Abstract
Oesophageal squamous cell carcinoma (OSCC) has a high incidence in southern Africa and a poor prognosis. Limited information is available on the contribution of genetic variants in susceptibility to OSCC in this region. However, recent genome-wide association studies have identified multiple susceptibility loci in Asian and European populations. In this study, we investigated genetic variants from seven OSCC risk loci identified in non-African populations for association with OSCC in the South African Black population. We performed association studies in a total of 1471 cases and 1791 controls from two study sample groups, which included 591 cases and 852 controls from the Western Cape and 880 cases and 939 controls from the Johannesburg region in the Gauteng province. Thereafter, we performed a meta-analysis for 11 variants which had been genotyped in both studies. A single nucleotide polymorphism in the CHEK2 gene, rs1033667, was significantly associated with OSCC [P = 0.002; odds ratio (OR) = 1.176; 95% confidence interval (CI): 1.06–1.30]. However, single nucleotide polymorphisms in the CASP8/ALS2CR12, TMEM173, PLCE1, ALDH2, ATP1B2/TP53 and RUNX1 loci were not associated with the disease (P > 0.05). The lack of association of six of these loci with OSCC in South African populations may reflect different genetic risk factors in non-African and African populations or differences in the genetic architecture of African genomes. The association at CHEK2, a gene with key roles in cell cycle regulation and DNA repair, in an African population provides further support for the contribution of common genetic variants at this locus to the risk of oesophageal cancer.
Single nucleotide polymorphisms (SNPs) associated with oesophageal squamous cell carcinoma (OSCC) in Asian populations were genotyped in OSCC cases and controls from the South African Black population. A SNP in CHEK2 was associated with an increased risk of OSCC.
Introduction
Oesophageal cancer (OC) is the eighth most common cancer worldwide (1), with oesophageal squamous cell carcinoma (OSCC) accounting for nearly 90% of all cases of OC worldwide (2). Particularly high incidence rates are observed in regions such as southern Africa, China and Japan. In South Africa, OC was the third most common cancer in males of African ancestry and the fourth most common cancer in females of African ancestry in 2014, with age standardized incidence rates of 4.79 and 3.16 per 100 000, respectively (3). In addition to the recognized risk factors for OSCC of alcohol consumption and tobacco use, polycyclic aromatic hydrocarbons exposure, high food temperatures and other factors such as diet and consumption of Fusarium-contaminated maize may also be involved in the development of the disease (reviewed in (4–7)).
Over the past 9 years, several genome-wide association studies (GWAS) have been performed in non-African populations to determine the genetic risk factors involved in the development of OSCC. In 2009, a GWAS for OSCC in the Japanese population found that single nucleotide polymorphisms (SNPs) at ADH1B (rs1229984) and ALDH2 (rs671) were associated with the disease (8), and a European GWAS of upper aerodigestive tract cancers detected genome-wide significant association with OSCC at ALDH2 (rs4767364) (9). Since 2010, four OSCC GWAS and a meta-analysis have been performed in the Chinese population, which identified multiple additional risk loci (10–14). These included SNPs at PLCE1, HEATR3, HAP1, CHEK2/XBP1, ST6GAL1, SMG6, PTPN2, SLC52A3 (C20orf54), PDE4D, RUNX1, UNC5CL and CYP26B1. However, in a subsequent analysis, the associations at HAP1, SMG6, SLC52A3 (C20orf54) and PTPN2 were not confirmed (14).
We previously tested several of these variants for association with OSCC in the South African Black (SAB) population recruited in the Western Cape region (15,16). Coding SNPs in ADH1B and ALDH2 were absent in the SAB population (15), and none of the lead SNPs from the Chinese GWAS studies were associated with OSCC (PLCE1 rs2274223, SLC52A3/C20orf54 rs13042395, PDE4D rs10052657, RUNX1 rs2014300, UNC5CL rs10484761). However, further analysis of the PLCE1 locus identified a significant association with the SNP Arg548Leu (rs17417407).
The Johannesburg Cancer Study (JCS) was established in 1995, with the aim of recruiting Black, treatment naïve, patients diagnosed with cancer from the greater Johannesburg area in Gauteng province (17). The JCS recruited all cancer cases, including OSCC, as well as non-cancer, ethnically matched controls from three tertiary academic hospitals in the greater Johannesburg region. The University of Cape Town (UCT) cancer study was established to recruit OSCC patients from the Western Cape region of South Africa through tertiary hospitals (15,18). Here, we investigate the association of seven main-effect susceptibility loci (CASP8/ALS2CR12, TMEM173, PLCE1, ALDH2, ATP1B2/TP53, RUNX1 and CHEK2/XBP1) identified by GWAS (9–13) with OSCC in SAB populations recruited in these two regions of South Africa.
Materials and methods
Study subjects
The UCT study sample consisted of 591 SAB OSCC cases and 852 ethnically matched non-cancer controls that were available for genotyping. The cases and controls were mainly Xhosa-speakers (97% and 95%, respectively) from the Western or Eastern Cape provinces of South Africa. The JCS samples consisted of 880 SAB OSCC cases and 939 ethnically matched non-cancer controls available for genotyping. The JCS cases and controls were recruited from the greater Johannesburg area in Gauteng province, with self-reported African ancestry. The major linguistic groups in the JCS in both cases and controls were the Nguni (Zulu and Xhosa speakers, 37.8%) and Sotho-Tswana (Southern Sotho, Tswana and Northern Sotho speakers, 39.2%). Control individuals for both study samples had no history of cancer, lived in the same residential areas and had a similar socioeconomic status to the cases. The JCS controls were recruited mainly from the cardiovascular unit at the Charlotte Maxeke Academic Hospital in Johannesburg, a high proportion of whom consumed alcohol.
