Abstract
Recent studies have identified FAT tumour suppressor homologue 4 (FAT4), an essential component of adherents junctions, involved in several cancers. However, its role in endometrial cancer (EC) remains unclear. In this study, we first analyzed the association between FAT4 expression and tumour stage, tumour type, and patient prognosis in 552 tumour samples and 35 non-tumour samples from The Cancer Genome Atlas (TCGA) database. The association of decreased FAT4 expression with advanced signature (lymph node metastasis, lymphovascular invasion and muscular infiltration) in EC patients was also confirmed by our own dataset. Stable FAT4 Knockdown promoted EC cell lines proliferation and invasion. FAT4 overexpression inhibited the parental cell phenotype. FAT4 silencing resulted in decreased phosphorylation of the LATS1/2 and YAP while increased YAP nuclear translocation which was associated with the promotion of proliferation and invasion. PCR array analysis of the negative control and shFAT4 HEC-1B cell lines revealed that the deubiquitinating enzyme USP51 was a FAT4 interacting target gene. Ablating USP51 by shRNA decreased cellular FAT4 protein level while overexpression of USP51 increased FAT4 protein level. Coimmunoprecipitation confirmed the direct binding of FAT4 and USP51 which was essential for FAT4’s function in EC. The growth inhibitory effect of FAT4 was also attenuated by USP51 down-regulation. In conclusion, suppression of FAT4 by inactivation of deubiquitinating enzyme USP51 promoted proliferation and invasion of EC cells via inhibiting Hippo pathway.
Keywords: FAT4, USP51, Hippo pathway, endometrial cancer
Introduction
Endometrial cancer (EC) is one of the most common gynaecologic malignancies in China, with 63,400 newly diagnosed cases and 21,800 estimated deaths [1]. Although the mortality rate among EC patients has remained steady, the incidence rate of EC is increasing, and a shift towards a younger affected population has occurred in recent decades [2]. Since 1983, EC has been classified into two major pathogenic types, defined by Doctor Bokhman [3], that differ in clinical behaviour and histological appearance. Type I tumours, which include endometrioid carcinoma, account for almost 80% of EC cases, are generally oestrogen receptor (ER) and progesterone receptor (PR) positive and have a relatively excellent prognosis. Conversely, type II tumours are often poorly differentiated and are represented primarily by clear cell carcinoma and serous carcinoma, which are ER- and PR-negative and are associated with a poor prognosis [4]. As treatment of metastatic and recurrent EC remains challenging, novel markers of progression and treatment response are needed for better clinical management of EC patients.
The vertebrate FAT cadherin family consists of four members, FAT1, FAT2, FAT3 and FAT4, all of which are closely related in structure to Drosophila ft and ft2 and encode large proteins with extracellular cadherin repeats, EGF-like domains, laminin G-like domains and one intracellular domain [5,6]. Sequence analysis has revealed that the orthologue of vertebrate FAT4 is structurally similar to Drosophila ft in flies and that the FAT family is well conserved in function and structure in humans [7]. FAT4 is a single-stranded transmembrane receptor that acts as a tumour suppressor and regulates cell proliferation, motility and differentiation. As FAT genes encode very large proteins, this family has been termed “sleeping giants”. New technologies in recent years have allowed for greater insight into the FAT family. As an orthologue of Drosophila ft, the FAT4 gene is involved in the maintenance of planar cell polarity (PCP) and the Hippo signalling pathway, which have been reported to have roles in tumourigenesis and cancer metastasis [8,9]. In various types of cancer, such as breast cancer, gastric cancer and melanoma, FAT4 mRNA expression is downregulated by promoter hypermethylation, point mutations and copy number aberrations. Through whole-exome sequencing (WES) technology, nonsynonymous mutations and missense mutations in the human FAT4 gene have been detected in several cancers such as melanoma (40%) [10], pancreatic cancer (8%) [11], head and neck squamous cell carcinoma (6%) [12], and gastric cancer (5%) [13].
The hippo signaling pathway plays an essential role in controlling organ size and suppressing tumorigenesis. The core comments of the Hippo pathway are a series of kinase cascades driven by MST1/2 kinases and their downstream kinases LATS1/2. MST1/2 activate LATS1/2 through direct phosphorylation of one adaptor protein of MST1/2 named SAV1 and also through direct phosphorylation one adaptor protein of LATS1/2 named MOB1 [14]. YAP transcription coactivator, a candidate human oncogene, is inhibited by the Hippo pathway kinase cascades LATS1/2 via its phosphorylation, which results in YAP/14-3-3 (the scaffold protein) binding and cytoplasmic retention. Cytoplasmic YAP can be also degraded by the E3 ligase SCFβ-TRCP [15]. Inactivation of the Hippo pathway results in the YAP nuclear translocation and interaction with a series of transcription factors such as TEAD1-4 proteins and Smad family proteins which leading to the pro-proliferation and pro-metastasis [16]. In Drosophila, multiple positive regulators of the Hippo pathway have been confirmed including WWC1, Kibra, Merlin and FRMD6 [17-20]. In addition to the above regulators, numerous studies have indicated that ft acts as an upstream regulator involved in cell growth via the Hippo pathway in Drosophila [21-23]. In mammals, suppression of FAT4 led to the YAP nuclear accumulation indicating that FAT4 is associated with the Hippo pathway [24]. Collectively, these reports reveal the importance of the FAT4, in particular its cytoplasmic tail, as a central hub for regulating downstream partners.
In this study, we aimed to investigate the tumour-suppressive function of FAT4 in EC and whether it is involved in the Hippo pathway. Furthermore, we report a novel interaction between the FAT4 cytoplasmic tail and USP51, a deubiquitinating enzyme, that modulates the cellular levels of FAT4.
Materials and methods
Clinical specimens
The present study was approved by the ethics committee of the Obstetrics and Gynecology Hospital of Fudan University. The 116 EC tissues [3 clear cell carcinoma, 5 serious carcinoma and 108 endometrioid endometrial carcinoma (46 G1, 48 G2 and 14 G3)] and 86 para-carcinoma tissues and 34 normal endometrium samples examined Immunohistochemically underwent surgical resection at Fudan University Obstetrics and Gynecology Hospital between 2013 and 2017. Among those tissues, we extracted RNA of 33 normal endometrium and 65 EC tissues for the next qRT-PCR validation. Normal epithelial endometrial cells extracted from 10 samples during hysteroscopy. The pathology of all 10 samples was proliferative endometrium.
Establishment of FAT4 stable knockdown and overexpressing cell lines and USP51 transient transfection
The FAT4 stable knockdown cell lines (HEC-1A and HEC-1B) were established by lentiviral-based stable shRNA (CCGGGCGCATTGTTAGATAGGGAAActcgagTTTCCCTATCTAACAATGCGCTTTTTG). Non-target control shRNA served as a negative control (NC). The final stable cell line named HEC-1A-NC, HEC-1A-shFAT4, HEC-1B-NC and HEC-1B-shFAT4. The plasmids Lenti-dCas9-VP64-Puro and U6-sgRNAs-SV40-MS2-P65-HSF1-T2A-Neo were used to simultaneously express wild-type dCas9 and single guide RNA (sgRNA). Three guide RNA sequences, GAGGTTCTTTGAAATAGCAG (sgRNA1), GCTACTTGCTTTTGCCGGAC (sgRNA2), and TCTAGGTAGCCAGTTGAACG (sgRNA3) were used to target the FAT4 promoter region to promote the expression FAT4 expression. Stable cell lines RL95-2+dCas9+sgRNA1, RL95-2+dCas9+sgRNA2 and RL95-2+dCas9+sgRNA3 were established. RL95-2+dCas9+NC served as a control. The shUSP51 plasmid (ShUSP51 sequence: AAAGACATAGAACAGATTGCCAA) was cloned into RNAi pLenti hU6-MCS-CMV-zsGreen1-PGK-Puro vector.
