Abstract
Irrawaddy squirrel (Callosciurus pygerythrus) may play an important role in the transmission of zoonotic bacteria, but little is known about the carriage of zoonotic bacteria in this common frugivorous rodent in Bangladesh. We aimed to investigate the presence of common zoonotic bacterial pathogens in Irrawaddy squirrel in the southeast part of Bangladesh. A total of 27 rectal and 27 oro‐nasal swabs were collected from 27 healthy wild Irrawaddy squirrels. Four common zoonotic bacteria were isolated following routine laboratory procedures, and were identified based on colony morphology, and biochemical and staining properties. The pathogenic potential of the identified bacteria was confirmed by detection of virulence genes by PCR. All isolates were subjected to antimicrobial susceptibility test against seven antibiotics from six generic groups which are commonly used in human and veterinary medicine in Bangladesh. The prevalence of Escherichia coli, Salmonella spp., Yersinia spp. and Staphylococcus spp. was 44.4% (95% CI, 32.0–57.6), 13% (95% CI, 6.1–24.7), 44.4% (95% CI, 32.0–57.6), and 72.2% (95% CI, 59.0–82.5), respectively. We identified potential zoonotic virulence genes in all of these four bacterial species. Antimicrobial susceptibility testing revealed the presence of several multidrug resistant bacterial strains in squirrels. To the best of our knowledge, this is the first report in Bangladesh of the detection of antibiotic resistant zoonotic bacteria in Irrawaddy squirrels. The findings underpin the role of Irrawaddy squirrel as a source of pathogenic antibiotic resistant bacteria, consequently, fruit rejected because of squirrel consumption and squirrel‐bites deserve more concern than previously.
Keywords: antibiotic resistance, Irrawaddy squirrel, PCR, zoonotic bacteria
Introduction
Irrawaddy squirrel (Callosciurus pygerythrus), a small‐ to medium‐size rodent in the family Sciuridae, is widely distributed in Asian countries including Bangladesh (20°34′N; 92°41′E) (Adhri et al. 2015). Squirrels are one of the most common wild rodents that live near human habitants. Recently, they have become a major cause of nuisance in orchards and fruit gardens in Bangladesh; they eat or destroy both green and ripened fruits. Generally, children and many adults directly consume fruits half‐eaten by squirrels. Little is known about the microbial risk of consuming squirrel contaminated half‐eaten fruits. On the other hand, squirrels are also reared as pets in different parts of the world. Furthermore, the ground squirrel animal model is a long‐standing model for the studies of hibernation biochemistry and physiology (Vaughan et al. 2006) and it is an emerging model for other medical experimentation (Arendt et al. 2003). Thus, squirrel bites are a common health hazard for pet lovers and researchers. However, it is not known whether squirrel carry any antibiotic resistant zoonotic bacteria or not. Therefore, we aimed to investigate the presence of common zoonotic bacteria in Irrawaddy squirrels, and the antibiotic susceptibility of any isolates.
Materials and methods
Study design and sample collection
On the basis of a preliminary survey on the abundance of Irrawaddy squirrel, the study was conducted in different environments (e.g. forest, orchards and fruits gardens etc.) of the Cox's bazar (area 1) and the Bandarban district (area 2) of Bangladesh during January 2016 to June, 2017. The live squirrels were captured using steel wire traps (30 cm × 15 cm × 15 cm) and nets, balancing animal welfare and personal safety. Animals were captured in three stages. The first stage was during post‐fruiting period (May, 2016); the second stage was performed in a period of the year when there are relatively fewer fruits available in the study area (October–November, 2016); and the third stage was carried out during fruiting season (April, 2017). The captured squirrels were anaesthetized using cotton wool soaked in Forane (Isoflurane; Abbott Laboratories Ltd.) to avoid any hazard related to squirrel bites. One rectal swab and one oro‐nasal swab samples were collected using sterile cotton tipped swab stick in sterile 50 mL Falcon Tubes containing 10 mL Buffered Peptone Water (Oxoid Ltd., PH: 6.2 ± 0.0) and/or in 1.5 mL Eppendorf tube containing 1 mL Stuart's Transport Media. Samples were immediately transported to the laboratory of Department of Microbiology and Veterinary Public Health (DMVPH), Chittagong Veterinary and Animal Sciences University (CVASU), Chittagong, Bangladesh, for microbiological analysis. After sample collection, the squirrels were fed and kept in a comfortable cage until being able to normally move, and then they were released within their natural habitat.
