive effects [59].
Tumor suppressor
The expression of vascular endothelial growth factor (VEGF), a stimulator of angiogenesis, is inversely correlated with that of ZNF191 in human breast and colon carcinoma cells [61]. This indicates that ZNF24 may act as a negative regulator in cancer by inhibiting angiogenesis. In vitro, electrophoretic mobility shift assays and chromatin immunoprecipitation assays showed that ZNF24 binds to the VEGF promoter to repress its transcription. In vivo, a transgenic zebrafish model confirmed the inhibitory effect of ZNF24 on tumor angiogenesis [62]. Another transcriptional target of ZNF24 is the receptor protein tyrosine kinase platelet-derived growth factor receptor-b (PDGFR-b), which affects cellular proliferation, migration, and survival [63]. In gastric cancer, ZNF24 acts as a potential suppressor in the microRNA-940 mediated epithelial mesenchymal transition process [64].
Tumor promoter
In hepatocellular carcinoma, ZNF24 binds directly to the promoter of β-catenin, activating its transcription and promoting cell proliferation [65]. Similarly, a positive role of ZNF24 in the regulation of proliferation and angiogenesis was reported in human primary microvascular endothelial cells [60]. Investigation of the underlying mechanism showed that silencing of ZNF24 upregulates cell cycle inhibitors such as cyclin dependent kinase inhibitor 3 and downregulates cell cycle activators such as cyclin D2. Furthermore, knockdown of ZNF24 decreased the expression of migration promoters such as vascular endothelial growth factor receptor 2 (VEGFR2) and MMP2 [60].
ZKSCAN domain 1 (ZKSCAN1)
ZKSCAN1, also known as ZNF139, is closely associated with gastric cancer. A group from China reported that ZNF139 is overexpressed in gastric cancer tissues and cells, especially in metastatic tissues. Immunohistochemical analysis of 108 gastric cancer tissue samples and paired adjacent tissues showed that ZNF139 was negatively related to prognosis [66]. This finding revealed its role as an independent prognostic factor in gastric cancer [66]. In vitro and in vivo experiments indicated that ZNF139 may promote cell viability and proliferation by modulating the apoptosis pathway [67]. This result was confirmed by another group [68]. Furthermore, small interfering RNA mediated knockdown of ZNF139 expression effectively reduced gastric cancer cell invasion and migration ability [69]. Regarding its role in chemotherapy resistance, this group showed that the expression of ZNF139 was positively correlated with resistance to therapy [66]. In melanoma, a new fusion protein, ZKSCAN1-MET, was predicted through DNA sequence and confirmed by RT-PCR. This fusion transcript includes a scan domain at the N-terminal of the kinase domain of mesenchymal epithelial transition (MET), which is integral for MET to become constitutively active. It was identified that ZKSCAN1-MET may active the mitogen-activated protein kinase (MAPK)/PI3K signaling pathway and may play important roles in tumor progression [70]. However, its inhibition effect on cell growth, migration and invasion is demonstrated in hepatocellular carcinoma [71].
ZKSCAN domain 3 (ZKSCAN3)
ZKSCAN3, a novel “driver” of cancer progression, is located on chromosome 6p22.1, which is amplified in colorectal cancer. Its overexpression in tumor tissues compared with adjacent nonmalignant mucosa was confirmed by qPCR and immunohistochemistry [72]. A recent study demonstrated a direct relationship between ZKSCAN3 and carcinoembryonic antigen, which plays an active role in the development of colorectal cancer liver metastasis. The results indicated that ZKSCAN3 may facilitate hepatic metastasis [73]. Unbiased screening identified many candidate downstream genes of ZKSCAN3, including several genes favoring tumor progression such as integrin β4, which plays an important role in breast and colorectal cancer tumorigenicity by activating phosphatidic 3-kinase [74]. Datasets from Oncomine and the multiple myeloma genomics portal (MMGP) reveal that the mRNA level of ZKSCAN3 is increased in primary patient samples. It may bind directly to the promoter of cyclin D2, inducing its expression and promoting myeloma proliferation [75]. ZKSCAN3 is also highly expressed in prostate cancer, especially in metastatic prostate cancer. Exogenous expression of ZKSCAN3 promotes prostate cancer cell migration and tumor progression [76]. ZKSCAN3 may also act as a transcriptional repressor of autophagy in bladder cancer cells. It transcriptionally modulates the expression of more than 60 genes involved in various steps of autophagy and lysosome biogenesis [77]. In a research of uterine cervical cancer, clinicopathological analysis and immunohistochemistry shows that ZKSCAN3 is strongly overexpressed in cancer samples [78]. What’s more, ZKSCAN3 also promotes cell proliferation, migration and invasion in breast cancer [79].
Other family members
In addition to these well-reported ZSCAN transcription factors, other family members involved in tumor progression have been identified.
ZSCAN4, an important gene expressed in embryo development [80], regulates genome maintenance and telomere extension in embryonic stem cells [81]. The interaction between ZSCAN4 and Rap1 or TRF1, telomere-associated proteins, is also confirmed in cancer cells [82,83]. In a recent report about resistance to anticancer treatments, ZSCAN4 was found to take part in telomeric DNA damage repair [84].
ZSCAN10, also known as ZNF206, functions as a differentiation repressor in mouse embryonic stem cells [85], and it is one of the important factors associated with human neuroblastoma genesis [86].
In basal cell carcinoma, ZNF396 prevents tumor cell differentiation and promotes cell proliferation [87].
Low expression caused by promoter DNA methylation of ZSCAN18 is found in most gastrointestinal cancers, such as colorectal, gastric, and pancreatic cancer [88]. This finding suggests that ZSCAN18 serves as a biomarker in gastrointestinal cancers.
A study on cervical cancer that analyzed microarray gene expression profiles from patients treated with radiotherapy and chemoradiotherapy showed that ZNF449 is involved in the regulation of differentially expressed genes in response to radiotherapy and chemoradiotherapy [89].
ZKSCAN4 is a transcription factor that was identified approximately 10 years ago. ZKSCAN4 inhibits the transcriptional activities of p53 and p21 [90], and it was shown to interact with the glucocorticoid receptor and play a role in cell proliferation [91].
