Abstract
We delineate a KMT2E-related neurodevelopmental disorder on the basis of 38 individuals in 36 families. This study includes 31 distinct heterozygous variants in KMT2E (28 ascertained from Matchmaker Exchange and three previously reported), and four individuals with chromosome 7q22.2-22.23 microdeletions encompassing KMT2E (one previously reported). Almost all variants occurred de novo, and most were truncating. Most affected individuals with protein-truncating variants presented with mild intellectual disability. One-quarter of individuals met criteria for autism. Additional common features include macrocephaly, hypotonia, functional gastrointestinal abnormalities, and a subtle facial gestalt. Epilepsy was present in about one-fifth of individuals with truncating variants and was responsive to treatment with anti-epileptic medications in almost all. More than 70% of the individuals were male, and expressivity was variable by sex; epilepsy was more common in females and autism more common in males. The four individuals with microdeletions encompassing KMT2E generally presented similarly to those with truncating variants, but the degree of developmental delay was greater. The group of four individuals with missense variants in KMT2E presented with the most severe developmental delays. Epilepsy was present in all individuals with missense variants, often manifesting as treatment-resistant infantile epileptic encephalopathy. Microcephaly was also common in this group. Haploinsufficiency versus gain-of-function or dominant-negative effects specific to these missense variants in KMT2E might explain this divergence in phenotype, but requires independent validation. Disruptive variants in KMT2E are an under-recognized cause of neurodevelopmental abnormalities.
Keywords: KMT2E, global developmental delay, intellectual disability, epilepsy, epileptic encephalopathy, autism, neurodevelopmental disorder, H3K4 methylation
Main Text
KMT2E (GenBank: NM_182931.2, MIM: 608444) encodes a member of the lysine N-methyltransferase 2 (KMT2) family. This family of enzymes plays a vital role in regulating post-translational histone methylation of histone 3 on lysine 4 (H3K4).1 Proper H3K4 methylation is required to maintain open chromatin states for regulation of transcription. There are at least eight known monogenic disorders that impair regulation of H3K4 methylation and that present with neurodevelopmental syndromes2, 3, 4, 5, 6, 7, 8 (Table S1). In addition to these Mendelian disorders, dysregulated H3K4 methylation is believed to play a role in the pathogenesis of schizophrenia and autism.9 Truncating variants in KMT2E have previously been reported in three unrelated males in a large sequencing study of non-syndromic autism, but phenotypic data were limited.10, 11, 12 In this report, we present 35 additional individuals with heterozygous variants in KMT2E in an effort to define a KMT2E-related neurodevelopmental disorder.
New cases were ascertained from GeneMatcher through the Matchmaker Exchange Network and MyGene2 between September 2016 and August 2018.13, 14 Microdeletions were detected by chromosomal microarrays in some individuals, whereas all other individuals were found to have variants in KMT2E via exome or genome sequencing. Written consent for publication of photographs was provided from the individuals’ parents or legal guardians. Additional phenotype data and genetic findings for individuals are summarized in Table S2.
KMT2E is constrained for protein-truncating variation in the general population. The Genome Aggregation Database (gnomAD) is a large-scale reference database with high-quality, jointly processed exome or genome data from more than 140,000 individuals.15 Constraint analysis performed on the gnomAD dataset shows that KMT2E is a candidate haploinsufficient gene. KMT2E is very depleted (presumably as a result of negative selection) for protein-truncating variants; there is a probability of loss-of-function intolerance (pLI) score of 1.0 and an observed/expected ratio of 0.01 (showing 1% [0–0.06 95% CI] of expected loss-of-function variation in gnomAD).
We reviewed the 28 loss-of-function variants present in gnomAD v2.1 (Table S3). The majority of these variants are not expected to result in protein truncation for a variety of reasons, including annotation artifacts (n = 8), sequence errors at a simple repeat (n = 5), somatic mosaicism (n = 1), and a splice-site rescue (n = 1). Four variants are part of a complex variant found in one individual; this complex variant, when resolved, is not expected to result in truncation. Four variants found in eight individuals in gnomAD are in the last exon; two are expected to result in truncation of the last exon, and two will result in protein extension. Of note, the two protein-extension variants are located close to the variant in individual 28 (c.5453_5460delTGGCCCTG [p.Val1818Alafs∗48]). The inheritance of this variant is unknown because the father is not available for testing, although it is not present in his mother, so this remains a variant of uncertain significance.
After review, we found that five variants in gnomAD appear to result in protein truncation. These are found in three males and two females between the ages of 30 and 70. All five are absent from the control-only subset in gnomAD (although it should be noted that gnomAD does not contain cohorts recruited for severe, pediatric-onset disease; rather, it contains cohorts recruited for common adult-onset diseases such as cardiovascular disease and type II diabetes). By reviewing the data subsets, we found two variants that appear to be from neurologic cohorts and three from non-neurologic and non-cancer cohorts. Overall, very few variants present in this large general population reference database are likely to result in protein truncation of KMT2E.
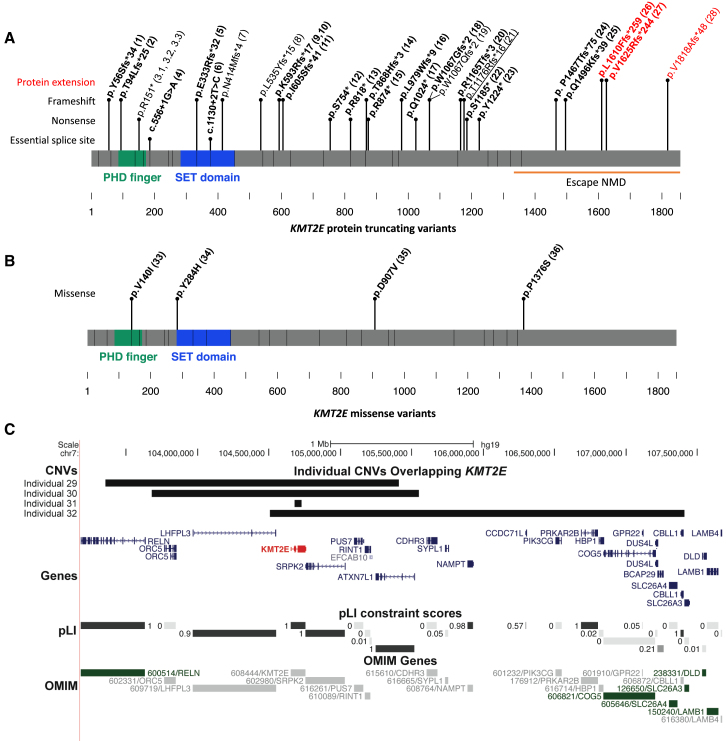
We identified 38 individuals with KMT2E variants in association with a neurodevelopmental phenotype. Including the three previously reported cases,10, 11, 12 34 individuals from 32 families were found to have single-nucleotide or indel variants in KMT2E, and four additional individuals had copy-number variants encompassing KMT2E (Figure 1, Table 1, Table S2). The KMT2E variants arose de novo in 26 individuals in our cohort. The variant was maternally inherited in one previously reported individual (maternal phenotype unknown).12 Inheritance of the variant was unknown in four families where neither parent was available for testing. In one family, a variant was found in three affected male siblings. The variant was not found in their mother. The father was not available for testing but was reported to have an intellectual disability. 30 variants were protein-truncating variants: 24 were indels, four were nonsense variants, and two were variants at essential splice sites (Figure 1A). Only one variant was seen in two independent families (c.1776_1780delAAAGA [p.Lys593Argfs∗17]); it was found in a male (individual 9) and a female (individual 10). 23 of these variants are predicted to produce transcripts that would be subject to nonsense-mediated decay. Five of the protein-truncating variants fall in the terminal exon of the gene, potentially escaping nonsense-mediated decay; three (found in individuals 26, 27, and 28) of these five variants extend the open reading frame. The last exon in individuals 26 and 27 in our cohort has a frameshift variant that alters the last 244–259 amino acids of KMT2E, whereas individual 28 has an alteration in the last 48 amino acids. We evaluated the impact of this on protein structure. Wild-type KMT2E has a very disordered C terminus, but these upstream frameshifts result in increased stability and the formation of a predicted homeodomain (Figure S1). CADD scores are summarized in Table 1.
Figure 1.
KMT2E Variants in 38 Individuals
(A) 28 protein-truncating variants in KMT2E identified in 30 individuals. Variants in bold are de novo in the proband, whereas the underlined variant was inherited. In some cases, both parents are unavailable and the de novo status is unknown (non-bold). Variants in the last exon are predicted to escape nonsense-mediated decay (individuals 24–28), whereas the last three variants (red) also result in protein extension (individuals 26–28).