Isolation and identification of bacteria
Four selective bacteria (Escherichia coli, Salmonella spp., Yersinia spp. and Staphylococcus spp.) were isolated from swab samples, following standard laboratory protocols. The mentioned bacteria were identified based on colony morphology on selective media, biochemical and staining properties (Carter et al. 1995), and Polymerase Chain Reaction (PCR) of species‐specific pathogenic genes (Table 1). Identified bacterial isolates were preserved at −80°C in brain heart infusion broth with 50% glycerine for further use.
Table 1.
Sequences of primers used to detect species‐specific pathogenic gene(s) in Escherichia coli, Salmonella spp., Yersinia spp. and Staphylococcus spp
| Organism | Target Gene | Primer Sequence | Annealing Temp (°C) | Amplicon (bp) | Reference |
|---|---|---|---|---|---|
| E. coli | hly | F: ACGATGTGGTTTATTCTG GA | 58 | 165 | DesRosiers et al. 2001; |
| R:CTTCACGTGACCATACAT AT | |||||
| stx1 | F:ACACTG GATGATCTCAGTGG | 58 | 614 | DesRosiers et al. 2001; | |
| R:CTGAATCCCCCTCCATTATG | |||||
| stx2 | F:CCATGACAACGGACAGCAGTT | 58 | 779 | Islam et al. 2015; | |
| R:CCTGTCAACTGAGCAGCACTTT | |||||
| eae | F:CCCGAATTCGGCACAAGCAT | 59 | 881 | Oswald 2000; | |
| R:CCCGGATCCGTCTCGCCAGTA | |||||
| Salmonella spp. | – | ST11:AGCCAACCATTGCTAAATTGGCGCA | 60 | 429 | Aabo et al. 1993; |
| ST15:GGTAGAAATTCCCAGCGGGTACTG | |||||
| Yersinia spp. | pla | F:ATCTTACTTTCCGTGAGAAG | 55 | 478 | Hinnebusch & Schwan 1993; |
| R:CTTGGATGTTGAGCTTCCTA | |||||
| ail | F:TAATGTGTACGCTGCGAG | 57 | 351 | Thoerner et al. 2003; | |
| R:GACGTCTTACTTGCACTG | |||||
| Staphylococcus spp. | fnbA | F:GCGGAGATCAAAGACAA | 50 | 1279 | Signas et al. 1989 |
| R:CCATCTATAGCTGTGTGG |
Temp, temperature; °C, degree centigrade; bp, base pair; F, Forward primer; R, Reverse primer.
Polymerase chain reaction (PCR)
Bacterial colonies grown overnight on blood agar plates were used for DNA extraction by boiling lysates method, according to previously described protocol (Islam et al. 2015). PCR was performed to detect species specific pathogenic gene(s) in bacteria using previously reported standard primer sequences and protocols (Table 1). In brief, 25 μL PCR mix was prepared by mixing 12.5 μL ready to use master mix (Thermo Scientific Ltd., USA), 9.5 μL nuclease‐free water, 1 μL of each primer, and 1 μL DNA template. PCR amplification was performed on a thermo‐cycler (Applied Biosystem, 2720 thermal cycler, Singapore) using 94–95°C denaturation temperature, 50–60°C annealing temperature and 72°C extension temperature for 30–35 cycles.
Antimicrobial susceptibility test
All cultured isolates were tested for antimicrobial susceptibility profile using a panel of seven antibiotics within six generic groups by standard disc diffusion according to Kirby‐Bauer method (Bauer et al. 1966). Zone diameters were measured and interpreted following Clinical Laboratory and Standards Institute guideline (CLSI, 2016).
Data analysis
All attributed data were recorded in an Excel spread sheet (Excel 2007, Microsoft Corporation, USA) and subjected to statistical analysis. Logistic regression and Pearson's Chi‐squared (two tailed) test was performed in R (version 3.3.2). The 95% confidence interval of the prevalence values were calculated by the modified Wald method using QuickCalcs in the Graph Pad software.