ZSCAN7, also known as ZNF165, is highly expressed in several tumors, such as hepatocellular carcinoma (HCC), gastric cancer, colon cancer, non-small cell lung carcinoma, and urinary bladder cancer [92-94]. In soft tissue myoepithelial carcinoma, a novel fusion gene EWSR1-ZNF444 was identified and shown to contribute to tumorigenesis [95].
Genome-wide methylation analysis shows that ZSCAN12 is hypermethylated in bladder cancer, especially in high-grade disease. This conclusion is validated in other data sets from TCGA [96].
An expression profile signature analysis of oral squamous cell carcinoma (OSCC) using data derived from two different sources shows that ZSCAN16 is differentially expressed between node-positive OSCCs with and without extra-capsular spread [97].
A single-nucleotide polymorphism (SNP) analysis of Thyrotropin (TSH)-secreting pituitary adenomas detects a novel DNA candidate driver mutation of ZSCAN23 [98].
To identify novel mutations in endometrial cancer patients, whole-exome sequencing is performed. The results show ZSCAN29 may act as a potential passenger gene [99].
A genome-wide DNA methylation study in lung cancer suggests ZSCAN31 as a novel gene target [100].
Weighted gene co-expression network analysis shows ZNF215 can be used as a biomarker for diagnosis and prognosis of the basal like breast cancer [101].
ZNF496 is found to be an interactor of ERα in breast cancer cells, this interaction selectively represses target genes transcription and so that inhibits cell proliferation [102].
Potential mechanisms underlying the dual function of ZSCAN transcription factors
General patterns of transcriptional regulation by transcription factors
Transcription factors are critical determinants of gene expression. Their transcriptional activities are multivariate, and many factors such as nuclear-plasma transposition, post-translational modifications, and interaction with other cofactors can change transcriptional output. Furthermore, several transcription factors may bind to the same or independent cis element, including promoter, intron, or enhancer, and work together to regulate transcription.
Diverse roles of ZSCAN transcription factors in transcriptional regulation
ZSCAN transcription factors have different functions and may even elicit opposing effects on cancer progression. The differential transcriptional regulation of various genes may account for the controversial function of these transcription factors. A wide variety of downstream genes can be regulated by ZSCAN transcription factors, and their transcriptional function may be context-dependent. Binding of ZSCAN transcription factors to specific promoter sequences is mediated by their zinc finger motif. By binding to different target sequences, these transcription factors may favor distinct cofactors, thereby activating or repressing transcription in different cell types [103,104]. The oncogene of gastric cancer, ZNF139, may increase the expression of a large set of genes related to cell migration and invasion, such as MMP-2, MMP-9, and intercellular adhesion molecule 1 (ICAM-1) [68]. The suppressive role of ZNF307 was confirmed by its effect on p53 expression [90].
Many family members function as both transcriptional activator and repressor, such as MZF1 and ZKSCAN3 [5,105]. ZKSCAN3 transcriptionally activates integrin β4 and VEGF during colorectal tumorigenesis [74]. It can also transcriptionally repress Map1lC3b and Wipi2, which are involved in autophagy and lysosome biogenesis [77]. Four and a half Lim domain protein-3 (FHL3) is a co-repressor of MZF1 and participates in the regulation of genes related to cancer development [106], whereas in osteosarcoma, MZF1 binds to the YAP1 enhancer and upregulates its expression [38].
Elucidating the potential mechanisms underlying the different effects of these transcription factors on transcriptional regulation is important. Many studies show that protein interactions and post-translational modifications play important roles in the modulation of the transcriptional activity.
Potential mechanisms of transcriptional regulation by ZSCAN transcription factors
Interaction through the SCAN domain: homodimer or heterodimer
Several studies indicated that the SCAN domain acts as a protein interaction domain to form homodimers. A yeast two-hybrid assay showed that ZNF174 forms homodimers via the SCAN domain 13. This self-oligomerization can promote DNA binding affinity and specificity, because PEG3 binds to certain target genes as a homodimer [107].
Many family members of the ZSCAN transcription factors have different protein isoforms, among which longer ones have the SCAN domain and DNA-recognition element zinc finger domain, whereas the smaller ones have only the SCAN domain, such as ZNF202 [108] and MZF1. Interestingly, the smaller isoforms interact with the intact form and change its transcriptional capacity [5,109].
In addition to these interactions, the SCAN domain can also regulate heterodimer formation between different family members, and this interaction affects the activation of transcription factors. For example, co-localization of MZF1 and ZNF24 was found in the nucleus by immunofluorescence, and GST pull-down assays confirmed this interaction. However, the mutant MZF1 without the SCAN domain shows disrupted co-localization with ZNF24 in the nucleus [110]. Moreover, SDP1, which encodes an isolated SCAN domain, abolishes the expressional capacity of ZNF202 by preventing binding of its co-repressor [108]. SDP1 was also reported to interact with a co-activator of peroxisome proliferator activated receptor gamma 2 (PPARg2) [111]. These SCAN-SCAN associations are selective, and not all the family members can interact with each other [5].
Although the interaction is important for transcriptional activity, the mechanisms have not been totally explored.
Interaction with epigenetic chromatin modification enzymes
During the regulation of transcription, transcription factors may recruit many co-regulators to form transcriptional complexes, including histone-modifying enzymes (histone acetyltransferases and histone methyltransferases) and chromatin remodeling factors, among others. ZNF274 recruits the histone methyltransferase SET domain bifurcated 1 (SETDB1) to H3K9me3 sites and represses transcription [112].
The recruitment of nuclear receptor binding SET domain protein 1 (NSD1), a SET-domain histone lysine methyltransferase, may be partly associated with the ZNF496 repressor properties [113].
H2A.Z, which is involved in chromosome segregation, centromeric function, and transcriptional regulation, was identified as a novel interacting protein of ZNF24 by yeast two-hybrid, GST pull-down, co-immunoprecipitation, and co-localization assays [114].
Post-translational modifications
The post-translational modifications of transcription factors, especially phosphorylation, may also affect the transcription into different outcomes. The SCAN domain is defined as a leucine rich region, and as a result, phosphorylation mediates transcriptional activation. For example, TGFβ-receptor II-associated ErbB2 may activate downstream MZF1 in breast cancer [27]. ErbB2 activation results in phosphorylation of MZF1-S27, which so that results in increased transcriptional activity of MZF1.