Subcutaneous tumour implantation model
In total, 1 × 106 (0.2 ml) HEC-1B-NC and HEC-1B-shFAT4 cells, RL95-2+dCas9+NC and RL95-2+dCas9+sgRNA2 cells were injected subcutaneously into 4-week-old immunodeficient BALB/c-nu mice (Shanghai, JSJ Company). Tumour formation was measured every 3 days, and tumour volume was calculated as 1/2 × length × width2 for 4 weeks.
EC cell lines
KLE, AN3-CA, Ishikawa, HEC-1B, ECC-1, and HEC-293T were kindly provided by JieZhu professor, Obstetrics and Gynecology hospital of Fudan University. HEC-1A and RL95-2 cells were provided by Chinese Academy of Science (www.cellbank.org.cn.). The certificate of STR analysis was included for all cell lines. All cells were maintained in RPMI-1640 medium supplemented with 10% foetal bovine serum (FBS, Gibco Inc., NY, USA), 100 μg/ml penicillin/streptomycin and 2 mM L-glutamine in a humidified incubator at 37°C and 5% CO2. All tested negative for mycoplasma contamination.
Western blotting
EC cell proteins were collected and extracted with RIPA lysis buffer containing PMSF (#89900, Thermo Fisher Scientific Inc., MA, USA). For subcellular fractionation, protein was extracted using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, P0027). HaltTM Phosphatase inhibitor cocktail was added to preserve protein phosphorylation. Proteins were separated on 4% or 10% SDS-PAGE gels (Beyotime, China) and transferred to PVDF membranes (0.22 μm). Primary antibodies against the following proteins were used: FAT4 (1:1000, ab130076, Abcam), YAP (1:1000, #12395, CST), phospho-YAP (1:1000, #13008, CST), LATS1 (1:1000, #3477, CST), phospho-LATS1 (1:1000, #8654, CST), LATS2 (1:1000, #5888, CST), phospho-LATS2 (1:1000, ab111344, Abcam), cyclin D1 (1:1000, ab131475, Abcam), CDK2 (1:1000, ab32147, Abcam), cyclin A1+A2 (1:1000, ab185619, Abcam), CDK3 (1:1000, ab191503, Abcam), CDK4 (1:1000, ab108357, Abcam), E-cadherin (1:1000, #3195, CST), N-cadherin (1:1000, #13116, CST), Smad2 (1:1000, #5339, CST), phospho-Smad2 (1:1000, #9510, CST), Smad3 (1:1000, #9523, CST), phospho-Smad3 (1:1000, #9520, CST), GAPDH (1:3000, Cat: 10494-1-AP, Proteintech), histone H3 (1:1000, ab1791, Abcam), HA-tag (1:1000, #3724, CST), and 3 × Flag-tag (1:1000, #8146, CST). Western blotting bands were visualized using an ECL kit (Merck Millipore Corp, MA, USA).
Reverse transcription-quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted from EC cells and patient tissue cultures using TRIzol (Thermo Fisher Scientific Inc., MA, USA) according to the manufacturer’s protocol. The PrimeScriptTM Reagent Kit and SYBR Premix Ex TaqTM II (Takara Bio Inc., Shiga, Japan) were used to reverse transcribe mRNA to cDNA and to perform real-time PCR. The associated primers are shown in the Table 2.
Table 2.
The primer sequences in the article
| Primer | Sequence (5’ to 3’) |
|---|---|
| FAT4-F | CAA ATG CTG TGA TTG CGT AT |
| FAT4-R | AAC AGT GGC AAA GCT ACA CCT |
| GAPDH-F | GGG AAG GTG AAG GTC GGA GT |
| GAPDH-R | GGG GTC ATT GAT GGC AAC A |
| YAP-F | CAA TAG CTC AGA TCC TTT CCT |
| YAP-R | TAG TAT CAC CTG TAT CCA TCT C |
| LATS1-F | ACG AGG GAA AAC AAT AAG GG |
| LATS1-R | GAC AGC AAA AAT CCC CTG AG |
| LATS2-F | AAG AGC TAC TCG CCA TAC GCC TTT |
| LATS2-R | AGC TTT GGC CAT TTC TTG CTC CAG |
| Mst1-F | GAA CAC AGA CCT GTG GAT TG |
| Mst1-R | CGC CTT GAT ATC TCG GTG TA |
| Mst2-F | TCT CCT CAA TAC AGA AGG AC |
| Mst2-R | AGA AGT AAT GCC AAG GGA CC |
| TAZ-F | GGT GCT ACA GTG TCC CCA CAA |
| TAZ-R | TTT CTC CTG TAT CCA TCT CAT CCA |
Immunohistochemistry (IHC)
In total, 116 EC tissues, 86 paired adjacent noncancerous tissues and 34 normal tissues were formalin-fixed and paraffin-embedded. Antigen retrieval was performed at 99°C for 30 min. The sections were incubated with primary antibodies against FAT4 (1:100, HPA052819, Sigma-Aldrich), YAP (1:400, #12395, CST), Ki67 (1:200, Wuhan Servicebio Technology Co., Ltd), LATS1/2 (1:200, ab70565, Abcam), and USP51 (1:100, ab121147, Abcam). Expression was scored based on intensity (0, no staining; 1+, weak positive staining; 2+, moderate positive staining; and 3+, intense positive staining) and percentage of stained cells (0, no cells stained; 1+, <10% positive cells; 2+, 10%-50% positive cells; and 3+, >50% positive cells). The final scores for IHC images were recorded based on a 4-point scale: 1 or 2, low staining; and 3 or 4, high staining. All pathological sections were reviewed by 2 pathologists.
Colony formation assay
HEC-1A-NC, HEC-1A-shFAT4, HEC-1B-NC, HEC-1B-shFAT4, RL95-2+dCas9+NC, and RL95-2+dCas9+sgRNA1/2/3 cells were seeded in six-well plates at 600 cells/well respectively and then incubated for 10 days. The detailed steps were acquired in our previous study [25].
Immunofluorescence (IF)
EC cells were incubated overnight with an anti-FAT4 antibody (1:100, #ab130076, Abcam) in PBS at 4°C and then with an Alexa Fluor 488-conjugated donkey anti-goat secondary antibody (1:500, Abcam). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (Beyotime, China). Images were captured by confocal microscopy (Leica, Germany).
