Abstract
This study aimed to identify specific microRNAs (miRNAs) related to postmenopausal osteoporosis (OP) in human. A total of 67 conserved miRNAs, including 50 miRNAs significantly up-regulated and 17 miRNAs significantly downregulated, showed differential expression between OP group and control group. 180 hairpin structures were predicted and 199 potential novel miRNA candidates with 18 to 25 nt in length, which will greatly enrich the human miRBase. 4 miRNAs (miR-518b, miR-582-3p, miR-148a-3p and miRNA-223-3p) had upregulated expression and 4 (miR-7d-5p, miR-210-3p, miR-324-5p and miR-654-3p) showed down-regulated expression. Target genes of these miRNAs were involved in bone development, cell proliferation in bone marrow, osteoblast development, negative regulation of osteoblast differentiation, and negative regulation of osteoclast development, as well as several osteogenesis related pathways. Canonical Wnt signaling pathway was selected for verification and function analysis. The expression of Wnt1, FZD10, LRP5, DVL2 and LEF1 was down-regulated significantly, while that of SFRP1, DKK1, and CHD8 was up-regulated markedly. In conclusion, these genes play important roles in OP, which improves our understanding of pathogenesis of OP.
Keywords: microRNAs, osteoporosis, bone marrow, Solexa sequencing
Introduction
Osteoporosis (OP) is a systemic skeletal disorder characterized by bone mass reduction and microarchitecture deterioration which are related to the increased bone fragility and elevated risk of fractures [1-3]. Bone homeostasis is a dynamic equilibrium between bone formation regulated by osteoblasts and bone resorption regulated by osteoclasts [4]. Many factors, such as hormones, cytokines and mechanical stimulation, mediate the complex regulation of bone homeostasis [4,5]. Postmenopausal OP (PMOP), which is considered to be resulted from estrogen deficiency, is a type of primary OP (POP) in clinical practice [2,3,6].
MicroRNAs (miRNAs) are a class of endogenous, small, single-stranded, noncoding RNA molecules with 18-25 nucleotides. miRNAs can regulate the target genes at the posttranscriptional level. In brief, miRNAs specifically bind to the 3’-untranslational region (3’-UTR) of target mRNAs, subsequently preventing the translation of target mRNAs and/or simulating the degradation of target mRNAs [5-7]. Increasing evidence indicates that miRNAs play important roles in the bone homeostasis, such as osteoblast differentiation, osteoclast differentiation, and cell fate.
Over the past several years, a variety of studies have been conducted to investigate the differentially expressed miRNAs between OP patients and various controls. Li et al. found three miRNAs (miR-21, miR-133a and miR-146) presented differential expression in the plasma between osteopenia/OP patients and controls [8]. Bedene et al. found the expression of miR-148a-3p was significantly up-regulated in plasma of OP patients as compared to controls [9], and the expression of miR126-3p and miR423-5p was related to the bone quality. In the study of Jian et al. miR-30b-5p expression was significantly decreased in osteopenia/OP patients, while the expression of miR-103-3p, miR-142-3p and miR-328-3p was significantly down-regulated in only OP patients [10]. Ding et al. showed miR-194-5p in the serum was a potential biomarker for postmenopausal OP [11]. Cao et al. investigated miRNA biomarkers in human circulating monocytes related to PMOP and found that miR-422a expression was markedly higher in OP group than in control group [12]. Garmilla et al. detected the expression of miRNAs in trabecular bone samples of osteoporotic patients with hip fractures and found that miR-187 and miR-518f were differentially expressed in the samples from OP patients [13]. Chen et al. found the expression of miR125b was significantly higher in osteoporotic hBMSCs. In addition, miR125b over-expression was found to suppress the proliferation and osteogenic differentiation of hBMSCs, while inhibition of miR125b promoted the proliferation and osteogenic differentiation of hBMSCs [14].
Despite increasing studies have investigated the role of miRNAs in OP and different samples (e.g., serum, BMSCs, circulating monocytes, and trabecular bone samples) have been employed for investigations in different conditions (i.e., osteopenia or OP patients and a normal group), none has been conducted to focus on the miRNAs in the bone marrow of OP patients. The imbalanced homeostasis of bone marrow, in which osteoblasts and osteoclasts develop, may firstly and directly result in imbalanced bone homeostasis. Therefore, this study was initiated to investigate the differentially expressed miRNAs in the bone marrow of OP patients.
Microarray analysis and Solexa sequencing are tow common, high-throughput methods used to screen differentially expressed miRNAs between paired samples. Microarray analysis was only used to identify the obtained target because of lack of corresponding probes [15]. Compared with microarray, Solexa sequencing can be used to explore and identify a new miRNA which can be further validated. Therefore, Solexa sequencing was also employed to identify the differentially expressed miRNAs in the bone marrow of OP patients.
In this study, to further investigate the regulation of miRNAs in the bone marrow of postmenopausal OP and explore the underlying mechanisms, the differentially expressed miRNAs in bone marrow of postmenopausal OP were examined by comparing to those in controls via Solexa sequencing. The putative target genes were predicted with bioinformatics analysis. The target genes of selected differentially expressed miRNAs were also analyzed in the gene ontology (GO) and KEGG biological pathway. Our study will deepen our understanding of the regulatory role of miRNAs in postmenopausal OP.
Methods and materials
Patients and sample collection
This study was conducted according to the World Medical Association Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects and approved by the Institutional Ethical Committee. Written informed consent was obtained from patients before sample collection. In this study, a total of 56 patients with hip fractures were recruited into present study, including 33 women with PMOP (age: 43-88.2 years) and 23 postmenopausal control women (age: 45.5-78.1 years old), from Shanghai Minhang Hospital between February 2016 and June 2018. The inclusion criteria were as follows: natural menopause after 40 years of age and a bone mineral density (BMD) of at least 2.5 standard deviation (SD) below the peak mean BMD of healthy young women (-2.5 T-score) at the lumbar spine, total hip or femoral neck. Patients with a medical history of OP treatment, hormone replacement therapy, early menopause (< 40 years), abnormal menopause, acute gastrointestinal inflammation, or chronic renal failure were excluded. Bone marrow samples were collected during the implantation of a total endoprosthesis or gamma nail into the proximal femur.
BMD measurement
Dual-energy X-ray absorptiometry (DXA; GE Healthcare, Madison, Wisconsin, USA) was performed to evaluate the BMD of the lumbar spine, total hip and femoral neck. The X-ray absorptiometer was calibrated and reference values were obtained according to previously study [16]. BMD measurements were used to exclude the regions of severe scoliosis, fracture, and operated sites.
Preparation of small RNA libraries and Solexa sequencing
Six small RNA (sRNA) library was prepared on the basis of bone marrow samples from OP group (OP, n = 3) and control group (Ctrl, n = 3). According to the manufacturer’s instructions, total RNA was extracted with Trizol reagent (Invitrogen, USA) from bone marrow. The RNA concentration was quantified using the 2100 Bioanalyzer (Agilent Technologies). For each group, 10 mg of total RNA from each sample was used for constructing library and sRNA sequencing. The 18- to 30-nt fraction of total RNA was purified through 15% Tris-Borate-EDTA (TBE) denaturing polyacrylamide gel electrophoresis (PAGE) and ligated to 3’ and 5’ RNA adaptors. The adaptor-ligated sRNAs were subjected to RT-PCR with 15 cycles of amplification. The PCR products were purified via 4% agarose gels and then used for sequencing analysis using an Illumina/Solexa G1 sequencer (Shanghai Oebiotech Co. Ltd, China).