(B) De novo missense variants in KMT2E in individuals 33–36.
(C) De novo deletions overlapping KMT2E were identified in individuals 29–32. All OMIM gene-disease associations (green) are for recessive disease.
Table 1.
Summary of KMT2E Variants Found in 38 Individuals with Neurodevelopmental Phenotypes
| Individual | Sex, Age | Variant, GenBank: NM_182931.2 | Consequence | Inheritance | CADD | ID | Autism | Delay | Epilepsy | Macrocephalya |
|---|---|---|---|---|---|---|---|---|---|---|
| 111 | male, 11 y | c.167delA, (p.Tyr56Serfs∗34) | frameshift, expect NMD | de novo | 30 | mild | yes | NA | no | no |
| 2 | female, 12 y | c.280delA, (p.Thr94Leufs∗25) | frameshift, expect NMD | de novo | 33 | moderate | no | yes | no | yes |
| 3.1 | male, 9 y, 6 m | c.450dupT, (p.Arg151∗) | nonsense, expect NMD | unknown | 34 | NA | yes | yes | NA | no |
| 3.2 | male, 7 y | c.450dupT, (p.Arg151∗) | nonsense, expect NMD | unknown | 34 | NA | yes | yes | NA | no |
| 3.3 | male, 6 y | c.450dupT, (p.Arg151∗) | nonsense, expect NMD | unknown | 34 | NA | yes | yes | NA | no |
| 4 | male, 5 y, 9 m | c.556+1G>A | essential splice site, expect NMD | de novo | 34 | NA | no | yes | yes | no |
| 5 | male, 12 y, 2 m | c.997delG, (p.Glu333Argfs∗32) | frameshift, expect NMD | de novo | 33 | NA | no | yes | no | yes |
| 6 | male, 3 y, 1 m | c.1130+2T>C | essential splice site, expect NMD | de novo | 33 | yes | no | yes | no | yes |
| 7 | female, 21 y | c.1239delC (p.Asn414Metfs∗4) | frameshift, expect NMD | unknown | 34 | moderate | no | yes | yes | yes |
| 8 | female, 8 y | c.1603delC (p.Leu535Tyrfs∗15) | frameshift, expect NMD | unknown | 25 | NA | no | yes | NA | relative |
| 9 | male, 11 y, 4 m | c.1776_1780delAAAGA, (p.Lys593Argfs∗17) | frameshift, expect NMD | de novo | 34 | yes | no | yes | no | yes |
| 10 | female, 3 y, 6 m | c.1776_1780delAAAGA, (p.Lys593Argfs∗17) | frameshift, expect NMD | de novo | 34 | yes | no | yes | yes | no |
| 11 | female, 1 y, 10 m | c.1812delG, (p.Ile605Serfs∗41) | frameshift, expect NMD | de novo | 26 | NA | NA | yes | no | no |
| 12 | male, 3 y, 7 m | c.2261delC, (p.Ser754∗) | nonsense, expect NMD | de novo | 34 | low-normal | no | yes | no | no |
| 13 | male, 4 y, 3 m | c.2452C>T, (p.Arg818∗) | nonsense, expect NMD | de novo | 37 | mild | no | yes | no | no |
| 14 | male, 8 y | c.2602_2605delACTA, (p.Thr868Hisfs∗3) | frameshift, expect NMD | de novo | 35 | NA | yes | yes | no | no |
| 15 | male, 1 y, 7 m | c.2620C>T, (p.Arg874∗) | nonsense, expect NMD | de novo | 39 | NA | no | yes | no | no |
| 16 | female, 3 y, 6 m | c.2936delT, (p.Leu979Trpfs∗9) | frameshift, expect NMD | de novo | 23 | NA | no | yes | no | yes |
| 17 | male, 4 y, 8 m | c.3070C>T, (p.Gln1024∗) | nonsense, expect NMD | de novo | 38 | NA | no | no | no | yes |
| 1810 | male, 12 y | c.3198delC, (p.Trp1067Glyfs∗2) | frameshift, expect NMD | de novo | 35 | mild | yes | NA | no | yes |
| 19 | female, 6 y, 5 m | c.3198_3234del, (p.Trp1067Glnfs∗2) | frameshift, expect NMD | unknown | 35 | mild | no | yes | no | yes |
| 20 | male, 5 y, 10 m | c.3494_3495delGA, (p.Arg1165Thrfs∗3) | frameshift, expect NMD | de novo | 34 | NA | no | yes | no | yes |
| 2112 | male, NA | c.3527_3530delCAGA, (p.Thr1176Argfs∗16) | frameshift, expect NMD | maternally inherited | 20 | NA | yes | NA | no | NA |
| 22 | female, 9 y | c.3554C>G, (p.Ser1185∗) | nonsense, expect NMD | de novo | 35 | mild | no | yes | yes | no |
| 23 | male, 6 y | c.3672_3673delTA, (p.Tyr1224∗) | frameshift, expect NMD | de novo | 24 | NA | no | yes | no | yes |
| 24 | male, 5 y | c.4397_4398ins19, (p.Pro1467Thrfs∗75) | frameshift, last exon, escape NMD | de novo | NA | mild | no | yes | no | yes |
| 25 | male, 12 y, 10 m | c.4485_4486delTC, (p.Gln1496Lysfs∗39) | Frameshift, last exon, escape NMD | de novo | 24 | mild | NA | yes | no | no |
| 26 | male, 6 y, 7 m | c.4829dupT, (p.Leu1610Phefs∗259) | frameshift, protein extension | de novo | 34 | low-normal | NA | NA | no | yes |
| 27 | male, 8 y, 8 m | c.4872dupC, (p.Val1625Argfs∗244) | frameshift, protein extension | de novo | 24 | yes | no | yes | no | yes |
| 28 | male, 24 y | c.5453_5460delTGGCCCTG, (p.Val1818Alafs∗48) | frameshift, protein extension | unknown | 35 | moderate | no | yes | no | relative |
| 29 | female, 12 y, 11 m | 7:103354482-105407628x1, 2.05 Mb | microdeletion | de novo | NA | moderate | yes | yes | no | yes |
| 30 | female, 18 y | 7:104678742-104730547x1, 0.052 Mb | microdeletion | de novo | NA | moderate | no | yes | yes | no |
| 31 | male, 22 y | 7:103679146-105547471x1, 1.87 Mb | microdeletion | de novo | NA | mild/moderate | no | yes | yes | no |
| 3226 | male, 7 y | 7:104099959-107002808x1, 2.9 Mb | microdeletion | de novo | NA | mild | no | yes | yes | yes |
| 33 | male, 16 y, 3 m | c.418G>A (p.Val140Ile) | missense | de novo | 25 | NA | yes | yes | yes | NA |
| 34 | male, 2 y, 5 m | c.850T>C (p.Tyr284His) | missense | de novo | 24 | severe | NA | yes | yes | no |
| 35 | female, 2 y, 11 m | c.2720A>T (p.Asp907Val) | missense | de novo | 24 | severe | no | yes | yes | microcephaly |
| 36 | female, 36 y | c.4126C>T (p.Pro1376Ser) | missense | de novo | 11 | mild | no | yes | yes | microcephaly |
NA = not available; NMD = nonsense-mediated decay.
Macrocephaly is defined here as a head circumference >2 standard deviations (SD) above mean for age and microcephaly as >−2 SD below mean for age. Relative macrocephaly is defined here as a head circumference 1 SD above the SD of the height.
Four of the individuals had de novo missense variants, three of which occurred at highly conserved positions and/or regions of the gene (Figure 1B). Pro1376 is not well conserved, and serine is present in some mammalian species. None of the KMT2E variants are reported in public databases (gnomAD, Exome Variant Server, or 1000 Genomes),15, 16, 17 although another missense change is seen at Pro1376 in gnomAD (p.Pro1376Leu, allele frequency 0.015%).