SUMOylation is another kind of post-translational modification [115], and it is considered as an inhibitor of gene transcription [116,117]. The potential mechanism may be that the SUMO interacting motifs (SIM) occur in the co-repressors, such as the HDAC1 complex [117]. Recruitment of these co-repressors to the SUMOylated transcription factors leads to gene repression [106]. SUMOylation also modulates ZSCAN transcription factors. For example, SUMOylated MZF1 is accumulated in promyelocytic leukemia nuclear bodies through this SUMO/SIM interaction, resulting in gene repression [110,115,118].
Regulation of the expression of ZSCAN transcription factors
In view of their essential roles in cancer, many studies focused on the regulation of the expression of ZSCAN transcription factors. Studies showed that gene mutation or alterations of gene copy number lead to different expressional levels of the ZSCAN transcription factors. This was reported for EWSR1-ZNF444 in soft tissue myoepithelial carcinoma [95] and ZKSCAN1-MET in melanoma [70]. Moreover, low expression of several members caused by promoter DNA methylation was also reported. For example, hypermethylation of the PEG3-CpG island was detected in many types of cancer [46]. Similarly, ZSCAN18 was found to be frequently methylated in gastrointestinal cancers [88]. Cancer related miRNA regulation or transcriptional regulation also accounts for expression differences. For instance, miRNA-940 may downregulate ZNF24 in gastric cancer [64]. Alternative promoters of PEG3 may change the potential cis-regulatory motifs involved in regulating the expression patterns of PEG3 [119]. Sequence analysis identified alternatively spliced transcripts of ZNF215 involved in congenital growth alterations because its five alternatively spliced transcripts were imprinted in a tissue-specific manner [120]. In summary, gene mutation, DNA methylation, miRNA regulation, and alternative splicing account for various expression levels of the ZSCAN transcription factors.
Discussion
Transcription factors dominate specific gene expression dynamically by recognizing and binding to short DNA sequence motifs. Recently, an increasing number of studies have shown that the ZSCAN transcription factors play important roles in cancer progression. Gene mutation, DNA methylation, alternative splicing, and miRNA regulation result in various expression levels of the ZSCAN transcription factors. ZSCAN transcription factors differentially regulate downstream genes, and therefore have different functions in cancer progression. These target genes may be involved in angiogenesis, cell apoptosis, cell differentiation, cell migration and invasion, cell proliferation, chemotherapy sensitivity, and stem cell properties. And their efforts involve different kinds of cancer, such as breast cancer, colorectal cancer, gastric cancer, glioma, and prostate cancer. In this article, we summarized 54 expressed members of the human ZSCAN family and their effects on cancer, among them several members such as MZF1, PEG3, ZNF24, ZKSCAN1, and ZKSCAN3 are well-reported. In addition to these known effects, the roles of many family members remain to be explored.
Regarding the potential underlying mechanisms, regulation of transcriptional activity seems to be quite important for the ZSCAN transcription factors. Protein-protein interactions are important for transcriptional regulation. The SCAN box, which mediates dimerization, is the key domain and contributes to the mystery surrounding the ZSCAN transcription factors. It can form a homodimer with the transcription factor itself or a heterodimer with other proteins with SCAN box. Moreover, the recruitment of co-regulating molecules and post-translational modifications including phosphorylation and SUMOylation, may determine the different effects of these factors on transcription. A better exploration of the structure and the potential mechanisms underlying their selection of interacting partners is necessary. Moreover, elucidating the widespread regulatory mode of the ZSCAN transcription factors is essential.
In summary, a better understanding of the ZSCAN transcription factors will provide further insight into the mechanism of transcriptional regulation and suggest novel therapeutic strategies against tumor progression.
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript. This work was supported by the grants from the National Natural Science Foundation of China (NSFC nos. 81572457 and 81702442) and the Natural Science Foundation of Jiangsu province, China (no. BK20170623).
Disclosure of conflict of interest
None.
References
- 1.Bemer M, van Dijk ADJ, Immink RGH, Angenent GC. Cross-family transcription factor interactions: an additional layer of gene regulation. Trends Plant Sci. 2017;22:66–80. doi: 10.1016/j.tplants.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA-dependent formation of transcription factor pairs alter their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- 3.Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 4.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Bellefroid E, Poncelet D, Lecocq P, Revelant O, Martial J. The evolutionarily conserved Krüppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci U S A. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 8.Chung HR, Schafer U, Jackle H, Bohm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in drosophila. EMBO Rep. 2002;3:1158–1162. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams A, Khachigian L, Shows T, Collins T. Isolation and characterization of a novel zinc-finger protein with transcriptional repressor activity. J Biol Chem. 1995;270:22143–22152. doi: 10.1074/jbc.270.38.22143. [DOI] [PubMed] [Google Scholar]