Flow cytometry (FCM)
To analyse the cell cycle, EC cells were fixed in 4% paraformaldehyde and permeabilized with 75% ethanol. After the cells were centrifuged at 150 × g for 10 min at room temperature (RT), the precipitate was resuspended in 1 ml 0.9% physiological saline and centrifuged at 150 × g for 10 min at RT. Cell cycle progression was examined after propidium iodide (PI) staining in the dark for 30 min. Cells transfected with shFAT4 or shCtrl were seeded in six-well culture plates and cultured to 90% confluence.
Plasmid construction, transfection and immunoprecipitation
HA-tagged truncated FAT4 (NM_054582) and Flag-USP51 (NM_201286) were subcloned into the pcDNA3.1 vector by LncBio Co., Shanghai, China. HEK293T cells were cultured in RPMI-1640 medium (Invitrogen) containing 10% FBS. Cells were transfected with pcDNA3.1-HA-FAT4 (small-scale tandem epitope) and pcDNA3.1-Flag-USP51 for FAT4 and USP51 binding analysis. After 72-96 h of incubation, the transfected HEK293T cells were washed three times with Modified Dulbecco’s PBS, and IP Lysis/Wash Buffer was added, as recommended in the CoIP kit instructions (Thermo Scientific). The following primers were used: USP51 plasmid sequencing primers, F: CTTGGTACCGAGCTCGGATCCGCCACCATGGCCCAGGTC and R: GAAGGGCCCTCTAGACTCGAGGTCCTTCTCCAGGCCCTGC; truncated-FAT4 sequencing primers, F: CTTGGTACCGAGCTCGGATCCGCCACCATGAACCAGTGCAGGGGGAA and R: GAAGGGCCCTCTAGACTCGAGCACATACTGTTCTGCTTCCCCA.
Screening for selected ubiquitin and deubiquitin enzymes using a PCR array experiment
An RT2 Profiler Custom PCR array was used to examine the mRNA levels of 70 genes FAT4 associated with ubiquitin and deubiquitin enzymes and five housekeeping genes in 384 well plates according to the manufacturer’s protocol (Bio TNT, Shanghai, China). Real-time PCR was performed using the RT2 SYBR Green qPCR Master Mix (SABiosciences) on an ABI 7500 Fast 96-well realtime PCR machine (Applied Biosystems, Foster City, CA, USA). Results were analyzed using the PCR Array Data Analysis Web Portal (SABiosciences). Relative mRNA levels of target genes were calculated by the 2-ΔΔCt method.
Statistical analysis
Statistical analysis was performed using IBM SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). The data are all presented as mean ± standard deviation (Sd). Student’s t-test and one-way analysis of variance were applied to assess the differences. χ2 test or Fisher exact test was used to evaluate the enumeration data. Kaplan-Meier method was used to draw the cumulative survival curves. Log rank test was used to perform the difference of the survival between two groups (FAT4-low expression and FAT4-high expression). P<0.05 was considered statistically significant.
Results
Frequent downregulation of FAT4 in human EC
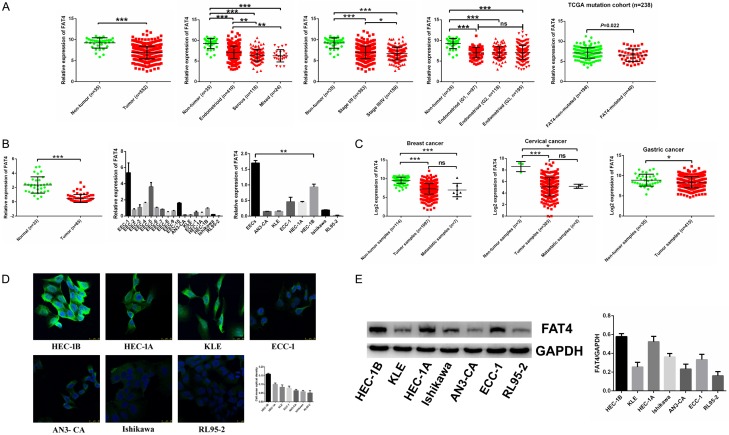
We downloaded and normalized the TCGA EC RNA-Seq data (35 non-tumor and 552 EC tumor samples) (https://cancergenome.nih.gov/). Subsequent analysis of the TCGA database confirmed that FAT4 expression was decreased in 35 non-tumour tissues and 552 tumour tissues. FAT4 mRNA expression correlated with the probability of progression to high stage (III/IV) disease and with a highly invasive tumour type (serous type vs. endometrioid type) (*P<0.05, **P<0.01, Figure 1A). We also analyzed TCGA mutation data and RNA-Sequence data in endometrial cancer and found that the FAT4 mRNA expression was relative lower in FAT4 mutated group compared with FAT4 non-mutated group (P = 0.022, Figure 1A). We used qRT-PCR to validate this initial expression observation using the Fudan University cohort consisting of 33 normal endometrial tissues and 65 EC tissues; we also extracted original normal epithelial endometrial cells from 10 samples during hysteroscopy and compared these samples with 7 EC cell lines by qRT-PCR. As expected, FAT4 was significantly downregulated in EC (P<0.001, Figure 1B). These findings suggest that downregulation of FAT4 expression is a general phenomenon in human EC. More strikingly, FAT4 expression was also significantly downregulated in multiple cancer types according to the TCGA database, which indicates that FAT4 deregulation is a common alteration in human cancer (P<0.01, Figure 1C). Immunofluorescence and Western blotting were also employed to evaluate FAT4 expression in 7 EC cell lines and revealed that FAT4 had relatively high expression in HEC-1A and HEC-1B cells and low expression in RL95-2 cells (Figure 1D and 1E).
Figure 1.
The frequent down-regulation of FAT4 in human EC in the Fudan cohort and in the TCGA database. A: FAT4 mRNA expression was significantly decreased in 552 endometrial cancer (EC) samples compared with their non-tumorous samples. FAT4 downregulated mRNA expression was associated with advanced stage (*P<0.05, t test) and histological type (**P<0.01, t test) while no association with grade. Analysis of TCGA mutation cohort, the mRNA expression of FAT4 in FAT4-mutated group was relative lower compared with FAT4-non-mutated group (P = 0.022, t test). B: FAT4 mRNA expression was significantly decreased in 65 EC samples compared with 33 normal samples in the Fudan cohort (***P<0.0001, t test). FAT4 mRNA expression was also decreased in original normal epithelial endometrial cells extracted from 10 patients during hysteroscopy compared with 7 EC cell lines (***P<0.0001, t test). C: FAT4 expression was also strikingly downregulated in multiple tumour types available in the TCGA (*P<0.05, ***P<0.0001, t test). D, E: FAT4 protein expression in 7 EC cell lines was examined by immunofluorescence and Western blot. FAT4 protein expression was relatively high in the HEC-1A and HEC-1B cell lines and was low in the RL95-2 cell line.