All patients had a histologically confirmed primary invasive OSCC and were recruited between 1995 and 2016. Smoking status was subdivided into ever-smokers (those who had smoked at some point in their lives) or never-smokers. Drinkers were defined as subjects who consumed alcohol at least once every week or non-drinkers. Whole blood samples were collected, with informed consent, from all subjects and DNA was extracted at the University of Cape Town and the University of the Witwatersrand as previously described (15,19). Ethical approval for the study was obtained from the joint University of Cape Town/Groote Schuur Hospital Research Ethics Committee and the University of Stellenbosch/Tygerberg Hospital Ethics Committee (UCT HREC 040/2005) and the Human Research Ethics Committee (Medical) of the University of the Witwatersrand (M140271, M160807).
SNP selection and genotyping
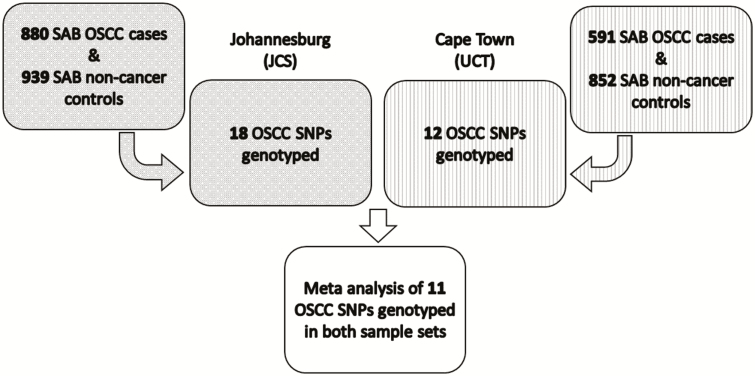
For the UCT samples, 12 SNPs were selected from the 7 OSCC susceptibility loci described above and genotyped. The list of these SNPs and loci are shown in Table 2, the majority of which were the lead SNPs from published GWAS (9–14). The index SNP for the TMEM173 locus on chromosome 5q31.2, rs7447927 (14) failed TaqMan assay design; a proxy for this SNP, rs13153461, was genotyped which is in strong linkage disequilibrium (LD) with rs7447927 in both Chinese (r2 = 0.98) and African (r2 = 0.69 in Yoruban) populations in the 1000 Genome Project Phase 3. The functional SNP in the TP53 gene (rs1800371, p.P47S), which has only been detected in African populations (20), was also genotyped. For the JCS samples, the lead SNPs from the seven OSCC susceptibility loci and one or more tagging SNPs in LD with the lead SNP were genotyped to provide additional coverage in case of genotype assay failure. Tag SNP selection was based on allele frequencies and LD in the Yoruban (YRI) and Chinese (CHB) populations from the 1000 Genomes Consortium (21), using LDLink 3.0 (22), which resulted in selection of 18 SNPs for genotyping (Table 2). An overview of the study design is shown in Figure 1.
Table 2.
Case-control association study analysis for SNPs genotyped in seven known risk loci for OSCC in the SAB populations
| UCT | JCS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene | SNP | Alleles | MAF case | MAF control | P-value | OR (95% CI) | MAF case | MAF control | P-value | OR (95% CI) |
| 2 | CASP8 | rs10931936 | C/T | 0.19 | 0.20 | 0.391 | 0.90 (0.71–1.15) | 0.22 | 0.20 | 0.271 | 1.09 (0.93–1.29) |
| 2 | ALS2CR12 | rs13016963a | G/A | 0.35 | 0.35 | 0.994 | 1.00 (0.82–1.22) | 0.39 | 0.38 | 0.515 | 1.05 (0.91–1.20) |
| 2 | ALS2CR12 | rs10201587a | A/G | – | – | – | – | 0.38 | 0.39 | 0.783 | 0.98 (0.86–1.12) |
| 5 | TMEM173 | rs13181561 | A/G | – | – | – | – | 0.48 | 0.49 | 0.275 | 0.93 (0.82–1.06) |
| 5 | TMEM173 | rs13153461 | G/A | 0.04 | 0.05 | 0.357 | 0.83 (0.56–1.23) | – | – | – | – |
| 10 | PLCE1 | rs17417407 | G/T | 0.17 | 0.21 | 0.014* | 0.76 (0.60–0.95) | 0.19 | 0.19 | 0.881 | 0.99 (0.84–1.17) |
| 10 | PLCE1 | rs7084339 | G/A | – | – | – | – | 0.48 | 0.46 | 0.178 | 1.09 (0.96–1.25) |
| 10 | PLCE1 | rs3765524a | T/C | 0.47 | 0.47 | 0.854 | 1.02 (0.83–1.26) | 0.48 | 0.46 | 0.186 | 1.09 (0.96–1.24) |
| 10 | PLCE1 | rs2274223a | A/G | 0.42 | 0.40 | 0.508 | 1.06 (0.89–1.26) | 0.41 | 0.43 | 0.334 | 0.94 (0.82–1.07) |
| 10 | PLCE1 | rs11187850 | A/G | – | – | – | – | 0.21 | 0.19 | 0.204 | 1.11 (0.94–1.31) |
| 12 | ALDH2 | rs4767364a | A/G | – | – | – | – | 0.12 | 0.11 | 0.138 | 1.17 (0.95–1.44) |
| 17 | ATP1B2/TP53 | rs1642764a | C/T | 0.21 | 0.20 | 0.527 | 1.07 (0.86–1.33) | 0.18 | 0.18 | 0.982 | 1.00 (0.85–1.19) |