- 10.Lu D, Searles MA, Klug A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. doi: 10.1038/nature02088. [DOI] [PubMed] [Google Scholar]
- 11.Pengue G, Calabrò V, Bartoli P, Pagliuca A, Lania L. Repression of transcriptional activity at a distance by the evolutionarily conserved KRAB domain present in a subfamily of zinc finger proteins. Nucleic Acids Res. 1994;22:2908–2914. doi: 10.1093/nar/22.15.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander T, Haas A, Peterson M, Morris J. Identification of a novel SCAN box-related protein that interacts with MZF1B. J Biol Chem. 2000;275:12857–12867. doi: 10.1074/jbc.275.17.12857. [DOI] [PubMed] [Google Scholar]
- 13.Williams A, Blacklow S, Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol Cell Biol. 1999;19:8526–8535. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis CM, Wang LL, Ballinger D, Bowen BR, Wagner S. The SCAN domain mediates selective oligomerization. J Biol Chem. 2000;275:17173–17179. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- 15.Eguchi T, Prince T, Wegiel B, Calderwood SK. Role and regulation of myeloid zinc finger protein 1 in cancer. J Cell Biochem. 2015;116:2146–2154. doi: 10.1002/jcb.25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hromas R, Collins SJ, Hickstein D, Raskind W, Deaven LL, O’Hara P, Hagen FS, Kaushansky K. A retinoic acid-responsive human zinc finger gene, MZF-1, preferentially expressed in myeloid cells. J Biol Chem. 1991;266:14183–14187. [PubMed] [Google Scholar]
- 17.Peterson MJ, Morris JF. Human myeloid zinc finger gene MZF produces multiple transcripts and encodes a scan box protein. Gene. 2000;254:105–118. doi: 10.1016/s0378-1119(00)00281-x. [DOI] [PubMed] [Google Scholar]
- 18.Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, Broxmeyer HE, Hromas R, Pandolfi PP. Mzf1 controls cell proliferation and tumorigenesis. Genes Dev. 2001;15:1625–1630. doi: 10.1101/gad.902301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, Lei Q, Yu Z, Xu G, Tang H, Wang W, Wang Z, Li G, Wu M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget. 2015;6:7930–7943. doi: 10.18632/oncotarget.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen PM, Cheng YW, Wang YC, Wu TC, Chen CY, Lee H. Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus-associated tumorigenesis. Neoplasia. 2014;16:961–971. doi: 10.1016/j.neo.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee EH, Ji KY, Kim EM, Kim SM, Song HW, Choi HR, Chung BY, Choi HJ, Bai HW, Kang HS. Blockade of Axl signaling ameliorates HPV16E6-mediated tumorigenecity of cervical cancer. Sci Rep. 2017;7:5759. doi: 10.1038/s41598-017-05977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mudduluru G, Vajkoczy P, Allgayer H. Myeloid zinc finger 1 induces migration, invasion, and in vivo metastasis through Axl gene expression in solid cancer. Mol Cancer Res. 2010;8:159–169. doi: 10.1158/1541-7786.MCR-09-0326. [DOI] [PubMed] [Google Scholar]
- 23.Yue CH, Huang CY, Tsai JH, Hsu CW, Hsieh YH, Lin H, Liu JY. MZF-1/Elk-1 complex binds to protein kinase calpha promoter and is involved in hepatocellular carcinoma. PLoS One. 2015;10:e0127420. doi: 10.1371/journal.pone.0127420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jou YC, Chiu YW, Chen YH, Hwang JM, Chao PY, Shiu JJ, Hwang WH, Liu JY, Hsu LS. Expression of protein kinase Calpha and the MZF-1 and elk-1 transcription factors in human bladder transitional cell carcinoma cells. Chin J Physiol. 2012;55:75–81. doi: 10.4077/CJP.2012.AMM121. [DOI] [PubMed] [Google Scholar]
- 25.Yue CH, Chiu YW, Tung JN, Tzang BS, Shiu JJ, Huang WH, Liu JY, Hwang JM. Expression of protein kinase C alpha and the MZF-1 and Elk-1 transcription factors in human breast cancer cells. Chin J Physiol. 2012;55:31–36. doi: 10.4077/CJP.2012.AMM109. [DOI] [PubMed] [Google Scholar]
- 26.Weber CE, Kothari AN, Wai PY, Li NY, Driver J, Zapf MA, Franzen CA, Gupta GN, Osipo C, Zlobin A, Syn WK, Zhang J, Kuo PC, Mi Z. Osteopontin mediates an MZF1-TGF-beta1-dependent transformation of mesenchymal stem cells into cancer-associated fibroblasts in breast cancer. Oncogene. 2015;34:4821–4833. doi: 10.1038/onc.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rafn B, Nielsen CF, Andersen SH, Szyniarowski P, Corcelle-Termeau E, Valo E, Fehrenbacher N, Olsen CJ, Daugaard M, Egebjerg C, Bottzauw T, Kohonen P, Nylandsted J, Hautaniemi S, Moreira J, Jaattela M, Kallunki T. ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol Cell. 2012;45:764–776. doi: 10.1016/j.molcel.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 28.Luan H, Mohapatra B, Bielecki TA, Mushtaq I, Mirza S, Jennings TA, Clubb RJ, An W, Ahmed D, El-Ansari R, Storck MD, Mishra NK, Guda C, Sheinin YM, Meza JL, Raja S, Rakha EA, Band V, Band H. Loss of the nuclear pool of ubiquitin ligase CHIP/STUB1 in breast cancer unleashes the MZF1-cathepsin pro-oncogenic program. Cancer Res. 2018;78:2524–2535. doi: 10.1158/0008-5472.CAN-16-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brix DM, Tvingsholm SA, Hansen MB, Clemmensen KB, Ohman T, Siino V, Lambrughi M, Hansen K, Puustinen P, Gromova I, James P, Papaleo E, Varjosalo M, Moreira J, Jäättelä M, Kallunki T. Release of transcriptional repression via ErbB2-induced, SUMO-directed phosphorylation of myeloid zinc finger-1 serine 27 activates lysosome redistribution and invasion. Oncogene. 2019;38:3170–3184. doi: 10.1038/s41388-018-0653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosell R, Skrzypski M, Jassem E, Taron M, Bartolucci R, Sanchez JJ, Mendez P, Chaib I, Perez-Roca L, Szymanowska A, Rzyman W, Puma F, Kobierska-Gulida G, Farabi R, Jassem J. BRCA1: a novel prognostic factor in resected nonsmall-cell lung cancer. PLoS One. 2007;2:1129. doi: 10.1371/journal.pone.0001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reymann S, Borlak J. Transcription profiling of lung adenocarcinomas of c-myc-transgenic mice: identification of the c-myc regulatory gene network. BMC Syst Biol. 2008;2:46. doi: 10.1186/1752-0509-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai LH, Wu JY, Cheng YW, Chen CY, Sheu GT, Wu TC, Lee H. The MZF1/c-MYC axis mediates lung adenocarcinoma progression caused by wild-type lkb1 loss. Oncogene. 2015;34:1641–1649. doi: 10.1038/onc.2014.118. [DOI] [PubMed] [Google Scholar]
