Abstract
Increasing evidences demonstrate that long noncoding RNAs (lncRNAs) play an important role in the tumorigenesis of hepatocellular carcinoma (HCC). LncRNA RMRP (RNA component of mitochondrial RNA processing endoribonuclease) has been proved to involve in the tumorigenesis of several human cancers. However, the role and molecular mechanism of RMRP in HCC remain largely unknown. In current study, we compared the expression profiles of RMRP in 52 paired HCC specimens and corresponding adjacent non-tumor tissues. The clinicopathological characteristics of HCC patients in relation to RMRP expression were analyzed, and the prognostic value of RMRP expression was examined. RNAi technology was performed to knockdown RMRP expression in HCC cells, and the effects of RMRP on cell proliferation, cell cycle, migration and invasion of HCC cells were assessed by using MTT, flow cytometry, wound scratch and transwell invasion assays. The potential binding site of RMRP to miR-613 was confirmed by using dual luciferase reporter gene and qRT-PCR assays. Here, we found that RMRP expression was significantly upregulated in the HCC tissues and cells, and high RMRP expression was positively correlated with tumor aggressive phenotypes and poor overall survival of patients with HCC. Furthermore, RMRP knockdown suppressed cell proliferation, migration, invasion and induced cell cycle arrest at G0/G1 phase. The expression of miR-613 was dramatically down-regulated in HCC tissues and cells, and RMRP could negatively regulate miR-613 expression by acting as a ceRNA. In mechanism, RMRP exerted an oncogenic role in HCC via downregulation of miR-613. In vivo experiment also revealed that RMRP knockdown inhibited HCC tumorigenesis, while miR-613 silencing could partly reversed the inhibitory effect of RMRP knockdown on HCC tumorigenesis. In conclusion, our data demonstrated that RMRP plays an oncogenic role in regulating HCC tumorigenesis by acting as a ceRNA of miR-613, indicating RMRP/miR-613 axis may serve as a novel molecular target for the treatment of HCC.
Keywords: HCC, RMRP, ceRNA, miR-613, tumorigenesis
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common and aggressive malignancy, and is the third leading cause of cancer-related death worldwide [1,2]. Emerging evidences have reported that the morbidity of HCC is mainly caused by infection of hepatitis B/C virus, alcohol addiction, liver cirrhosis and environmental pollution [3,4]. Despite curative improvements have made in HCC diagnostic and therapeutic approaches recent years, the 5-year survival rate of HCC patients still remains poor due to tumour invasion, metastasis and high frequency of recurrence [5]. Although great advances in uncovering many aberrantly expressed genes and altered pathway signals involved in the carcinogenesis of HCC over the past decades, the precise molecular mechanisms for HCC tumorigenesis are not entirely clear [6]. Therefore, it is crucial and urgent to unravel novel regulating mechanisms for hepatocarcinogenesis, and to provide new strategies for the diagnosis and treatment of HCC.
Long non-coding RNAs (lncRNAs) are a class of non-protein coding RNA transcripts with longer than 200 bp nucleotides that modulate gene expression at post-transcriptional and/or transcriptional levels, including chromatin remodeling, epigenetic modification, histone modification and sponge effects [7,8]. LncRNAs occupy a large proportion of transcriptomes in eukaryotes, and they have been shown to emerge as essential regulators in a diverse range of cellular processes, such as development, differentiation, metastasis and apoptosis as well as tumorigenesis [9]. Mounting evidences have indicated that dysregulation of lncRNAs can serve as oncogenes or tumour suppressor genes involving in tumour initiation, proliferation and metastasis [10]. For instance, lncRNA XIST (X inactive specific transcript) regulates PTEN (Phosphatase and tensin homolog) expression by sponging miR-181a and promotes the progression of HCC [11]. LncRNA HOTAIR (HOX transcript antisense RNA) promotes tumour migration and invasion by regulating MKL1 via inhibition miR-206 expression in cervical cancer [12]. LncRNA MALAT1 (Metastasis associated lung adenocarcinoma transcript 1) promotes tumour growth and metastasis by targeting miR-124/foxq1 axis in bladder transitional cell carcinoma [13]. Although large numbers of lncRNAs have been annotated, the roles and molecular regulatory mechanisms of lncRNAs in HCC tumorigenesis still require further investigation.
The RNA component of mitochondrial RNA processing endoribonuclease (RMRP), a new lncRNA, was first identified in cartilage-hair hypoplasia (CHH) [14]. RMRP is primarily located in the nucleus, nucleolus and mitochondria, and is highly expressed in a wide range of human tissues and is essential for early embryonic development in murine [15]. RMRP can help endonuclease to cleave mitochondrial RNA at a priming site of mitochondrial DNA replication, and can interact with the TERT (telomerase-associated reverse transcriptase) catalytic subunit to form a complex and produce dsRNAs (double-stranded RNAs) involving in small interfering RNA [16]. In recent years, RMRP has been proved to exhibit dysregulated expression in lung adenocarcinoma and gastric cancer, and ectopic expression of RMRP promotes tumorigenesis by acting as a miR-206 sponge [17,18]. However, the biological role and molecular mechanism of RMRP in HCC remain largely unknown.
In the current study, we found RMRP was highly expressed in HCC tissues and cells, and miR-613 was down-regulated in HCC tissues and cells. Upregulated RMRP and downregulated miR-613 expression were correlated with tumor aggressive phenotypes and poor overall survival of patients with HCC. RMRP knockdown inhibited cell proliferation, invasion, and migration of HCC cells in vitro, and suppressed HCC tumorigenesis in vivo. In mechanism, RMRP acted as a miRNA sponge of miR-613, and exerted an oncogenic role in HCC tumorigenesis via downregulation of miR-613. In summary, this study showed a new axis of RMRP/miR-613 that promotes HCC tumorigenesis by impacting oncogenic phenotypes and may serve as a novel molecular target for HCC treatment.
Materials and methods
Patient samples
52 pairs of HCC tissues and matched adjacent tissues (more than 5 cm laterally from the edge of the cancerous region) were collected from patients with clinically and pathologically confirmed HCC between June 2006 and December 2012 from Department of Hepatobiliary Surgery, Hunan Provincial People’s Hospital (Changsha, China). None of patients received chemotherapy, radiotherapy and percutaneous ablation before operation. Medical records of clinicopathological characteristics in HCC patients, and all patients after surgery were followed up for 60 months. The clinicopathological characteristics in patients with HCC were shown in Table 1. The study was obtained with written informed consent of all patient, and was approved by Ethics Committee of The First Affiliated Hospital of Hunan Normal University. All tissues were immediately immersed in RNAlater (Qiagen, Hilden, Germany) for 30 min and then stored at -80°C until RNA extraction.