Clinicopathological significance of FAT4 downregulation in human EC
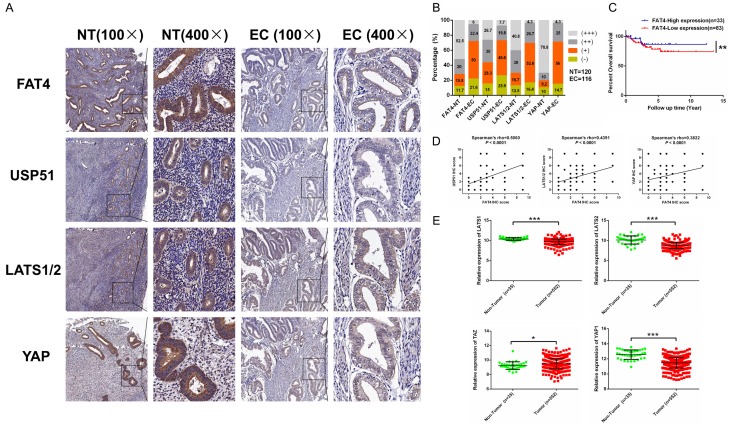
We next examined the clinicopathological significance of FAT4 protein expression in 116 EC samples, 86 adjacent noncancerous samples and 34 normal endometrial tissues by IHC (Figure 2A). High FAT4 expression was defined as a score of 2-3, while low FAT4 expression was defined as a score of 0-1. The proportion of FAT4-low EC samples was significantly higher than that of FAT4-high noncancerous samples (71.6% vs. 27.5%, P<0.01) (Figure 2B). Next, we investigated the clinical implications of FAT4 downregulation in human EC by correlating FAT4 expression changes with various clinicopathological features. We found that low FAT4 expression was significantly associated with aggressive and metastatic features of EC, such as the presence of lymph-vascular invasion and local lymph node metastasis (Table 1). Furthermore, We divided the 116 EC patients into two groups as mentioned in Table 1 and collected followed-up data. We analyzed the survival difference between two groups and found EC patients with low expression of FAT4 exhibited worse overall survival compared with patients with high expression of FAT4 (Figure 2C).
Figure 2.
FAT4 protein expression in EC tissues. A: Immunohistochemical staining of FAT4, USP51, LATS1/2 and YAP in EC tissues, noncancerous tissues and normal tissues. Original magnification, × 100 and × 400, scale bar = 100 μm and 50 μm, respectively. B: Proportions of samples negative (- or +) or positive (++ or +++) for FAT4, USP51, LATS1/2 and YAP among 116 EC tissues (EC) and 120 normal tissues (NT). C: EC patients with low expression of FAT4 exhibited worse overall survival compared with patients with high expression of FAT4 (**P<0.01, Log rank test). Kaplan-Meier method was used to draw the cumulative survival curves. D: The correlations of the scores for the four molecules, FAT4, USP51, LATS1/2 and YAP, were measured by Pearson’s coefficient. E: The core components of the Hippo signalling pathway, including LATS1, LATS2, TAZ and YAP. LATS1, LATS2 and YAP mRNA expression was downregulated in 552 tumour samples compared with 35 noncancerous samples, as revealed by the TCGA RNA-Seq analysis, while TAZ showed the opposite trend (*P<0.05, ***P<0.0001, t test).
Table 1.
Clinicopathological characteristics of Test trial patients according to FAT4 expression
| Characteristics | Patients | FAT4 expression | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| No. | % | High | Low | χ2 | P-Valuea | |
| All patients | 116 | 100 | 33 | 83 | ||
| Age (years) | 0.834 | 0.361 | ||||
| <60 | 64 | 55.2 | 16 | 48 | ||
| ≥60 | 52 | 44.8 | 17 | 35 | ||
| Tumor size (cm) | 0.478 | 0.489 | ||||
| <4 | 68 | 58.6 | 21 | 47 | ||
| ≥4 | 48 | 41.4 | 12 | 36 | ||
| Lymphovascular invasion | 7.068 | 0.008 | ||||
| Positive | 21 | 18.1 | 1 | 20 | ||
| Negative | 95 | 81.9 | 32 | 63 | ||
| Lymph node metastasis | 6.226 | 0.013 | ||||
| Metastasis | 29 | 25 | 3 | 26 | ||
| No metastasis | 87 | 75 | 30 | 57 | ||
| Histology Diagnosis | 1.117 | 0.290 | ||||
| Endometrioid type | 91 | 78.4 | 28 | 63 | ||
| Serious type or others | 25 | 21.6 | 5 | 20 | ||
| FIGO stage | 2.527 | 0.112 | ||||
| I/II | 95 | 81.9 | 30 | 65 | ||
| III/IV | 21 | 18.1 | 3 | 18 | ||
| Muscular infiltration | 6.766 | 0.009 | ||||
| <1/2 | 86 | 74.1 | 30 | 56 | ||
| ≥1/2 | 30 | 25.9 | 3 | 27 | ||
| YAP nuclear location | 12.158 | 0.000 | ||||
| Positive | 68 | 58.6 | 11 | 57 | ||
| Negative | 48 | 41.4 | 22 | 26 | ||
P-value <0.05 marked in bold font shows statistical significant.
FAT4 silencing promoted EC cell proliferation and invasion in vitro
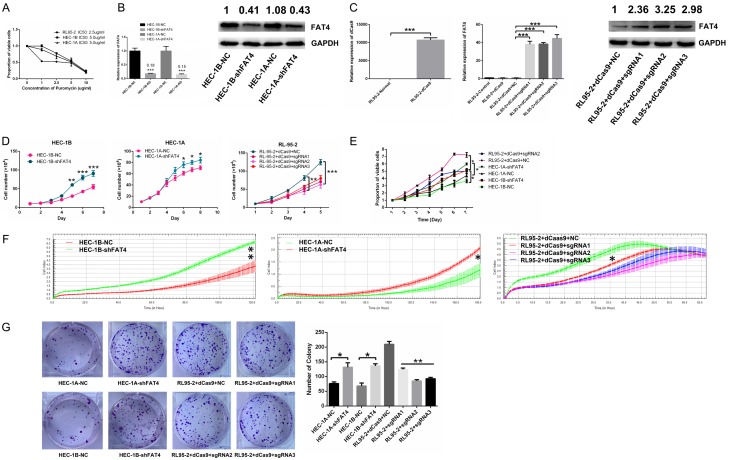
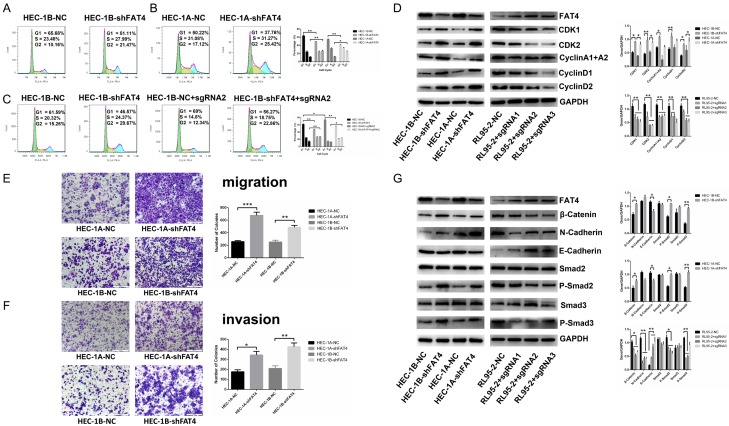
We performed Cell Counting Kit-8 (CCK-8) assays, Real-time Cell Analysis (RTCA), and colony formation to test the proliferative effects of FAT4 on EC cells growth. The results showed that FAT4 knockdown significantly prohibited the proliferation in both HEC-1A-shFAT4 and HEC-1B-shFAT4 cells, while FAT4 overexpression in RL95-2+dCas9+sgRNA cells significantly inhibited proliferation. Moreover, rescued FAT4 expression in HEC-1B-shFAT4 cells inhibited its proliferation ability (Figure 3D-G). In addition, cell cycle analysis showed that FAT4 knockdown in ECs decreased the proportion of cells in G0/G1 phase and increased the proportion of cells in S and G2/M phases (Figure 4A and 4B). Then, we transfected FAT4 sgRNA2 into HEC-1B-shFAT4 cells, which rescued proliferation (Figure 4C). Western blotting was used to examine proliferation-associated markers, including CDK1, CDK2, cyclin A1+A2, cyclin D1 and cyclin D2. These proteins were upregulated in HEC-1A-shFAT4 and HEC-1B-shFAT4 cells but downregulated in RL95-2+sgRNA cells (Figure 4D). Collectively, these results suggest that FAT4 critically regulates EC proliferation.