Results
Overall prevalence
We collected 54 swabs (27 rectal, 27 oro‐nasal) from 27 squirrels. E. coli, Salmonella spp., Yersinia spp. and Staphylococcus spp. were isolated from 24 (3 oro‐nasal, 21 rectal), 7 (0 oro‐nasal, 7 rectal), 24 (9 oro‐nasal, 15 rectal) and 39 (19 oro‐nasal, 20 rectal) samples, respectively. A comprehensive result of bacteriological analysis is shown in Table 2. The overall prevalence of these four bacteria was relatively higher in rectal swab samples (58.3%) compared to oro‐nasal swabs (28.7%). The prevalence of E. coli, Salmonella spp., Yersinia spp. and Staphylococcus spp. in oro‐nasal swabs were 11.1%, 0%, 33.3% and 74.1%, respectively. Conversely, the prevalence of E. coli, Salmonella spp., Yersinia spp. and Staphylococcus spp. in rectal swabs were 77.8%, 25.9%, 55.6% and 74.1% respectively. Although, the presence of E. coli and Salmonella spp. was significantly (P < 0.05) related to type of sample, the presence of Yersinia spp. and Staphylococcus spp. was not related to this variable. The prevalence of Yersinia spp. and Staphylococcus spp. in Irrawaddy squirrel was significantly (P < 0.05) related to the season in the study area, while the isolation of E. coli and Salmonella spp. showed no significant relationship with seasons. There was no significant effect of sex on the prevalence of any of the four species of bacteria, although male squirrels had relatively higher loads of Staphylococcus spp., but with a borderline significance (P = 0.083).
Table 2.
Relationship between different categorical variables and prevalence of bacteria identified, based on colony morphology, staining and biochemical properties
| Variables | Categories | N | Proportionate Prevalence | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | Salmonella spp. | Yersinia spp. | Staphylococcus spp. | |||||||
| PP (95% CI) | P | PP (95% CI) | P | PP (95% CI) | P | PP (95% CI) | P | |||
| Season | May, 2016 | 8 | 37.5 (13.5–69.6) | 0.533 | 0.0 (0.0–37.2) | 0.352 | 0.0 (0.0–37.2) | 0.016* | 12.5 (0.1–49.2) | 0.000* |
| Oct–Nov, 2016 | 14 | 57.1 (32.6–78.7) | 21.4 (6.8–48.3) | 42.9 (21.3–67.5) | 50.0 (26.8–73.2) | |||||
| April, 2017 | 32 | 40.6 (25.5–57.8) | 12.5 (4.4–28.7) | 56.2 (39.3–71.9) | 100 (87.3–100) | |||||
| Sex | M | 30 | 43.3 (27.4–60.8) | 0.854 | 13.3 (4.7–30.4) | 0.928 | 46.7 (30.2–63.9) | 0.713 | 83.3 (66.0–93.1) | 0.083 |
| F | 24 | 45.8 (27.9–64.9) | 12.5 (3.5–31.8) | 41.7 (24.4–61.2) | 62.5 (42.6–78.9) | |||||
| Sample | ON | 27 | 11.1 (3.0–28.9) | 0.000* | 0.0 (0.0–14.8) | 0.005* | 33.3 (18.5–52.3) | 0.100 | 74.1 (55.1–87.1) | 1.000 |
| R | 27 | 77.8 (58.9–89.7) | 25.9 (12.9–44.9) | 55.6 (37.3–72.4) | 74.1 (55.1–87.1) | |||||
| Total | 54 | 44.4 (32.0–57.6) | 13.0 (6.1–24.7) | 44.4 (32.0–57.6) | 72.2 (59.0–82.5) | |||||
*, significant P‐value; ON, Oro‐nasal swab; R, rectal swab; M, male squirrel; F, female squirrel; %, percentage; N, number of samples; PP, prevalence; P, probability value; CI, confidence interval.
Detection of virulence genes
Among 24 culture and biochemically positive E. coli isolates, eight isolates (33.3%; 95% CI, 17.8–53.4) carried shiga toxin producing gene (stx2); one (4.2%; 95% CI, 0.01–21.8) isolate carried the intimin encoding gene (eae). Furthermore, one isolate harboured both stx2 and eae genes. Two (28.6%; 95% CI, 7.6–64.8) of seven culture‐positive Salmonella spp. were positive for ST DNA fragment. One (4.2%; 95% CI, 0.01–21.8) of 24 tentatively identified Yersinia spp. was carrying the plasminogen activator protein (pla) encoding gene while another two (8.3%; 95% CI, 1.2–27.0) were harbouring the attachment invasion locus protein (ail) producing gene. Although Staphylococcus spp. was the most prevalent bacteria isolated from squirrel, only one (2.6%; 95% CI, 0.01–20.6) of 39 culture‐positive isolates was positive for the fibrinogen binding protein A (fnbA) encoding gene (Table 3).