Table 1.
Association of RMRP expression with clinicopathological features of HCC patients
| Features | Number of cases (n = 52) | Low RMRP expression (n = 23) | High RMRP expression (n = 29) | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 41 | 16 | 25 | 0.182 |
| Female | 11 | 7 | 4 | |
| Age (y) | ||||
| < 60 | 24 | 12 | 12 | 0.438 |
| ≥ 60 | 28 | 11 | 17 | |
| Tumor size (cm) | ||||
| < 5 | 30 | 14 | 16 | 0.680 |
| ≥ 5 | 22 | 9 | 13 | |
| Liver cirrhosis | ||||
| Without | 7 | 2 | 5 | 0.444 |
| With | 45 | 21 | 24 | |
| AFP levels (ng/ml) | ||||
| < 20 | 13 | 5 | 8 | 0.629 |
| ≥ 20 | 39 | 18 | 21 | |
| TNM stage | ||||
| I + II | 26 | 16 | 10 | 0.012* |
| III + IV | 26 | 7 | 19 | |
| Venous invasion | ||||
| Absent | 34 | 20 | 14 | 0.007** |
| Present | 18 | 3 | 15 | |
| Lymph node metastasis | ||||
| No | 31 | 18 | 13 | 0.015* |
| Yes | 21 | 5 | 16 |
HCC: hepatocellular carcinoma;
P < 0.05;
P < 0.01.
Cell culture and transfection
Two HCC cell lines (Hep3B and HCCLM3) and one normal liver cell line (HL-7702) were obtained from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, Rockford, IL, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Rockford, IL, USA), 100 U/ml penicillin and 100 μg/ml streptomycin in a humidified chamber with 95% air and 5% CO2 at 37°C.
The small interfering RNA (siRNA) specifically targeting RMRP (siRNA-RMRP) and siRNA-control were synthesized from GenePharma Co., Ltd (Shanghai, China). The sequences were listed as follows: siRNA-RMRP: 5’-CCUAGGCUACACACUGAGGACUTT-3’, siRNA-control: 5’-UUCUCCGAACGUGUCACGUTT-3’. miR-613 mimic, the corresponding mimic control (mimic control), miR-613 inhibitor and the corresponding inhibitor control (inhibitor control) were obtained from RiboBio Co., Ltd (Guangzhou, China). The sequences were shown as follows: miR-613 mimic: 5’-AGGAAUGUUCCUUCUUUGCC-3’; mimic control: 5’-UUCUCCGAACGUGUCACGUTT-3’; miR-613 inhibitor: 5’-GGCAAAGAAGGAACAUUCCT-3’ and inhibitor control: 5’-AUCCGUAGGCGUUAGCCUAU-3’. For cell transfection, Hep3B and HCCLM3 cells were dissociated into single cells and plated in 6-well plates at a density of 6 × 105 cells/well, and were transfected with siRNA-RMRP or siRNA-control, and miR-613 inhibitor or inhibitor control by using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Rockford, IL, USA) and Opti-MEM® I Reduced Serum Medium (Thermo Fisher Scientific, Rockford, IL, USA) in accordance with the manufacturer’s instructions, respectively. At 48 h post-transfection, Hep3B and HCCLM3 cells were harvested for further analysis.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from the prepared tissues and cells by using TRIzol reagent (Thermo Fisher Scientific, Rockford, IL, USA). NanoDrop 2000/2000c (Thermo Fisher Scientific, Rockford, IL, USA) was utilized to quantify the density of each RNA. Reverse transcription (RT) was conducted by using a PrimeScript RT reagent Kit (TaKaRa, Dalian, China) following to manufacturer’s information. qRT-PCR was conducted to determine the RMRP and miR-613 expression by using a GoTaq® qPCR Master Mix (Promega, Madison, WI, USA) on the ABI PRISM® 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The PCR primers for RMRP, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), miR-613 and U6 were as follows: RMRP-F, 5’-ACTCCAAAGTCCGCCAAGA-3’ and RMRP-R, 5’-TGCGTAACTAGAGGGAGCTGAC-3’; GAPDH-F, 5’-GATTCCACCCATGHCCAAATTC-3’ and GAPDH-R, 5’-CTGGAAGATGGTGATGGGATT-3’; miR-613-F, 5’-GTGAGTGCGTTTCCAAGTGT-3’ and miR-613-R, 5’-TGAGTGGCAAAGAAGGAACAT-3’; U6-F, 5’-CTCGCTTCGGCAGCACA-3’ and U6-R, 5’-ACGCTTCACGAATTTGCGT-3’. Their relative expression levels of RMRP and miR-613 were calculated by using the 2-ΔΔCt method with GAPDH and U6 as the control, respectively.
Cell proliferation assay
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to determine the cell proliferation. 24 h after transfection, Hep3B and HCCLM3 cells were dissociated into single cells and plated in plated in 96-well plates at a density of 6000 cells/well for 0, 24, 48 and 72 hours. At the indicated time points, 15 μl MTT (5 mg/ml; Sigma, St. Louis, MO, USA) was added to each well and incubated for another 4 h in a humidified chamber at 37°C. Then, the medium was discarded, and 200 μl DMSO (Sigma, St. Louis, MO, USA) was added into each well to dissolve formazan crystals. The absorbance at the 540 nm was determined by using a microplate reader (MultiskanEX, Lab systems, Helsinki, Finland).
Cell cycle analysis
Flow cytometry (FACS) was used for cell cycle detection. The transfected Hep3B and HCCLM3 cells were seeded in 6 well plates with 6 × 105 cell/well. Then, these cells were collected at 48 h and washed twice with 1 × PBS, and resuspended in 2 ml 75% ethanol at -20°C for 60 min. After that, cells were stained with 2 μl PI (Propidium iodide) by using a BD Cycletest™ Plus DNA Reagent Kit (Franklin Lakes, NJ, USA) at 37°C for 30 min. Finally, the stained cells were analyzed by using a BD FACSCalibur™ Flow Cytometer (Franklin Lakes, NJ, USA) followed as the manufacture’s protocols. The percentage of the cells in G0/G1, S, and G2/M phase was analyzed.
Cell migration assay
Wound scratch assay was used to assess the migratory ability of HCC cells. Approximately 6 × 105 cell/well transfected Hep3B and HCCLM3 cells were seed in 6 well plates. After 24 h of cell culture, vertical and horizontal wounds were made by using 10 μl sterile pipette tips, and the cells were rinsed with PBS and cultured in a humidified chamber at 37°C for another 24 h. Cell migration was observed by using an inverted microscope (Leica Microsystems, Tokyo, Japan) and images were captured with a digital camera system at 0 and 24 h after the wounds were made.