Analysis of sequencing data and identification of miRNAs
Clean reads were obtained via removing the low-quality reads, such as reads without the 3’ adaptor, 5’ adaptor-contaminant reads, reads without the insert fragment, reads containing poly (A) stretches, and reads of less than 18 nt. Clean reads were aligned with the human genome using SOAP (http://soap. genomics.org.cn). Clean reads were also mapped to sRNA in the GenBank and Rfam to analyze their distribution and annotate sRNA sequences. Clean reads were aligned against known miRNA precursors and mature miRNAs in the miRBase 21.0 to identify conserved miRNAs.
After identification of the conserved miRNAs, the hairpin structures of two libraries, which are the characteristics of miRNA precursors, were used to predict novel miRNAs by mapping with the integrated human transcriptome. Structural features of potentially novel miRNAs were analyzed via miRDeap 2 software, and the resulting structures were retained as novel miRNA candidates only if they met the criteria described by Allen et al. [17] and Friedlander et al. [18]. Then, the stem-loop structures of potentially novel miRNAs were predicted and checked using RNA fold software (http://rna.tbi.univie.ac.at). The sequences could be candidate miRNA precursors only if the number of base pairs in a stem was ≥ 18 nt; the number of errors in one bulge was ≤ 18; the free energy of secondary structures of the hairpins was less than -20 kcal/mol; the percentage of miRNA in the stem was ≥ 80%; the hairpin length was ≥ 53 nt; the hairpin loop length was ≤ 22 nt; the percentage of A and U in the mature miRNA was 30%-70%.
Differential expression assay
To identify differentially expressed miRNAs, the expression of known miRNAs was compared between two libraries. miRNA expression in two libraries (OP and Ctrl) was normalized to obtain transcripts per million with following formula: normalized expression (NE) = actual miRNA count/total count of clean reads*1,000,000. Then, the fold change was calculated from the normalized expression as follow: Fold change = log2 (OP - NE/Ctrl - NE) and then generated the log2 ratio plot and scatter plot. P-value was calculated based on the normalized expression as follow (Equation):
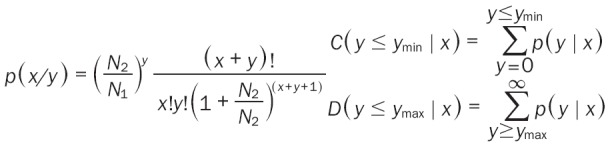 |
where x and y represent normalized expression, and N1 and N2 represent total count of clean reads of a given miRNA in a small RNA library of Ctrl and OP, respectively.
Validation and expression detection of human miRNAs via qPCR
qPCR was used to validate the differentially expressed miRNAs. miRNA was extracted from the OP and Ctrl bone marrow according to the miRcute miRNA Isolation Kit (TIANGEN, China, DP501). 1 mg of total RNA from each sample was subjected to reverse-transcription using the NCodeTM EXPRESS SYBR® GreenERTM miRNA qPCR Kit (Invitrogen). qPCR was performed using the GeneAmp PCR system 9600 (Perkin-Elmer). The miRNA sequences were obtained from the miRBase 21.0 database (http://www.mirbase.org/). The primer sequences of miRNAs are shown in Table 1. U6 served as an endogenous control gene for miRNAs. The primer sequences of target mRNAs used are shown in Table 2. β-actin served as an endogenous control gene for mRNAs. Detection was performed thrice, and the relative expression level was calculated with the 2-ΔΔCt method. The differential expression of miRNA in OP and Ctrl was measured using the fold change (log2 ratio).
Table 1.
The probe sequences of miRNAs for qRT-PCR in the experiment
| Gene | Primer Sequence |
|---|---|
| hsa-let-7d-5p | AGAGGTAGTAGGTTGCATAGTT |
| hsa-miR-148a-3p | TCAGTGCACTACAGAACTTTGT |
| hsa-miR-210-3p | CTGTGCGTGTGACAGCGGCT |
| hsa-miR-223-3p | TGTCAGTTTGTCAAATACCCCA |
| hsa-miR-324-5p | CGCATCCCCTAGGGCATTGG |
| hsa-miR-518b | CAAAGCGCTCCCCTTTAGAGG |
| hsa-miR-582-3p | TAACTGGTTGAACAACTGAACC |
| hsa-miR-654-3p | TATGTCTGCTGACCATCACCTT |
Table 2.
Primers sequence of target mRNA for qRT-PCR in the experiment
| Gene | Primers (F = forward; R = reverse) |
|---|---|
| WNT1 | F: 5’-CGGCGTTTATCTTCGCTA TC-3’ |
| R: 5’-GGGCGATTTCTCGAAGTA GA-3’ | |
| FZD10 | F: 5’-TCGAAGCCAACAGCAGCTAC-3’ |
| R: 5’-AGGATCAGGATGGTCTTCACC-3’ | |
| LRP5 | F: 5’-TGCCACTGGTGAGATTGAC-3’ |
| R: 5’-ACTGCTGCTTGATGAGGAC-3’ | |
| DVL2 | F: 5’-TAGCTCTCACAGCCACCACTGA-3’ |
| R: 5’-CTGATCCGACACACCGTCAA-3’ | |
| LEF1 | F: 5’-CAGAGGTCAACCCCAAGCAA-3’ |
| R: 5’-TTTCAGGAGCTGGTGGGTGTD-3’ | |
| SFRP1 | F: 5’-TTTGAGGAGAGCACCCTAGGC-3’ |
| R: 5’-TGTGTATCTGCTGGCAACAGG-3’ | |
| DKK1 | F: 5’-GGGTCTTTGTCGCGATGGTA-3’ |
| R: 5’-CTGGTACTTATTCCCGCCCG-3’ | |
| CHD8 | F: 5’-GGTCACACAAGACCCCATTGA-3’ |
| R: 5’-AGGGACCAGTTCACTTGCTG-3’ |
GO and KEGG (Kyoto encyclopedia of genes and genomes) pathway analysis
GO and KEGG analyses were used to assess the functional enrichment of mRNA in the miRNA-mRNA regulatory network. InterProScan [19] and Blast2go [20] were used for GO annotation and enrichment analysis for molecular function, cellular component, and biological process. Cytoscape software V2.8.2 (http://www.cytoscape.org/) [21] and the ClueGO plug-in (http://apps.cytoscape.org/apps/cluego) [22] were used to decipher the KEGG (http://www.genome.jp/kegg/) [23] pathway, in which the miRNAs and target genes were enriched and their biological functions were determined. In all the tests, the significant GO terms and pathways were identified by the Benjamini-corrected modified Fisher’s exact test, and a value of P ≤ 0.05 was considered statistically significant.
Statistical analysis
Data are presented as mean ± standard deviation (SD). Student’s t test was used for comparison between two groups. For multiple comparisons, analysis of variance (ANOVA) was used. A value of P < 0.05 were considered statistically significant.
Results
Sequencing data
The clinical characteristics of patients included are shown in Table 3. There was no significant difference in the age between OP patients and patients without OP. The mean values of height, weight, and BMI were higher in patients without OP (P < 0.05). In addition, the osteoporotic patients have significantly lower BMD at all measured sites (P < 0.05) as compared to patients without OP.
Table 3.
Clinical characteristics of participants
| Osteoporosis (n = 33) | Control (n = 23) | P value | |
|---|---|---|---|
| Age, year | 69.4 ± 10.6 | 66.6 ± 8.0 | > 0.05 |
| Height, cm | 152.3 ± 6.7 | 156.5 ± 5.9 | < 0.05 |
| Weight, kg | 50.5 ± 6.5 | 58.2 ± 8.9 | < 0.001 |
| BMI, kg/m2 | 21.8 ± 2.7 | 23.8 ± 3.3 | < 0.05 |
| Lumbar spine BMD, g/cm2 | 0.81 ± 0.10 | 1.00 ± 0.13 | < 0.001 |
| Femoral neck BMD, g/cm2 | 0.64 ± 0.11 | 0.83 ± 0.15 | < 0.001 |
| Total hip BMD, g/cm2 | 0.67 ± 0.09 | 0.85 ± 0.15 | < 0.001 |
Notes: Abbreviations: BMI, body mass index; BMD, bone mineral density.