Table 3.
Presence of virulence genes in isolated bacteria
| Organism | N | Target genes | Positive samples | Proportion | 95% CI |
|---|---|---|---|---|---|
| E. coli | 24 | hly | 0 | 0.0 | 0.0–16.3 |
| stx1 | 0 | 0.0 | 0.0–16.3 | ||
| stx2 | 8 | 33.3 | 17.8–53.4 | ||
| eae | 1† | 4.2 | 0.01–21.8 | ||
| Salmonella spp. | 7 | ST * | 2 | 28.6 | 7.6–64.8 |
| Yersinia spp. | 24 | pla | 1 | 4.2 | 0.01–21.8 |
| ail | 2 | 8.3 | 1.2–27.0 | ||
| Staphylococcus spp. | 39 | fnbA | 1 | 2.6 | 0.01–20.6 |
N, No. of culturally and biochemically positive samples; CI, Confidence interval.
*ST is a genus‐specific DNA fragment of Salmonella.
One sample carried both eae and stx2 genes.
Antimicrobial resistance pattern of isolated bacteria
All culturally positive bacterial isolates were tested for antimicrobial susceptibility for a panel of seven commonly used antibiotics under six generic groups. Test results are represented in Fig. 1. During this study, E. coli showed highest resistance to colistin sulphate (53.8%) and sensitive to gentamicin (100%). About 71.4% Salmonella spp. were resistant to colistin sulphate while most of the Salmonella isolates were sensitive to gentamycin (100%), sulphamethoxazole‐ trimethoprim (85.7%). Yersinia isolates were resistant to amoxicillin (69.2%) but sensitive to gentamicin (84.6%). Staphylococcus spp. isolates were most likely sensitive to ciprofloxacin (92.3%). Bivariate logistic regression analysis revealed that Yersinia spp. and Staphylococcus spp. were relatively more likely to be resistant to the antibiotics tested, but Salmonella spp. isolates also showed almost similar resistance pattern of E. coli (Table 4)
Figure 1.
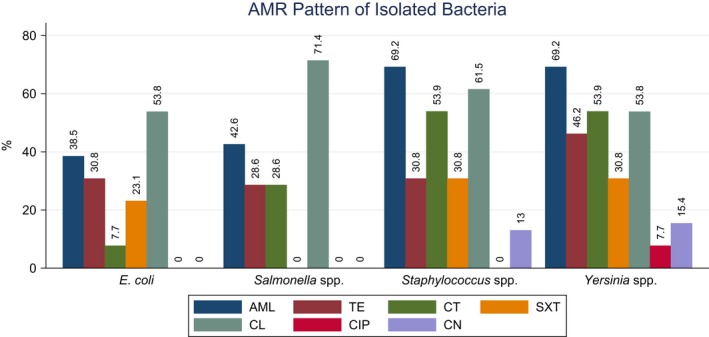
Antibiotic resistance pattern (expressed in percentage) of Escherichia coli, Salmonella spp., Staphylococcus spp. and Yersinia spp., isolated from Irrawaddy Squirrel, against AML, amoxicillin; CIP, ciprofloxacin; CL, cephalexin; CN, gentamycin; CT, colistin sulphate; SXT, sulphamethoxazole‐ trimethoprim; TE, tetracycline.
Table 4.