Cell invasion assay
To assess cell invasive ability, transfected HCC cells were seeded on Matrigel-coated transwell inserts with 500 μl of serum-free DMEM medium. The lower chamber was filled with 500 μl of DMEM medium containing 10% FBS. After incubation for 24 h in a humidified chamber at 37°C, the cells inside the upper chamber were obliterated with cotton swabs, and the cells on the lower membrane surface were fixed with 100% methanol for 30 min at 37°C and then stained with 0.5% crystal violet solution for 20 min at 37°C. The invade cells were photographed and counted by using an inverted microscope (Leica Microsystems, Tokyo, Japan).
Dual luciferase reporter gene assay
To corroborate whether RMRP directly binds to miR-613, Hep3B and HCCLM3 cells were chosen for dual luciferase reporter gene assay. The wild-type (WT) RMRP transcript contained binding site of miR-613 was subcloned into pGL3 basic vector (Promega, Madison, WI, USA) to produce WT pGL3-RMRP reporter plasmid. Similarly, the mutant (MUT) RMRP transcript contained no binding site of miR-613 was also inserted into pGL3 vector to produce MUT pGL3-RMRP reporter plasmid. pRL-TK vector was transfected and used as an internal control plasmid. The Hep3B and HCCLM3 cells were seeded in 6 well plates with 6 × 105 cell/well. After 24 h of incubation, the cells were co-transfected with the WT or MUT pGL3-RMRP reporter plasmid, pRL-TK plasmid, and miR-613 mimic or mimic control. At 24 h post-transfection, the luciferase activities were detected by using a Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) in accordance with the manufacturer’s protocols.
Xenograft assays
Six-week-old male athymic Balb/c nude mice (weighing 20-25 g) were obtained from Model Animal Research Center of Nanjing University (Nanjing, China). The mice were housed in specific pathogen-free (SPF) conditions and randomly assigned to the siRNA-RMRP, siRNA-control, siRNA-RMRP + miR-613 inhibitor, and siRNA-RMRP + inhibitor control groups (ten mice in each group). The animal study protocol was approved by the Institutional Animal Care and Use Committee of The First Affiliated Hospital of Hunan Normal University.
For in vivo tumorigenicity, A total of 6 × 106 transfected Hep3B cells were subcutaneously injected into the left flanks of male nude mice. Tumor volumes were detected every 5 days, and was calculated based on the formula: 0.5 × length × width2. All mice were sacrificed after 35 days, and the tumors were surgically removed, weighed and stored in liquid nitrogen for further experiments.
Statistical analysis
Data were exhibited as mean ± SD (Standard Deviation) of three independent experiments, and statistical analyses were processed by using SPSS 17.0 statistical software (SPSS Inc. Chicago, IL, USA). Survival curve was calculated by using Kaplan-Meier method and was assessed by using log-rank test. For comparisons, one-way analysis of variance, chi-squared test, and two-tailed Student’s t-test were performed, as appropriate. P < 0.05 was regarded as statistically significant.
Results
RMRP is frequently up-regulated in HCC tissues and cell lines
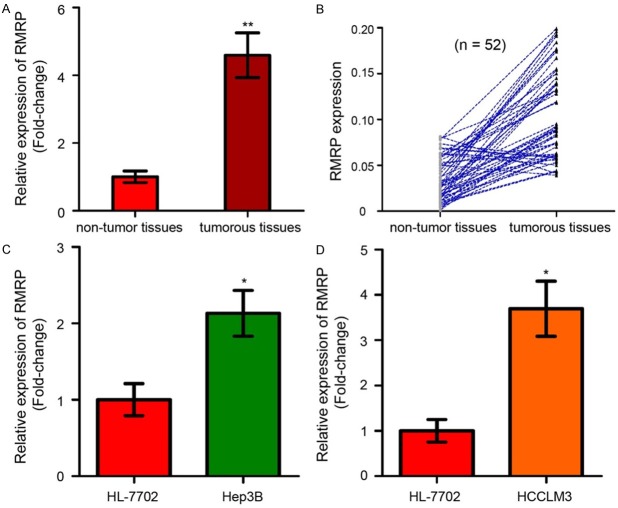
To analyze the physiological expression of RMRP, the levels of RMRP in 52 paired HCC tissues and corresponding adjacent non-tumor tissues were examined by using qRT-PCR assay. As shown in Figure 1A, RMRP expression were significantly higher in tumorous tissues than that in the adjacent non-tumor tissues (P < 0.01). Of the 52 HCC patients, RMRP was up-regulated in 45 HCC tissues (45/52, 86.54%) compared with adjacent normal tissues (Figure 1B). Moreover, the expression of RMRP was up-regulated in Hep3B and HCCLM3 cells compared with that in HL-7702 cells (Figure 1C and 1D, P < 0.05).
Figure 1.
LncRNA RMRP is frequently up-regulated in HCC tissues and cell lines. A. The expression levels of RMRP in tumorous tissues and non-tumor tissues were determined by qRT-PCR analysis and normalized with the control U6. RMRP was higher expressed in tumorous tissues than non-tumor tissues. B. Of the 52 HCC patients, RMRP was up-regulated in 45 HCC tissues (45/52, 86.54%) compared with adjacent normal tissues. C. RMRP was dramatically upregulated in Hep3B cell line compared with a normal liver cell line, HL-7702. D. RMRP expression was increased in HCCLM3 cell line compared to HL-7702 cell line. LncRNA, long non-coding RNA; qRT-PCR, quantitative real-time PCR; RMRP: RNA component of mitochondrial RNA processing endoribonuclease; HCC: hepatocellular carcinoma. Mean of three independent experiments with SD (Standard Deviation) were shown. * and ** represents P value < 0.05 and < 0.01, respectively.