We sequenced all small RNAs (10 to 30 nt) in the bone marrow of OP group and Ctrl group by Solexa sequencing. The total number of clean reads in OP patients was 29160833 to 46611231. The total number of clean reads in Ctrl group was 35397695 to 35964204. There was a large amount of small RNAs with 21-23 nt in length from both libraries, which was consistent with the miRNAs size.
To assess the efficiency of high-throughput sequencing for sRNA detection, total population of clean sRNAs was annotated and classified via alignment with GenBank and Rfam databases. A total of 35.68 ± 0.41 M reads in control library (85.68%) and 37.89 ± 3.73 M reads in OP library (81.38%) were mapped to the human genome. The classification of RNA in each library is shown in Figure 1. The classification annotation revealed that 4.89 M and 1.73 M reads in control library (13.67%) and OP library (4.79%) were annotated as known miRNAs, respectively, while 3.02 M and 1.66 M reads in control library (8.47%) and OP library (4.68%) were unannotated, respectively, and required further analysis for novel miRNA candidates (Figure 1).
Figure 1.
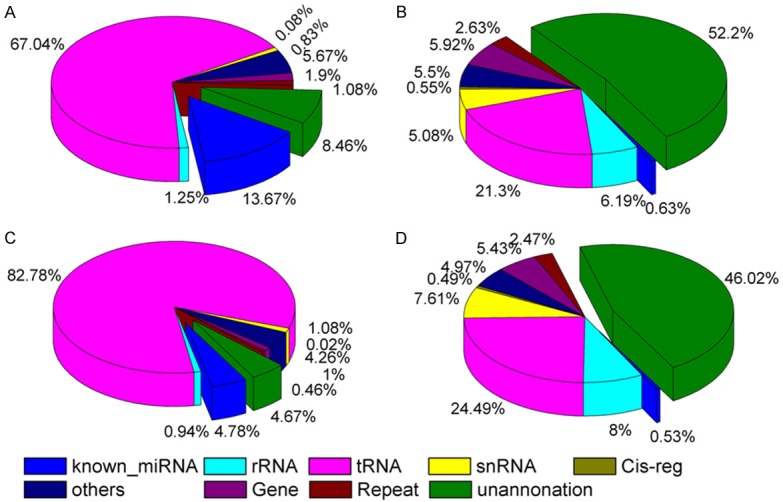
Distribution of sRANs among different categories in the Ctrl and OP library. 4.89 M and 1.73 M reads in the control (13.67%) and OP (4.79%) libraries, respectively, were classified as know miRNAs, while 3.02 M and 1.66 M reads in the control (8.47%) and OP (4.68%) libraries, respectively, were unannotated and required further analysis for novel miRNA candidates. A: Total number of reads in Ctrl. B: Total number of unique sequences in Ctrl. C: Total number of reads in OP. D: Total number of unique sequences in OP.
Identification of known conserved miRNAs
The clean reads were aligned to the precursor/mature miRNAs in the miRBase 21.0 database. The sequence and count of families were obtained (Table S1).
A total of 1091 conserved miRNAs were identified. 967 of these were present in control library and 851 in OP library. In addition, 364 miRNAs were identified in only one library, while 727 miRNAs were found in both libraries. For example, miR-1-5p, miR-1287-3p, and miR-612 were found in only control library, while miR-141-5p, miR-3591-5p, miR-498 and miR-765 were found in only OP library.
The differential expression profiles of all known miRNAs in OP and Ctrl libraries are shown in Table S2. The expression of known miRNAs was presented using a Log2-ratio and visualized in scatter plot (Figure 2). A total of 67 conserved miRNAs showed significantly differential expression (P < 0.05) between two groups. Compared with control library, 50 miRNAs in OP library showed significant up-regulation (P < 0.05), while 17 miRNAs exhibited significant down-regulation (P < 0.05) (Figure 2 and Table S2).
Figure 2.
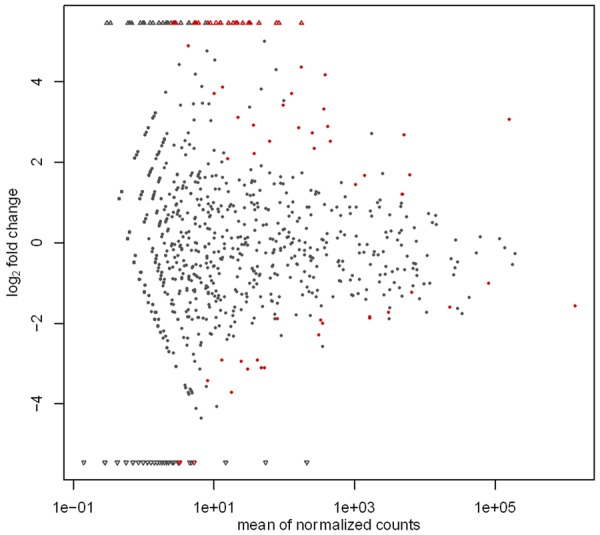
Differential expression of conversed miRNAs between OP and control library. Each point in the figure represents a miRNA. Dots represent miRNAs presented in both libraries. Regular triangle represents miRNAs detected in only OP library. Inverted triangles represent miRNAs detected in only control library. Red points represent significant differentially expressed miRNAs.
Prediction of novel miRNAs
Furthermore, the potential novel miRAN candidates were predicted via aligning the remaining sequences of two libraries with human integrated transcriptome. The miRDeep2 software was used to predict potential novel miRNA candidates. RNA fold software was also used to predict the typical secondary structures of miRNA precursors and remove pseudo-pre-miRNAs. A total of 180 hairpin structures were predicted and 199 potential novel miRNA candidates were identified with the length ranging from 18 nt to 25 nt (Table S3).
Validation of miRNAs by qRT-PCR
The Solexa results from control group and OP group were further validated individually by qPCR. 4 miRNAs with up-regulated expression (miR-518b, miR-582-3p, miR-148a-3p and miRNA-223-3p) and 4 miRNAs with down-regulated expression (miR-7d-5p, miR-210-3p, miR-324-5p and miR-654-3p) were selected in OP group for validation by qPCR. Consistent with sequencing findings, the expression of miR-518b, miR-582-3p, miR-148a-3p and miRNA-223-3p was up-regulated, while that of miR-7d-5p, miR-210-3p, miR-324-5p and miR-654-3p down-regulated in OP group (Figure 3). Thus, qPCR findings supported the results from Solexa sequencing.
Figure 3.
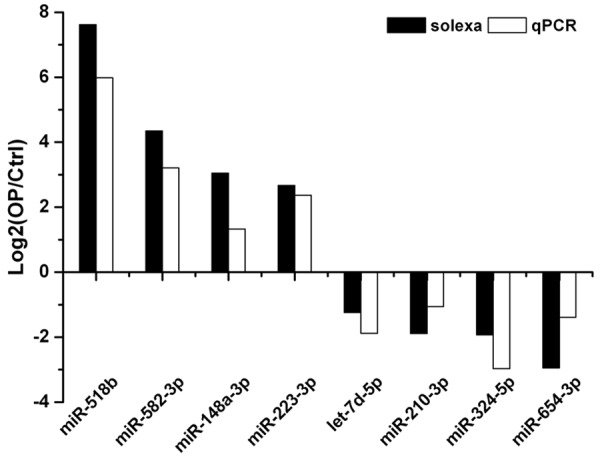
qPCR validation of miRNAs identified in bone marrow using Solexa sequencing technology. miR-518b, miR-582-3p, miR-148a-3p, miRNA-223-3p were all upregulated, whereas miR-7d-5p, miR-210-3p, miR-324-5p, miR-654-3p were all downregulated in OP group.
