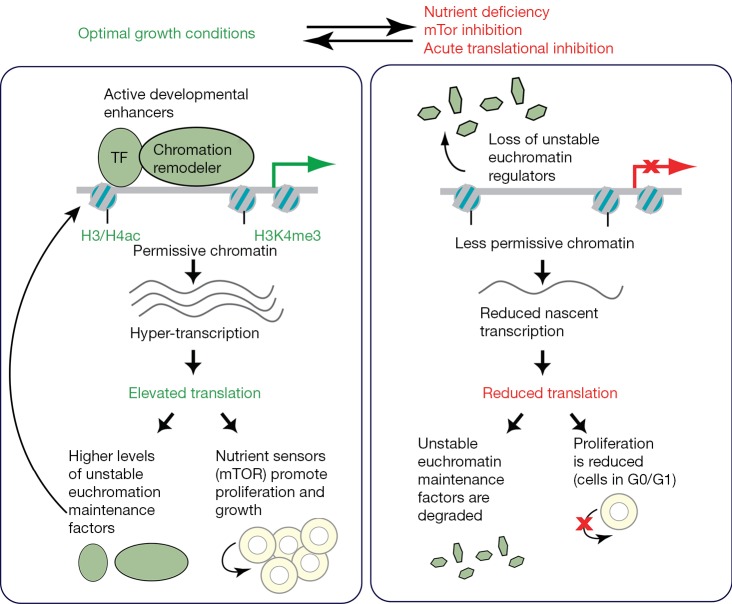
A transcriptionally permissive chromatin state is a characteristic of both the early blastocyst embryo and of embryonic stem cells (ESCs). Recent work from the laboratory of Miguel Ramalho-Santos, described in this Editorial, demonstrates that this state is very sensitive to translational output, with a positive feedback loop linking high rates of translation with maintenance of a stem cell state of hyper-transcription and elevated chromatin accessibility (1). The authors demonstrate that such tight control of translational output acts as a rheostat coordinating modulation of transcription and chromatin state with stem cell proliferation and growth, to enable the rapid growth of the embryo during early development (Figure 1).
Figure 1.
Schematic depicts the interdependence between euchromatin maintenance by unstable regulators, transcriptional and translational status, and proliferation in stem cells, and demonstrates how levels of translation under optimal versus non-optimal growth conditions can act as a cellular rheostat to influence these cellular parameters. TF, transcription factor.
Cells of the mouse pre-implantation embryo, and mouse ESCs derived from these early embryos, have a distinct chromatin landscape which promotes high levels of transcription. Chromatin is maintained in a decondensed, permissive, euchromatic state, with very low levels of compact heterochromatin and high levels of histone modifications associated with transcriptional activity, such as acetylation of histone H3 and H4 and tri-methylation of histone H3 lysine 4 (H3K4me3) (2-8). Accordingly, ESCs are in a transcriptionally hyperactive state, with global elevation of nascent transcript output. A number of chromatin remodeling activities were previously shown to influence stem cell euchromatin maintenance, including histone acetyltransferases such as Tip60/p400 and ATP-dependent chromatin remodelers such as Ino80 and Chd1 (9,10). Chd1 binds the ESC genome broadly at transcribed regions and promotes RNA polymerase I- and II-dependent hypertranscription, and this is required for the rapid proliferation and growth of ESCs and epiblast cells of the embryo prior to implantation (10).
While this state of permissive chromatin and global hypertranscription appears to be required for developmental transitions involving rapid proliferation of stem cells, it was not clear how stem cells sense and regulate their state of transcription to accommodate the demands of rapid proliferation and growth (4,5,7). Bulut-Karslioglu et al. recently addressed this question, defining a relationship between growth capacity of ESCs, their translational output, regulation of the permissive chromatin state, and dynamic control of the levels of unstable regulators of euchromatin maintenance (1). The authors began by constructing an ESC line expressing a Chd1 chromodomain protein fusion to the enhanced green fluorescent protein (EGFP) reporter, which was highly responsive to euchromatin state. They used this reporter to perform a genome-wide RNAi screen, identifying genes whose knockdown was associated with reduced reporter fluorescence. This work defined multiple regulators of transcription and of the ESC euchromatin state but also, surprisingly, strongly selected for regulators of cell growth and protein synthesis, including RNA polymerase (Pol I) complex components, ribosomal proteins, and translation factors including mTor, a key nutrient sensor and regulator of translation (2,10).
Pursuing these findings, the authors studied the relationship between translational regulation and euchromatin maintenance. They found that acutely inhibiting protein synthesis, mTor, or Myc/Max activity diminished EGFP reporter expression and euchromatin maintenance, both delocalizing the Chd1-EGFP reporter from chromatin and promoting its protein turnover. Therefore, Chd1-EGFP stabilization and chromatin association reflects a cell state with high levels of euchromatin, nascent transcription, and translation, all characteristics of undifferentiated stem cells. The authors next looked at the connection between acute inhibition of translation and this euchromatic state: a brief translational block reduced active chromatin modifications at enhancers, without affecting overall levels of histone H3 or repressive histone modifications. In the mouse blastocyst stage embryo, reductions of active histone modifications were likewise seen after a brief translational block, while mTor inhibition had similar effects on active histone modifications in both ESCs and in the early embryo. Therefore, this work demonstrated that euchromatin maintenance in stem cells is acutely sensitive to translational status.