miR-613 is significantly lowly expressed in HCC tissues and cell lines
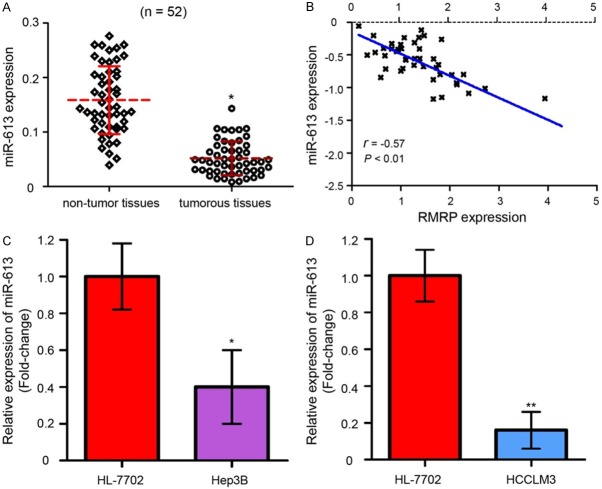
The expression of miR-613 in HCC specimens was also determined. Downregulated miR-613 were found in HCC tissues when compared with adjacent non-tumor tissues as detected by qRT-PCR assay (Figure 2A, P < 0.05). Furthermore, we indicated that the level of miR-613 was reversely related with the RMRP expression in HCC specimens (Figure 2B, P < 0.01). In addition, the expression of miR-613 at cellular levels were detected in a normal liver cell line, HL-7702, and two HCC cell lines, Hep3B and HCCLM3. As the data showed in Figure 2C and 2D, decreased miR-613 expression were presented in the Hep3B and HCCLM3 cells as comparing with HL-7702 cells (P < 0.05).
Figure 2.
miR-613 is significantly lowly expressed in HCC tissues and cell lines. A. qRT-PCR assay showed that miR-613 expression was significantly down-regulated in HCC tissues compared with adjacent normal tissues. B. The level of miR-613 was negatively related with RMRP expression in HCC samples. C. miR-613 expression was lower in Hep3B cells compared with HL-7702 cells. D. miR-613 was lower expressed in HCCLM3 cells compared to HL-7702 cells. The experiments were performed in triplicate. miR: microRNA. * and ** represents P value < 0.05 and < 0.01, respectively.
Upregulated RMRP and downregulated miR-613 are correlated with the tumor aggressive phenotypes and poor prognosis of HCC patients
Subsequently, the 52 HCC patients were subdivided into low RMRP expression group (n = 23) and high RMRP expression group (n = 29) according to the median expression of RMRP. We further investigated the associations between RMRP expression and clinicopathological characteristics in patients with HCC. As shown in Table 1, high RMRP expression was significantly correlated with lymph node metastasis (P = 0.015), TNM stage (P = 0.012) and venous invasion (P = 0.007). However, up-regulation of RMRP was not correlated with other factors, including gender, age, liver cirrhosis, tumor size and AFP levels in patients with HCC. Furthermore, Kaplan-Meier survival analysis indicated that HCC patients with higher levels of RMRP had a shorter overall survival rates compared with those patients with lower RMRP expression (Figure 3A, P < 0.05). Simultaneously, we found that low miR-613 expression was significantly correlated with TNM stage (P = 0.023) and lymph node metastasis (P = 0.004, Table 2). HCC patients with lower levels of miR-613 had a worse overall survival rates compared with those patients with higher miR-613 expression (Figure 3B, P < 0.05). In a word, high RMRP and low miR-613 expression are correlated with tumor malignances and poor overall survival of patients with HCC.
Figure 3.
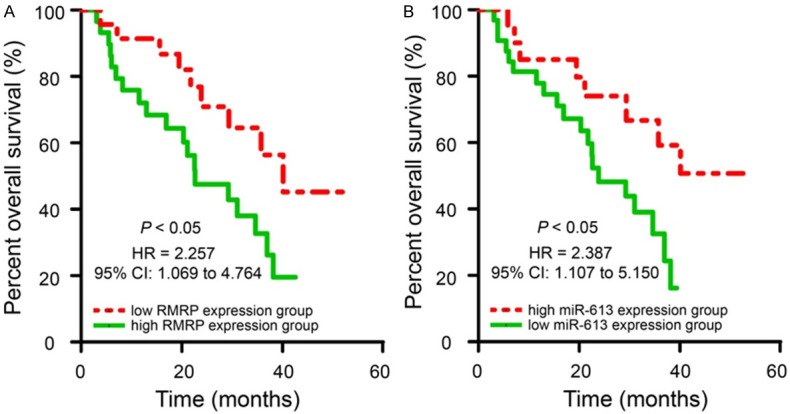
Upregulated RMRP and downregulated miR-613 expression in HCC patients are significantly correlated with poor prognosis. A. The correlation between RMRP expression and HCC prognosis. The overall survival (OS) rates in HCC patients with high RMRP levels were significantly shorter than that in the patients with low RMRP expression as determined by Kaplan-Meier survival curves. B. The correlation between miR-613 expression and HCC prognosis. HCC patients with lower levels of miR-613 had a worse OS rates than those patients with high miR-613 levels.
Table 2.
Associations between miR-613 expression and patients’ clinicopathological features in HCC
| Features | Number of cases (n = 52) | Low miR-613 expression (n = 32) | High miR-613 expression (n = 20) | P-value |
|---|---|---|---|---|
| Gender | ||||
| Male | 41 | 24 | 17 | 0.497 |
| Female | 11 | 8 | 3 | |
| Age (y) | ||||
| < 60 | 24 | 15 | 9 | 0.895 |
| ≥ 60 | 28 | 17 | 11 | |
| Tumor size (cm) | ||||
| < 5 | 30 | 20 | 10 | 0.375 |
| ≥ 5 | 22 | 12 | 10 | |
| Liver cirrhosis | ||||
| Without | 7 | 4 | 3 | 1.000 |
| With | 45 | 28 | 17 | |
| AFP levels (ng/ml) | ||||
| < 20 | 13 | 11 | 2 | 0.057 |
| ≥ 20 | 39 | 21 | 18 | |
| TNM stage | ||||
| I + II | 26 | 12 | 14 | 0.023* |
| III + IV | 26 | 20 | 6 | |
| Venous invasion | ||||
| Absent | 34 | 19 | 15 | 0.249 |
| Present | 18 | 13 | 5 | |
| Lymph node metastasis | ||||
| No | 31 | 14 | 17 | 0.004** |
| Yes | 21 | 18 | 3 |
P < 0.05;
P < 0.01.
RMRP functions as an oncogene in the development of HCC
To further investigate the role of RMRP in HCC, we transfected siRNA-RMRP and siRNA-control into Hep3B and HCCLM3 cells to knockdown RMRP expression. qRT-PCR results revealed that the siRNA-RMRP for RMRP knockdown were workable in Hep3B and HCCLM3 cells (Figure 4A, P < 0.01). MTT assay showed that knockdown of RMRP could markedly decrease the cell viability in Hep3B (Figure 4B, P < 0.05) and HCCLM3 cells (Figure 4C, P < 0.05). Flow cytometry showed that inhibition of RMRP increased an accumulation of cells in the G0/G1 phase, and this increase was accompanied with a concomitant decrease of cell number in S phase (Figure 4D and 4E, P < 0.05). To further investigate whether RMRP affects cell migration and invasion, wound scratch and transwell invasion assays were performed in our study. Results showed that RMRP inhibition significantly decreased the abilities of cell migration (Figure 4F, P < 0.05) and invasion (Figure 4G, P < 0.01) in Hep3B and HCCLM3 cells. All of these results indicated that RMRP acts as an oncogenic role in the development of HCC that promoting cell proliferation, migration and invasion.