To understand the biophysical consequence of KMT2E protein sequence changes, we used structural-prediction programs (HMMER,18 PHYRE2,19 InterProScan,20 and NetPhos21) that evaluate the presence of protein domains and major secondary structure elements (e.g., helices, strands, loops, disorder, post-translational modification sites, etc.). A large protein of 1,858 amino acids, KMT2E has two N-terminal domains: a SET enzymatic domain (aa 282–445), which is predicted to be inactive, and a Zn-finger PHD domain (aa 120–165), and most of the protein has few scattered helices and strands, as well as a disordered C terminus. There was no clustering of the missense variants; one is in the SET domain, one is in the PHD domain, and two are not in identified domains. KMT2E is not significantly constrained for missense variation in the general population (Z score +1.42, observed/expected ratio of 0.87 [0.82–0.92 95% CI] for missense variation in gnomAD). All four missense variants might significantly change local structure by introducing rotamers (c.418G>A [p.Val104Ile]),22 or by changing the charge and hydrophobicity of local sequences (c.850T>C [p.Tyr284His], c.2720A>T [p.Asp907Val], and c.4126C>T [p.Pro1376Ser]). Additionally, p.Tyr284His abolishes and p.Pro1376Ser creates potential phosphorylation sites. Changing rotamers, electrical charge, and hydrophobicity might alter KMT2E binding properties.
For the four individuals with chromosome microdeletions encompassing KMT2E, all deletions occurred de novo. Deletion sizes range from 0.052 to 3.2 Mb. The 0.052 Mb deletion in individual 30 involves only KMT2E, whereas the other three deletions include additional genes.23 Figure 1C illustrates the genes included in these deletions. Median maternal and paternal age across the cohort was 30 and 36 years, respectively. There were phenotypic differences between individuals with protein-truncating, missense, and copy-number variants, as summarized below.
For the 30 individuals with protein-truncating variants in KMT2E, 22 were male and eight were female (Figure 2). Age at most recent evaluation ranged from 19 months to 24 years. Prenatal and neonatal courses were largely uncomplicated for most individuals with protein-truncating variants. One individual was born prematurely at 35 weeks. Several individuals had neonatal jaundice, one had hypoglycemia, one had sinus tachycardia, and two had neonatal feeding difficulties. Individual 10 developed respiratory arrest at 14 h of life and had a hypoxic-ischemic injury with typical sequelae seen on neuroimaging. She has spastic quadriplegia and epilepsy, and she is not included in the analysis below because her acquired injury significantly influences her phenotype and is most likely not representative of the disorder itself (although it cannot be excluded that the genetic disorder predisposed her to the injury).
Figure 2.
Photos of Individuals with KMT2E Variants
Each individual is noted with the corresponding number used throughout the manuscript. Included on the bottom right of each cluster is the individual’s sex.
(A) Individual 9, 11 years old
(B) Individual 11, 1 year, 10 months old
(C) Individual 12, 4.5 years old
(D) Individual 13, 6 years old
(E) Individual 15, 1 year, 7 months old
(F) Individual 20, 6 years old
(G) Individual 24, 5 years old
(H) Individual 25, 12 years old
(I) Individual 30, 18 years old
(J) Individual 31, 22 years old
(K) Individual 32, 7 years old
(L) Individual 33, 16 years old
Consistent facial features include dolichocephaly, large foreheads, and deep-set eyes, often with down-slanting palpebral fissures, periorbital fullness, prominent cheeks, and prominent nasolacrimal folds.
Of the remaining 29 individuals in this group (i.e., excluding individual 10), 24 had early developmental delay documented. For three individuals without documented developmental delay, these are cases previously reported from autism studies where only limited clinical information is available.10, 11, 12 The mean age of independent walking in this group was 20 months (range 12 to 48 months, Figure 3). All individuals are currently able to walk independently. 12 of the 29 individuals have hypotonia. Individual 15 had normal initial motor development but developed progressive spastic diplegia at 14 months of age. Neuroimaging in this individual demonstrated cerebral white-matter abnormalities.
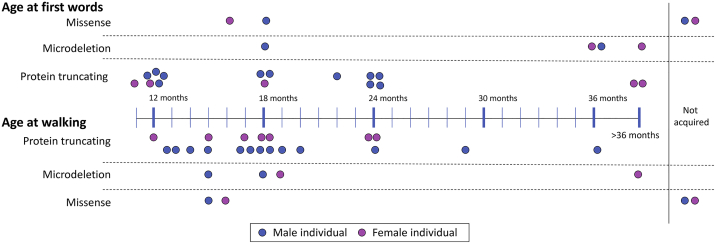
Figure 3.
Developmental Milestones in Individuals with Variants in KMT2E
Most children with protein-truncating variants acquire first words and walking by 24 months of age, though a minority are more significantly delayed. Only individual 12, who experienced a cardiac arrest and injury, did not acquire these skills. A majority of individuals with a microdeletion had significant delay in speech development but walked at a similar time to individuals with protein-truncating variants. Of those with missense variants, those with severe infantile epilepsy had significant delays.
The mean age of acquired first word in this group was 20 months (range 12 to 48 months, Figure 3). Although this information is not available for all individuals, 14 (out of 17) individuals are verbal, but seven are noted to speak poorly or to have articulation problems. Three of the individuals were reported to have speech regression. Intelligence quotient (IQ) data were available for only seven out of the 29 individuals: the mean IQ was 74 (range 62–98). Seven of the individuals have been diagnosed with autism. One additional individual was diagnosed with a sensory integration disorder, and another diagnosed with difficulty in social interaction but did not meet the criteria for autism. At least two of the individuals have been diagnosed with attention-deficit/hyperactivity disorder (ADHD). Additional behavioral concerns were reported in 11 of the individuals; these included stereotypies, skin-picking behavior, self-injurious behavior, aggression, and anxiety.
14 of the 30 individuals had macrocephaly, defined by a head circumference equal to two or more standard deviations above the mean, or in the 95th percentile or greater. An additional two individuals have relative macrocephaly, defined here as head circumference one standard deviation higher than the standard deviation for the height. Individual 6 also had a de novo pathogenic PTEN (GenBank: NM_000314.6, MIM: 601728) c.493G>A (p.Gly165Arg) variant, which can also account for his macrocephaly. Other growth parameters were variable for individuals in this group, but most were in the normal range for height and weight.
Excluding individual 10, who had a hypoxic-ischemic injury, only four of the individuals in this group (4, 7, 8, and 22) had epilepsy (two or more unprovoked seizures); an additional individual (9) had a history of just one seizure at eight years of age. There was no consistent seizure semiology or epilepsy syndrome described across the individuals. Only one of the four individuals with epilepsy (7) had treatment-resistant epilepsy. 19 of the individuals had undergone at least one brain MRI. MRI findings were normal or non-specific, and there were no consistent abnormalities (Table S2). Noted abnormalities included thinning or partial agenesis of the corpus callosum (individuals 5, 12, and 15); various cysts including pineal, epidermoid, arachnoid, and ependymal (individuals 6, 7, 9, and 19, respectively); increased white-matter signal (individuals 8 and 17); hyperintense signal in the basal ganglia (individual 10); decreased volume (individuals 5, 10, 12, and 15); delayed myelination (individual 19); small areas of heterotopia (individual 20); and Chiari I malformation (individual 14).
Many of the individuals were reported to have gastrointestinal symptoms, including reflux, vomiting, or bowel motility issues; these are issues commonly seen in individuals with hypotonia. All individuals tested had normal hearing. There were no significant ophthalmological findings. There were no other recurrent health complications noted in this group. When we compared individuals with truncating variants in the terminal exon of KMT2E to those with earlier-truncating variants, there were no clear phenotypic differences, although the number of individuals available for comparison is small.
It is notable that 22 out of the 30 individuals with protein-truncating variants were male. It is possible that decreased penetrance or variable expressivity of the condition in females means that fewer female individuals with de novo protein-truncating variants come to diagnostic attention. Additionally, the expressivity of certain aspects of the phenotype is variable between males and females (Table 2). Although the rates of intellectual disability and macrocephaly were similar, interestingly, epilepsy was seen in 43% of females but in only 5% of males (p = 0.047, Fisher’s exact test), whereas autism was seen in 35% of males and in none of the females (p = 0.14, Fisher’s exact test) with protein-truncating variants in KMT2E. These sex-related differences in phenotype parallel differences in the epidemiology of autism and epilepsy: autism is four times more common in males than in females,24 whereas polygenic idiopathic generalized epilepsies are more common in females.25
Table 2.