The authors next demonstrated that this translational output positively feeds back to regulate levels of nascent transcription in ESCs. An acute translational block strongly diminished production of both RNA polymerase I- and II-generated nascent transcripts, while steady state transcript levels were much less affected. This translational block also rapidly diminished RNA polymerase II occupancy at transcription start sites and in the body of highly transcribed genes, both in ESCs and in cells of the blastocyst embryo. Therefore, inhibiting translation had an unexpected and rapid effect on nascent transcription. They also assessed whether these effects were specific to stem cells. By contrast with the effects in stem cells, several types of differentiating cells had a lower basal level of nascent transcripts and had a much more muted response than pluripotent cells to acute translational inhibition. These data suggest that the permissive, hypertranscribing ESC chromatin state is particularly sensitive to acute translational inhibition.
Finally, they analyzed the proteomic changes that accompanied a brief (1 or 3 hours) translational block. Proteins depleted in ESCs following this block included cell cycle regulators, while there was relative enrichment in regulators of the translational and RNA processing machinery. Cells exhibited a correspondingly altered cell cycle, with a diminished percentage of cells in S phase, and increased numbers of G0/G1 cells. The unstable proteins detected here included both some regulators of euchromatin maintenance obtained in prior reporter screening, including Chd1 and Tip60/p400, as well as many sequence-specific transcription factors that bind enhancer and promoter regions. Accordingly, this translational block rapidly deactivated developmental enhancers, reducing their accessibility, potentially due to increased turnover of both euchromatin maintenance factors and sequence-specific transcription factors that bind these regions. By contrast, the translational block increased the accessibility of regions of chromatin that encode transposable elements, potentially priming their expression.
Conclusions
This study combines data from transcriptomic, epigenomic, and proteomic analysis to define translational status as a key feedback regulator that integrates control of both cell growth and of the euchromatic and hypertranscriptional state of stem cells. The transcriptionally permissive chromatin state that characterizes mouse ESCs and cells of the early mouse embryo is particularly sensitive to levels of translational output, with acute inhibition of translation rapidly delocalizing RNA Polymerase II, reducing nascent transcription, and diminishing chromatin accessibility at developmental enhancers, while increasing accessibility at histone genes and transposable elements. Accordingly, a number of euchromatin regulators are unstable proteins, which are only maintained under conditions of high translational output. Thus, this positive feedback loop establishes an inter-dependence between the level of translation and the maintenance of euchromatin and high levels of transcription and translation, which may facilitate the rapid rate of cell proliferation and growth that accompanies early embryogenesis. In vivo, translational regulators that sense nutrients, such as mTor, may act as a rheostat, modulating translation levels in response to environmental conditions of optimal versus limited nutrition. Feedback from this translational sensor may be used to promote and integrate control of multiple central properties of pluripotent stem cells, including their maintenance of a euchromatic and hypertranscriptional and hypertranslational state, and rapid cell growth.
Acknowledgements
Funding: This work supported by NIH/NIGMS (GM66815), March of Dimes (1-FY13-413), WUSM Institute for Clinical and Translational Sciences (ICTS) (JIT-NOA_619), and grants from Washington University (WU) McDonnell Center for Cellular and Molecular Neurobiology and WU Center for Regenerative Medicine to KL Kroll.
Provenance: This is an invited Editorial Commentary commissioned by the Editorial Office, Stem Cell Investigation.
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Bulut-Karslioglu A, Macrae TA, Oses-Prieto JA, et al. The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output. Cell Stem Cell 2018;22:369-83.e8. 10.1016/j.stem.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaspar-Maia A, Alajem A, Meshorer E, et al. Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 2011;12:36-47. 10.1038/nrm3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efroni S, Duttagupta R, Cheng J, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008;2:437-47. 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development 2017;144:365-73. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed K, Dehghani H, Rugg-Gunn P, et al. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 2010;5:e10531. 10.1371/journal.pone.0010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Percharde M, Wong P, Ramalho-Santos M. Global Hypertranscription in the Mouse Embryonic Germline. Cell Rep 2017;19:1987-96. 10.1016/j.celrep.2017.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Percharde M, Bulut-Karslioglu A, Ramalho-Santos M. Hypertranscription in Development, Stem Cells, and Regeneration. Dev Cell 2017;40:9-21. 10.1016/j.devcel.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 2011;145:183-97. 10.1016/j.cell.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 2008;134:162-74. 10.1016/j.cell.2008.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman-Ayala M, Sachs M, Koh FM, et al. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development 2015;142:118-27. 10.1242/dev.114843 [DOI] [PMC free article] [PubMed] [Google Scholar]