Target gene prediction for miRNAs
miRNA target gene prediction was further performed based on miRNA/mRNA interactions, and results indicated these differentially expressed miRNAs played important roles in physiological functions and biologic processes during OP. In total, 5968 potential target genes were identified for 67 differentially expressed miRNAs (Table S4).
GO enrichment and KEGG pathway analyses of target genes
The biological functions of target genes were further defined via GO and KEGG pathway analyses. In total, 2618 significantly enriched GO terms (corrected P < 0.05) were identified (Table S5). The top 10 enriched GO terms in biological process, cellular component and molecular function are shown in Figure 4. GO analysis showed a wide range of transcription-DNA-templated (GO: 0006351), negative regulation of transcription from RNA polymerase II promoter (GO: 0000122), signal transduction (GO: 0007165) in biological process, cytoplasm (GO: 0005737), nucleus (GO: 0005634), plasma membrane (GO: 0005886) in cellular component, protein binding (GO: 0005515), and metal ion binding (GO: 0046872) in molecular function.
Figure 4.
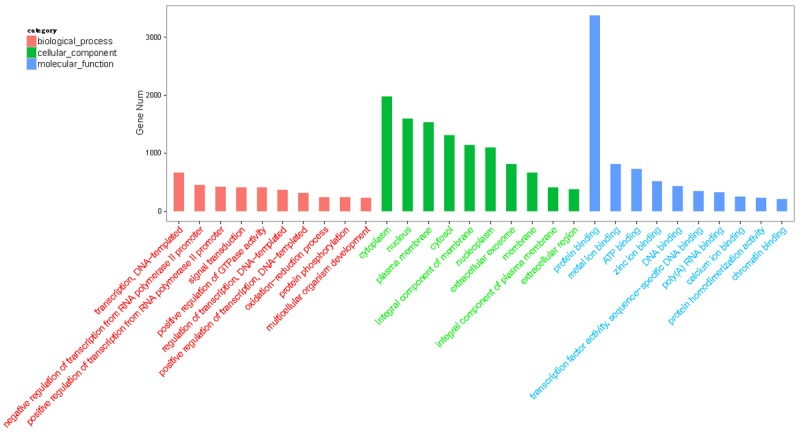
Gene ontology classification annotated by gene2go for target genes of differentially expressed miRNAs. The figure shows top 10 most enriched GO for the predicted target genes in ontologies in ontologies of biological processes, cellular component, and molecular function.
Moreover, 274 enriched KEGG pathways were identified (Table S6). The top 20 enriched KEGG pathways, including MAPK signaling pathway, Ras signaling pathway, Notch signaling pathway and calcium signaling pathway were involved in osteogenic differentiation of stem cells, osteoclast differentiation and bone formation (Figure 5). In addition, Wnt signaling pathway and TGFβ signaling pathway, which were reported to play important roles in bone formation, skeletal development and adult skeletal homeostasis, were significantly enriched in KEGG pathways (Table S6). The GO and KEGG analysis indicate that the target genes of differentially expressed miRNAs are involved in a wide range of regulatory functions in OP.
Figure 5.
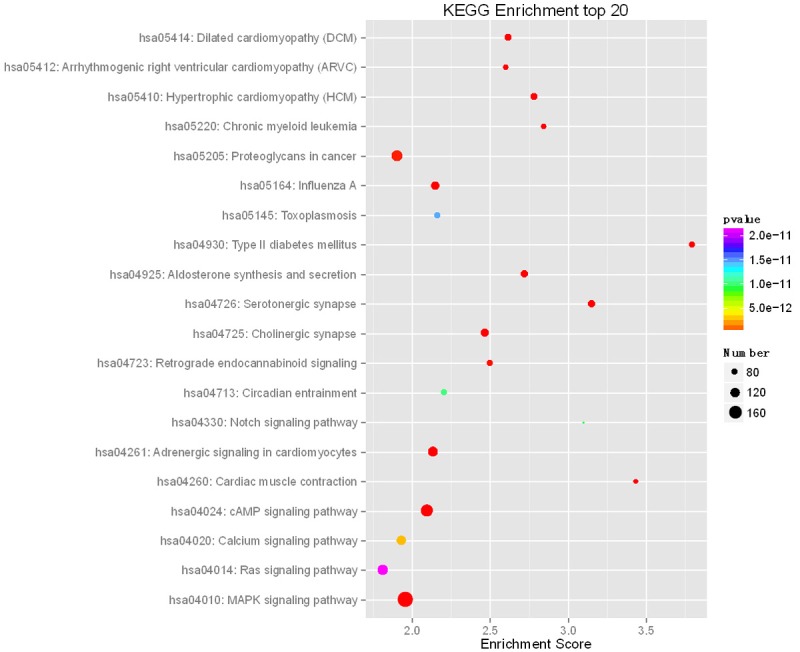
Top 20 significantly enriched Kyoto Encyclopedia of Genes and Genomes pathways from target genes, including MAPK signaling pathway, Ras signaling pathway, Notch signaling pathway, and Calcium signaling pathway.
Molecules in Wnt signaling pathway were differentially expressed in the bone marrow of OP patients
Wnt signaling pathways are major signaling cascades responsible for OP. Our results indicated that Wnt signaling pathway was significantly enriched in the bioinformatics analysis. Furthermore, a number of OP related genes in the canonical Wnt signaling pathway, such as Wnt1, Wnt4, FZD10, LRP5, DVL2, GSK3B, LEF1, SFRP1, DKK1 and CHD8, were predicted to be target genes of differentially expressed miRNAs (Table 4). Among them, the expression of Wnt1, FZD10, LRP5, DVL2 and LEF1, which are positive regulators of Wnt signaling pathway, was significantly down-regulated in OP, which was confirmed by qPCR; while the expression of SFRP1, DKK1, and CHD8, negative regulators of Wnt signaling pathway, was significantly up-regulated (Figure 6).
Table 4.
Predicted target genes of differentially expressed miRNAs in the “canonical Wnt signaling pathway” gene ontology group
| Target genes | Function | miRNAs |
|---|---|---|
| Wnt1 | Wnt ligand protein, osteogenesis | hsa-miR-148a-3p, hsa-let-7d-5p, hsa-let-7a-5p |
| Wnt4 | Wnt ligand protein, Osteogenesis | hsa-miR-424-5p, hsa-miR-9-5p, hsa-miR-4676-3p |
| FZD10 | Wnt receptor, osteogenesis, osteoporosis | hsa-miR-522-3p, hsa-miR-524-5p, hsa-miR-520g-3p, hsa-miR-512-3p, hsa-miR-450b-5p, hsa-miR-424-5p, hsa-miR-654-5p |
| LRP5/6 | Wnt receptor, osteogenesis, osteoporosis | hsa-miR-381-3p, hsa-miR-499a-5p, hsa-miR-582-5p, hsa-miR-424-5p, hsa-miR-128-3p |
| DVL2 | Signal transduction, upregulation of Wnt signaling | hsa-miR-381-3p, hsa-miR-128-3p |
| GSK3B | Regulation of Wnt signaling, osteogenesis | hsa-miR-424-5p, hsa-miR-26b-5p, hsa-miR-582-5p, hsa-miR-142-5p, hsa-miR-128-3p, hsa-miR-9-5p |
| LEF1 | Binding factor, activates transcription of genes with β-catenin | hsa-miR-372-3p, hsa-miR-381-3p, hsa-miR-520a-3p, hsa-miR-26b-5p |
| SFRP1 | Negatively regulates Wnt signaling, osteogenesis, osteoporosis | hsa-miR-128-3p |
| DKK1 | Negatively regulates Wnt signaling, osteogenesis, osteoporosis | hsa-miR-372-3p, hsa-miR-520a-3p |
| CHD8 | Negatively regulates Wnt signaling | miR-194-5p |
Figure 6.