Summarized Phenotypes by Variant Type
| Subset | # | Intellectual Disability | Autism | Epilepsy | Macrocephaly | Microcephaly |
|---|---|---|---|---|---|---|
| Protein-Truncating Variants | ||||||
| Total | 30 | 81% (13/16) | 26% (7/27) | 15% (4/26) | 55% (16/29) | 0% (0/29) |
| Male | 22 | 82% (9/11) | 35% (7/20) | 5% (1/19) | 52% (11/21) | 0% (0/21) |
| Female | 8 | 80% (4/5) | 0% (0/7) | 43% (3/7) | 63% (5/8) | 0% (0/8) |
| Microdeletion | ||||||
| Total | 4 | 100% (4/4) | 25% (1/4) | 75% (3/4) | 50% (2/4) | 0% (0/4) |
| Missense | ||||||
| Total | 4 | 100% (3/3) | 33% (1/3) | 100% (4/4) | 0% (0/3) | 66% (2/3) |
Of the four individuals with de novo 7q22.2-22.3 chromosome deletions including KMT2E, two were male and two were female (Figure 2). The age at most recent evaluation ranged from 7 to 22 years. Clinically, individuals with deletions presented similarly to those with truncating variants. Although the sample size is small, there appear to be more severe developmental delays in this group. The average age of first words was 34.5 months (range 18 to 48 months, Figure 3). Only two of the four individuals are verbal. Walking was delayed in all; age of walking ranged from 15 to 42 months. Three of the four individuals in this group have epilepsy (30, 31, and 32). Two of the four individuals in this group have macrocephaly (29 and 32).
Individual 32 has been previously reported.26 He presented with global developmental delay, overgrowth, macrocephaly, delayed bone age, and treatment refractory generalized epilepsy. MRI of the brain demonstrated reduction of cerebral white matter, corpus callosum hypoplasia, right cerebellar hypoplasia, and an enlarged cisterna magna. Brain imaging was also performed in individuals 30 and 31. The MRI of individual 31 demonstrated global cerebral atrophy, and the MRI of individual 30 demonstrated a possible focal cortical dysplasia.
Of the four individuals with de novo missense variants in KMT2E, two were male and two were female (Figure 2). The age at most recent evaluation ranged from 29 months to 36 years. All four of the individuals with missense variants had epilepsy. Individual 33 had five generalized tonic-clonic seizures, starting at the age of 15 years. Individuals 34, 35, and 36 all presented with infantile epileptic encephalopathy. Individual 34 developed seizures at 6 months of age, and individuals 35 and 36 both developed seizures in the neonatal period. Reported seizure semiologies include generalized tonic-clonic, tonic, atonic, and myoclonic seizures and epileptic spasms. The initial EEG in individual 35 showed burst suppression and subsequently evolved into hypsarrhythmia. The EEG in individual 36 also showed hypsarrhythmia. The EEG in individual 34 demonstrated background disorganization and multifocal and generalized epileptiform discharges. All three individuals have treatment-resistant epilepsy. Individual 34 was started on the ketogenic diet at 14 months of age, but this diet did not improve seizure control.
In our cohort, individuals with missense variants also had more severe developmental delays than did the individuals with truncating variants. Only two of the four individuals can walk independently, and none of the individuals were verbal at most recent follow-up (Figure 3). Two of the four individuals in this category have microcephaly, and the other two are normocephalic. Three of these individuals have had a brain MRI: one individual had delayed myelination, one had cerebral atrophy, and one had an incidental abnormality in the right cerebral peduncle.
Comparison of the facial features of eleven of the individuals in our cohort suggests some commonalities, including macrocephaly, dolichocephaly, high forehead, deep-set eyes, periorbital fullness, prominent cheeks, and prominent nasolabial folds (Figures 2 and 4). Utilizing Face2Gene (FDNA, Boston, MA) facial recognition software, we created a composite image from frontal photographs of these 11 individuals (excluding individual 30, who wore glasses in the photograph) to represent the common facial gestalt.
Figure 4.
Composite Photo from Face2Gene
Individuals in Figure 2 were used in this analysis, excluding individual 30, who is wearing glasses.
KMT2E encodes a histone methyltransferase protein, a transcriptional regulator reported to play key roles in diverse biological processes, including cell-cycle progression, maintenance of genomic stability, adult hematopoiesis, and spermatogenesis. The gene is highly expressed in the brain, particularly during fetal development.11 KMT2E appears to be distinct from other members of the KMT2 family. Most KMT2 proteins contain an enzymatically active SET domain that possesses methyltransferase function.9, 27 Although the KMT2E protein contains a SET domain, its sequence and location within the protein are different from those of other members of the KMT2 family, and studies suggest that it might lack intrinsic methyltransferase activity.28 However, the SET domain is still highly conserved in KMT2E, and it has been proposed that KMT2E might have an indirect effect on H3K4 methylation, possibly through transcriptional regulation of additional histone-modifying enzymes. Most members of the KMT2 family contain multiple PHD finger domains that function as H3K4 methylation readers. In contrast, KMT2E contains a single PHD finger domain. PHD fingers typically bind to specific epigenetic histone marks in order to recruit transcription factors and nucleosome-associated complexes to chromatin. Finally, whereas most members of the KMT2 family function as global activators of open chromatin, KMT2E is believed to be a repressor, although the precise mechanisms involved in KMT2E regulation of gene transcription have not yet been elucidated.29
Of the individuals in our cohort, those who have protein-truncating KMT2E variants present with syndromic intellectual disability. Most individuals are functioning in the low normal to mild range of intellectual disability. Seven of the male individuals (including three of the previously reported individuals10, 11, 12) have also been formally diagnosed with autism. There appears to be a subtle common facial gestalt among the individuals whose images were available for review. Additional features, albeit not obligate or specific, include macrocephaly, hypotonia, and GI dysmotility. Neuroimaging is normal or non-specific. Epilepsy was not common among individuals with protein-truncating variants. There were no significant phenotypic differences between individuals with truncating variants in the terminal exon of the gene and those with earlier-truncating variants, suggesting a probable common pathophysiology of haploinsufficiency.
Whereas, in our cohort, only approximately 14% of the individuals with protein-truncating variants have epilepsy, all of the individuals we report as having missense variants have epilepsy. This association met statistical significance (p = 0.0026, Fisher’s exact test). Three of the individuals with missense variants fall in the category of an infantile-onset epileptic encephalopathy. In addition, these individuals have more severe developmental delays, and two have microcephaly. We hypothesize that the phenotype of epileptic encephalopathy could be variant specific and might relate to an alternate mechanism such as a gain-of-function or dominant-negative effect. Recently, distinct developmental disorder phenotypes have been identified to result from PTVs and missense variants in the same gene.30, 31 Additional cases and further functional studies are required to clarify this.
Overall, the individuals with chromosome 7q22.2-22.3 microdeletions encompassing KMT2E presented similarly to those with truncating variants, further supporting haploinsufficiency as the disease mechanism. Although the sample size was small, these individuals appeared to have more severe developmental delays than did those individuals with truncating variants, which is likely explained by the influence of additional genes included in their deletions. The 7q22.2-22.3 region contains multiple additional genes involved in the regulation of the cell cycle; such genes include SRPK2 (MIM: 602980), RINT1 (MIM: 610089), and LHFPL3 (MIM: 609719).26 In particular, SRPK2 and LHFPL3 show depletion of loss-of-function variation from expectation in the gnomAD database (pLI of 1.0 and 0.9, respectively) and are expressed in the central nervous system. SRPK2 encodes a cell-cycle-regulated protein kinase that phosphorylates serine and arginine domain-containing proteins and modulates pre-mRNA splicing in neurons.32 LHFPL3 is a transmembrane protein, but little is known about its function to date.
Several Kmt2e (Mll5)-deficiency mouse models have been created and characterized.29, 33, 34, 35, 36 These mice present with growth restriction and increased mortality, as well as impaired hematopoiesis. A neurological phenotype in these mice has not been reported. Both homozygous and heterozygous loss of Kmt2e in mice results in DNA damage and elevated levels of reactive oxygen species (ROS).36 The cellular effects were effectively reversed by supplementation with the glutathione precursor N-acetylcysteine (NAC).36 This has interesting therapeutic implications for humans because NAC supplementation has been used in the treatment of glutathione depletion in acetaminophen overdose as well as rare inborn errors of metabolism associated with increased free-radical damage. Further studies are required if we are to establish whether humans haploinsufficient for KMT2E are also vulnerable to increased ROS and whether there might be a benefit in treating with NAC or other antioxidants. This evaluation could include clinically measuring urine F2 isoprostanes and blood glutathione levels.37
In this report, we define a KMT2E-related neurodevelopmental disorder, which adds to the growing list of KMT2 gene family disorders. Most individuals with protein-truncating variants appear to present with generally mild developmental delay and intellectual disability. Autism is also relatively common. Additional common, but not obligate, features include relative macrocephaly, hypotonia, and functional gastrointestinal disturbances. There appears to be a subtle facial gestalt. Epilepsy was not common among individuals with protein-truncating variants. We suspect haploinsufficiency as the disease mechanism. The similar phenotype seen in individuals with microdeletions of this region is consistent with this hypothesis. In contrast, individuals with missense variants all presented with epilepsy, including infantile-onset epileptic encephalopathy, and more severe developmental delays. Variant-specific alterations in KMT2E function, possibly even gain-of-function alterations, might explain this divergence in phenotype. Further studies are required if we are to further understand the correlation between genotype and phenotype. There is no established therapy for KMT2E-related disorders, although based on animal data, there might be a role for NAC or other antioxidant treatments.