Figure 3.
Silencing FAT4 promoted EC proliferation, and overexpressing FAT4 suppressed EC proliferation. A: IC50 values of puromycin in HEC-1A, HEC-1B and RL95-2 cells. B: Knockdown (shFAT4) efficiency of FAT4 at the mRNA and protein levels in HEC-1A and HEC-1B cells. The data are presented mean ± SD, t-test, *P<0.05, **P<0.01. C: Efficiency of upregulating dCas9, FAT4 mRNA and FAT4 protein levels in RL95-2 cells. The data are presented as mean ± SD, t-test, ***P<0.001. D-F: Knockdown (shFAT4) of FAT4 in both the HEC-1B and HEC-1A cell lines significantly promoted EC cell proliferation compared with the negative (non-target) control (NC); upregulating (sgRNA) FAT4 in RL95-2 cells significantly inhibited EC cell proliferation compared with NC. The data are presented mean ± SD, t-test, *P<0.05, **P<0.01. G: EC cell colony formation was significantly increased by knockdown (shFAT4) of FAT4 in both HEC-1B and HEC-1A cells and significantly decreased upon upregulating (sgRNA) FAT4 in the RL95-2 cell line, The data are presented mean ± SD, t-test, *P<0.05, **P<0.01.
Figure 4.
FAT4 inhibited EC cell cycle progression, cell invasion and altered cell cycle-associated protein markers and invasion-associated protein markers in the HEC-1A, HEC-1B and RL95-2 cell lines. A, B: Flow cytometry assay showed a decrease in the percentage of cells in G1 phase and a concomitant increase in that in G2 phase, while the percentage in S phase remained stable in FAT4-silenced HEC-1A and HEC-1B cells. The data are presented as mean ± SD, t-test, *P<0.05, **P<0.01. C: Flow cytometry assay revealed that upregulating FAT4 in HEC-1B-NC cells increased the G1 phase population and that upregulating FAT4 in HEC-1B-shFAT4 cells rescued the increase in proliferation upon FAT4 silencing. The data are presented mean ± SD, t-test, *P<0.05, **P<0.01. D: Western blotting showed that CDK1, CDK2, cyclin A1+A2, cyclin D1 and cyclin D2 expression was increased in HEC-1A-shFAT4 and HEC-1B-shFAT4 cells compared with HEC-1A-NC and HEC-1B-NC cells. The expression of these markers was decreased in RL95-2-sgRNA cells with upregulated FAT4 expression compared with RL95-2 negative control cells, The data are presented as the mean ± SD, t test. *P<0.05, **P<0.01. E, F: Knockdown of FAT4 in both the HEC-1A and HEC-1B cell lines significantly promoted EC cell migration and invasion, as shown by adhesion and transwell cell migration and invasion assays. The data are reported as the average cell count in five random areas in each transwell membrane. Each sample was tested in triplicate. **P<0.01, ***P<0.001, t test. G: FAT4 modulated the expression of migration-associated markers. HEC-1A and HEC-1B cells were infected with NC or shFAT4, and the expression of β-catenin, N-cadherin, P-Smad2 and P-Smad3 was significantly decreased, E-cadherin expression increased, and Smad2 and Smad3 showed stable expression. RL95-2 cells were infected with NC or sgRNA1-3, and the expression of β-catenin, N-cadherin, P-Smad2 and P-Smad3 was significantly upregulated, E-cadherin was downregulated, and Smad2 and Smad3 were stably expressed. GAPDH served as a loading control. The data are presented as the mean ± SD, t test. *P<0.05, **P<0.01.
To examine the effect of FAT4 on EC cell invasion, Transwell cell migration and invasion assays were performed, and the results revealed that FAT4 silencing promoted HEC-1A and HEC-1B cells migration and invasion (Figure 4E and 4F). We also found that in HEC-1A-shFAT4 and HEC-1B-shFAT4 cells β-catenin and N-cadherin were up-regulated while E-cadherin, P-smad2, and P-smad3 were down-regulated compared with HEC-1A-NC and HEC-1B-NC cells, respectively. RL95-2+sgRNA cells obtained the opposite results (Figure 4G), which corresponded with our observations in the clinicopathological analysis of EC patients and highlighted the essential regulation of EC cell motility and invasion by FAT4.
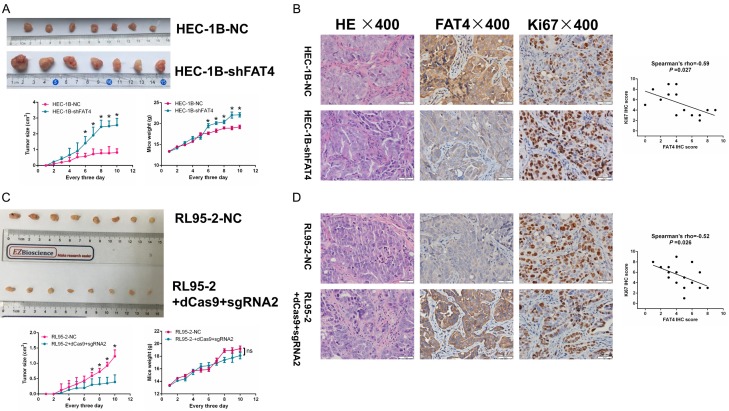
FAT4 suppressed EC tumourigenicity in vivo
To support the in vitro findings, we evaluated the tumour-promoting role of FAT4 in EC using a nude mouse models. Mice were injected subcutaneously with 1 × 106 (0.2 ml) viable HEC-1B-NC and HEC-1B-shFAT4 cells. Compared with the mice injected with HEC-1B-NC cells, those injected with HEC-1B-shFAT4 cells displayed higher rates of tumour growth. In addition, the tumours derived from the shFAT4 group were significantly larger than those derived from the NC group, whereas body weight showed the opposite trend (Figure 5A). Moreover, injection of RL95-2+dCas9+sgRNA2 cells yielded the opposite results (Figure 5C). IHC was applied to examine FAT4 expression in tumours derived from nude mice, and we confirmed that FAT4 expression was lower in HEC-1B-shFAT4 cells and was correlated negatively with Ki67 expression (Figure 5B and 5D). Taken together, the above data indicate that FAT4 functions as a tumour suppressor gene and is essential for EC cell growth.
Figure 5.