Bivariate relationship between different categorical variables and AMR of bacteria. E. coli, rectal swab and area‐1 were used as reference value for organisms, sample type and location, respectively, to determine the association of these categorical variables with the resistance of bacteria to different antibiotics
| Categories | Antimicrobials | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SXT | CT | AML | TE | CIP | CL | CN | ||||||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| (A) Organisms | ||||||||||||||
| E. coli | Ref | Ref | Ref | Ref | Ref | Ref | Ref | |||||||
| Salmonella spp. | 0.6 (0.0–6.6) | 0.642 | 4.8 (0.4–65.8) | 0.240 | 2.1 (0.3–15.3) | 0.448 | 0.5 (0.1–3.3) | 0.448 | 4.8 (0.3–65.8) | 0.240 | 1.0 | NA | 1.0 | NA |
| Yersinia spp. | 1.4 (0.3–8.5) | 0.659 | 14 (1.4–141.5) | 0.025* | 4.7 (0.7–30.3) | 0.102 | 1.4 (0.3–6.4) | 0.695 | 5.3 (0.5–56.2) | 0.164 | 0.1 (0.0–1.4) | 0.090 | 0.3 (0.0–1.9) | 0.197 |
| Staphylococcus spp. | 2.1 (0.4–11.5) | 0.399 | 14 (1.4–141.5) | 0.025* | 1.9 (0.4–9.6) | 0.423 | 0.7 (0.1–3.5) | 0.692 | 1.0 (0.1–17.9) | 1.000 | 0.5 (0.0–5.8) | 0.547 | 1.0 | NA |
| (B) Sample type | ||||||||||||||
| R | Ref | Ref | Ref | Ref | Ref | Ref | Ref | |||||||
| ON | 0.5 (0.1–2.0) | 0.302 | 1.6 (0.4–5.4) | 0.487 | 0.4 (0.1–1.4) | 0.157 | 2.2 (0.6–7.6) | 0.206 | 4.1 (0.8–20.1) | 0.083 | 0.2 (0.0–1.3) | 0.083 | 3 (0.6–15.5) | 0.190 |
| (C) Location | ||||||||||||||
| Area 1 | Ref | Ref | Ref | Ref | Ref | Ref | Ref | |||||||
| Area 2 | 1.0 (0.3–4.1) | 0.975 | 0.9 (0.3–3.4) | 0.908 | 1.9 (0.4–8.4) | 0.384 | 0.4 (0.1–1.5) | 0.184 | 0.3 (0.0–2.5) | 0.250 | 0.7 (0.1–3.3) | 0.634 | 0.3 (0.0–3.1) | 0.332 |
*, Significant P‐value; ON, oro‐nasal swab; OR, odd ratio; NA, not applicable; R, rectal swab; P, probability value; Ref, reference value.
Multi‐drug resistance (MDR) of isolated bacteria
The drug resistance pattern of isolated bacteria shows that the Irrawaddy squirrel can carry multi‐drug resistant bacteria. From 27 Irrawaddy squirrel, four E. coli, four Salmonella spp., six Yersinia spp. and six Staphylococcus spp. were isolated that showed resistance against three or more unrelated groups of antibiotics (Table S1).
Discussion
The result of this study revealed that Irrawaddy squirrels can carry Escherichia coli, Salmonella spp., Yersinia spp. and Staphylococcus spp. Bacterial load was higher in rectal swabs (58.3%) in comparison to oral swabs (28.7%). E. coli is the predominant facultative anaerobic organism among the aerobic commensal flora in gut of human and animal (Chart 2002; Russo & Johnson 2003).
Among all strains, a group of E. coli, the Shiga toxin producing E. coli (STEC), have recently received particular attention due to the serious clinical implications associated with large scale epidemics in humans attributed to this pathogen. Along with many other virulence markers such as enterohemolysin, catalase‐peroxidase, adhesion, intimin etc. (Etcheverria & Padola 2013), STEC produces two major groups of Shiga toxins, Stxl and Stx2 (Kuntz & Kuntz 1999) which are responsible for severe diseases, such as haemolytic uremic syndrome, in humans (Griffin & Tauxe 1991; Friedrich et al. 2002). In this study, the presence of stx2 gene in eight (33.3%, 95% CI, 17.8–53.4) E. coli isolates was confirmed by PCR; which indicates the potential significance of squirrels carrying the virulent strain as the strain producing Stx2 alone is considered more virulent than those producing Stx1 alone, or Stx1 and Stx2 in combination (Donohue‐Rolfe et al. 2000). Furthermore, the presence of the intimin encoding gene (eae gene) in the same isolate enhanced the probability of the existence of virulent and zoonotic strains of E. coli in squirrels. However, there was no isolate showing positivity for haemolytic properties on blood agar or hly and/or Stx1 gene(s) in PCR. This suggests that this wild species may not carry E. coli O157 strain. E. coli isolated from squirrels showed resistance to many commonly used antibiotics in human and veterinary medicine such as‐ colistin sulphate (53.8%), amoxicillin (38.5%) and tetracycline (30.8%).