Consortia
The members of the Deciphering Developmental Disorders (DDD) Study are Jeremy F. McRae, Stephen Clayton, Tomas W. Fitzgerald, Joanna Kaplanis, Elena Prigmore, Diana Rajan, Alejandro Sifrim, Stuart Aitken, Nadia Akawi, Mohsan Alvi, Kirsty Ambridge, Daniel M. Barrett, Tanya Bayzetinova, Philip Jones, Wendy D. Jones, Daniel King, Netravathi Krishnappa, Laura E. Mason, Tarjinder Singh, Adrian R. Tivey, Munaza Ahmed, Uruj Anjum, Hayley Archer, Ruth Armstrong, Jana Awada, Meena Balasubramanian, Siddharth Banka, Diana Baralle, Angela Barnicoat, Paul Batstone, David Baty, Chris Bennett, Jonathan Berg, Birgitta Bernhard, A. Paul Bevan, Maria Bitner-Glindzicz, Edward Blair, Moira Blyth, David Bohanna, Louise Bourdon, David Bourn, Lisa Bradley, Angela Brady, Simon Brent, Carole Brewer, Kate Brunstrom, David J. Bunyan, John Burn, Natalie Canham, Bruce Castle, Kate Chandler, Elena Chatzimichali, Deirdre Cilliers, Angus Clarke, Susan Clasper, Jill Clayton-Smith, Virginia Clowes, Andrea Coates, Trevor Cole, Irina Colgiu, Amanda Collins, Morag N. Collinson, Fiona Connell, Nicola Cooper, Helen Cox, Lara Cresswell, Gareth Cross, Yanick Crow, Mariella D’Alessandro, Tabib Dabir, Rosemarie Davidson, Sally Davies, Dylan de Vries, John Dean, Charu Deshpande, Gemma Devlin, Abhijit Dixit, Angus Dobbie, Alan Donaldson, Dian Donnai, Deirdre Donnelly, Carina Donnelly, Angela Douglas, Sofia Douzgou, Alexis Duncan, Jacqueline Eason, Sian Ellard, Ian Ellis, Frances Elmslie, Karenza Evans, Sarah Everest, Tina Fendick, Richard Fisher, Frances Flinter, Nicola Foulds, Andrew Fry, Alan Fryer, Carol Gardiner, Lorraine Gaunt, Neeti Ghali, Richard Gibbons, Harinder Gill, Judith Goodship, David Goudie, Emma Gray, Andrew Green, Philip Greene, Lynn Greenhalgh, Susan Gribble, Rachel Harrison, Lucy Harrison, Victoria Harrison, Rose Hawkins, Liu He, Stephen Hellens, Alex Henderson, Sarah Hewitt, Lucy Hildyard, Emma Hobson, Simon Holden, Muriel Holder, Susan Holder, Georgina Hollingsworth, Tessa Homfray, Mervyn Humphreys, Jane Hurst, Ben Hutton, Stuart Ingram, Melita Irving, Lily Islam, Andrew Jackson, Joanna Jarvis, Lucy Jenkins, Diana Johnson, Elizabeth Jones, Dragana Josifova, Shelagh Joss, Beckie Kaemba, Sandra Kazembe, Rosemary Kelsell, Bronwyn Kerr, Helen Kingston, Usha Kini, Esther Kinning, Gail Kirby, Claire Kirk, Emma Kivuva, Alison Kraus, Dhavendra Kumar, V. K. Ajith Kumar, Katherine Lachlan, Wayne Lam, Anne Lampe, Caroline Langman, Melissa Lees, Derek Lim, Cheryl Longman, Gordon Lowther, Sally A. Lynch, Alex Magee, Eddy Maher, Alison Male, Sahar Mansour, Karen Marks, Katherine Martin, Una Maye, Emma McCann, Vivienne McConnell, Meriel McEntagart, Ruth McGowan, Kirsten McKay, Shane McKee, Dominic J. McMullan, Susan McNerlan, Catherine McWilliam, Sarju Mehta, Kay Metcalfe, Anna Middleton, Zosia Miedzybrodzka, Emma Miles, Shehla Mohammed, Tara Montgomery, David Moore, Sian Morgan, Jenny Morton, Hood Mugalaasi, Victoria Murday, Helen Murphy, Swati Naik, Andrea Nemeth, Louise Nevitt, Ruth Newbury-Ecob, Andrew Norman, Rosie O’Shea, Caroline Ogilvie, Kai-Ren Ong, Soo-Mi Park, Michael J. Parker, Chirag Patel, Joan Paterson, Stewart Payne, Daniel Perrett, Julie Phipps, Daniela T. Pilz, Martin Pollard, Caroline Pottinger, Joanna Poulton, Norman Pratt, Katrina Prescott, Sue Price, Abigail Pridham, Annie Procter, Hellen Purnell, Oliver Quarrell, Nicola Ragge, Raheleh Rahbari, Josh Randall, Julia Rankin, Lucy Raymond, Debbie Rice, Leema Robert, Eileen Roberts, Jonathan Roberts, Paul Roberts, Gillian Roberts, Alison Ross, Elisabeth Rosser, Anand Saggar, Shalaka Samant, Julian Sampson, Richard Sandford, Ajoy Sarkar, Susann Schweiger, Richard Scott, Ingrid Scurr, Ann Selby, Anneke Seller, Cheryl Sequeira, Nora Shannon, Saba Sharif, Charles Shaw-Smith, Emma Shearing, Debbie Shears, Eamonn Sheridan, Ingrid Simonic, Roldan Singzon, Zara Skitt, Audrey Smith, Kath Smith, Sarah Smithson, Linda Sneddon, Miranda Splitt, Miranda Squires, Fiona Stewart, Helen Stewart, Volker Straub, Mohnish Suri, Vivienne Sutton, Ganesh Jawahar Swaminathan, Elizabeth Sweeney, Kate Tatton-Brown, Cat Taylor, Rohan Taylor, Mark Tein, I. Karen Temple, Jenny Thomson, Marc Tischkowitz, Susan Tomkins, Audrey Torokwa, Becky Treacy, Claire Turner, Peter Turnpenny, Carolyn Tysoe, Anthony Vandersteen, Vinod Varghese, Pradeep Vasudevan, Parthiban Vijayarangakannan, Julie Vogt, Emma Wakeling, Sarah Wallwark, Jonathon Waters, Astrid Weber, Diana Wellesley, Margo Whiteford, Sara Widaa, Sarah Wilcox, Emily Wilkinson, Denise Williams, Nicola Williams, Louise Wilson, Geoff Woods, Christopher Wragg, Michael Wright, Laura Yates, Michael Yau, Chris Nellåker, Michael Parker, Helen V. Firth, Caroline F. Wright, David R. FitzPatrick, Jeffrey C. Barrett, and Matthew E. Hurles.
Declaration of Interests
A.T., C.R., H.M.M., I.M.W., K.M., R.H., and R.P. are employees of GeneDx, a wholly-owned subsidiary of OPKO Health. F.K. and L.L.P.R. are employees of Mendelics Genomics Analysis.
Acknowledgments
We thank the families who participated in this study, GeneMatcher, MyGene2, the gnomAD team, and the Deciphering Developmental Disorders study. We appreciate guidance from Nicole Fleisher for using the Face2Gene software and Steven Harrison for guidance with curation and ClinVar submission.
Support was provided by the National Institutes of Health’s National Institute of Child Health and Human Development (NICHD) (K12HD052896) and the Boston Children’s Hospital Faculty Development Fellowship to A.H.O.L.; the National Human Genome Research Institute-funded Broad Center for Mendelian Genomics (UM1HG008900) to L.S.P., A.S., and S.B.; by NICHD (R01 HD073104 and R01 HD091846) to V.L.; by Chile’s National Commission for Scientific and Technological Research (CONICYT) (72160007) to V.F.; by the German Research Society (DFG WE4896/3-1, WE4896/4-1, HE5415/3-1, HE5415/5-1, HE5415/6-1, HE5415/7-1) to Y.W. and I.H.; by intramural funds from the Children's Hospital of Philadelphia and the University of Kiel to I.H.; by Dietmar-Hopp-Stiftung (1DH1813319) to S.S.; by the Health Innovation Challenge Fund (R6-388/WT 100127) and the National Institute for Health Research through the Comprehensive Clinical Research Network to J.C.T.; and by a Skaggs-Oxford Scholarship to J.Z. The DDD Study (Cambridge South REC approval 10/H0305/83 and the Republic of Ireland REC GEN/284/12) presents independent research commissioned by the Health Innovation Challenge Fund (HICF-1009-003), a parallel funding partnership between the Department of Health and the Wellcome Sanger Institute (WT098051). Views expressed here are those of the authors and not necessarily those of the Wellcome or the Department of Health.