FAT4 inhibited EC carcinogenicity in vivo. A: HEC-1B cells were tranfected with either NC or shFAT4 and injected into BALB/c nude mice subcutaneously (0.2 mL, 1 × 106 cells). Tumor size and weight were monitored every 3 days. Silenced FAT4 significantly promoted EC tumor growth, the data are mean ± SD, *P<0.05, t test. C: RL95-2 cells were tranfected with either NC or sgRNA2 and injected into BALB/c nude mice subcutaneously (0.2 mL, 1 × 106 cells). Tumor size and weight were monitored every 3 days. Upregulated FAT4 significantly inhibited EC tumor growth, the data are mean ± SD, *P<0.05, t test. B and D: IHC staining for FAT4 and Ki67 in sections of transplanted tumors. *P<0.05, t test. Person’s correlation.
FAT4 repression induced dysregulation of the Hippo pathway and increased the nuclear translocation of YAP
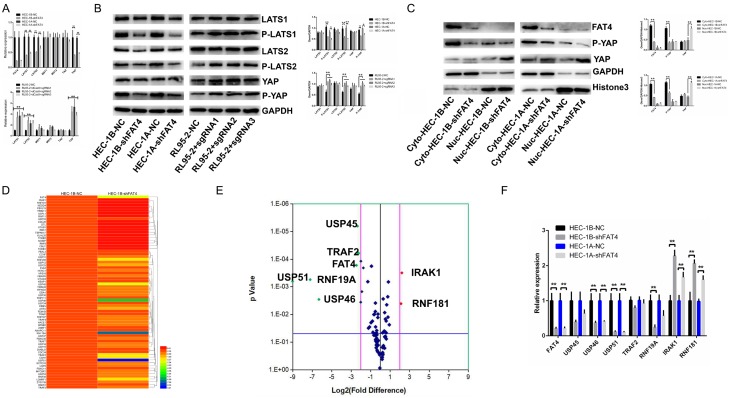
Sequence analysis revealed that the orthologue of vertebrate FAT4 is structurally most similar to ft in Drosophila, which is involved in the maintenance of the PCP and Hippo signalling pathways. However, as the underlying dysregulated pathways involved in EC remain unclear, we investigated the Hippo signalling pathway and related signalling components. The effect of FAT4 on the Hippo pathway was evaluated by measuring changes in mRNA and protein expression of critical elements of the Hippo pathway (LATS1, LATS2, MST1, MST2, YAP and TAZ). qRT-PCR analysis showed that FAT4 knockdown significantly downregulated LATS1/2 and YAP expression, while FAT4 overexpression upregulated LATS1/2, YAP and TAZ expression; in contrast, MST1 and MST2 expression remained stable (Figure 6A). Similar results were obtained at the protein level as based on Western blotting (Figure 6B). Therefore the Hippo pathway in EC cells in vitro is inhibited by FAT4 downregulation and promoted by FAT4 upregulation. Furthermore, we also found that FAT4 silencing induced the nuclear accumulation of the YAP protein which is associated with the promotion of proliferation and metastasis (Figure 6C).
Figure 6.
Altered FAT4 levels regulated the Hippo signalling pathway and the PCR-array analysis of HEC-1B-NC and HEC-1B-shFAT4 including 70 common ubiquitin and deubiquitin enzymes. A: The mRNA expression of the core modulators of the Hippo pathway (LATS1, LATS2, MST1, MST2, TAZ and YAP) in HEC-1A and HEC-1B cells infected with either NC or shFAT4 and in RL95-2 cells infected with NC or sgRNA, The data are presented as the mean ± SD, t test. *P<0.05, **P<0.01. B: LATS1, P-LATS1, LATS2, P-LATS2, YAP and P-YAP protein expression in HEC-1A and HEC-1B cells infected with either NC or shFAT4 and in RL95-2 cells infected with NC or sgRNA, The data are presented as the mean ± SD, t test. *P<0.05, **P<0.01. C: FAT4 repression increased the nuclear accumulation of YAP. Cytoplasmic P-YAP expression decreased, nuclear YAP levels increased, and cytoplasmic total YAP levels remained stable in FAT4-silenced cells compared with NC cells (HEC-1A and HEC-1B). GAPDH and histone 3 were used as loading controls. The data are presented as the mean ± SD, t test. *P<0.05, **P<0.01. D, E: Cluster analysis and volcano plot of the difference genes regulated by FAT4, red point represents upregualted genes and green point downregulated genes; F: Validating the significantly regulated genes (USP45, USP46, USP51, TRAF2, RNF19A, IRAK1 and RNF181) by FAT4 in HEC-1A and HEC-1B infected with either NC or shFAT4. The data are presented as the mean ± SD, t test. *P<0.05, **P<0.01.
Suppressing USP51 attenuated the tumour suppressor function of FAT4
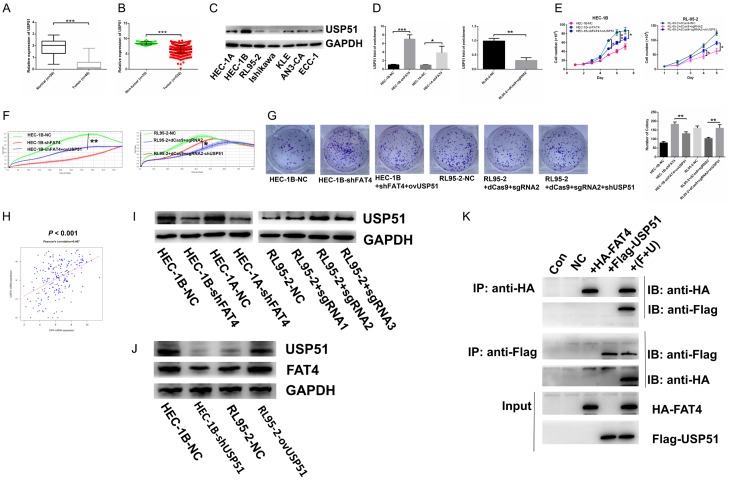
The data presented above indicated that FAT4 inhibited EC cell proliferation and invasion in a Hippo signalling pathway-dependent manner. However, low expression of FAT4 in EC remains unclear and a novel interacting partner of FAT4’s cytoplasmic tail would further allow for dissection. To identify possible interacting cellular contexts of FAT4, we performed a PCR array containing 70 common ubiquitinating and deubiquitinating enzymes associated with FAT4 based on TCGA-RNA-Seq database to investigate changes upon stable FAT4 Knockdown in HEC-1B cells. The USP45, USP46, USP51, TRAF2 and RNF19A genes were significantly down-regulated and IRAK1 and RNF181 were up-regulated (>2-fold) (Figure 6D and 6E). We next validated these genes’ changes in mRNA level (Figure 6F). We confirmed that USP51 was the most significantly altered target gene. In our EC cell models, USP51 expression was significantly downregulated by FAT4 knockdown in HEC-1A and HEC-1B cells and significantly upregulated by FAT4 overexpression in RL95-2 cells (Figure 7G).
Figure 7.