Antimicrobial resistance (AMR) was relatively, but not significantly, higher in squirrels captured from area 1, which has previous history of poultry farms in the area. Three out of four multi‐drug resistant E. coli were isolated from samples collected from area 1. So, it can be hypothesized that squirrels acquired their antibiotic‐resistant bacteria from environmental sources contaminated by drug resistant E. coli strain from poultry farms, although further research needs to be done.
Water‐borne and enteric bacteria Salmonella spp. can cause diseases in humans, mammals and poultry. It is an important zoonotic bacteria and in this study, seven (25.9%; 95% CI, 12.9–44.9) rectal swab samples were found culture‐positive for Salmonella spp. Among them, the presence of genus‐specific DNA portion ST (ST11 and ST15) in two samples confirmed the isolates as pathogenic Salmonella spp. (Aabo et al. 1993). However, a further study is required to identify them fully. Salmonella spp., also, were resistant to colistin sulphate (71.4%) but sensitive to gentamicin (100%). The almost similar antimicrobial resistance of both E. coli and Salmonella spp. isolates may be due to their phylogenetic relatedness and/or their emergence from similar source. It may be a consequence of inter‐species resistance gene transfer although this needs to be proved by further research.
Yersinia is a genus of important and zoonotic significant bacteria that includes Y. pestis, Y. enterocolitica and Y. pseudotuberculosis. In this study, 24 (9 oro‐nasal, 15 rectal) samples were found to be culture positive for Yersinia spp. One isolate (4.2%; 95% CI, 0.01–21.8) was confirmed as Y. pestis through detection of pla gene with an amplicon size 478 bp. pla gene encoding plasminogen activating proteins (coagulase and fibrinolysin) which is found in pathogenic strains of Y. pestis (Kukkonen et al. 2001). However, recent findings suggest that pla gene is not specific to Y. pestis as it is also found in E. coli and Citrobacter koseri (Hansch et al. 2015). The ail gene encodes Attachment invasion locus protein (an outer membrane protein that helps bacteria to invade host cells) and is found in pathogenic strains of Y. enterocolitica (Sihvonen et al. 2011). In this study, two (8.3%; 95% CI, 1.2–27.0) isolates were confirmed as Y. enterocolitica as they were positive for ail gene in PCR assay (Miller et al. 1989). So, the present findings indicate that Irrawaddy squirrel can carry pathogenic and zoonotic Yersinia bacteria. However, we found no isolate that fully coincided with the cultural and biochemical properties of Y. pseudotuberculosis. Yersinia spp. showed the highest resistance to amoxicillin (69.2%) and highest sensitivity to gentamicin (84.6%). This may be due to the long‐term use of β‐lactam antibiotics in human and veterinary medicine, through gene transfer or mutation or selection pressure, and spread of amoxicillin‐resistant Yersinia spp. within the squirrel population. Staphylococcus spp. is one of the most prevalent bacteria isolated from animals (including squirrel) and human bite wounds (Goldstein et al. 1978). During this study, Staphylococcus spp. was isolated from 39 (72.2%) samples. Presence of fnbA gene in one sample (2.6%; 95% CI, 0.01–20.6) is an indication of pathogenic Staphylococcal carriage by Irrawaddy squirrels. The isolated staphylococci in this study were mostly resistant to amoxicillin (69.2%) and sensitive to ciprofloxacin (92.3%). This may reflect contamination of the natural environment with bacteria that have already acquired resistance against β‐lactam antibiotics as a consequence of their long term and widespread use in human and veterinary medicine.
During this study, multi‐drug resistance (MDR) was observed in all four types of bacteria. In present study, four (16.7%; 95% CI, 6.1–36.5) E. coli isolates, three (42.9%; 95% CI, 15.8–75.0) Salmonella spp. isolates, six (25%; 95% CI, 11.7–45.2) Yersinia spp. isolates and six (15.4%; 95% CI, 6.9–30.1) Staphylococcus spp. isolates were found resistant to three or more unrelated groups of antibiotics at a time, in vitro. This finding supports the existence of MDR in wild‐caught squirrels (Cloud‐Hansen et al. 2007), that challenges the treatment of any illness resulting from the consumption of squirrel rejected half‐eaten fruits or squirrel dung contaminated water or from squirrel bites during handling, in both humans and livestock. The bacterial load in squirrel was significantly related to the seasons. According to findings of this study, more bacteria were isolated during the fruiting period (April) compared to post‐fruiting (May) or off fruiting (October to November) periods. So, it can be assumed that there is a probability of transmission of pathogenic bacteria from squirrels to humans, if people eat squirrel rejected or half‐eaten fruits.