| 17 | ATP1B2/TP53 | rs1641511 | A/G | – | – | – | – | 0.39 | 0.42 | 0.081 | 0.89 (0.78–1.02) |
| 17 | TP53 | rs1800371a | G/A | 0.02 | 0.03 | 0.145 | 0.67 (0.38–1.16) | 0.03 | 0.02 | 0.117 | 1.38 (0.92–2.07) |
| 21 | RUNX1 | rs2014300a | A/G | 0.38 | 0.40 | 0.376 | 0.92 (0.77–1.10) | 0.36 | 0.36 | 0.795 | 1.02 (0.89–1.17) |
| 21 | RUNX1 | rs2834718 | T/A | – | – | – | – | 0.33 | 0.33 | 0.667 | 0.97 (0.84–1.11) |
| 22 | CHEK2 | rs4822983a | C/T | 0.46 | 0.39 | 0.001*** | 1.32 (1.12–1.56) | 0.43 | 0.42 | 0.836 | 1.01 (0.89–1.16) |
| 22 | CHEK2 | rs1033667a | C/T | 0.44 | 0.38 | 0.002** | 1.30 (1.10–1.53) | 0.42 | 0.39 | 0.145 | 1.10 (0.97–1.26) |
| 22 | XBP1 | rs2239815a | C/T | 0.21 | 0.16 | 0.001*** | 1.41 (1.15–1.74) | 0.16 | 0.18 | 0.162 | 0.88 (0.74–1.05) |
MAF, minor allele frequency; C/T, major allele/minor allele; OR, odds ratio, for each minor allele carried; 95% CI, 95% confidence interval.
aPreviously published lead SNPs (9–14); other SNPs tag the lead SNPs.
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Figure 1.
Schematic of the overall study design.
The Agena MassARRAY iPLEX genotyping assay (Agena Bioscience, San Diego) was used to genotype 18 SNPs in the JCS samples. The assay was performed using the iPLEX assay protocol (23). Oligonucleotide primers were designed using the Agena Assay Design Suite (Agena Bioscience). Reactions for the PCR assays were carried out in 5.0 µl volumes in 96-well plates. Each reaction contained 25 ng DNA, 0.5 µl 10X PCR buffer, 2 mM MgCl2, 500 µM deoxynucleoside triphosphate mix, 100 nM primer mix and 1.0 U Taq polymerase, and distilled water (dH20) made up the remaining volume. Reactions for the shrimp alkaline phosphatase (SAP) post-PCR clean-up were carried out by adding 2.0 µl SAP mix, each containing 0.17 µl 10X SAP buffer, 0.51U SAP enzyme and dH20 to each reaction well. Reactions for the iPLEX single-base extension were carried out by adding 4.0 µl iPLEX mix, each containing 0.2 µl 10X iPLEX buffer plus, 0.2 µl iPLEX termination mix, 8 µM primer mix, 0.041 µl iPLEX enzyme and dH20 to each reaction well. Primer extension products were spotted onto the SpectroCHIPs and detection of the primer extension products by mass spectrometry was done on the Agena MassARRAY Compact mass spectrometer. All but one of 18 SNPs (rs1642764) produced discrete genotype clusters in the iPLEX assay.
The 12 SNPs for the UCT samples were genotyped using TaqMan SNP assays (Life Technologies, Carlsbad). A TaqMan SNP assay was also used to genotype the SNP (rs1642764) in the JCS samples that had failed in the iPLEX assay. Reactions for the TaqMan SNP assays were carried out in 2.5 µl volumes in 96-well plates. Each reaction contained 20 ng DNA, 1.25 µl ABsolute QPCR ROX mix (Thermo Scientific, Massachusetts) and 0.03 µl 40X TaqMan SNP assay mix (Life Technologies), with the PCR performed on a PTC-0225 DNA Engine (MJ Research, Hampton) according to manufacturer’s protocol. Fluorescent levels at the PCR endpoint were determined using a 7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City) and genotypes assigned using SDS 2.2.2 software (Applied Biosystems), with additional manual checks for discrete genotype clusters. High levels of concordance of genotyping results between MassARRAY chemistry and TaqMan chemistry have been described previously, including for clinical implementation (24,25).
Pearson’s chi-square (χ2) test was used to determine deviations from the Hardy–Weinberg equilibrium in controls only, using a cut-off of P < 0.001. Call rates for all other SNPs genotyped by either method were >95%.