Suppression of USP51 contributed to promoting tumorigenic function of FAT4 and FAT4 directly connected with USP51 to regulate mutual expression. A: mRNA expression of USP51 downregulated in EC tissues compared with normal tissues in Fudan cohort (***P<0.001, t test); B: mRNA expression of USP51 decreased in EC tissues compared with non-cancerous tissues in TCGA database (***P<0.001, t test); C: Protein expression of USP51 in 7 EC cell lines; D: To overexpress USP51 in HEC-1B-shFAT4 and HEC-1A-shFAT4 cell lines and knockdown USP51 in RL95-2+dCas9+sgRNA2 cell line (**P<0.01, ***P<0.001, t test); E-G: Increasing USP51 rescued the promoting proliferation function of silenced-FAT4 in HEC-1B cell line while HEC-1A presented no significance. Decreasing USP51 rescued the inhibiting proliferation function of upregulated-FAT4 in RL95-2 cell line (*P<0.05, **P<0.01, One-way ANOVA); H: mRNA expression of FAT4 and USP51 association based on TCGA database (Person’s correlation = 0.467); I: Validating the protein expression of USP51 in HEC-1A and HEC-1B infected with either NC or shFAT4 and RL95-2 infected with either NC or sgRNA. GAPDH was a loading control; J: Detection the protein expression of USP51 and FAT4 in HEC-1B cells transfected either with NC or shUSP51; RL95-2 cells transfected either with NC or ovUSP51 plasmid. K: CoIP assay was used to test the direct connection between FAT4 and USP51 in HEK293T cell line.
USP51 is a possible interacting target of FAT4 in EC
Considering the role of FAT4 in EC cells, we aimed to determine whether USP51 functions as a tumour suppressor DUB. Silencing USP51 increased the proliferation rate of both HEC-1A and HEC-1B cells, which conflicts with a previous report [26]. According to the TCGA database, USP51 expression was downregulated in tumour tissue compared with its expression in noncancerous tissue, which corresponded with our Fudan cohort data (Figure 7A and 7B). We also analysed USP51 protein levels in EC tissues and normal tissues by IHC and found that they were downregulated in tumour samples compared with normal samples (Figure 2A). In HEC-1A-shFAT4 and HEC-1B-shFAT4 cells, we rescued USP51 expression using the USP51 overexpression plasmid, while in RL95-2+sgRNA2 cells, we downregulated USP51 expression using shUSP51 (Figure 7D). Cell proliferation was rescued in HEC-1B and RL95-2 cells (Figure 7E-G). In addition, a positive correlation was observed between USP51 and FAT4 in the TCGA cohort and in our own EC samples (Figures 7H and 2D). Furthermore, in HEC-1B cells we decreased USP51 expression by shUSP51 plasmid and in RL95-2 cells we increased USP51 expression by ovUSP51 plasmid and found that FAT4 expression downregulated in HEC-1B-shUSP51 cells and upregulated in RL95-2-ovUSP51 cells (Figure 7J). To investigate whether FAT4 directly interact with USP51, we performed an in vitro binding assay. We transfected HA-Flag-truncated FAT4 alone or with Flag-USP51 into HEK293T cells and found that purified HA-FAT4 bound to purified Flag-USP51, which demonstrates a direct interaction between FAT4 and USP51 (Figure 7K). These results collectively suggest that USP51 is a direct interacting partner of FAT4 and that USP51 suppression significantly contributes to the tumour suppressor function of FAT4 in human EC.
Discussion
In this study, frequent downregulation of FAT4 was a common characteristic of EC compared with normal samples that was validated in the TCGA EC samples and in our Fudan sample cohort. According to the TCGA database, FAT4 expression decreased as the EC stage progressed and in serous-type EC compared with endometrioid-type EC, revealing that FAT4 downregulation might facilitate EC progression. Clinicopathological analysis in our Fudan cohort showed that low FAT4 expression was significantly associated with aggressive features of EC, such as advanced lymphovascular invasion, lymph node metastasis and muscular infiltration, showing FAT4’s suppressive impact in EC carcinogenesis and progression.
We further investigated the molecular mechanisms responsible. Previous studies have shown that FAT4 may be involved in the Hippo signalling pathway and planar cell polarity (PCP) in vitro and in vivo [27]. Ma et al. [24] found that FAT4 silencing in gastric cancer cells induced nuclear accumulation of YAP, promoting proliferation and migration. In human triple-negative breast cancer [28], FAT4 was found to inhibit breast cancer cell proliferation, migration and invasion. There are several explanations for the suppression of FAT4. Low FAT4 expression in gastric cancer is associated with mutations, and promoter methylation is also involved [29,30]. In breast cancer cells, transient activation of the Src oncoprotein repressed FAT4 mRNA expression through actin depolymerization [29,30]. In our previous study, our exon sequencing panel revealed that FAT4 was frequently mutated in EC (41%, 12/29). In addition, according to the TCGA mutation cohort, we found that the FAT4 mRNA expression was relative lower in FAT4 mutated group compared with FAT4 non-mutated group in EC (Figure 1A). Thus, mutation may be one factor involving suppression of FAT4 expression. In the present study, we examined the deregulation of the Hippo signalling pathway in HEC-1A and HEC-1B cells after FAT4 silencing and in RL95-2 cells upon FAT4 overexpression. The results revealed that FAT4 silencing downregulated the Hippo core components p-LATS1/2 and p-YAP while LATS1/2 and YAP type remained unchanged. Furthermore, cellular fraction experiments showed that FAT4 silencing could increase the nuclear accumulation of YAP in the shFAT4 cells while decrease the nuclear YAP accumulation in the FAT4-overexpression RL95-2 cells, which might explain why FAT4 exhibites the function of inhibiting cell proliferation and invasion.
Ectopic FAT4 cadherin expression is implicated in various cancer types. The regulation of FAT4 and its downstream signalling pathways remain incompletely understood. Previous study demonstrated that SH3RF1, E3 ubiquitin-protein ligase, as a new regulator of FAT1 (one FAT family member) protein levels [32]. Therefore, we speculated that the FAT4 cytoplasmic tail could delineate its regulation and function though binding a serious ubiquitinating and deubiquitinating enzymes. Thus we designed a PCR array including 70 ubiquitinating and deubiquitinating enzymes which had strong correlations with FAT4 based on the TCGA database. We found three deubiquitinating enzymes (USP45, USP46, and USP51) and three ubiquitinating enzymes (RNF19A, IRAK1 and RNF181) were differentially expressed between the HEC-1B-NC and HEC-1B-shFAT4 groups. Among which, USP51 was the most downregulated gene after FAT4 silencing. USP51, a member of ubiquitin-specific protease, exert different functions in different tumour types. In human breast cancer, USP51 promotes the deubiquitination and stabilization of ZEB1, an EMT-inducing transcription factor that plays an essential role in cancer cell metabolism, metastasis and chemotherapy resistance [33,34]. In addition, Jing Zou et al. [35] found that USP51 was downregulated in the SKOV3/DDP and A2780/DDP drug-resistant cell lines. In our study, the expression of USP51 was decreased in EC tissues and positively correlated with FAT4 expression, suggesting USP51 acting as a tumour suppressor in EC. We speculated that USP51 is an interacting partner of FAT4 since USP51 re-expression in HEC-1B-shFAT4 cells effectively rescued cell proliferation and invasion. CoIP confirmed that FAT4 binded directly to USP51. Taken together, our results suggest that FAT4 silencing is positively regulated by USP51, downregulating p-LATS1/2 expression and thus inducing the nuclear accumulation of YAP and ultimately contributing to EC proliferation and invasion.