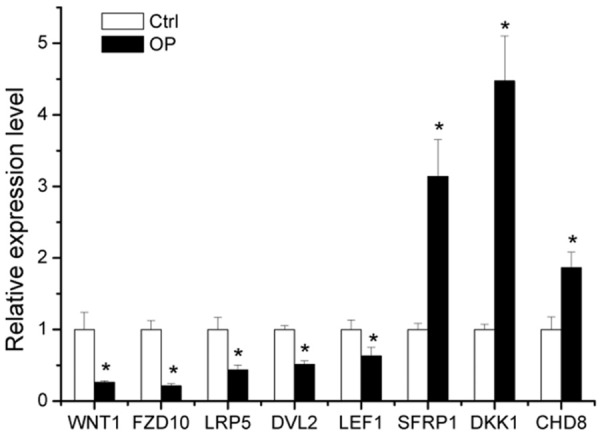
Eight osteoporosis-related genes in the canonical Wnt signaling pathway which were predicted to be target genes of the differentially expressed miRNAs were confirmed to be significantly regulated in osteoporosis by qPCR. Data are expressed as mean ± SE of each group of bone marrow from three separate experiments. *P < 0.05, compared to control group.
Discussion
PMOP is a systemic skeletal disease characterized by low bone mass, impaired bone microstructure and increased skeletal fragility, with increased incidence of fractures. OP results from imbalance between bone formation and bone resorption due to the interactions of genetic, epigenetic and environmental factors [24]. Numerous studies have indicated that epigenetic mechanisms, including miRNAs, are key regulators of gene expression [5,25]. Postmenopausal women usually have multiple independent predisposing factors, such as estrogen deficiency, continuous calcium loss, and aging, and thus have a high risk for OP [7,10,11]. In the present study, Solexa sequencing was used to profile miRNA expression and screen differentially expressed microRNAs (P < 0.05) between PMOP and control group.
Studies have shown that a number of miRNAs are involved in the pathogenesis of PMOP, but most of studies are performed in samples of serum or by microarray assay. Meng et al. analyzed pooled blood samples of PMOP using miRNA microarray and found that five miRNAs (has-miR-130b-3p, has-miR-151a-3p, has-miR-151b, has-miR-194-5p, and has-miR-590-5p) were upregulated [17]. Mandourah et al. identified fifteen differentially expressed miRNAs in OP, of which two miRNAs had up-regulated expression (has-miR-21-3p and has-miR-1231) and thirteen miRNAs showed down-regulated expression (has-miR-100-5p, has-miR-122-5p, has-miR-215-5p, has-miR-3911, has-miR-1290, has-miR-194-5p, has-miR-145-3p, has-let-7a-3p, has-miR-4306, has-miR-10b-5p, has-miR-365b-5p, has-miR-200-b-3p and has-miR-99a-5p) [26]. You et al. employed miRNA microarray to investigate differentially expressed miRNAs in the serum of PMOP and their results showed the expression of 33 miRNAs was down-regulated by > 2 folds in the osteoporotic patients as compared to controls [27]. In the study of Maria et al. the expression of 5 miRNAs was significantly deregulated in the serum of patients with low bone mass as compared to controls, among which miR-124 and miR-2861 were significantly higher, whereas miR-21, miR-23 and miR-29 were lower [20].
In this study, the differentially expressed miRNAs were assayed in bone marrow of PMOP patients. Among the 67 miRNAs identified in the present study, 17 miRNAs showed significantly down-regulated expression, whereas 50 miRNAs had significantly up-regulated expression. Moreover, only 11 miRNAs were observed during osteogenesis, adipogenesis and/or OP. miR-26 family members mediated formation and degradation of collagens and extracellular matrix (ECM) proteins [28]. Li et al. showed the miR-26b-5p expression was significantly up-regulated in the osteoarthritic anterior cruciate ligament (ACL) [29]. Sartori et al. showed miR-26b-5p was differentially expressed in the osteoblasts on the nanotopography surface [30]. miR-142-3p was an important osteogenesis related miRNA and its expression significantly decreased in the serum of OP patients [4]. miR-142-3p expression was markedly up-regulated during osteogenic differentiation of hFOB1.19 cells. miR-142-3p can stimulate osteoblast differentiation via targeting APC and activating Wnt signaling pathway [31]. Furthermore, over-expression of miR-142-3p prevents the transformation of monocytes to macrophage and inhibits osteoclast differentiation [32]. miR-142-5p has been proven to be the most significantly induced miRNA in the bone marrow mesenchymal stem cells (BMSCs), it can bind to the 3’UTR of vascular cell adhesion molecule 1 (VCAM-1) to inhibit VCAM-1 expression in rats with ovariectomy- (OVX-) induced OP, and down-regulated VCAM-1 inhibits the migration of BMSCs in OP [33]. On the contrary, Tu et al. found miR-142-5p showed up-regulated expression in osteoblasts and could promote bone repair by maintaining osteoblast activity and matrix mineralization via targeting WWdomain-containing E3 ubiquitin protein ligase 1 [34]. miR-148a-3p can regulate adipogenesis and osteogenesis of marrow stromal cells through directly targeting Kdm6b [35]. Overexpression of miR-148a-3p induces adipocyte differentiation of ST2 and blunts osteoblast differentiation [35]. The expression of miR-148a-3p, a potential new plasma-based biomarker for OP, is significantly up-regulated in OP patients as compared to controls [9,36]. Kelch et al. found the expression of miR-148a-3p was significantly higher in osteoporotic osteoclasts [36]. Cheng et al. reported miR-148a promoted the differentiation of peripheral blood mononuclear cells (PBMCs) into osteoclasts via repressing V-maf musculoaponeurotic fibrosarcoma oncogene homolog B (MAFB) protein expression by binding to the 3’-UTR of MAFB mRNA [37]. In vivo, silencing of miR-148a using a specific antagomir increased bone mass in ovariectomized mice. miR-194-5p is highly expressed in the blood and was identified as a potential biomarker for OP [17]. miR-210-3p can stimulate the expression of osteogenic genes in BMSCs in vitro and miR-210-3p/β-tricalcium phosphate (β-TCP)/bone mesenchymal stem cell (BMSC) construct could enhance the regeneration of critical-sized bone defects in vivo [38]. In studies conducted in humans or mice, more than 20 genes have been validated as the targets of miR-223, such as C/EBPβ, IKKα, NFIA and FGFR2 [39,40]. FGFR2 and NFIA as miR-223 targets can regulate the osteoblasts differentiation, adipogenic differentiation and osteoclast differentiation [41,42]. miR-223 can suppress the monocyte and macrophage differentiation via binding the 3’UTR and inhibiting the protein expression of IKKα, a critical regulator of NF-κB pathway [43]. As we know, osteoclasts are monocyte and macrophage-derived cells [44], IKKα is involved in the regulatory effects of miR-223 on osteoclast differentiation. Sun et al. indicated that miR-375-3p directly bound to the 3’UTR of LRP5 and β-catenin to decreased their expression, which suppressed the osteogenic differentiation and bone formation in vivo [45], while Chen et al. found that miR-375 expression was up-regulated during the osteogenesis of human adipose derived mesenchymal stem cells (hASCs) [46]. Over-expression of miR-375 significantly enhanced the osteogenesis of hASCs via the YAP1/DEPTOR/AKT network. miR-451a expression was significantly up-regulated in standard healing fractures as compared to non-healing fractures [47]. miR-451a was reported to reduce inflammation via targeting 14-3-3ζ and Rab5a, resulting in suppressed phosphorylation of p38 mitogen activated protein kinase (MAPK) [48], which plays an important role in orchestrating injury or stress-induced responses in bone formation [49]. miR-542-3p induces osteoblast differentiation and bone formation by inhibiting SFRP1 [50]. On the contrary, Kureel et al. reported that miR-542-3p suppressed the proliferation and differentiation of osteoblasts and inhibited bone formation by targeting BMP-7 signaling pathway [51]. miR-618 expression was down-regulated during osteogenic differentiation of RAW264.7 cells. In addition, over-expression of miR-618 attenuated osteoclast differentiation of RAW264.7 cells in vitro, whereas inhibition of miR-618 promoted this progress [52]. miR-654-3p expression was reported to be up-regulated in the osteogenic differentiation of human periodontal ligament stem cells [53]. However, the remaining 56 miRNAs were identified for the first time in the bone marrow of OP.