Published: May 9, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.03.021.
Contributor Information
Anne H. O’Donnell-Luria, Email: Anne.ODonnell@childrens.harvard.edu.
Lance H. Rodan, Email: Lance.Rodan@childrens.harvard.edu.
Deciphering Developmental Disorders (DDD) Study:
Jeremy F. McRae, Stephen Clayton, Tomas W. Fitzgerald, Joanna Kaplanis, Elena Prigmore, Diana Rajan, Alejandro Sifrim, Stuart Aitken, Nadia Akawi, Mohsan Alvi, Kirsty Ambridge, Daniel M. Barrett, Tanya Bayzetinova, Philip Jones, Wendy D. Jones, Daniel King, Netravathi Krishnappa, Laura E. Mason, Tarjinder Singh, Adrian R. Tivey, Munaza Ahmed, Uruj Anjum, Hayley Archer, Ruth Armstrong, Jana Awada, Meena Balasubramanian, Siddharth Banka, Diana Baralle, Angela Barnicoat, Paul Batstone, David Baty, Chris Bennett, Jonathan Berg, Birgitta Bernhard, A. Paul Bevan, Maria Bitner-Glindzicz, Edward Blair, Moira Blyth, David Bohanna, Louise Bourdon, David Bourn, Lisa Bradley, Angela Brady, Simon Brent, Carole Brewer, Kate Brunstrom, David J. Bunyan, John Burn, Natalie Canham, Bruce Castle, Kate Chandler, Elena Chatzimichali, Deirdre Cilliers, Angus Clarke, Susan Clasper, Jill Clayton-Smith, Virginia Clowes, Andrea Coates, Trevor Cole, Irina Colgiu, Amanda Collins, Morag N. Collinson, Fiona Connell, Nicola Cooper, Helen Cox, Lara Cresswell, Gareth Cross, Yanick Crow, Mariella D’Alessandro, Tabib Dabir, Rosemarie Davidson, Sally Davies, Dylan de Vries, John Dean, Charu Deshpande, Gemma Devlin, Abhijit Dixit, Angus Dobbie, Alan Donaldson, Dian Donnai, Deirdre Donnelly, Carina Donnelly, Angela Douglas, Sofia Douzgou, Alexis Duncan, Jacqueline Eason, Sian Ellard, Ian Ellis, Frances Elmslie, Karenza Evans, Sarah Everest, Tina Fendick, Richard Fisher, Frances Flinter, Nicola Foulds, Andrew Fry, Alan Fryer, Carol Gardiner, Lorraine Gaunt, Neeti Ghali, Richard Gibbons, Harinder Gill, Judith Goodship, David Goudie, Emma Gray, Andrew Green, Philip Greene, Lynn Greenhalgh, Susan Gribble, Rachel Harrison, Lucy Harrison, Victoria Harrison, Rose Hawkins, Liu He, Stephen Hellens, Alex Henderson, Sarah Hewitt, Lucy Hildyard, Emma Hobson, Simon Holden, Muriel Holder, Susan Holder, Georgina Hollingsworth, Tessa Homfray, Mervyn Humphreys, Jane Hurst, Ben Hutton, Stuart Ingram, Melita Irving, Lily Islam, Andrew Jackson, Joanna Jarvis, Lucy Jenkins, Diana Johnson, Elizabeth Jones, Dragana Josifova, Shelagh Joss, Beckie Kaemba, Sandra Kazembe, Rosemary Kelsell, Bronwyn Kerr, Helen Kingston, Usha Kini, Esther Kinning, Gail Kirby, Claire Kirk, Emma Kivuva, Alison Kraus, Dhavendra Kumar, V. K. Ajith Kumar, Katherine Lachlan, Wayne Lam, Anne Lampe, Caroline Langman, Melissa Lees, Derek Lim, Cheryl Longman, Gordon Lowther, Sally A. Lynch, Alex Magee, Eddy Maher, Alison Male, Sahar Mansour, Karen Marks, Katherine Martin, Una Maye, Emma McCann, Vivienne McConnell, Meriel McEntagart, Ruth McGowan, Kirsten McKay, Shane McKee, Dominic J. McMullan, Susan McNerlan, Catherine McWilliam, Sarju Mehta, Kay Metcalfe, Anna Middleton, Zosia Miedzybrodzka, Emma Miles, Shehla Mohammed, Tara Montgomery, David Moore, Sian Morgan, Jenny Morton, Hood Mugalaasi, Victoria Murday, Helen Murphy, Swati Naik, Andrea Nemeth, Louise Nevitt, Ruth Newbury-Ecob, Andrew Norman, Rosie O’Shea, Caroline Ogilvie, Kai-Ren Ong, Soo-Mi Park, Michael J. Parker, Chirag Patel, Joan Paterson, Stewart Payne, Daniel Perrett, Julie Phipps, Daniela T. Pilz, Martin Pollard, Caroline Pottinger, Joanna Poulton, Norman Pratt, Katrina Prescott, Sue Price, Abigail Pridham, Annie Procter, Hellen Purnell, Oliver Quarrell, Nicola Ragge, Raheleh Rahbari, Josh Randall, Julia Rankin, Lucy Raymond, Debbie Rice, Leema Robert, Eileen Roberts, Jonathan Roberts, Paul Roberts, Gillian Roberts, Alison Ross, Elisabeth Rosser, Anand Saggar, Shalaka Samant, Julian Sampson, Richard Sandford, Ajoy Sarkar, Susann Schweiger, Richard Scott, Ingrid Scurr, Ann Selby, Anneke Seller, Cheryl Sequeira, Nora Shannon, Saba Sharif, Charles Shaw-Smith, Emma Shearing, Debbie Shears, Eamonn Sheridan, Ingrid Simonic, Roldan Singzon, Zara Skitt, Audrey Smith, Kath Smith, Sarah Smithson, Linda Sneddon, Miranda Splitt, Miranda Squires, Fiona Stewart, Helen Stewart, Volker Straub, Mohnish Suri, Vivienne Sutton, Ganesh Jawahar Swaminathan, Elizabeth Sweeney, Kate Tatton-Brown, Cat Taylor, Rohan Taylor, Mark Tein, I. Karen Temple, Jenny Thomson, Marc Tischkowitz, Susan Tomkins, Audrey Torokwa, Becky Treacy, Claire Turner, Peter Turnpenny, Carolyn Tysoe, Anthony Vandersteen, Vinod Varghese, Pradeep Vasudevan, Parthiban Vijayarangakannan, Julie Vogt, Emma Wakeling, Sarah Wallwark, Jonathon Waters, Astrid Weber, Diana Wellesley, Margo Whiteford, Sara Widaa, Sarah Wilcox, Emily Wilkinson, Denise Williams, Nicola Williams, Louise Wilson, Geoff Woods, Christopher Wragg, Michael Wright, Laura Yates, Michael Yau, Chris Nellåker, Michael Parker, Helen V. Firth, Caroline F. Wright, David R. FitzPatrick, Jeffrey C. Barrett, and Matthew E. Hurles
Web Resources
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar
DECIPHER, https://decipher.sanger.ac.uk/
GeneMatcher, https://genematcher.org/
Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org
HMMER, http://hmmer.org/
InterProScan, https://www.ebi.ac.uk/interpro/search/sequence-search
MyGene2, NHGRI/NHLBI University of Washington-Center for Mendelian Genomics (UW-CMG), Seattle, WA, https://www.mygene2.org/MyGene2/
NetPhos 3.1, http://www.cbs.dtu.dk/services/NetPhos/
Online Mendelian Inheritance in Man (OMIM), https://omim.org/
Phyre2, http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index
UCSC Human Genome Browser, http://www.genome.ucsc.edu
Variant Validator, https://variantvalidator.org/variantvalidator/
Supplemental Data
References
- 1.Shen E., Shulha H., Weng Z., Akbarian S. Regulation of histone H3K4 methylation in brain development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:369. doi: 10.1098/rstb.2013.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shen, E., Shulha, H., Weng, Z., and Akbarian, S. (2014). Regulation of histone H3K4 methylation in brain development and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 369. [DOI] [PMC free article] [PubMed]
- 2.Jones W.D., Dafou D., McEntagart M., Woollard W.J., Elmslie F.V., Holder-Espinasse M., Irving M., Saggar A.K., Smithson S., Trembath R.C. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 2012;91:358–364. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jones, W.D., Dafou, D., McEntagart, M., Woollard, W.J., Elmslie, F.V., Holder-Espinasse, M., Irving, M., Saggar, A.K., Smithson, S., Trembath, R.C., et al. (2012). De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 91, 358-364. [DOI] [PMC free article] [PubMed]
- 3.Zech M., Boesch S., Maier E.M., Borggraefe I., Vill K., Laccone F., Pilshofer V., Ceballos-Baumann A., Alhaddad B., Berutti R. Haploinsufficiency of KMT2B, encoding the lysine-specific histone methyltransferase 2B, results in early-onset generalized dystonia. Am. J. Hum. Genet. 2016;99:1377–1387. doi: 10.1016/j.ajhg.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zech, M., Boesch, S., Maier, E.M., Borggraefe, I., Vill, K., Laccone, F., Pilshofer, V., Ceballos-Baumann, A., Alhaddad, B., Berutti, R., et al. (2016). Haploinsufficiency of KMT2B, encoding the lysine-specific histone methyltransferase 2B, results in early-onset generalized dystonia. Am. J. Hum. Genet. 99, 1377-1387. [DOI] [PMC free article] [PubMed]