Statistical analysis
In order to test for association with OSCC in the UCT and JCS study samples, allele frequencies in cases and controls were compared using Pearson’s χ2 test using Plink (version 1.9, https://www.cog-genomics.org/plink/1.9/). Allelic odds ratios (OR) and 95% confidence intervals (CI) were calculated using the common allele as the reference. In view of the differences in the ethno-linguistic composition of the subjects in the UCT and JCS samples and the different genotyping methodology used, a meta-analysis of the two data sets for fixed effects and random effects was performed using Plink (version 1.9, https://www.cog-genomics.org/plink/1.9/). A Bonferroni-corrected P-value of <0.0045 (0.05/11) was used as a significance threshold to account for the testing of 11 variants in the meta-analysis. The proportion of SNPs showing the same direction of effect in the meta-analysis as compared to the direction of effect observed in non-African studies was tested using the exact binomial test for difference from 0.5. As secondary analyses, dominant and recessive models were also tested for association with each SNP.
The power of the association tests was determined using Quanto (v1.2.4, http://hydra.usc.edu/gxe/). The average effect size of the loci tested was 1.26 and a range of minor allele frequency (MAF) for the SNPs was used for the power calculation. The power of the UCT study sample to detect an effect size of 1.26 for MAFs of 0.1–0.5 at alpha 0.05 was 48–86%. The power of the JCS samples to detect an effect size of 1.26 for MAFs of 0.05–0.45 at alpha 0.05 was 36–94%. The power of the combined study samples (1471 cases and 1791 controls) in the meta-analysis to detect an effect size of 1.26 was calculated for the range of MAFs observed in the combined study sample (0.17–0.46) and was 78–96% at alpha 0.0045.
Gene x environmental (GxE) interactions with tobacco smoking were investigated for SNPs with significant evidence of allelic association (P < 0.0045) by testing for association in cases and controls in the combined UCT/JCS study samples stratified by smoking status (ever versus never) in a χ2 analysis using Stata MP 15.1 (StataCorp, College Station). GxE interactions for alcohol consumption were investigated in a case-only analysis of the combined UCT/JCS study samples. Case-control interactions were not tested owing to the high level of alcohol use in the JCS controls.
Bioinformatic analysis of associated variants to look for potential functional effects was carried out using established tools and resources such as HaploReg (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), GTEx (https://gtexportal.org), RegulomeDB (http://www.regulomedb.org), ENCODE (https://www.encodeproject.org) and FuncPred (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html).
Results
The characteristics of the cases and controls in the JCS and UCT sample sets are shown in Table 1.
Table 1.
Characteristics of OSCC cases and controls in the SAB populations
| Study Samples | JCS | UCT | |||
|---|---|---|---|---|---|
| Case | Control | Case | Control | ||
| Total number | 880 | 939 | 591 | 852 | |
| Age, mean years (SD) | 58.2 (10.2) | 50.0 (15.5) | 60.2(11.3) | 48.9 (16.8) | |
| Sex, n (%) | Male | 545 (61.9) | 240 (25.6) | 284 (48.1) | 342 (40.1) |
| Female | 332 (37.7) | 698 (74.3) | 307 (51.9) | 507 (59.5) | |
| Unknown | 3 (0.3) | 1 (0.1) | 0 (0) | 3 (0.4) | |
| Smoking status, n (%) | Ever-smoker | 598 (68.0) | 333 (35.5) | 364 (61.6) | 338 (39.7) |
| Never-smoker | 276 (31.4) | 584 (62.2) | 227 (38.4) | 504 (59.1) | |
| Unknown | 6 (0.7) | 22 (2.3) | 0 (0) | 10 (1.2) | |
| Alcohol consumption, n (%) | Drinker | 473 (53.8) | 633 (67.4)a | 370 (62.6) | 458 (53.7) |
| Non-drinker | 404 (45.9) | 305 (32.5)a | 219 (37.1) | 390 (45.8) | |
| Unknown | 3 (0.3) | 1 (0.1)a | 2 (0.3) | 4 (0.5) |
aAscertainment bias of individuals with higher baseline alcohol consumption levels.
The mean age of diagnosis was similar in the JCS and UCT groups (58.2 and 60.2 years, respectively). The male to female ratio was 1.64 in the JCS cases but 0.93 in the UCT sample. Smoking rates were high (>60%) in both groups of cases. The proportion of cases reporting alcohol consumption was higher in the UCT group.
Three of the 12 variants tested, under the allelic additive model, in the SAB UCT study sample were significantly associated with OSCC after Bonferroni multiple testing correction (P < 0.0042) (Table 2). These were XBP1 rs2239815, CHEK2 rs4822983 and CHEK2 rs1033667. The other nine SNPs in CASP8, ALS2CR12, TMEM173, PLCE1, ATP1B2/TP53, TP53 and RUNX1 were not significantly associated with the disease, although rs17417407 at PLCE1 showed nominal evidence of association. In the secondary analysis of the SAB UCT study sample, results under a dominant and a recessive model of inheritance were similar to the additive model (Supplementary Table 1, available at Carcinogenesis Online), and all SNPs showing association with OSSC in the dominant or recessive models were also associated in the additive model. None of the 18 variants tested in the SAB JCS study sample showed evidence of association with OSCC (P > 0.05) under the allelic additive model (Table 2) or the dominant and recessive models (Supplementary Table 1, available at Carcinogenesis Online).