- 33.Ko H, Kim S, Yang K, Kim K. Phosphorylation-dependent stabilization of MZF1 upregulates N-cadherin expression during protein kinase CK2-mediated epithelial-mesenchymal transition. Oncogenesis. 2018;7:27. doi: 10.1038/s41389-018-0035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YK, Park UH, Kim EJ, Hwang JT, Jeong JC, Um SJ. Tumor antigen PRAME is up-regulated by MZF1 in cooperation with DNA hypomethylation in melanoma cells. Cancer Lett. 2017;403:144–151. doi: 10.1016/j.canlet.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Tian E, Borset M, Sawyer JR, Brede G, Vatsveen TK, Hov H, Waage A, Barlogie B, Shaughnessy JD Jr, Epstein J, Sundan A. Allelic mutations in noncoding genomic sequences construct novel transcription factor binding sites that promote gene overexpression. Genes Chromosomes Cancer. 2015;54:692–701. doi: 10.1002/gcc.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng Y, Wang J, Wang G, Jin Y, Luo X, Xia X, Gong J, Hu J. p55PIK transcriptionally activated by MZF1 promotes colorectal cancer cell proliferation. Biomed Res Int. 2013;2013:868131. doi: 10.1155/2013/868131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m 6 A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 38.Verma NK, Gadi A, Maurizi G, Roy UB, Mansukhani A, Basilico C. Myeloid zinc finger 1 and GA binding protein co-operate with Sox2 in regulating the expression of yes-associated protein 1 in cancer cells. Stem Cells. 2017;35:2340–2350. doi: 10.1002/stem.2705. [DOI] [PubMed] [Google Scholar]
- 39.Wu L, Han L, Zhou C, Wei W, Chen X, Yi H, Wu X, Bai X, Guo S, Yu Y, Liang L, Wang W. TGF-beta1-induced CK17 enhances cancer stem cell-like properties rather than EMT in promoting cervical cancer metastasis via the ERK1/2-MZF1 signaling pathway. FEBS J. 2017;284:3000–3017. doi: 10.1111/febs.14162. [DOI] [PubMed] [Google Scholar]
- 40.Tsai SJ, Hwang JM, Hsieh SC, Ying TH, Hsieh YH. Overexpression of myeloid zinc finger 1 suppresses matrix metalloproteinase-2 expression and reduces invasiveness of SiHa human cervical cancer cells. Biochem Biophys Res Commun. 2012;425:462–467. doi: 10.1016/j.bbrc.2012.07.125. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Kim SS, Lee HS, Hong S, Rajasekaran N, Wang LH, Choi JS, Shin YK. Upregulation of SMAD4 by MZF1 inhibits migration of human gastric cancer cells. Int J Oncol. 2017;50:272–282. doi: 10.3892/ijo.2016.3793. [DOI] [PubMed] [Google Scholar]
- 42.Lin S, Wang X, Pan Y, Tian R, Lin B, Jiang G, Chen K, He Y, Zhang L, Zhai W, Jin P, Yang L, Li G, Wu Y, Hu J, Gong W, Chang Z, Sheng J-q, Lu Y, Wang JM, Huang J. Transcription factor myeloid zinc-finger 1 suppresses human gastric carcinogenesis by interacting with metallothionein 2A. Clin Cancer Res. 2019;25:1050–1062. doi: 10.1158/1078-0432.CCR-18-1281. [DOI] [PubMed] [Google Scholar]
- 43.Ko CP, Yang LC, Chen CJ, Yeh KT, Lin SH, Yang SF, Chen MK, Lin CW. Expression of myeloid zinc finger 1 and the correlation to clinical aspects of oral squamous cell carcinoma. Tumour Biol. 2015;36:7099–7105. doi: 10.1007/s13277-015-3419-x. [DOI] [PubMed] [Google Scholar]
- 44.Horinaka M, Yoshida T, Tomosugi M, Yasuda S, Sowa Y, Sakai T. Myeloid zinc finger 1 mediates sulindac sulfide-induced upregulation of death receptor 5 of human colon cancer cells. Sci Rep. 2014;4:6000. doi: 10.1038/srep06000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain P, Clermont PL, Desmeules F, Zoubeidi A, Neveu B, Pouliot F. Development of a transcriptional amplification system based on the PEG3 promoter to target androgen receptor-positive and -negative prostate cancer cells. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nye MD, Hoyo C, Huang Z, Vidal AC, Wang F, Overcash F, Smith JS, Vasquez B, Hernandez B, Swai B, Oneko O, Mlay P, Obure J, Gammon MD, Bartlett JA, Murphy SK. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One. 2013;8:e56325. doi: 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otsuka S, Maegawa S, Takamura A, Kamitani H, Watanabe T, Oshimura M, Nanba E. Aberrant promoter methylation and expression of the imprinted PEG3 gene in glioma. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:157–165. doi: 10.2183/pjab.85.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Relaix F, Wei X, Li W, Pan J, Lin Y, Bowtell DD, Sassoon DA, Wu X. Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc Natl Acad Sci U S A. 2000;97:2105–2110. doi: 10.1073/pnas.040378897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson MD, Wu X, Aithmitti N, Morrison RS. Peg3/Pw1 is a mediator between p53 and Bax in DNA damage-induced neuronal death. J Biol Chem. 2002;277:23000–23007. doi: 10.1074/jbc.M201907200. [DOI] [PubMed] [Google Scholar]