Compared with microarray, Solexa sequencing can be used to explore and identify a new miRNA that can be further validated. Therefore, Solexa sequencing was exployred to identify distinct miRNAs in the bone marrow of OP patients. In the present study, 180 hairpin structures were predicted and 199 potential novel miRNA candidates were identified with the length ranging from 18 nt to 25 nt, which will greatly enrich the human miRBase.
In order to further investigate the function of these distinct miRNAs, bioinformatics analysis were used to explain the annotation of their functions. Abundant high-enrichment GOs of target genes regulated by these differentially expressed miRNAs might involve several biological processes such as bone development, cell proliferation in bone marrow, osteoblast development, negative regulation of osteoblast differentiation, negative regulation of osteoclast development, and MAPK cascade. In addition, specific KEGG pathways targeted by these differentially expressed miRNAs were found to be involved in MAPK signaling pathway, Ras signaling pathway, calcium signaling pathway, and Notch signaling pathway. Our previous study indicated that ERK and JNK cell signaling pathways, two distinct groups regulated by mitogen-activated protein kinases (MAPKs), played an important role in the osteogenic differentiation [54-56]. Activation of ERK and JNK is especially related to the sequential expression of osteogenesis-related genes, while blockage of them can inhibit the osteogenic differentiation of ASCs.
In addition, Wnt signaling pathway is known to play critical roles in bone formation, skeletal development and adult skeletal homeostasis [57,58]. Recently, increasing evidence shows that Wnt signaling pathway also plays important roles in the development of OP, possibly through regulating inflammation, bone resorption, bone remodeling and joint destruction [59-61]. In this study, our results indicated that some key molecules of OP (such as Wnt1, Wnt4, FZD10, LRP5, DVL2, GSK3B, LEF1, SFRP1, DKK1 and CHD8) which function as positive/negative regulators of OP and canonical Wnt signaling pathway mediators are included. In the canonical pathway, Wnt ligands (such as Wnt1 and Wnt4) bind to the LRP5-Frizzled (FZD) complex to activate the intracellular protein Dishevelled (DVL). DVL then inhibits the complex comprised of axin and GSK-3β. This inhibits the degradation of β-catenin, which accumulates and enters the nucleus to interact with TCF/LEF transcription factors mediating gene transcription [60,62]. SFRP1 was identified as an antagonist of canonical Wnt signaling pathway. Yao et al. reported that over-expression of SFRP1 suppressed bone formation and attenuated PTH anabolic action on the bone [63]. Wang et al. showed that SFRP1, in a high concentration of glucocorticoid, stimulated bone cell apoptosis and inhibitd bone formation activities, resulting in bone loss [64]. DKK1 is another inhibitor of canonical Wnt signaling pathway and plays a vital role in the occurrence and development of bone disease [65]. CHD8 is a negative regulator of canonical Wnt signaling pathway and can suppress β-catenin activity [66]. Our results showed the expression of Wnt1, FZD10, LRP5, DVL2 and LEF1 was significantly down-expressed in the bone marrow of OP, while that of SFRP1, DKK1 and CHD8 was markedly up-regulated. All these findings suggested that the differentially expressed miRNAs may regulate OP via inhibiting positive regulators and promoting negative regulators of canonical Wnt signaling pathway. Further study is currently ongoing in our group to explore the effect and mechanism of key miRNAs in regulating osteogenesis, bone formation and OP in vivo and in vitro in ovariectomy (OVX) mice.
Conclusion
Taken together, 17 miRNAs have significantly down-regulated expression in the bone marrow of OP, whereas 50 miRNAs display markedly up-regulated expression. 180 hairpin structures are predicted and 199 potential novel miRNA candidates identified with the length ranging from 18 nt to 25 nt, which will greatly enrich the human miRBase. Furthermore, the target genes of some miRNAs are found to be involved in bone development, cell proliferation in bone marrow, and canonical Wnt signaling pathway, playing critical roles in the pathogenesis of OP. These results will improve our understanding of the pathogenesis of OP, and subsequently provide potential novel biomarkers for further study.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 81301335 and 81772433), and Shanghai Municipal Commission of Health and Family Planning Foundation (No. 201440337).
Disclosure of conflict of interest
None.