Conclusion
Irrawaddy squirrel can carry, in oral or faecal material, pathogenic and zoonotic bacteria such as E. coli, Salmonella spp., Yersinia pestis, Y. enterocolitica, Staphylococcus aureus etc. As all these bacteria may be associated with illness in humans and domestic animals, it is important to be concerned about the consumption of squirrel rejected fruits and squirrel bite wound management. The bacteria may acquire transposon‐mediated resistance genes from environmental bacteria which have already acquired resistance from human or animal pathogens and/or from genetic mutations. Although bacteria carried by Irrawaddy squirrel can be resistant to many commonly used antibiotics, gentamicin or ciprofloxacin are, at present, relatively effective against carried bacteria.
Source of funding
Advanced Studies and Research, Chittagong Veterinary and Animal Sciences University.
Conflicts of interest
None.
Contribution
MSJ designed and carried out this project. ADu, PKD, ADa, MMH contributed in samples collection, data collection and lab works, in a team. MSJ and MZI analyzed data and wrote this article; and HB, PKB, AH supervised the project and checked the writing, of this article.
Ethics statement
We captured live squirrels from the selected study areas of Bangladesh with a formal permission from Bangladesh Forest Department, and Samples were collected whilst assuring animal welfare. This project was approved by the ethical committee of Chittagong Veterinary and Animal Sciences University, Bangladesh.
Supporting information
Table S1. Multi‐drug resistant pattern of isolated bacteria
Acknowledgement
We acknowledge Bangladesh Forest Department for endorsing permission to collect samples from their authorized area.
References
- Aabo S., Rasmussen O.F., Roseen L., Sørensen P.D. & Olsen J.E. (1993) Salmonella identification by the polymerase chain reaction. Molecular and Cellular Probes 7, 171–178. 10.1006/mcpr.1993.1026 [DOI] [PubMed] [Google Scholar]
- Adhri A.S., Sultana A. & Rahman S. (2015) Habit and habitat of squirrels in Bangladesh. International Journal of Recent Research in Life Sciences 2, 41–44. [Google Scholar]
- Arendt T., Stieler J., Strijkstra A.M., Hut R.A., Rudiger J., Van der Zee E.A. et al (2003) Reversible paired helical filament‐like phosphorylation of tau is an adaptive process associated with neuronal plasticity in hibernating animals. The Journal of Neuroscience 23, 6972–6981. doi: content/23/18/6972.short [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A.W., Kirby W.M., Sherris J.C. & Turck M. (1966) Antibiotic susceptibility testing by a standardized single disk method. American Journal of Clinical Pathology 45, 493. [PubMed] [Google Scholar]
- Carter G.R., Chengappa M.M., Roberts A.W. (1995). Enterobacteriaceae. Essentials of veterinary Microbiology. 5th edn, pp 151–165. Williams & Wilkins: Philadelphia. [Google Scholar]
- Chart H. (2002) Escherichia: urinary tract infection; travel lers'diarrhoea; haemorrhagic colitis; haemolytic uraemic syndrome In: Medical Microbiology: A guide to Microbial Infections: Pathogenesis, Immunity, Laboratory Diagnosis and Control (eds Greenwood D., Slack R., Peutherer L.), 16th edn, pp 265–274. Churchill Livingstone: Edinburgh. [Google Scholar]
- Cloud‐Hansen K.A., Villiard K.M., Handelsman J. & Carey H.V. (2007) Thirteen‐lined ground squirrels (Spermophilus tridecemlineatus) harbor multiantibiotic‐resistant bacteria. Journal of the American Association for Laboratory Animal Science 46, 17–20. doi: aalas/jaalas/2007/00000046/00000003/art00003 [PubMed] [Google Scholar]
- CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing. 26th edn, pp 52–74. CLSI supplement M100S. Clinical and Laboratory Standards Institute: Wayne, PA. doi: file:///C:/Users/HP/Downloads/CLSI‐2016%20(1).pdf [Google Scholar]
- DesRosiers A., Fairbrother J.M., Johnson R.P., Desautels C., Letellier A. & Quessy S. (2001) Phenotypic and genotypic characterization of Escherichia coli verotoxin‐producing isolates from humans and pigs. Journal of Food Protection 64, 1904–1911. 10.4315/0362-028X-64.12.1904. [DOI] [PubMed] [Google Scholar]