- 50.Neill T, Sharpe C, Owens RT, Iozzo RV. Decorin-evoked paternally expressed gene 3 (PEG3) is an upstream regulator of the transcription factor EB (TFEB) in endothelial cell autophagy. J Biol Chem. 2017;292:16211–16220. doi: 10.1074/jbc.M116.769950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torres A, Gubbiotti MA, Iozzo RV. Decorin-inducible Peg3 evokes beclin 1-mediated autophagy and thrombospondin 1-mediated angiostasis. J Biol Chem. 2017;292:5055–5069. doi: 10.1074/jbc.M116.753632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Relaix F, Wei X, Wu X, Sassoon DA. Peg3/Pw1 is an imprinted gene involved in the TNF-NFkappaB signal transduction pathway. Nat Genet. 1998;18:287–291. doi: 10.1038/ng0398-287. [DOI] [PubMed] [Google Scholar]
- 53.Jiang X, Yu Y, Yang HW, Agar NY, Frado L, Johnson MD. The imprinted gene PEG3 inhibits Wnt signaling and regulates glioma growth. J Biol Chem. 2010;285:8472–8480. doi: 10.1074/jbc.M109.069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohda T, Asai A, Kuroiwa Y, Kobayashi S, Aisaka K, Nagashima G, Yoshida MC, Kondo Y, Kagiyama N, Kirino T, Kaneko-Ishino T, Ishino F. Tumour suppressor activity of human imprinted gene PEG3 in a glioma cell line. Genes Cells. 2001;6:237–247. doi: 10.1046/j.1365-2443.2001.00412.x. [DOI] [PubMed] [Google Scholar]
- 55.Dowdy SC, Gostout BS, Shridhar V, Wu X, Smith DI, Podratz KC, Jiang SW. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol Oncol. 2005;99:126–134. doi: 10.1016/j.ygyno.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 56.Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast RC Jr. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112:1489–1502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 57.Huang JM, Kim J. DNA methylation analysis of the mammalian PEG3 imprinted domain. Gene. 2009;442:18–25. doi: 10.1016/j.gene.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bonfanti C, Rossi G, Tedesco FS, Giannotta M, Benedetti S, Tonlorenzi R, Antonini S, Marazzi G, Dejana E, Sassoon D, Cossu G, Messina G. PW1/Peg3 expression regulates key properties that determine mesoangioblast stem cell competence. Nat Commun. 2015;6:6364. doi: 10.1038/ncomms7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Chen X, Gong X, Liu Y, Feng H, Qiu L, Hu Z, Zhang J. A transcript profiling approach reveals the zinc finger transcription factor ZNF191 is a pleiotropic factor. BMC Genomics. 2009;10:241. doi: 10.1186/1471-2164-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia D, Huang L, Bischoff J, Moses MA. The endogenous zinc finger transcription factor, ZNF24, modulates the angiogenic potential of human microvascular endothelial cells. FASEB J. 2015;29:1371–1382. doi: 10.1096/fj.14-258947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harper J, Yan L, Loureiro RM, Wu I, Fang J, D’Amore PA, Moses MA. Repression of vascular endothelial growth factor expression by the zinc finger transcription factor ZNF24. Cancer Res. 2007;67:8736–8741. doi: 10.1158/0008-5472.CAN-07-1617. [DOI] [PubMed] [Google Scholar]
- 62.Jia D, Hasso SM, Chan J, Filingeri D, D’Amore PA, Rice L, Pampo C, Siemann DW, Zurakowski D, Rodig SJ, Moses MA. Transcriptional repression of VEGF by ZNF24: mechanistic studies and vascular consequences in vivo. Blood. 2013;121:707–715. doi: 10.1182/blood-2012-05-433045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Chen X, Liu Y, Ding L, Qiu L, Hu Z, Zhang J. The transcriptional repression of platelet-derived growth factor receptor-beta by the zinc finger transcription factor ZNF24. Biochem Biophys Res Commun. 2010;397:318–322. doi: 10.1016/j.bbrc.2010.05.110. [DOI] [PubMed] [Google Scholar]
- 64.Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu Z, Tang W, Gan L, Sun M, Li J. MicroRNA-940 promotes tumor cell invasion and metastasis by downregulating ZNF24 in gastric cancer. Oncotarget. 2015;6:25418–25428. doi: 10.18632/oncotarget.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu G, Jiang S, Wang C, Jiang W, Liu Z, Liu C, Saiyin H, Yang X, Shen S, Jiang D, Zhou P, Han D, Hu X, Yi Q, Yu L. Zinc finger transcription factor 191, directly binding to beta-catenin promoter, promotes cell proliferation of hepatocellular carcinoma. Hepatology. 2012;55:1830–1839. doi: 10.1002/hep.25564. [DOI] [PubMed] [Google Scholar]
- 66.Li Y, Zhao Q, Fan LQ, Wang LL, Tan BB, Leng YL, Liu Y, Wang D. Zinc finger protein 139 expression in gastric cancer and its clinical significance. World J Gastroenterol. 2014;20:18346–18353. doi: 10.3748/wjg.v20.i48.18346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan L, Tan B, Li Y, Zhao Q, Liu Y, Wang D, Zhang Z. Silencing of ZNF139-siRNA induces apoptosis in human gastric cancer cell line BGC823. Int J Clin Exp Pathol. 2015;8:12428–12436. [PMC free article] [PubMed] [Google Scholar]
- 68.Nie HF, Li Y, Li ZX, Mu JX, Wang JS. Effects of ZNF139 on gastric cancer cells and mice with gastric tumors. Oncol Lett. 2016;12:2550–2554. doi: 10.3892/ol.2016.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Tan BB, Zhao Q, Fan LQ, Wang D, Liu Y. ZNF139 promotes tumor metastasis by increasing migration and invasion in human gastric cancer cells. Neoplasma. 2014;61:291–298. doi: 10.4149/neo_2014_037. [DOI] [PubMed] [Google Scholar]
- 70.Yeh I, Botton T, Talevich E, Shain AH, Sparatta AJ, de la Fouchardiere A, Mully TW, North JP, Garrido MC, Gagnon A, Vemula SS, McCalmont TH, LeBoit PE, Bastian BC. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun. 2015;6:7174. doi: 10.1038/ncomms8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, Liu B, Yang Y. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang L, Hamilton SR, Sood A, Kuwai T, Ellis L, Sanguino A, Lopez-Berestein G, Boyd DD. The previously undescribed ZKSCAN3 (ZNF306) is a novel “driver” of colorectal cancer progression. Cancer Res. 2008;68:4321–4330. doi: 10.1158/0008-5472.CAN-08-0407. [DOI] [PubMed] [Google Scholar]