A fixed-effect meta-analysis was performed for the 11 SNPs genotyped in both study samples under the allelic additive model (Table 3). One SNP, CHEK2 rs1033667 [P = 0.002, OR = 1.18 (1.06–1.30)], was significantly associated with OSCC under this model. This was also the only SNP with evidence of association when analysed under dominant or recessive models (Supplementary Tables 2 and 3, available at Carcinogenesis Online). A random-effect meta-analysis was also performed to investigate potential heterogeneity between the two studies (Table 3). The only marker with any evidence of association with OSCC under the random-effect model was rs1033667 [P(R) = 0.032], and the direction of the effect was the same in both studies. Three variants showed evidence of heterogeneity (TP53 rs1800371, CHEK2 rs4822983 and XBP1 rs2239815), but none of these were associated with OSCC under the random-effect model. Of the 11 variants included in the meta-analysis, 9 were genotyped in the Chinese genome-wide scans and 5 had effects in the same direction as in the original Chinese studies (P = 0.556).
Table 3.
Meta-analysis of 11 variants genotyped in the SAB populations
| Chr | Gene | SNP | Major allele | Minor allele | P | P(R) | OR (95% CI) | Q | I 2 |
|---|---|---|---|---|---|---|---|---|---|
| 2 | CASP8 | rs10931936 | C | T | 0.663 | 0.886 | 1.030 (0.902–1.177) | 0.186 | 42.91 |
| 2 | ALS2CR12 | rs13016963 | G | A | 0.588 | 0.588 | 1.031 (0.923–1.152) | 0.726 | 0.00 |
| 10 | PLCE1 | rs17417407 | G | T | 0.116 | 0.310 | 0.898 (0.786–1.027) | 0.061 | 71.57 |
| 10 | PLCE1 | rs3765524 | T | C | 0.221 | 0.221 | 1.072 (0.959–1.197) | 0.588 | 0.00 |
| 10 | PLCE1 | rs2274223 | A | G | 0.708 | 0.780 | 0.980 (0.882–1.089) | 0.267 | 18.70 |
| 17 | ATP1B2/TP53 | rs1642764 | C | T | 0.682 | 0.682 | 1.028 (0.899–1.176) | 0.627 | 0.00 |
| 17 | TP53 | rs1800371 | G | A | 0.685 | 0.962 | 1.070 (0.772–1.483) | 0.037* | 77.10 |
| 21 | RUNX1 | rs2014300 | A | G | 0.740 | 0.740 | 0.982 (0.882–1.094) | 0.390 | 0.00 |
| 22 | CHEK2 | rs4822983 | C | T | 0.028* | 0.285 | 1.123 (1.013–1.245) | 0.015* | 83.03 |
| 22 | CHEK2 | rs1033667 | C | T | 0.002** | 0.032* | 1.176 (1.060–1.304) | 0.137 | 54.85 |
| 22 | XBP1 | rs2239815 | C | T | 0.313 | 0.648 | 1.071 (0.938–1.758) | 0.001*** | 91.30 |
OR, odds ratio, for each minor allele carried; 95% CI, 95% confidence interval; P(R), P-values for the random effect meta-analysis; Q, Cochrane’s Q-statistic P-values. I2, I2 heterogeneity index; P, P-value for the fixed effect meta-analysis.
*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
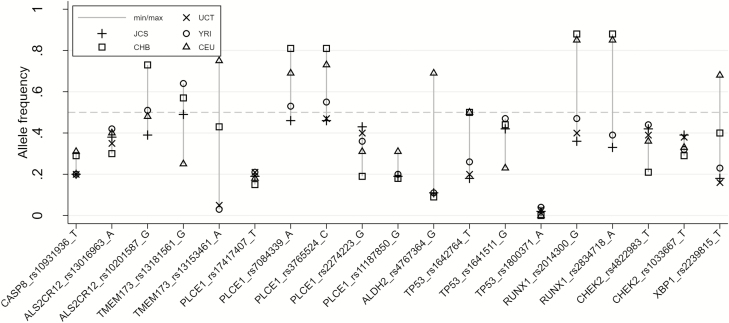
A comparison of allele frequencies of the 19 SNPs of interest in the UCT and JCS controls with their allele frequencies in the 1000 Genomes project phase 3 populations, Yoruban (YRI), Han Chinese (CHB) and Northern European (CEU), is shown in Figure 2 and Supplementary Table 4, available at Carcinogenesis Online. The majority of the SNPs were common in all these populations, but allele frequencies were very different for SNPs such as TMEM173 rs13153461, which had low frequencies in African populations. In such cases, there would be low power to detect an association of this marker with OSCC in Africans. Also, for SNPs such as ALDH2 rs4767364 and XBP1 rs2239815, the rarer allele in one population was the common allele in other populations. Notably, the SNP with the strongest evidence of association with OSCC, CHEK2 rs1033667, had similar frequencies in all these populations.
Figure 2.
Comparison of the allele frequencies of the 19 SNPs of interest by population.
The rs1033667 variant was investigated by bioinformatic analysis for evidence of a functional effect. The SNP is located in an intron of CHEK2, 90 bp downstream of a donor splice site. Analysis of this variant using a variety of predictive tools (see Materials and methods) did not provide any direct evidence for a functional effect. However, expression quantitative trait locus (eQTL) data show that it is associated with altered expression of several genes in this region in multiple tissue types, including increased expression of CHEK2 in liver and HSCB in oesophageal mucosa and reduced expression of TTC28 in transformed fibroblasts (26,27). The only SNP currently known to be in strong LD (r2 > 0.8) with rs1033667 in African populations is rs2078555, which is also an intronic SNP in CHEK2. Bioinformatic analysis of this variant using the same tools also failed to reveal any direct evidence for a functional effect and showed similar eQTLs to rs1033667 for CHEK2, HSCB and TTC28 both in terms of direction of effect and tissue type.