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
References
- 1.Marini F, Cianferotti L, Brandi ML. Epigenetic mechanisms in bone biology and osteoporosis: can they drive therapeutic choices? Int J Mol Sci. 2016;17 doi: 10.3390/ijms17081329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanis JA, Cooper C, Rizzoli R, Reginster JY Scientific Advisory Board of the European Society for Clinical and Economic Aspects of Osteoporosis and Osteoarthritis (ESCEO) and the Committees of Scientific Advisors and National Societies of the International Osteoporosis Foundation (IOF) European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int. 2019;30:3–44. doi: 10.1007/s00198-018-4704-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone HG, Wagman RB, Pannacciulli N, Papapoulos S. Denosumab treatment in postmenopausal women with osteoporosis - Authors’ reply. Lancet Diabetes Endocrinol. 2017;5:768–769. doi: 10.1016/S2213-8587(17)30288-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Li K, Pang Q, Yang C, Zhang H, Wu F, Cao H, Liu H, Wan Y, Xia W, Wang J, Dai Z, Li Y. Identification of suitable reference gene and biomarkers of serum miRNAs for osteoporosis. Sci Rep. 2016;6:36347. doi: 10.1038/srep36347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gennari L, Bianciardi S, Merlotti D. MicroRNAs in bone diseases. Osteoporos Int. 2017;28:1191–1213. doi: 10.1007/s00198-016-3847-5. [DOI] [PubMed] [Google Scholar]
- 6.Ge DW, Wang WW, Chen HT, Yang L, Cao XJ. Functions of microRNAs in osteoporosis. Eur Rev Med Pharmacol Sci. 2017;21:4784–4789. [PubMed] [Google Scholar]
- 7.Hendrickx G, Boudin E, Van Hul W. A look behind the scenes: the risk and pathogenesis of primary osteoporosis. Nat Rev Rheumatol. 2015;11:462–474. doi: 10.1038/nrrheum.2015.48. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Wang Z, Fu Q, Zhang J. Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers. 2014;19:553–556. doi: 10.3109/1354750X.2014.935957. [DOI] [PubMed] [Google Scholar]
- 9.Bedene A, Mencej Bedrac S, Jese L, Marc J, Vrtacnik P, Prezelj J, Kocjan T, Kranjc T, Ostanek B. MiR-148a the epigenetic regulator of bone homeostasis is increased in plasma of osteoporotic postmenopausal women. Wien Klin Wochenschr. 2016;128:519–526. doi: 10.1007/s00508-016-1141-3. [DOI] [PubMed] [Google Scholar]
- 10.Kranjc T, Ostanek B, Marc J. Bone microRNAs and Ageing. Curr Pharm Biotechnol. 2017;18:210–220. doi: 10.2174/1389201018666170203091828. [DOI] [PubMed] [Google Scholar]
- 11.Shao M. Construction of an mirna-regulated pathway network reveals candidate biomarkers for postmenopausal osteoporosis. Comput Math Methods Med. 2017;2017:9426280. doi: 10.1155/2017/9426280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Z, Moore BT, Wang Y, Peng XH, Lappe JM, Recker RR, Xiao P. MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One. 2014;9:e97098. doi: 10.1371/journal.pone.0097098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garmilla-Ezquerra P, Sanudo C, Delgado-Calle J, Perez-Nunez MI, Sumillera M, Riancho JA. Analysis of the bone microRNome in osteoporotic fractures. Calcif Tissue Int. 2015;96:30–37. doi: 10.1007/s00223-014-9935-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Yang L, Jie Q, Lin YS, Meng GL, Fan JZ, Zhang JK, Fan J, Luo ZJ, Liu J. MicroRNA125b suppresses the proliferation and osteogenic differentiation of human bone marrowderived mesenchymal stem cells. Mol Med Rep. 2014;9:1820–1826. doi: 10.3892/mmr.2014.2024. [DOI] [PubMed] [Google Scholar]
- 15.Cao Y, Lu X, Wang J, Zhang H, Liu Z, Xu S, Wang T, Ning S, Xiao B, Wang L. Construction of an miRNA-regulated drug-pathway network reveals drug repurposing candidates for myasthenia gravis. Int J Mol Med. 2017;39:268–278. doi: 10.3892/ijmm.2017.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao K, Zhao Q, Guo Z, Chen Z, Hu Y, Su J, Chen L, He Z, Cai X, Chen M, Zheng L, Wang W, Wang Q. Hsa_Circ_0001275: a potential novel diagnostic biomarker for postmenopausal osteoporosis. Cell Physiol Biochem. 2018;46:2508–2516. doi: 10.1159/000489657. [DOI] [PubMed] [Google Scholar]
- 17.Meng J, Zhang D, Pan N, Sun N, Wang Q, Fan J, Zhou P, Zhu W, Jiang L. Identification of miR-194-5p as a potential biomarker for postmenopausal osteoporosis. PeerJ. 2015;3:e971. doi: 10.7717/peerj.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan J, Li N, Gong X, He L. Serum 25-hydroxyvitamin D, bone turnover markers and bone mineral density in postmenopausal women with hip fractures. Clin Chim Acta. 2018;477:135–140. doi: 10.1016/j.cca.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 19.Kharroubi A, Saba E, Smoom R, Bader K, Darwish H. Serum 25-hydroxyvitamin D and bone turnover markers in Palestinian postmenopausal osteoporosis and normal women. Arch Osteoporos. 2017;12:13. doi: 10.1007/s11657-017-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yavropoulou MP, Anastasilakis AD, Makras P, Tsalikakis DG, Grammatiki M, Yovos JG. Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur J Endocrinol. 2017;176:169–176. doi: 10.1530/EJE-16-0583. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Guo Q, Zhang M, Song S, Quan T, Zhao T, Li H, Guo L, Jiang T, Wang G. Relationship of serum GDF11 levels with bone mineral density and bone turnover markers in postmenopausal Chinese women. Bone Res. 2016;4:16012. doi: 10.1038/boneres.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei QS, Huang L, Tan X, Chen ZQ, Chen SM, Deng WM. Serum osteopontin levels in relation to bone mineral density and bone turnover markers in postmenopausal women. Scand J Clin Lab Invest. 2016;76:33–39. doi: 10.3109/00365513.2015.1087045. [DOI] [PubMed] [Google Scholar]
- 23.Mukaiyama K, Kamimura M, Uchiyama S, Ikegami S, Nakamura Y, Kato H. Elevation of serum alkaline phosphatase (ALP) level in postmenopausal women is caused by high bone turnover. Aging Clin Exp Res. 2015;27:413–418. doi: 10.1007/s40520-014-0296-x. [DOI] [PubMed] [Google Scholar]
- 24.Bristow SM, Gamble GD, Stewart A, Horne L, House ME, Aati O, Mihov B, Horne AM, Reid IR. Acute and 3-month effects of microcrystalline hydroxyapatite, calcium citrate and calcium carbonate on serum calcium and markers of bone turnover: a randomised controlled trial in postmenopausal women. Br J Nutr. 2014;112:1611–1620. doi: 10.1017/S0007114514002785. [DOI] [PubMed] [Google Scholar]
- 25.Jeong TD, Lee W, Choi SE, Kim JS, Kim HK, Bae SJ, Chun S, Min WK. Relationship between serum total cholesterol level and serum biochemical bone turnover markers in healthy pre- and postmenopausal women. Biomed Res Int. 2014;2014:398397. doi: 10.1155/2014/398397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandourah AY, Ranganath L, Barraclough R, Vinjamuri S, Hof RV, Hamill S, Czanner G, Dera AA, Wang D, Barraclough DL. Circulating microRNAs as potential diagnostic biomarkers for osteoporosis. Sci Rep. 2018;8:8421. doi: 10.1038/s41598-018-26525-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You L, Pan L, Chen L, Gu W, Chen J. MiR-27a is Essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell Physiol Biochem. 2016;39:253–265. doi: 10.1159/000445621. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Bai L, Shen P, Sun Y, Chen Z, Wen Y. Identification of differentially expressed microRNAs in knee anterior cruciate ligament tissues surgically removed from patients with osteoarthritis. Int J Mol Med. 2017;40:1105–1113. doi: 10.3892/ijmm.2017.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sartori EM, Magro-Filho O, Silveira Mendonca DB, Li X, Fu J, Mendonca G. Modulation of Micro RNA expression and osteoblast differentiation by nanotopography. Int J Oral Maxillofac Implants. 2018;33:269–280. doi: 10.11607/jomi.5372. [DOI] [PubMed] [Google Scholar]