- 73.Kim CW, Roh SA, Tak KH, Koh BM, Ha YJ, Cho DH, Kim SY, Kim YS, Kim JC. ZKSCAN3 facilitates liver metastasis of colorectal cancer associated with CEA-expressing tumor. Anticancer Res. 2016;36:2397–2406. [PubMed] [Google Scholar]
- 74.Yang L, Zhang L, Wu Q, Boyd DD. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J Biol Chem. 2008;283:35295–35304. doi: 10.1074/jbc.M806965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang L, Wang H, Kornblau SM, Graber DA, Zhang N, Matthews JA, Wang M, Weber DM, Thomas SK, Shah JJ, Zhang L, Lu G, Zhao M, Muddasani R, Yoo SY, Baggerly KA, Orlowski RZ. Evidence of a role for the novel zinc-finger transcription factor ZKSCAN3 in modulating Cyclin D2 expression in multiple myeloma. Oncogene. 2011;30:1329–1340. doi: 10.1038/onc.2010.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, Jing Y, Qin Y, Hunsucker S, Meng H, Sui J, Jiang Y, Gao L, An G, Yang N, Orlowski RZ, Yang L. The zinc finger transcription factor ZKSCAN3 promotes prostate cancer cell migration. Int J Biochem Cell B. 2012;44:1166–1173. doi: 10.1016/j.biocel.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee S, Cho YE, Kim JY, Park JH. ZKSCAN3 upregulation and its poor clinical outcome in uterine cervical cancer. Int J Mol Sci. 2018;19:2859. doi: 10.3390/ijms19102859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chi Y, Xu H, Wang F, Chen X, Shan Z, Sun Y, Fan Q. ZKSCAN3 promotes breast cancer cell proliferation, migration and invasion. Biochem Biophys Res Commun. 2018;503:2583–2589. doi: 10.1016/j.bbrc.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 80.Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MS. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee K, Gollahon LS. Zscan4 interacts directly with human Rap1 in cancer cells regardless of telomerase status. Cancer Bio Ther. 2014;15:1094–1105. doi: 10.4161/cbt.29220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee K, Gollahon LS. ZSCAN4 and TRF1: a functionally indirect interaction in cancer cells independent of telomerase activity. Biochem Biophys Res Commun. 2015;466:644–649. doi: 10.1016/j.bbrc.2015.09.107. [DOI] [PubMed] [Google Scholar]
- 84.Portney BA, Khatri R, Meltzer WA, Mariano JM, Zalzman M. ZSCAN4 is negatively regulated by the ubiquitin-proteasome system and the E3 ubiquitin ligase RNF20. Biochem Biophys Res Commun. 2018;498:72–78. doi: 10.1016/j.bbrc.2018.02.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang ZX, Kueh JL, Teh CH, Rossbach M, Lim L, Li P, Wong KY, Lufkin T, Robson P, Stanton LW. Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells. 2007;25:2173–2182. doi: 10.1634/stemcells.2007-0085. [DOI] [PubMed] [Google Scholar]
- 86.Kawashima H, Sugito K, Yoshizawa S, Uekusa S, Furuya T, Ikeda T, Koshinaga T, Shinojima Y, Hasegawa R, Mishra R, Igarashi J, Kimura M, Wang X, Fujiwara K, Gosh S, Nagase H. DNA hypomethylation at the ZNF206-exon 5 CpG island associated with neuronal differentiation in mice and development of neuroblastoma in humans. Int J Oncol. 2012;40:31–39. doi: 10.3892/ijo.2011.1234. [DOI] [PubMed] [Google Scholar]
- 87.Bai J, Kito Y, Okubo H, Nagayama T, Takeuchi T. Expression of ZNF396 in basal cell carcinoma. Arch Dermatol Res. 2014;306:399–404. doi: 10.1007/s00403-014-1442-1. [DOI] [PubMed] [Google Scholar]
- 88.Vedeld HM, Andresen K, Eilertsen IA, Nesbakken A, Seruca R, Gladhaug IP, Thiis-Evensen E, Rognum TO, Boberg KM, Lind GE. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136:844–853. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferreira de Freitas R, Harding RJ, Franzoni I, Ravichandran M, Mann MK, Ouyang H, Lautens M, Santhakumar V, Arrowsmith CH, Schapira M. Identification and structure-activity relationship of HDAC6 zinc-finger ubiquitin binding domain inhibitors. Med Chem. 2018;61:4517–4527. doi: 10.1021/acs.jmedchem.8b00258. [DOI] [PubMed] [Google Scholar]
- 90.Li J, Wang Y, Fan X, Mo X, Wang Z, Li Y, Yin Z, Deng Y, Luo N, Zhu C, Liu M, Ma Q, Ocorr K, Yuan W, Wu X. ZNF307, a novel zinc finger gene suppresses p53 and p21 pathway. Biochem Biophys Res Commun. 2007;363:895–900. doi: 10.1016/j.bbrc.2007.08.180. [DOI] [PubMed] [Google Scholar]
- 91.Ecker K, Lorenz A, Wolf F, Ploner C, Bock G, Duncan T, Geley S, Helmberg A. A RAS recruitment screen identifies ZKSCAN4 as a glucocorticoid receptor-interacting protein. J Mol Endocrinol. 2009;42:105–117. doi: 10.1677/JME-08-0087. [DOI] [PubMed] [Google Scholar]
- 92.Singh PK, Srivastava AK, Dalela D, Rath SK, Goel MM, Bhatt ML. Frequent expression of zinc-finger protein ZNF165 in human urinary bladder transitional cell carcinoma. Immunobiology. 2015;220:68–73. doi: 10.1016/j.imbio.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 93.Dong XY, Yang XA, Wang YD, Chen WF. Zinc-finger protein ZNF165 is a novel cancer-testis antigen capable of eliciting antibody response in hepatocellular carcinoma patients. Br J Cancer. 2004;91:1566–1570. doi: 10.1038/sj.bjc.6602138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Atanackovic D, Blum I, Cao Y, Wenzel S, Bartels K, Faltz C, Hossfeld D, Hegewisch-Becker S, Bokemeyer C, Leuwer R. Expression of cancer-testis antigens as possible targets for antigen-specific immunotherapy in head and neck squamous cell carcinoma. Cancer Biol Ther. 2006;5:1218–1225. doi: 10.4161/cbt.5.9.3174. [DOI] [PubMed] [Google Scholar]