- 4.Koemans T.S., Kleefstra T., Chubak M.C., Stone M.H., Reijnders M.R.F., de Munnik S., Willemsen M.H., Fenckova M., Stumpel C.T.R.M., Bok L.A. Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLoS Genet. 2017;13:e1006864. doi: 10.1371/journal.pgen.1006864. [DOI] [PMC free article] [PubMed] [Google Scholar]; Koemans, T.S., Kleefstra, T., Chubak, M.C., Stone, M.H., Reijnders, M.R.F., de Munnik, S., Willemsen, M.H., Fenckova, M., Stumpel, C.T.R.M., Bok, L.A., et al. (2017). Functional convergence of histone methyltransferases EHMT1 and KMT2C involved in intellectual disability and autism spectrum disorder. PLoS Genet. 13, e1006864. [DOI] [PMC free article] [PubMed]
- 5.Van Laarhoven P.M., Neitzel L.R., Quintana A.M., Geiger E.A., Zackai E.H., Clouthier D.E., Artinger K.B., Ming J.E., Shaikh T.H. Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 2015;24:4443–4453. doi: 10.1093/hmg/ddv180. [DOI] [PMC free article] [PubMed] [Google Scholar]; Van Laarhoven, P.M., Neitzel, L.R., Quintana, A.M., Geiger, E.A., Zackai, E.H., Clouthier, D.E., Artinger, K.B., Ming, J.E., and Shaikh, T.H. (2015). Kabuki syndrome genes KMT2D and KDM6A: functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 24, 4443-4453. [DOI] [PMC free article] [PubMed]
- 6.Poeta L., Fusco F., Drongitis D., Shoubridge C., Manganelli G., Filosa S., Paciolla M., Courtney M., Collombat P., Lioi M.B. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 2013;92:114–125. doi: 10.1016/j.ajhg.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Poeta, L., Fusco, F., Drongitis, D., Shoubridge, C., Manganelli, G., Filosa, S., Paciolla, M., Courtney, M., Collombat, P., Lioi, M.B., et al. (2013). A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am. J. Hum. Genet. 92, 114-125. [DOI] [PMC free article] [PubMed]
- 7.Nakagawa T., Xiong Y. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol. Cell. 2011;43:381–391. doi: 10.1016/j.molcel.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nakagawa, T., and Xiong, Y. (2011). X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol. Cell 43, 381-391. [DOI] [PMC free article] [PubMed]
- 8.Brookes E., Laurent B., Õunap K., Carroll R., Moeschler J.B., Field M., Schwartz C.E., Gecz J., Shi Y. Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet. 2015;24:2861–2872. doi: 10.1093/hmg/ddv046. [DOI] [PMC free article] [PubMed] [Google Scholar]; Brookes, E., Laurent, B., Ounap, K., Carroll, R., Moeschler, J.B., Field, M., Schwartz, C.E., Gecz, J., and Shi, Y. (2015). Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum. Mol. Genet. 24, 2861-2872. [DOI] [PMC free article] [PubMed]
- 9.Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study. Deciphering Developmental Disorders (DDD) Study Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Faundes, V., Newman, W.G., Bernardini, L., Canham, N., Clayton-Smith, J., Dallapiccola, B., Davies, S.J., Demos, M.K., Goldman, A., Gill, H., et al.; Clinical Assessment of the Utility of Sequencing and Evaluation as a Service (CAUSES) Study; Deciphering Developmental Disorders (DDD) Study (2018). Histone lysine methylases and demethylases in the landscape of human developmental disorders. Am. J. Hum. Genet. 102, 175-187. [DOI] [PMC free article] [PubMed]
- 10.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.-H., Narzisi G., Leotta A. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Iossifov, I., Ronemus, M., Levy, D., Wang, Z., Hakker, I., Rosenbaum, J., Yamrom, B., Lee, Y.-H., Narzisi, G., Leotta, A., et al. (2012). De novo gene disruptions in children on the autistic spectrum. Neuron 74, 285-299. [DOI] [PMC free article] [PubMed]
- 11.Dong S., Walker M.F., Carriero N.J., DiCola M., Willsey A.J., Ye A.Y., Waqar Z., Gonzalez L.E., Overton J.D., Frahm S. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dong, S., Walker, M.F., Carriero, N.J., DiCola, M., Willsey, A.J., Ye, A.Y., Waqar, Z., Gonzalez, L.E., Overton, J.D., Frahm, S., et al. (2014). De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 9, 16-23. [DOI] [PMC free article] [PubMed]
- 12.Wang T., Guo H., Xiong B., Stessman H.A.F., Wu H., Coe B.P., Turner T.N., Liu Y., Zhao W., Hoekzema K. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 2016;7:13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, T., Guo, H., Xiong, B., Stessman, H.A.F., Wu, H., Coe, B.P., Turner, T.N., Liu, Y., Zhao, W., Hoekzema, K., et al. (2016). De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat. Commun. 7, 13316. [DOI] [PMC free article] [PubMed]
- 13.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sobreira, N., Schiettecatte, F., Valle, D., and Hamosh, A. (2015). GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 36, 928-930. [DOI] [PMC free article] [PubMed]
- 14.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]; Philippakis, A.A., Azzariti, D.R., Beltran, S., Brookes, A.J., Brownstein, C.A., Brudno, M., Brunner, H.G., Buske, O.J., Carey, K., Doll, C., et al. (2015). The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 36, 915-921. [DOI] [PMC free article] [PubMed]
- 15.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T., O’Donnell-Luria, A.H., Ware, J.S., Hill, A.J., Cummings, B.B., et al.; Exome Aggregation Consortium (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285-291. [DOI] [PMC free article] [PubMed]
- 16.Tennessen J.A., Bigham A.W., O’Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., Broad GO. Seattle GO. NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tennessen, J.A., Bigham, A.W., O’Connor, T.D., Fu, W., Kenny, E.E., Gravel, S., McGee, S., Do, R., Liu, X., Jun, G., et al.; Broad GO; Seattle GO; NHLBI Exome Sequencing Project (2012). Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 337, 64-69. [DOI] [PMC free article] [PubMed]
- 17.1000 Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]; 1000 Genomes Project Consortium, Auton, A., Brooks, L.D., Durbin, R.M., Garrison, E.P., Kang, H.M., Marchini, J.L., McCarthy, S., McVean, G.A., and Abecasis, G.R. (2015). A global reference for human genetic variation. Nature 526, 68-74. [DOI] [PMC free article] [PubMed]
- 18.Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eddy, S.R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. [DOI] [PMC free article] [PubMed]
- 19.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kelley, L.A., Mezulis, S., Yates, C.M., Wass, M.N., and Sternberg, M.J.E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845-858. [DOI] [PMC free article] [PubMed]
- 20.Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., McWilliam, H., Maslen, J., Mitchell, A., Nuka, G., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236-1240. [DOI] [PMC free article] [PubMed]
- 21.Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]; Blom, N., Sicheritz-Ponten, T., Gupta, R., Gammeltoft, S., and Brunak, S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633-1649. [DOI] [PubMed]