The CHEK2 variant rs1033667 was tested for GxE interactions with tobacco smoking in a case-control analysis. Tobacco smoking was an independent risk factor for OSCC, OR = 3.04 (2.36–3.90) for ever-smokers (P < 0.001) compared to non-smokers. No significant interaction with rs1033667 was observed for tobacco smoking (P = 0.60). A case-only analysis was carried out for interaction of rs1033667 with alcohol use (see Materials and methods), but no significant interaction was observed (P = 0.13). Although the ALDH2 SNP rs4767364 was not associated with OSCC in our study, this variant was tested for interaction with alcohol because of strong evidence for interaction between variants in this gene and alcohol use in other populations (8,13), but no interaction was detected (P = 0.30).
Discussion
This study investigated seven main-effect susceptibility loci that were previously reported to be associated with OSCC in non-African populations (9–14) for association with this disease in the SAB population. The meta-analysis included 1471 cases and 1791 controls from the Western Cape and Gauteng provinces, which is more than double the sample size of any previous African study of oesophageal cancer genetics. Only one SNP (CHEK2 rs1033667) was significantly associated with OSCC in the SAB population in the meta-analysis of the two study samples [P = 0.002, OR = 1.18 (1.06–1.30)]. This finding suggests that shared aetiology of the disease may exist between the Chinese populations and the SAB populations involving the CHEK2 gene. None of the other loci tested showed evidence of transference to the SAB population, even though this study was well powered to detect these associations except for two low-frequency SNPs.
CHK2 (encoded by CHEK2) is a serine-threonine kinase with a key role in the DNA damage response pathway. Upon activation by genotoxic stress, it can phosphorylate downstream proteins to lead the activation of DNA repair, cell-cycle arrest, senescence or apoptosis (28). Rare germline mutations in CHEK2 have been implicated in susceptibility to multiple cancers, including strong evidence of association with breast and prostate cancer (29). The CHEK2 gene lies within a 100 kb region of chromosome 22 which also contains other genes of interest in relation to cancer, XBP1, HSCB and TTC28. XPB1 encodes an X-box binding protein 1 which is involved in the unfolded protein response in the endoplasmic reticulum. The stress caused by the accumulation of unfolded proteins activates this response which leads to the restoration of normal protein folding, or in severe cases, apoptosis (30). XBP1 has been shown to have an important role in the development and progression of triple-negative breast cancer through its control of the HIF1alpha pathway (31). HSCB encodes a member of the heat shock cognate B (HscB) family of proteins involved in the synthesis of iron-sulphur clusters and redox reactions of mitochondrial electron transport. Expression of HSCB is elevated in breast cancer tissue, and downregulation of this gene has recently been shown to reduce cell proliferation (32). TTC28 encodes a tetratricopeptide repeat containing protein which may be involved in spindle formation in mitosis. Whole genome sequencing of OSCC tumour tissue has revealed frequent structural alterations in TTC28, the significance of which is not yet clear (33). The CHEK2 SNP rs1033667 is associated with altered expression of CHEK2 itself and also of HSCB and TTC28. A detailed fine-map of this region in large sample sizes will be required to address the genes and variants that are driving this association in more detail.
The variants from the other six loci of interest were not significantly associated with OSCC in either of our sample sets or in the meta-analysis. These included a low-frequency functional SNP in TP53, Pro47Ser, which is unique to African populations (20). However, the SNP has an allele frequency of only 2–3%, so it would require a very large sample size to resolve a possible association with this or any other cancers in African populations. Interestingly, this variant has recently been reported to be associated with pre-menopausal breast cancer in African-American women (34). In our previous study of the SAB population of the Western Cape, we reported evidence of a possible association of a coding SNP in PLCE1, rs17417407, with OSCC (16). However, this was not replicated in the JCS sample and the association was not significant in the meta-analysis. It is possible that the larger sample size is a more robust reflection of the effect size for this variant, or there may be inter-African population differences that will only be resolved by the analysis of very large sample sizes in these populations (35). As discussed previously, there are several possible reasons for a lack of association in our studies (15,16). First, there may be insufficient power to detect the effect sizes observed in the original GWAS studies. However, we had 78–96% power to detect an effect (OR) of 1.26 for the GWAS-derived SNPs at alpha 0.0046 in the meta-analysis, so detection of an association signal should have been possible at most of these loci.
Another possibility is that the actual causal variants in the Chinese population arose after the migration of humans out of Africa and are therefore not present in African populations. A recent exome-wide association study of OSCC reported six new susceptibility loci in the Chinese population (36). However, inspection of the 1000 Genome project phase 3 data for West and East African populations showed that two of the three low-frequency variants (rs117353193 and rs17848945) are monomorphic and one (rs138478634) has a MAF of 0.001; thus, these three loci are unlikely to make a significant contribution to OSCC risk in African populations. The three common associated variants from chromosome 6p21.3 in the exome study will be tested as part of an ongoing GWAS in the SAB population.