Figure 4.
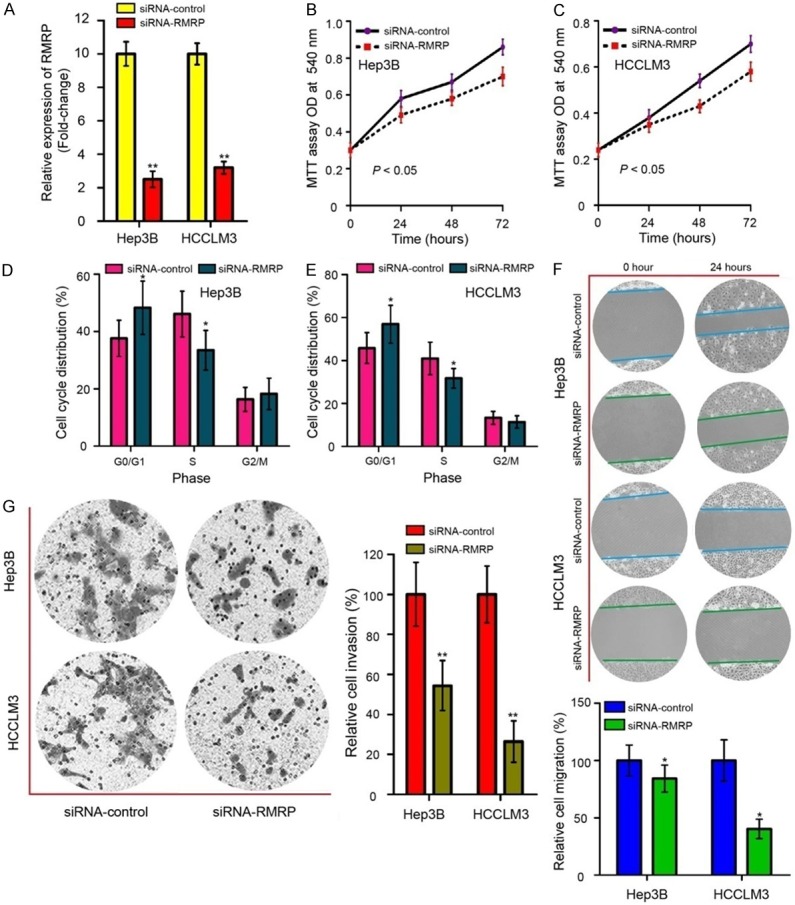
RMRP functions as an oncogene in the development of HCC. A. RMRP expression in Hep3B and HCCLM3 cells after transfected with siRNA-RMRP and siRNA-control was detected by qRT-PCR assay. B. MTT assay revealed that RMRP inhibition suppressed the growth of Hep3B cells. C. RMRP knockdown suppressed the growth of HCCLM3 cells. D. Knockdown of RMRP increased an accumulation of Hep3B cells in the G0/G1 phase, and this increase was accompanied with a concomitant decrease of cell number in S phase. E. RMRP knockdown converted HCCLM3 cells with lower capabilities of cell-cycle G1/S transition. F. Wound scratch assay (Magnification, × 200 times) was applied to determine the inhibitive effects due to depression of RMRP on Hep3B and HCCLM3 cells’ migration ability. G. Transwell invasion assay (Magnification, × 400 times) was used to measure cell invasion abilities in Hep3B and HCCLM3 cells after transfected with siRNA-RMRP and siRNA-control. siRNA-RMRP, small interfering RNA specifically targeting RMRP; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. Mean of three independent experiments with SD were shown. *P < 0.05 and **P < 0.01.
RMRP is directly targeting to miR-613
To assure whether RMRP regulates gene expression through competing endogenous RNA (ceRNA) mechanism, we then predicted the potential miRNA binding sites in RMRP by using miRcode (http://www.mircode.org/) and RegRNA 2.0 (http://regrna2.mbc.nctu.edu.tw/) algorithms. We found that RMRP sequence (ACATTCC) had a complementary binding site for miR-613 (UGUAAGG) (Figure 5A). Subsequently, miR-613 was chosen for further study because miR-613 ranked the top among all potential targets and the roles of miR-613 were well studied in HCC. To validate this prediction, we conducted WT and MUT pGL3-RMRP reporter plasmids (Figure 5B). As shown in Figure 5C, miR-613 mimic inhibited the luciferase activity in Hep3B and HCCLM3 cells transfected with WT pGL3-RMRP reporter plasmid (P < 0.01). When the binding site of miR-613 in RMRP transcript was mutated, miR-613 mimic had no effect on the luciferase activity. To further confirm the interaction between miR-613 and RMRP, we transfected Hep3B and HCCLM3 cells with siRNA-RMRP and found that RMRP inhibition significantly increased miR-613 expression (Figure 5D, P < 0.01). These results confirmed that RMRP directly targets miR-613.
Figure 5.