- Donohue‐Rolfe A., Kondova I., Oswald S., Hutto D. & Tzipori S. (2000) Escherichia coli 0157: H7 strains that express Shiga toxin (Stx) 2 alone are more neurotropic for gnotobiotic piglets than are isotypes producing only Stx1 or both Stx1 and Stx2. The Journal of Infectious Diseases 181, 1825–1829. 10.1086/315421. [DOI] [PubMed] [Google Scholar]
- Etcheverria A.I. & Padola N.L. (2013) Shiga toxin‐producing Escherichia coli: factors involved in virulence and cattle colonization. Virulence 4, 366–372. 10.4161/viru.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich A.W., Bielaszewska M., Zhang W.L., Pulz M., Kuczius T., Ammon A. & Karch H. (2002) Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. The Journal of Infectious Diseases 185, 74–84. 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- Goldstein E.J., Citron D.M., Wield B., Blachman U., Sutter V.L., Miller T.A. & Finegold S.M. (1978) Bacteriology of human and animal bite wounds. Journal of Clinical Microbiology 8, 667–672. https://doi.org/0095-1137/78/0008-0667$02.00/0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin P.M. & Tauxe R.V. (1991) The epidemiology of infections caused by Escherichia coli O157: H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiologic Reviews 13, 60–98. 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- Hansch S., Cilli E., Catalano G., Gruppioni G., Bianucci R., Stenseth N.C. et al (2015) The pla gene, encoding plasminogen activator, is not specific to Yersinia pestis . BMC Research Notes 8, 535 10.1186/s13104-015-1525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch J. & Schwan T.G. (1993) New method for plague surveillance using polymerase chain reaction to detect Yersinia pestis in fleas. Journal of Clinical Microbiology 31, 1511–1514. https://doi.org/0095-1137/93/061511-04$02.00/0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.Z., Christensen J.P. & Biswas P.K. (2015) Sorbitol non‐fermenting shiga toxin‐producing Escherichia coli in cattle on smallholdings. Epidemiology & Infection 143, 94–103. 10.1017/S0950268814000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen M., Lahteenmäki K., Suomalainen M., Kalkkinen N., Emody L., Lang H. & Korhonen T.K. (2001) Protein regions important for plasminogen activation and inactivation of α2‐antip lasmin in the surface protease Pla of Yersinia pestis . Molecular Microbiology 40, 1097–1111. 10.1046/j.1365-2958.2001.02451.x. [DOI] [PubMed] [Google Scholar]
- Kuntz T.B. & Kuntz S.T. (1999) Enterohemorrhagic E. coli infection. Primary Care Update for OB/GYNS 6, 192–196. [Google Scholar]
- Miller V.L., Farmer J.J., Hill W.E. & Falkow S. (1989) The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infection and Immunity 57, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald E. (2000) Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infection and Immunity 68, 64–71. 10.1128/IAI.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T.A. & Johnson J.R. (2003) Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes and Infection 5, 449–456. 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Signas C., Raucci G., Jonsson K., Lindgren P.E., Anantharamaiah G.M., Heoeok M. & Lindberg M. (1989) Nucleotide sequence of the gene for a fibronectin‐binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proceedings of the National Academy of Sciences of the United States of America 86, 699–703. https://doi.org/content/86/2/699.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihvonen L.M., Hallanvuo S., Haukka K., Skurnik M. & Siitonen A. (2011) The ail gene is present in some Yersinia enterocolitica biotype 1A strains. Foodborne Pathogens and Disease 8, 455–457. 10.1089/fpd.2010.0747. [DOI] [PubMed] [Google Scholar]
- Thoerner P., Kingombe C.B., Bogli‐Stuber K., Bissig‐Choisat B., Wassenaar T.M., Frey J. & Jemmi T. (2003) PCR detection of virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis and investigation of virulence gene distribution. Applied and Environmental Microbiology 69, 1810–1816. 10.1128/AEM.69.3.1810-1816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan D.K., Gruber A.R., Michalski M.L., Seidling J. & Schlink S. (2006) Capture, care, and captive breeding of 13‐lined ground squirrels, Spermophilus tridecemlineatus . Laboratory Animal 35, 33–40. https://doi.org/00b4952a2247e7a09d000000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Multi‐drug resistant pattern of isolated bacteria