An important challenge in the investigation of transference of genetic factors between African and non-African populations is that LD is generally lower across African genomes as compared to Asian and European genomes, with shorter haplotype blocks (37). Thus, the genotyped SNPs may be in high LD with the causal variant in the Chinese population but in low LD in the SAB populations, and hence the genotyped SNP would not ‘tag’ the causal SNP in an African population well and a disease association might not be observed. Finally, it is possible that differences in population structure between cases and controls could contribute to the weakening of genuine differences in allele frequencies of SNPs between cases and controls (37). This seems unlikely in the UCT sample since cases and controls were predominantly from the same ethno-linguistic group, but the JCS study sample included a broader ethnic mix of SAB participants. In general, one would expect differences in the population structure of cases and controls to increase false positive associations rather than reducing genuine disease associations, but this could only be resolved by high throughput genotyping of very large numbers of SNPs with appropriate statistical correction for any differences observed.
Tobacco smoking is a well-established risk factor for OSCC (reviewed in 4). We therefore investigated the only locus in our study associated with OSCC for interaction with smoking using a case-control analysis. We confirmed tobacco smoking as an independent risk factor for OSCC in this sample but did not find evidence for an interaction between the CHEK2 variant rs1033667 and smoking. A case-only analysis, rather than a case-control analysis, was done to investigate a gene-environment interaction for the rs1033667 variant with alcohol consumption owing to the very high alcohol consumption rate in the JCS control samples. No differences in allele frequencies of rs1033667 were observed when comparing drinkers with non-drinkers, which is consistent with a previous report that associations with OSCC at the chromosome 22q12 locus did not differ significantly between subgroups with different alcohol drinking status (13). Although the ALDH2 SNP rs4767364 was not associated with OSSC in our study, this variant was tested for interaction with alcohol use because of strong evidence for interaction between variants in this gene and alcohol in other populations (8,13), but no interaction was detected.
Encouragingly, our current study has identified a SNP in the CHEK2 locus that is associated with OSCC in the SAB population. This provides support for the hypothesis that at least some risk variants for OSCC will prove to be shared across multiple populations and for the prior evidence of association at CHEK2 in GWAS and DNA repair pathway analysis of OSCC in Chinese populations (13,14,38). Although most risk variants from other populations do not replicate in the SAB population, an African GWAS is needed to provide deep coverage of the known OSCC risk loci and to identify potential novel risk variants in African populations.
Funding
This work was supported by the Cancer Association of South Africa (to C.G.M., C.B.dV. and C.M.L.), the University of the Witwatersrand Research Council (to C.G.M.), the National Research Foundation of South Africa (to C.G.M.: incentive grant to a rated researcher; W.C.C.: German Academic Exchange Service-National Research Foundation Joint In-country Scholarship Programme and the National Research Foundation and Department of Science and Technology Thuthuka fund); Boehringer Ingelheim Fonds travel grant (to W.C.C.); The Grants, Innovation and Product Development Unit (GIPD) of the South African Medical Research Council (SAMRC) and the South African National Department of Health (to E.S.), King’s Health Partner’s Research and Development Challenge Fund, King’s College London (to C.G.M. and H.B.), the South African Medical Research Council Newton fund #045 to the ERICA-SA project (to C.G.M., C.B.dV., C.M.L. and E.S.) and the Wellcome Trust (grant 094491/Z/10/Z to N.J.P.). In addition, this paper represents independent research part-funded by the National Institute for Health Research Biomedical Research Centre at South London and Maudsley National Health Service Foundation Trust and King’s College London, and the National Institute for Health Research Biomedical Research Centre at Guy’s and St Thomas’ National Health Service Foundation Trust and King’s College London, United Kingdom. The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research or the Department of Health and Social Care. M.I.P., M.M. and C.G.M. were supported by a grant from the South African Medical Research Council, with funds received from the South African National Department of Health, the Medical Research Council (United Kingdom) and GlaxoSmithKline via the Newton Fund grant #046. M.M. was recipient of an International Centre for Genetic Engineering and Biotechnology Postdoctoral Fellowship.
Supplementary Material
Acknowledgements
We thank sisters Gloria Mokwatle, Patricia Rapoho and Pheladi Kale for assistance with sample collection, and Phillip Tombleson (National Institute for Health Research Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London), Margaret Urban and Tonicah Maphanga (National Cancer Registry, National Health Laboratory Service, South Africa) for assistance in the preparation and management of the sample database; Cassandra Soo and Natalie Smyth (Sydney Brenner Institute for Molecular Bioscience Biobank) and Ariella Rowe (International Centre for Genetic Engineering and Biotechnology) for DNA extractions; Dr Aron Abera and Felicity Azubuike (Inqaba Biotechnical Industries (Pty) Ltd. Southern Africa) for MassARRAY genotyping.