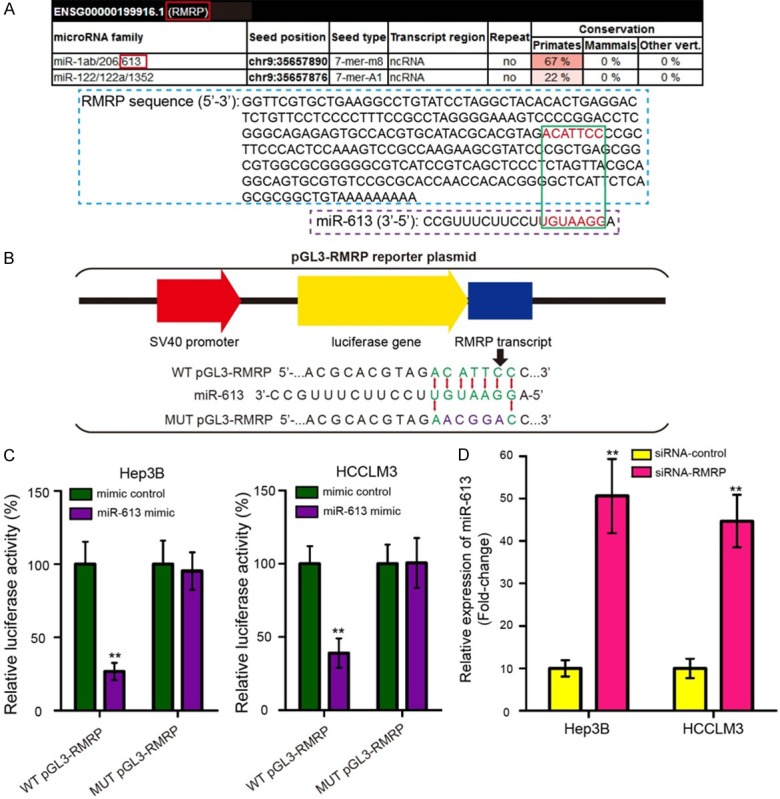
RMRP is directly targeting to miR-613. A. miRcode algorithm was used to predict the binding site of RMRP on miR-613. RMRP sequence (ACATTCC) had a complementary binding site for miR-613 (UGUAAGG). B. Diagram of the constructed RMRP reporter plasmid containing the wild-type (WT) and mutant (MUT) binding site for miR-613. C. The WT and MUT pGL3-RMRP reporter plasmids were co-transfected into Hep3B and HCCLM3 cells with miR-613 mimic or mimic control, and luciferase activities were measured by dual luciferase reporter gene assay. miR-613 mimic inhibited the luciferase activity in Hep3B and HCCLM3 cells transfected with WT pGL3-RMRP reporter plasmid. D. miR-613 expression was measured by qRT-PCR assay in Hep3B and HCCLM3 cells after transfection with siRNA-RMRP and siRNA-control. The experiments were performed in triplicate. *P < 0.05 and **P < 0.01.
RMRP sponges miR-613 to promote cell proliferation, migration, and invasion in HCC
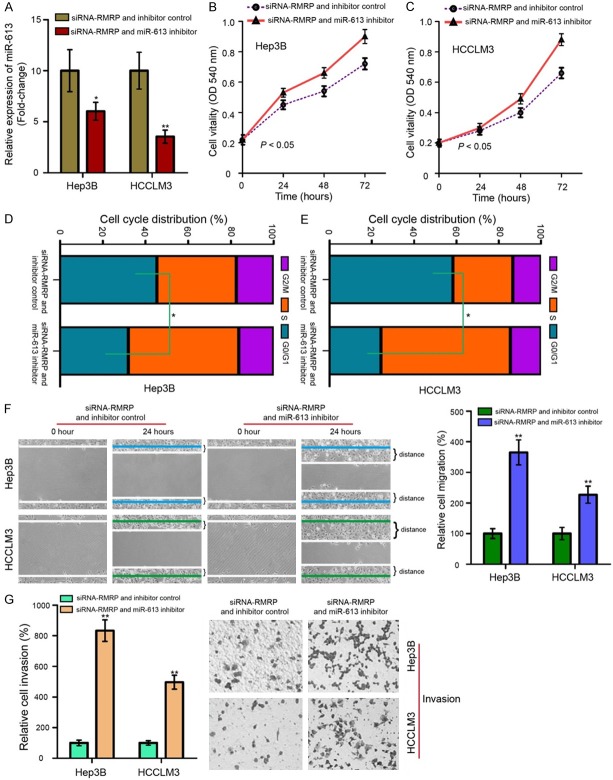
Rescue experiments were conducted to investigate whether RMRP exerted its roles in HCC cells via targeting miR-613. We re-executed cellular biological functional assays to demonstrate the ceRNA mechanism of the RMRP/miR-613 axial working on cell proliferation, migration and invasion in Hep3B and HCCLM3 cells. As shown in Figure 6A-G, transfection with miR-613 inhibitor could partly reversed the inhibitory effects of RMRP knockdown on cell proliferation, invasion and migration (P < 0.05 or P < 0.01). Our data strongly suggested that RMRP acts as a sponge for miR-613 by promoting cell proliferation, migration, and invasion in HCC.
Figure 6.
RMRP sponges miR-613 to promote cell proliferation, migration, and invasion in HCC. A. qRT-PCR assay was performed to measure the miR-613 expression in Hep3B and HCCLM3 cells treated with siRNA-RMRP and miR-613 inhibitor. The promotive effect of RMRP knockdown on miR-613 in HCC cells was reversed by miR-613 inhibitor. B. RMRP silence inhibited the growth of Hep3B cells, while addition of miR-613 inhibitor reversed this effect. C. The migration of HCCLM3 cells following transfection with siRNA-RMRP and miR-613 inhibitor. D. Proportion of cells in various phases of the cell cycle in Hep3B cells. Hep3B cells transfected with siRNA-RMRP and miR-613 inhibitor decreased an accumulation of cells in the G0/G1 phase, and this decrease was accompanied with a concomitant increase of cell number in S phase. E. Cotransfected with siRNA-RMRP and miR-613 inhibitor converted HCCLM3 cells with higher capabilities of cell-cycle G1/S transition. F. Migratory ability (Magnification, × 200 times) of Hep3B and HCCLM3 was suppressed by siRNA-RMRP, and the effect was attenuated by miR-613 inhibitor. G. siRNA-RMRP negatively affected cell invasion (Magnification, × 200 times), and the suppressive effect siRNA-RMRP presented was reversed by miR-613 inhibitor but not by inhibitor control in Hep3B and HCCLM3 cells, individually. Mean of three independent experiments with SD were shown. *P < 0.05 and **P < 0.01.