- 22.Watkins A.M., Bonneau R., Arora P.S. Side-chain conformational preferences govern protein-protein interactions. J. Am. Chem. Soc. 2016;138:10386–10389. doi: 10.1021/jacs.6b04892. [DOI] [PMC free article] [PubMed] [Google Scholar]; Watkins, A.M., Bonneau, R., and Arora, P.S. (2016). Side-chain conformational preferences govern protein-protein interactions. J. Am. Chem. Soc. 138, 10386-10389. [DOI] [PMC free article] [PubMed]
- 23.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. (2002). The human genome browser at UCSC. Genome Res. 12, 996-1006. [DOI] [PMC free article] [PubMed]
- 24.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]; Fombonne, E. (2009). Epidemiology of pervasive developmental disorders. Pediatr. Res. 65, 591-598. [DOI] [PubMed]
- 25.McHugh J.C., Delanty N. Epidemiology and classification of epilepsy: gender comparisons. Int. Rev. Neurobiol. 2008;83:11–26. doi: 10.1016/S0074-7742(08)00002-0. [DOI] [PubMed] [Google Scholar]; McHugh, J.C., and Delanty, N. (2008). Epidemiology and classification of epilepsy: gender comparisons. Int. Rev. Neurobiol. 83, 11-26. [DOI] [PubMed]
- 26.Uliana V., Grosso S., Cioni M., Ariani F., Papa F.T., Tamburello S., Rossi E., Katzaki E., Mucciolo M., Marozza A. 3.2 Mb microdeletion in chromosome 7 bands q22.2-q22.3 associated with overgrowth and delayed bone age. Eur. J. Med. Genet. 2010;53:168–170. doi: 10.1016/j.ejmg.2010.02.003. [DOI] [PubMed] [Google Scholar]; Uliana, V., Grosso, S., Cioni, M., Ariani, F., Papa, F.T., Tamburello, S., Rossi, E., Katzaki, E., Mucciolo, M., Marozza, A., et al. (2010). 3.2 Mb microdeletion in chromosome 7 bands q22.2-q22.3 associated with overgrowth and delayed bone age. Eur. J. Med. Genet. 53, 168-170. [DOI] [PubMed]
- 27.Rao R.C., Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rao, R.C., and Dou, Y. (2015). Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer 15, 334-346. [DOI] [PMC free article] [PubMed]
- 28.Mas-Y-Mas S., Barbon M., Teyssier C., Déméné H., Carvalho J.E., Bird L.E., Lebedev A., Fattori J., Schubert M., Dumas C. The Human Mixed Lineage Leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS ONE. 2016;11:e0165139. doi: 10.1371/journal.pone.0165139. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mas-Y-Mas, S., Barbon, M., Teyssier, C., Demene, H., Carvalho, J.E., Bird, L.E., Lebedev, A., Fattori, J., Schubert, M., Dumas, C., et al. (2016). The Human Mixed Lineage Leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS ONE 11, e0165139. [DOI] [PMC free article] [PubMed]
- 29.Zhang X., Novera W., Zhang Y., Deng L.-W. MLL5 (KMT2E): structure, function, and clinical relevance. Cell. Mol. Life Sci. 2017;74:2333–2344. doi: 10.1007/s00018-017-2470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang, X., Novera, W., Zhang, Y., and Deng, L.-W. (2017). MLL5 (KMT2E): structure, function, and clinical relevance. Cell. Mol. Life Sci. 74, 2333-2344. [DOI] [PMC free article] [PubMed]
- 30.Rivière J.-B., van Bon B.W.M., Hoischen A., Kholmanskikh S.S., O’Roak B.J., Gilissen C., Gijsen S., Sullivan C.T., Christian S.L., Abdul-Rahman O.A. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012;44:440–444. doi: 10.1038/ng.1091. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Riviere, J.-B., van Bon, B.W.M., Hoischen, A., Kholmanskikh, S.S., O’Roak, B.J., Gilissen, C., Gijsen, S., Sullivan, C.T., Christian, S.L., Abdul-Rahman, O.A., et al. (2012). De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 44, 440-444, S1-S2. [DOI] [PMC free article] [PubMed]
- 31.Cuvertino S., Stuart H.M., Chandler K.E., Roberts N.A., Armstrong R., Bernardini L., Bhaskar S., Callewaert B., Clayton-Smith J., Davalillo C.H., DDD Study ACTB loss-of-function mutations result in a pleiotropic developmental disorder. Am. J. Hum. Genet. 2017;101:1021–1033. doi: 10.1016/j.ajhg.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cuvertino, S., Stuart, H.M., Chandler, K.E., Roberts, N.A., Armstrong, R., Bernardini, L., Bhaskar, S., Callewaert, B., Clayton-Smith, J., Davalillo, C.H., et al.; DDD Study (2017). ACTB loss-of-function mutations result in a pleiotropic developmental disorder. Am. J. Hum. Genet. 101, 1021-1033. [DOI] [PMC free article] [PubMed]
- 32.Hong Y., Chan C.B., Kwon I.-S., Li X., Song M., Lee H.-P., Liu X., Sompol P., Jin P., Lee H.-G. SRPK2 phosphorylates tau and mediates the cognitive defects in Alzheimer’s disease. J. Neurosci. 2012;32:17262–17272. doi: 10.1523/JNEUROSCI.3300-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hong, Y., Chan, C.B., Kwon, I.-S., Li, X., Song, M., Lee, H.-P., Liu, X., Sompol, P., Jin, P., Lee, H.-G., et al. (2012). SRPK2 phosphorylates tau and mediates the cognitive defects in Alzheimer’s disease. J. Neurosci. 32, 17262-17272. [DOI] [PMC free article] [PubMed]
- 33.Heuser M., Yap D.B., Leung M., de Algara T.R., Tafech A., McKinney S., Dixon J., Thresher R., Colledge B., Carlton M. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113:1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]; Heuser, M., Yap, D.B., Leung, M., de Algara, T.R., Tafech, A., McKinney, S., Dixon, J., Thresher, R., Colledge, B., Carlton, M., et al. (2009). Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood 113, 1432-1443. [DOI] [PubMed]
- 34.Madan V., Madan B., Brykczynska U., Zilbermann F., Hogeveen K., Döhner K., Döhner H., Weber O., Blum C., Rodewald H.-R. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113:1444–1454. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]; Madan, V., Madan, B., Brykczynska, U., Zilbermann, F., Hogeveen, K., Dohner, K., Dohner, H., Weber, O., Blum, C., Rodewald, H.-R., et al. (2009). Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood 113, 1444-1454. [DOI] [PubMed]
- 35.Yap D.B., Walker D.C., Prentice L.M., McKinney S., Turashvili G., Mooslehner-Allen K., de Algara T.R., Fee J., de Tassigny Xd., Colledge W.H., Aparicio S. Mll5 is required for normal spermatogenesis. PLoS ONE. 2011;6:e27127. doi: 10.1371/journal.pone.0027127. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yap, D.B., Walker, D.C., Prentice, L.M., McKinney, S., Turashvili, G., Mooslehner-Allen, K., de Algara, T.R., Fee, J., de Tassigny, Xd., Colledge, W.H., and Aparicio, S. (2011). Mll5 is required for normal spermatogenesis. PLoS ONE 6, e27127. [DOI] [PMC free article] [PubMed]
- 36.Tasdogan A., Kumar S., Allies G., Bausinger J., Beckel F., Hofemeister H., Mulaw M., Madan V., Scharffetter-Kochanek K., Feuring-Buske M. DNA damage-induced HSPC malfunction depends on ROS accumulation downstream of IFN-1 signaling and bid mobilization. Cell Stem Cell. 2016;19:752–767. doi: 10.1016/j.stem.2016.08.007. [DOI] [PubMed] [Google Scholar]; Tasdogan, A., Kumar, S., Allies, G., Bausinger, J., Beckel, F., Hofemeister, H., Mulaw, M., Madan, V., Scharffetter-Kochanek, K., Feuring-Buske, M., et al. (2016). DNA damage-induced HSPC malfunction depends on ROS accumulation downstream of IFN-1 signaling and bid mobilization. Cell Stem Cell 19, 752-767. [DOI] [PubMed]
- 37.Il’yasova D., Scarbrough P., Spasojevic I. Urinary biomarkers of oxidative status. Clin. Chim. Acta. 2012;413:1446–1453. doi: 10.1016/j.cca.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; Il’yasova, D., Scarbrough, P., and Spasojevic, I. (2012). Urinary biomarkers of oxidative status. Clin. Chim. Acta 413, 1446-1453. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.