Glossary
Abbreviations
- eQTL
expression quantitative trait locus
- GxE
gene x environmental
- GWAS
genome-wide association studies
- JCS
Johannesburg Cancer Study
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OC
oesophageal cancer
- OSCC
oesophageal squamous cell carcinoma
- SAB
South African Black
- SAP
shrimp alkaline phosphatase
- SNP
single nucleotide polymorphism
- UCT
University of Cape Town
References
- 1. Ferlay J., et al. (2013) GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. International Agency for Research on Cancer, Lyon. [Google Scholar]
- 2. Arnold M., et al. (2015) Global incidence of oesophageal cancer by histological subtype in 2012. Gut, 64, 381–387. [DOI] [PubMed] [Google Scholar]
- 3. National Cancer Registry. Cancer in South Africa 2014 Full Report http://www.nioh.ac.za/assets/files/2014%20NCR%20tables.pdf (1 May 2018, date last accessed).
- 4. Abnet C.C., et al. (2018) Epidemiology of esophageal squamous cell carcinoma. Gastroenterology, 154, 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murphy G., et al. (2017) International cancer seminars: a focus on esophageal squamous cell carcinoma. Ann. Oncol., 28, 2086–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Matejcic M., et al. (2017) Alcohol metabolism and oesophageal cancer: a systematic review of the evidence. Carcinogenesis, 38, 859–872. [DOI] [PubMed] [Google Scholar]
- 7. Matejcic M., et al. (2015) Gene-environment interactions in esophageal cancer. Crit. Rev. Clin. Lab. Sci., 52, 211–231. [DOI] [PubMed] [Google Scholar]
- 8. Cui R., et al. (2009) Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology, 137, 1768–1775. [DOI] [PubMed] [Google Scholar]
- 9. McKay J.D., et al. (2011) A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet., 7, e1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abnet C.C., et al. (2010) A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet., 42, 764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L.D., et al. (2010) Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat. Genet., 42, 759–763. [DOI] [PubMed] [Google Scholar]
- 12. Wu C., et al. (2011) Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations. Nat. Genet., 43, 679–684. [DOI] [PubMed] [Google Scholar]
- 13. Wu C., et al. (2012) Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat. Genet., 44, 1090–1097. [DOI] [PubMed] [Google Scholar]
- 14. Wu C., et al. (2014) Joint analysis of three genome-wide association studies of esophageal squamous cell carcinoma in Chinese populations. Nat. Genet., 46, 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bye H., et al. (2011) Population-specific genetic associations with oesophageal squamous cell carcinoma in South Africa. Carcinogenesis, 32, 1855–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bye H., et al. (2012) Distinct genetic association at the PLCE1 locus with oesophageal squamous cell carcinoma in the South African population. Carcinogenesis, 33, 2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urban M., et al. (2012) Injectable and oral contraceptive use and cancers of the breast, cervix, ovary, and endometrium in black South African women: case-control study. PLoS Med., 9, e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matejcic M., et al. (2011) Association of a deletion of GSTT2B with an altered risk of oesophageal squamous cell carcinoma in a South African population: a case-control study. PLoS One, 6, e29366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen W.C., et al. (2018) The integrity and yield of genomic DNA isolated from whole blood following long-term storage at -30°C. Biopreserv. Biobank., 16, 106–113. [DOI] [PubMed] [Google Scholar]
- 20. Jennis M., et al. (2016) An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev., 30, 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. The 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Machiela M.J., et al. (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics, 31, 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gabriel S., et al. (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet., Chapter 2, Unit 2.12. [DOI] [PubMed] [Google Scholar]
- 24. Goh L.L., et al. (2017) Analysis of genetic variation in CYP450 genes for clinical implementation. PLoS One, 12, e0169233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Syrmis M.W., et al. (2011) Comparison of a multiplexed MassARRAY system with real-time allele-specific PCR technology for genotyping of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect., 17, 1804–1810. [DOI] [PubMed] [Google Scholar]
- 26. Westra H.J., et al. (2013) Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet., 45, 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. GTEx Consortium (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multi-tissue gene regulation in humans. Science, 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zannini L., et al. (2014) CHK2 kinase in the DNA damage response and beyond. J. Mol. Cell Biol., 6, 442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Southey M.C., et al. ; Australian Ovarian Cancer Study Group (2016) PALB2, CHEK2 and ATM rare variants and cancer risk: data from COGS. J. Med. Genet., 53, 800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hetz C., et al. (2013) Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov., 12, 703–719. [DOI] [PubMed] [Google Scholar]
- 31. Chen X., et al. (2014) XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature, 508, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S., et al. (2018) ChIP-seq analysis reveals alteration of H3K4 trimethylation occupancy in cancer-related genes by cold atmospheric plasma. Free Radic. Biol. Med., 126, 133–141. [DOI] [PubMed] [Google Scholar]
- 33. Chang J., et al. (2017). Genomic analysis of oesophageal squamous cell carcinoma identifies alcohol drinking related mutation signature and genomic alterations. Nat. Comm., 8, 15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murphy M.E., et al. (2017) A functionally significant SNP in TP53 and breast cancer risk in African-American women. NPJ Breast Cancer, 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gurdasani D., et al. (2015) The African genome variation project shapes medical genetics in Africa. Nature, 517, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang J., et al. (2018) Exome-wide analyses identify low-frequency variant in CYP26B1 and additional coding variants associated with esophageal squamous cell carcinoma. Nat. Genet., 50, 338–343. [DOI] [PubMed] [Google Scholar]
- 37. Teo Y.Y., et al. (2010) Methodological challenges of genome-wide association analysis in Africa. Nat. Rev. Genet., 11, 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wen-Qing L., et al. (2013). Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis, 34, 1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.