- 31.Lu C, Wan Y, Cao J, Zhu X, Yu J, Zhou R, Yao Y, Zhang L, Zhao H, Li H, Zhao J, He L, Ma G, Yang X, Yao Z, Guo X. Wnt-mediated reciprocal regulation between cartilage and bone development during endochondral ossification. Bone. 2013;53:566–574. doi: 10.1016/j.bone.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Fordham JB, Guilfoyle K, Naqvi AR, Nares S. MiR-142-3p is a RANKL-dependent inducer of cell death in osteoclasts. Sci Rep. 2016;6:24980. doi: 10.1038/srep24980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng Z, Xie X, Zhu Y, Liu J, Hu X, Na Q, Zhang X, Wei G, Xu S, Liu Y, Xian CJ. miR-142-5p in bone marrow-derived mesenchymal stem cells promotes osteoporosis involving targeting adhesion molecule VCAM-1 and inhibiting cell migration. Biomed Res Int. 2018;2018:3274641. doi: 10.1155/2018/3274641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu M, Tang J, He H, Cheng P, Chen C. MiR-142-5p promotes bone repair by maintaining osteoblast activity. J Bone Miner Metab. 2017;35:255–264. doi: 10.1007/s00774-016-0757-8. [DOI] [PubMed] [Google Scholar]
- 35.Tian L, Zheng F, Li Z, Wang H, Yuan H, Zhang X, Ma Z, Li X, Gao X, Wang B. miR-148a-3p regulates adipocyte and osteoblast differentiation by targeting lysine-specific demethylase 6b. Gene. 2017;627:32–39. doi: 10.1016/j.gene.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Kelch S, Balmayor ER, Seeliger C, Vester H, Kirschke JS, van Griensven M. miRNAs in bone tissue correlate to bone mineral density and circulating miRNAs are gender independent in osteoporotic patients. Sci Rep. 2017;7:15861. doi: 10.1038/s41598-017-16113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY, Luo XH. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res. 2013;28:1180–1190. doi: 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 38.Hu B, Li Y, Wang M, Zhu Y, Zhou Y, Sui B, Tan Y, Ning Y, Wang J, He J, Yang C, Zou D. Functional reconstruction of critical-sized load-bearing bone defects using a Sclerostin-targeting miR-210-3p-based construct to enhance osteogenic activity. Acta Biomater. 2018;76:275–282. doi: 10.1016/j.actbio.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Xie Y, Zhang L, Gao Y, Ge W, Tang P. The multiple roles of Microrna-223 in regulating bone metabolism. Molecules. 2015;20:19433–19448. doi: 10.3390/molecules201019433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haneklaus M, Gerlic M, O’Neill LA, Masters SL. miR-223: infection, inflammation and cancer. J Intern Med. 2013;274:215–226. doi: 10.1111/joim.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 42.Guan X, Gao Y, Zhou J, Wang J, Zheng F, Guo F, Chang A, Li X, Wang B. miR-223 regulates adipogenic and osteogenic differentiation of mesenchymal stem cells through a C/EBPs/miR-223/FGFR2 regulatory feedback loop. Stem Cells. 2015;33:1589–1600. doi: 10.1002/stem.1947. [DOI] [PubMed] [Google Scholar]
- 43.Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol. 2010;11:799–805. doi: 10.1038/ni.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang P, Xiong Q, Ge W, Zhang L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol. 2014;11:1355–1363. doi: 10.1080/15476286.2014.996462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun T, Li CT, Xiong L, Ning Z, Leung F, Peng S, Lu WW. miR-375-3p negatively regulates osteogenesis by targeting and decreasing the expression levels of LRP5 and beta-catenin. PLoS One. 2017;12:e0171281. doi: 10.1371/journal.pone.0171281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Zheng Y, Zhang S, Jia L, Zhou Y. Promotion effects of miR-375 on the osteogenic differentiation of human adipose-derived mesenchymal stem cells. Stem Cell Reports. 2017;8:773–786. doi: 10.1016/j.stemcr.2017.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waki T, Lee SY, Niikura T, Iwakura T, Dogaki Y, Okumachi E, Oe K, Kuroda R, Kurosaka M. Profiling microRNA expression during fracture healing. BMC Musculoskelet Disord. 2016;17:83. doi: 10.1186/s12891-016-0931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murata K, Yoshitomi H, Furu M, Ishikawa M, Shibuya H, Ito H, Matsuda S. MicroRNA-451 down-regulates neutrophil chemotaxis via p38 MAPK. Arthritis Rheumatol. 2014;66:549–559. doi: 10.1002/art.38269. [DOI] [PubMed] [Google Scholar]
- 49.Zhou FH, Foster BK, Zhou XF, Cowin AJ, Xian CJ. TNF-alpha mediates p38 MAP kinase activation and negatively regulates bone formation at the injured growth plate in rats. J Bone Miner Res. 2006;21:1075–1088. doi: 10.1359/jbmr.060410. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Zhu Y, Zhang C, Liu J, Sun T, Li D, Na Q, Xian CJ, Wang L, Teng Z. miR-542-3p prevents ovariectomy-induced osteoporosis in rats via targeting SFRP1. J Cell Physiol. 2018;233:6798–6806. doi: 10.1002/jcp.26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kureel J, Dixit M, Tyagi AM, Mansoori MN, Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A, Singh D. miR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014;5:e1050. doi: 10.1038/cddis.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang WW, Yang L, Wu J, Gao C, Zhu YX, Zhang D, Zhang HX. The function of miR-218 and miR-618 in postmenopausal osteoporosis. Eur Rev Med Pharmacol Sci. 2017;21:5534–5541. doi: 10.26355/eurrev_201712_13989. [DOI] [PubMed] [Google Scholar]
- 53.Hao Y, Ge Y, Li J, Hu Y, Wu B, Fang F. Identification of MicroRNAs by microarray analysis and prediction of target genes involved in osteogenic differentiation of human periodontal ligament stem cells. J Periodontol. 2017;88:1105–1113. doi: 10.1902/jop.2017.170079. [DOI] [PubMed] [Google Scholar]
- 54.Gu H, Guo F, Zhou X, Gong L, Zhang Y, Zhai W, Chen L, Cen L, Yin S, Chang J, Cui L. The stimulation of osteogenic differentiation of human adipose-derived stem cells by ionic products from akermanite dissolution via activation of the ERK pathway. Biomaterials. 2011;32:7023–7033. doi: 10.1016/j.biomaterials.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 55.Gu H, Huang Z, Yin X, Zhang J, Gong L, Chen J, Rong K, Xu J, Lu L, Cui L. Role of c-Jun N-terminal kinase in the osteogenic and adipogenic differentiation of human adipose-derived mesenchymal stem cells. Exp Cell Res. 2015;339:112–121. doi: 10.1016/j.yexcr.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Liu Q, Cen L, Zhou H, Yin S, Liu G, Liu W, Cao Y, Cui L. The role of the extracellular signal-related kinase signaling pathway in osteogenic differentiation of human adipose-derived stem cells and in adipogenic transition initiated by dexamethasone. Tissue Eng Part A. 2009;15:3487–3497. doi: 10.1089/ten.TEA.2009.0175. [DOI] [PubMed] [Google Scholar]
- 57.Rudnicki MA, Williams BO. Wnt signaling in bone and muscle. Bone. 2015;80:60–66. doi: 10.1016/j.bone.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lerner UH, Ohlsson C. The WNT system: background and its role in bone. J Intern Med. 2015;277:630–649. doi: 10.1111/joim.12368. [DOI] [PubMed] [Google Scholar]
- 59.Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol. 2013;9:575–583. doi: 10.1038/nrendo.2013.154. [DOI] [PubMed] [Google Scholar]
- 60.Baron R, Gori F. Targeting WNT signaling in the treatment of osteoporosis. Curr Opin Pharmacol. 2018;40:134–141. doi: 10.1016/j.coph.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 61.Weitzmann MN. Bone and the immune system. Toxicol Pathol. 2017;45:911–924. doi: 10.1177/0192623317735316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19:179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 63.Yao W, Cheng Z, Shahnazari M, Dai W, Johnson ML, Lane NE. Overexpression of secreted frizzled-related protein 1 inhibits bone formation and attenuates parathyroid hormone bone anabolic effects. J Bone Miner Res. 2010;25:190–199. doi: 10.1359/jbmr.090719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang FS, Lin CL, Chen YJ, Wang CJ, Yang KD, Huang YT, Sun YC, Huang HC. Secreted frizzled-related protein 1 modulates glucocorticoid attenuation of osteogenic activities and bone mass. Endocrinology. 2005;146:2415–2423. doi: 10.1210/en.2004-1050. [DOI] [PubMed] [Google Scholar]
- 65.Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, Niehrs C, Izpisua Belmonte JC, Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1:423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- 66.Thompson BA, Tremblay V, Lin G, Bochar DA. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes. Mol Cell Biol. 2008;28:3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