In summary, FAT4 inactivation accelerated EC cell line proliferation and invasion both in vitro and in vivo. FAT4 exerted its tumour suppressor function partly by downregulating expression of the deubiquitinating enzyme USP51, which in return could promote deubiquitination and stabilization of FAT4. Inactivation of FAT4 led to the decreased p-LATS1/2 and increased nuclear translocation of YAP, which bound various nuclear transcription factors to promote cancer proliferation and metastasis. Thus, targeting FAT4 may be a novel strategy for EC treatment.
Acknowledgements
This work was supported by grants from Science and Technology Commission of Shanghai Municipality (grant number 16411953500), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant, the Shanghai Committee on Science and Technology (Key project 164-11953500), and the National Natural Science Foundation of China (81572836).
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–7. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 4.Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, Wolk A, Wentzensen N, Weiss NS, Webb PM, van den Brandt PA, van de Vijver K, Thompson PJ, Strom BL, Spurdle AB, Soslow RA, Shu XO, Schairer C, Sacerdote C, Rohan TE, Robien K, Risch HA, Ricceri F, Rebbeck TR, Rastogi R, Prescott J, Polidoro S, Park Y, Olson SH, Moysich KB, Miller AB, McCullough ML, Matsuno RK, Magliocco AM, Lurie G, Lu L, Lissowska J, Liang X, Lacey JJ, Kolonel LN, Henderson BE, Hankinson SE, Hakansson N, Goodman MT, Gaudet MM, Garcia-Closas M, Friedenreich CM, Freudenheim JL, Doherty J, De Vivo I, Courneya KS, Cook LS, Chen C, Cerhan JR, Cai H, Brinton LA, Bernstein L, Anderson KE, Anton-Culver H, Schouten LJ, Horn-Ross PL. Type I and II endometrial cancers: have they different risk factors? J Clin Oncol. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoeng JC, Ivanov NV, Hodor P, Xia M, Wei N, Blevins R, Gerhold D, Borodovsky M, Liu Y. Identification of new human cadherin genes using a combination of protein motif search and gene finding methods. J Mol Biol. 2004;337:307–317. doi: 10.1016/j.jmb.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 6.Katoh M. Function and cancer genomics of FAT family genes (review) Int J Oncol. 2012;41:1913–8. doi: 10.3892/ijo.2012.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanoue T, Takeichi M. New insights into fat cadherins. J Cell Sci. 2005;118:2347–2353. doi: 10.1242/jcs.02398. [DOI] [PubMed] [Google Scholar]
- 8.Alders M, Al-Gazali L, Cordeiro I, Dallapiccola B, Garavelli L, Tuysuz B, Salehi F, Haagmans MA, Mook OR, Majoie CB, Mannens MM, Hennekam RC. Hennekam syndrome can be caused by FAT4 mutations and be allelic to Van Maldergem syndrome. Hum Genet. 2014;133:1161–1167. doi: 10.1007/s00439-014-1456-y. [DOI] [PubMed] [Google Scholar]
- 9.Mao Y, Francis-West P, Irvine KD. Fat4/Dchs1 signaling between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development. 2015;142:2574–2585. doi: 10.1242/dev.122630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, Michielin O, Muehlethaler K, Speiser D, Beckmann JS, Xenarios I, Halazonetis TD, Jongeneel CV, Stevenson BJ, Antonarakis SE. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2011;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 11.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Trevino L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 14.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF (beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mesrouze Y, Bokhovchuk F, Meyerhofer M, Fontana P, Zimmermann C, Martin T, Delaunay C, Erdmann D, Schmelzle T, Chene P. Dissection of the interaction between the intrinsically disordered YAP protein and the transcription factor TEAD. Elife. 2017:6. doi: 10.7554/eLife.25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 20.Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 22.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of Expanded in the Hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 24.Ma L, Cui J, Xi H, Bian S, Wei B, Chen L. Fat4 suppression induces Yap translocation accounting for the promoted proliferation and migration of gastric cancer cells. Cancer Biol Ther. 2016;17:36–47. doi: 10.1080/15384047.2015.1108488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Che X, Yan H, Sun H, Dongol S, Wang Y, Lv Q, Jiang J. Grifolin induces autophagic cell death by inhibiting the Akt/mTOR/S6K pathway in human ovarian cancer cells. Oncol Rep. 2016;36:1041–1047. doi: 10.3892/or.2016.4840. [DOI] [PubMed] [Google Scholar]
- 26.Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, McIvor E, Li W, Zhang Y, Florens L, Byrum SD, Mackintosh SG, Calhoun-Davis T, Koutelou E, Wang L, Tang DG, Tackett AJ, Washburn MP, Workman JL, Dent SY. ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell. 2016;62:558–571. doi: 10.1016/j.molcel.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai J, Feng D, Hu L, Chen H, Yang G, Cai Q, Gao C, Wei D. FAT4 functions as a tumour suppressor in gastric cancer by modulating Wnt/beta-catenin signalling. Br J Cancer. 2015;113:1720–1729. doi: 10.1038/bjc.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou L, Chen M, Zhao X, Li J, Deng S, Hu J, Yang H, Jiang J. FAT4 functions as a tumor suppressor in triple-negative breast cancer. Tumour Biol. 2016 doi: 10.1007/s13277-016-5421-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Pilehchian LM, Nikbakhsh N, Samadani AA, Fattahi S, Taheri H, Shafaei S, Amirbozorgi G, Pilehchian LR, Akhavan-Niaki H. FAT4 hypermethylation and grade dependent downregulation in gastric adenocarcinoma. J Cell Commun Signal. 2017;11:69–75. doi: 10.1007/s12079-016-0355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida S, Yamashita S, Niwa T, Mori A, Ito S, Ichinose M, Ushijima T. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer. 2017;20:136–145. doi: 10.1007/s10120-016-0593-5. [DOI] [PubMed] [Google Scholar]
- 31.Ito T, Taniguchi H, Fukagai K, Okamuro S, Kobayashi A. Inhibitory mechanism of FAT4 gene expression in response to actin dynamics during Src-induced carcinogenesis. PLoS One. 2015;10:e118336. doi: 10.1371/journal.pone.0118336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bock CE, Hughes MR, Snyder K, Alley S, Sadeqzadeh E, Dun MD, McNagny KM, Molloy TJ, Hondermarck H, Thorne RF. Protein interaction screening identifies SH3RF1 as a new regulator of FAT1 protein levels. FEBS Lett. 2017;591:667–678. doi: 10.1002/1873-3468.12569. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z, Zhang P, Hu X, Kim J, Yao F, Xiao Z, Zeng L, Chang L, Sun Y, Ma L. USP51 promotes deubiquitination and stabilization of ZEB1. Am J Cancer Res. 2017;7:2020–2031. [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou J, Yin F, Wang Q, Zhang W, Li L. Analysis of microarray-identified genes and microRNAs associated with drug resistance in ovarian cancer. Int J Clin Exp Pathol. 2015;8:6847–6858. [PMC free article] [PubMed] [Google Scholar]