RMRP/miR-613 axis enhances the tumorigenesis of HCC in vivo
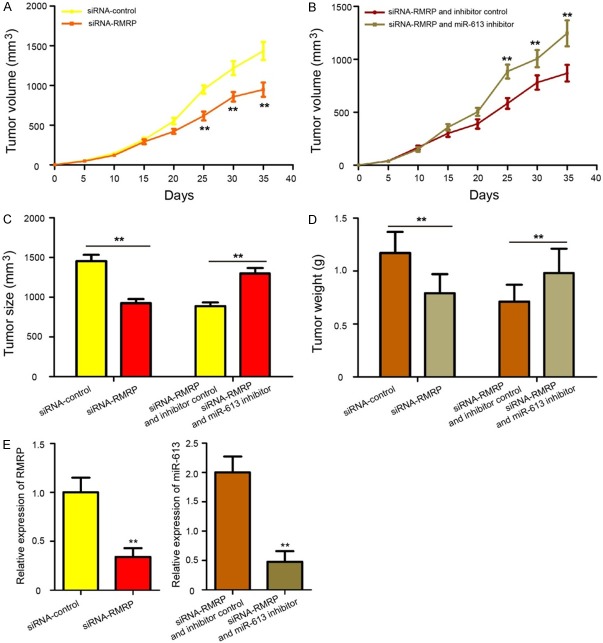
To determine the effects of RMRP/miR-613 axis in HCC tumorigenesis in vivo, transfected Hep3B cells were subcutaneously injected into the left flanks of male athymic Balb/c nude mice, and HCC tumorigenesis was measured. The results showed that tumor growth was slower in the siRNA-RMRP group than in the siRNA-control group (Figure 7A, P < 0.01), while tumor growth in the siRNA-RMRP + miR-613 inhibitor was faster compared with the siRNA-RMRP + inhibitor control group (Figure 7B, P < 0.01). At 35 days after injection, all mice were killed, and the xenograft tumors were removed and weighed. We found that knockdown RMRP decreased tumor size and weight, while miR-613 silencing could partly reversed the inhibitory effects of RMRP knockdown on HCC tumorigenesis (Figure 7C and 7D, P < 0.01). Additionally, RMRP expression was downregulated in tumor tissues from the siRNA-RMRP group compared with that in the siRNA-control group, and the expression of miR-613 was decreased in the siRNA-RMRP + miR-613 inhibitor group compared with that in the siRNA-RMRP + inhibitor control group (Figure 7E, P < 0.01). Summarily, these data strong demonstrated that RMRP/miR-613 axis promotes the tumorigenesis of HCC.
Figure 7.
RMRP/miR-613 axis promotes the tumorigenesis of HCC. A. The transfected Hep3B cells were subcutaneously injected into the left flanks of male athymic Balb/c nude mice and HCC tumorigenesis was measured. The tumor size was monitored every five days. The growth curves generated for tumor volume in the nude mice demonstrated that knockdown of RMRP significantly inhibited tumor growth in vivo. B. Co-transfected with siRNA-RMRP and miR-613 inhibitor increased tumor growth compared to cells treated with siRNA-RMRP and inhibitor control. C. Knockdown RMRP decreased tumor size, while miR-613 silencing could partly reversed the inhibitory effect of RMRP knockdown. D. The tumors were isolated and weighed after 35 days. RMRP knockdown decreased tumor weight, and tumor weight in siRNA-RMRP and miR-613 inhibitor group was increased compared to siRNA-RMRP and inhibitor control group. E. The expression of RMRP and miR-613 in xenograft from the nude mice was determined by qRT-PCR assay. The experiments were performed in triplicate. **P < 0.01.
Discussion
Owing to difficult early diagnosis and poor prognosis, HCC has become the rapidest growing cause of cancer death globally. Mounting evidences have highlighted the critical roles of lncRNAs in the carcinogenesis of HCC, and indicating lncRNAs may serve as an effective therapeutic targets for HCC treatment [19]. For example, Guo et al. found that lncRNA PVT1 (Pvt1 oncogene) promotes cell propagation and inhibited apoptotic cells in HCC by restraining P53 protein expression [20]. Mo et al. elucidated that LINC01287 plays an oncogenic role in tumour growth and metastasis and serves as a novel molecular target for the treatment of HCC [21]. Huang et al. demonstrated that lncRNA PTTG3P (Pituitary tumor-transforming 3, pseudogene) promotes tumour growth and metastasis via up-regulating PTTG1 (Pituitary tumor-transforming 1) and activating PI3K/Akt signaling in HCC [22]. In addition, Tang et al. showed that lncRNA CRNDE (Colorectal neoplasia differentially expressed) promotes tumour cell proliferation by regulating PI3K/Akt/β-catenin signaling in HCC [23]. In the current study, we showed a novel lncRNA, RMRP, which was significantly up-regulated in HCC tissues and cell lines, given the clue that RMRP might play an important role in the development of HCC.
RMRP, also known as NME1 or RRP2, is firstly identified and named by Rogler LE and his colleagues in human cartilage-hair hypoplasia [14]. RMRP is located at chromosome 9p13.3, and is abundantly expressed in various human tissues [15]. Previous study has shown that RMRP expression is upregulated in lung adenocarcinoma tissues compared to the adjacent normal tissues [18]. However, Shao et al. revealed that the expression levels of RMRP were downregulated in gastric cancer tissues and gastric dysplasia. Moreover, RMRP suppressed the expression of miR-206 and modulated cell cycle by regulating Cyclin D2 expression in gastric cancer [17]. Our findings were similarity with these previous reports that RMRP was elevated in HCC tissues and cell lines compared to corresponding adjacent non-tumor tissues and a normal liver cell line, respectively. We also noticed that highly expressed RMRP in HCC patients was significantly correlated with lymph node metastasis, TNM stage, venous invasion and poor prognosis, and is not correlated with other factors, including gender, age, liver cirrhosis, tumor size and AFP levels. These clues suggested that highly expressed RMRP are associated with hepatocarcinogenesis.
To further study the biological functions of RMRP in HCC, we firstly performed loss-of-function experiment in Hep3B and HCCLM3 cells in vitro. Our results showed that decreased expression of RMRP converted HCC cells into less aggressive cells, with lower capabilities of cell-cycle G1/S transition, proliferation, migration and invasion. These findings clearly suggested that RMRP functions as an oncogene in HCC development. Similar to our results, normal human gastric mucosa epithelial cells and gastric cancer cells transfected with RMRP siRNA represented significant G0/G1 arrest [17]. Moreover, Meng et al. reported that ectopic expression of RMRP promoted lung adenocarcinoma cell proliferation, colony formation and invasion [18]. Consistently, RMRP has been demonstrated to promote gastric tumor cell proliferation in vitro and in vivo [17].
Recent studies revealed that lncRNAs have been demonstrated to regulate gene expression through functioning as miRNA sponge or competing endogenous RNA (ceRNA) [24,25]. To elucidate further molecular mechanism by which RMRP exerts its oncogenic function in HCC development, we predicted its putative miRNA binding sites by using the prediction algorithms of miRcode and RegRNA 2.0. Of the miRNAs listed, miR-613 was chosen for further investigation because miR-613 ranked the top among all potential targets and the roles of miR-613 were well studied in HCC. It is reported that miR-613 functions as tumor suppressor gene in many human cancers including HCC. For example, miR-613 acts as a tumor suppressor in glioma cells by directly targeting sex-determining region Y-box 9 [26]. miR-613 inhibits cell migration and invasion by downregulating Daam1 (Dishevelled associated activator of morphogenesis 1) in triple-negative breast cancer [27]. miR-613 suppresses the laryngeal squamous cell carcinoma progression through regulating pyruvate dehydrogenase kinase 1 [28]. In addition, miR-613 functions as tumor suppressor in HCC by targeting YWHAZ (Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta) [29]. miR-613 has been reported to be a ceRNA of lncRNA NEAT1 (Nuclear paraspeckle assembly transcript 1) in HCC [30] and lncRNA HOTAIR in pancreatic cancer [31]; however, the interaction between RMRP and miR-613 has not been reported in HCC.