- 95.Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009;48:1051–1056. doi: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- 96.Olkhov-Mitsel E, Savio AJ, Kron KJ, Pethe VV, Hermanns T, Fleshner NE, van Rhijn BW, van der Kwast TH, Zlotta AR, Bapat B. Epigenome-wide DNA methylation profiling identifies differential methylation biomarkers in high-grade bladder cancer. Translational Oncology. 2017;10:168–177. doi: 10.1016/j.tranon.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang W, Lim WK, Leong HS, Chong FT, Lim TK, Tan DSW, Teh BT, Iyer NG. An eleven gene molecular signature for extra-capsular spread in oral squamous cell carcinoma serves as a prognosticator of outcome in patients without nodal metastases. Oral Oncology. 2015;51:355–362. doi: 10.1016/j.oraloncology.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 98.Sapkota S, Horiguchi K, Tosaka M, Yamada S, Yamada M. Whole-exome sequencing study of thyrotropin-secreting pituitary adenomas. J Clin Endocrinol Metab. 2017;102:566–575. doi: 10.1210/jc.2016-2261. [DOI] [PubMed] [Google Scholar]
- 99.Chang YS, Huang HD, Yeh KT, Chang JG. Identification of novel mutations in endometrial cancer patients by whole-exome sequencing. Int J Oncol. 2017;50:1778–1784. doi: 10.3892/ijo.2017.3919. [DOI] [PubMed] [Google Scholar]
- 100.Kettunen E, Hernandez-Vargas H, Cros MP, Durand G, Le Calvez-Kelm F, Stuopelyte K, Jarmalaite S, Salmenkivi K, Anttila S, Wolff H, Herceg Z, Husgafvel-Pursiainen K. Asbestos-associated genome-wide DNA methylation changes in lung cancer. Int J Cancer. 2017;141:2014–2029. doi: 10.1002/ijc.30897. [DOI] [PubMed] [Google Scholar]
- 101.Maind A, Raut S. Identifying condition specific key genes from basal-like breast cancer gene expression data. Comput Biol Chem. 2019;78:367–374. doi: 10.1016/j.compbiolchem.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 102.Wang J, Zhang X, Ling J, Wang Y, Xu X, Liu Y, Jin C, Ju J, Yuan Y, He F, Zhao C, Wang J, Tian C. KRAB-containing zinc finger protein ZNF496 inhibits breast cancer cell proliferation by selectively repressing ERalpha activity. Biochim Biophys Acta Gene Regul Mech. 2018 doi: 10.1016/j.bbagrm.2018.07.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 103.Arvey A, Agius P, Noble WS, Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012;22:1723–1734. doi: 10.1101/gr.127712.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hutchins AP, Diez D, Takahashi Y, Ahmad S, Jauch R, Tremblay ML, Miranda-Saavedra D. Distinct transcriptional regulatory modules underlie STAT3’s cell type-independent and cell type-specific functions. Nucleic Acids Res. 2013;41:2155–2170. doi: 10.1093/nar/gks1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tanaka N, Saitoh H. A real-time SUMO-binding assay for the analysis of the SUMO-SIM protein interaction network. Biosci Biotechnol Biochem. 2010;74:1302–1305. doi: 10.1271/bbb.100082. [DOI] [PubMed] [Google Scholar]
- 107.Rimsa V, Eadsforth TC, Hunter WN. Structure of the SCAN domain of human paternally expressed gene 3 protein. PLoS One. 2013;8:e69538. doi: 10.1371/journal.pone.0069538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Porsch-Ozcurumez M, Langmann T, Heimerl S, Borsukova H, Kaminski WE, Drobnik W, Honer C, Schumacher C, Schmitz G. The zinc finger protein 202 (ZNF202) is a transcriptional repressor of ATP binding cassette transporter A1 (ABCA1) and ABCG1 gene expression and a modulator of cellular lipid efflux. J Biol Chem. 2001;276:12427–12433. doi: 10.1074/jbc.M100218200. [DOI] [PubMed] [Google Scholar]
- 109.Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi: 10.1016/s0378-1119(03)00509-2. [DOI] [PubMed] [Google Scholar]
- 110.Noll L, Peterson FC, Hayes PL, Volkman BF, Sander T. Heterodimer formation of the myeloid zinc finger 1 SCAN domain and association with promyelocytic leukemia nuclear bodies. Leukemia Res. 2008;32:1582–1592. doi: 10.1016/j.leukres.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 111.Babb R, Bowen BR. SDP1 is a peroxisome-proliferator-activated receptor gamma 2 co-activator that binds through its SCAN domain. Biochem J. 2003;370:719–727. doi: 10.1042/BJ20021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frietze S, O’Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 recruits the histone methyltransferase SETDB1 to the 3’ ends of ZNF genes. PLoS One. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nielsen AL, Jorgensen P, Lerouge T, Cervino M, Chambon P, Losson R. Nizp1, a novel multitype zinc finger protein that interacts with the NSD1 histone lysine methyltransferase through a unique C2HR motif. Mol Cell Biol. 2004;24:5184–5196. doi: 10.1128/MCB.24.12.5184-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li J, Chen X, He J, Li M, Liu Y, Zi H, Hu Z, Zhang J. A yeast two-hybrid screen identifies histone H2A. Z as a transcription factor ZNF24 interactor. J Cell Biochem. 2012;113:3411–3418. doi: 10.1002/jcb.24217. [DOI] [PubMed] [Google Scholar]
- 115.Heun P. SUMOrganization of the nucleus. Curr Opin Cell Biol. 2007;19:350–355. doi: 10.1016/j.ceb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 116.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 117.Stielow B, Kruger I, Diezko R, Finkernagel F, Gillemans N, Kong-a-San J, Philipsen S, Suske G. Epigenetic silencing of spermatocyte-specific and neuronal genes by SUMO modification of the transcription factor Sp3. PLoS Genet. 2010;6:e1001203. doi: 10.1371/journal.pgen.1001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Bio. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 119.Perera BP, Kim J. Alternative promoters of Peg3 with maternal specificity. Sci Rep. 2016;6:24438. doi: 10.1038/srep24438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Alders M, Ryan A, Hodges M, Bliek J, Feinberg AP, Privitera O, Westerveld A, Little PF, Mannens M. Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann syndrome. Am J Hum Genet. 2000;66:1473–1484. doi: 10.1086/302892. [DOI] [PMC free article] [PubMed] [Google Scholar]