In our study, we found that the expression of miR-613 was down-regulated in HCC tissues and cell lines. And the expression of miR-613 was negatively correlated with that of RMRP in HCC tissues. Furthermore, the constructed dual luciferase reporter gene assay strongly confirmed that RMRP was directly targeting to miR-613. Thus, we hypothesized that RMRP could promote cell proliferation, migration, and invasion in HCC by acting as a ceRNA of miR-613. A subsequent experiments demonstrated that knockdown of miR-613 could partly reversed the inhibitory effects of RMRP inhibition on cell proliferation, invasion and migration. Xenograft assays also revealed that RMRP knockdown inhibited HCC tumorigenesis, while miR-613 silencing could partly reversed the inhibitory effects of RMRP knockdown on HCC tumorigenesis. Our data strongly suggested that RMRP acts as a sponge for miR-613 by promoting HCC tumorigenesis.
Overall, our data provided the first evidence that the RMRP/miR-613 axis is associated with poor prognosis and promotes the tumorigenesis of HCC by impacting oncogenic phenotypes. Therapeutics that targeting RMRP/miR-613 axis may therefore improve the treatment of HCC.
Acknowledgements
This work was supported by the Hunan Provincial Natural Science Foundation (no. 2017JJ3176) and the Youth PhD Fund of Hunan Provincial People’s Hospital (BSJJ201808).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Marquardt JU, Thorgeirsson SS. SnapShot: hepatocellular carcinoma. Cancer Cell. 2014;25:550, e1. doi: 10.1016/j.ccr.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 4.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tejeda-Maldonado J, Garcia-Juarez I, Aguirre-Valadez J, Gonzalez-Aguirre A, Vilatoba-Chapa M, Armengol-Alonso A, Escobar-Penagos F, Torre A, Sanchez-Avila JF, Carrillo-Perez DL. Diagnosis and treatment of hepatocellular carcinoma: an update. World J Hepatol. 2015;7:362–376. doi: 10.4254/wjh.v7.i3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Umeda S, Kanda M, Kodera Y. Emerging evidence of molecular biomarkers in hepatocellular carcinoma. Histol Histopathol. 2018;33:343–355. doi: 10.14670/HH-11-936. [DOI] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 9.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. doi: 10.1186/s12885-017-3216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng P, Yin Z, Wu Y, Xu Y, Luo Y, Zhang TC. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun Signal. 2018;16:5. doi: 10.1186/s12964-018-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao D, Li Z, Zhu M, Wang Y, Wu G, Han X. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC) Am J Cancer Res. 2018;8:748–760. [PMC free article] [PubMed] [Google Scholar]
- 14.Rogler LE, Kosmyna B, Moskowitz D, Bebawee R, Rahimzadeh J, Kutchko K, Laederach A, Notarangelo LD, Giliani S, Bouhassira E, Frenette P, Roy-Chowdhury J, Rogler CE. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum Mol Genet. 2014;23:368–382. doi: 10.1093/hmg/ddt427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbluh J, Nijhawan D, Chen Z, Wong KK, Masutomi K, Hahn WC. RMRP is a non-coding RNA essential for early murine development. PLoS One. 2011;6:e26270. doi: 10.1371/journal.pone.0026270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K, Smagula CS, Parsons WJ, Richardson JA, Gonzalez M, Hagler HK, Williams RS. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X, Yang Y, Xiao B, Guo J. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812–37824. doi: 10.18632/oncotarget.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng Q, Ren M, Li Y, Song X. LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One. 2016;11:e0164845. doi: 10.1371/journal.pone.0164845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Guo J, Hao C, Wang C, Li L. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18:98. doi: 10.1186/s12935-018-0582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo Y, He L, Lai Z, Wan Z, Chen Q, Pan S, Li L, Li D, Huang J, Xue F, Che S. LINC01287/miR-298/STAT3 feedback loop regulates growth and the epithelial-to-mesenchymal transition phenotype in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2018;37:149. doi: 10.1186/s13046-018-0831-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang JL, Cao SW, Ou QS, Yang B, Zheng SH, Tang J, Chen J, Hu YW, Zheng L, Wang Q. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer. 2018;17:93. doi: 10.1186/s12943-018-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Q, Zheng X, Zhang J. Long non-coding RNA CRNDE promotes heptaocellular carcinoma cell proliferation by regulating PI3K/Akt/beta-catenin signaling. Biomed Pharmacother. 2018;103:1187–1193. doi: 10.1016/j.biopha.2018.04.128. [DOI] [PubMed] [Google Scholar]
- 24.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kartha RV, Subramanian S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front Genet. 2014;5:8. doi: 10.3389/fgene.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sang Q, Liu X, Sun D. Role of miR-613 as a tumor suppressor in glioma cells by targeting SOX9. Onco Targets Ther. 2018;11:2429–2438. doi: 10.2147/OTT.S156608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong H, Yan T, Zhang W, Shi F, Jiang X, Wang X, Li S, Chen Y, Chen C, Zhu Y. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell Signal. 2018;44:33–42. doi: 10.1016/j.cellsig.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Yang S, Ge W, Wang Y, Han C, Li M. MiR-613 suppressed the laryngeal squamous cell carcinoma progression through regulating PDK1. J Cell Biochem. 2018;119:5118–5125. doi: 10.1002/jcb.26468. [DOI] [PubMed] [Google Scholar]
- 29.Jiang X, Wu J, Zhang Y, Wang S, Yu X, Li R, Huang X. MiR-613 functions as tumor suppressor in hepatocellular carcinoma by targeting YWHAZ. Gene. 2018;659:168–174. doi: 10.1016/j.gene.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Zou Q, Song M, Chen J. NEAT1 promotes cell proliferation and invasion in hepatocellular carcinoma by negative regulating miR-613 expression. Biomed Pharmacother. 2017;94:612–618. doi: 10.1016/j.biopha.2017.07.111. [DOI] [PubMed] [Google Scholar]
- 31.Cai H, Yao J, An Y, Chen X, Chen W, Wu D, Luo B, Yang Y, Jiang Y, Sun D, He X. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR-613 in pancreatic cancer. Oncotarget. 2017;8:32905–32917. doi: 10.18632/oncotarget.16462. [DOI] [PMC free article] [PubMed] [Google Scholar]