Abstract
Background
Codeletion of chromosome arms 1p and 19q (1p/19q codeletion) highly benefits diagnosis and prognosis in gliomas. In this study, we investigated the effect of 1p/19q codeletion on cancer cell metabolism and evaluated possible metabolic targets for tailored therapies.
Methods
We combined in vivo 1H (proton) magnetic resonance spectroscopy (MRS) measurements in human gliomas with the analysis of a series of standard amino acids by liquid chromatography–mass spectroscopy (LC-MS) in human glioma biopsies. Sixty-five subjects with low-grade glioma were included in the study: 31 underwent the MRI/MRS examination, 47 brain tumor tissue samples were analyzed with LC-MS, and 33 samples were analyzed for gene expression with quantitative PCR. Additionally, we performed metabolic tracer experiments in cell models with 1p deletion.
Results
We report the first in vivo detection of cystathionine by MRS in 1p/19q codeleted gliomas. Selective accumulation of cystathionine was observed in codeleted gliomas in vivo, in brain tissue samples, as well as in cells harboring heterozygous deletions for serine- and cystathionine-pathway genes located on 1p: phosphoglycerate dehydrogenase (PHGDH) and cystathionine gamma-lyase (CTH). Quantitative PCR analyses showed 40–50% lower expression of both PHGDH and CTH in 1p/19q codeleted gliomas compared with their non-codeleted counterparts.
Conclusions
Our results provide strong evidence of a selective vulnerability of codeleted gliomas to serine and glutathione depletion and point to cystathionine as a possible noninvasive marker of treatment response.
Keywords: cystathionine, MRS, brain glioma, 1p/19q codeletion, MEGA-PRESS
Key Points.
1. This study represents the first in vivo detection of cystathionine by MRS.
2. Cystathionine accumulates selectively in 1p/19q codeleted gliomas.
3. Cystathionine is a candidate marker for tailored therapies in codeleted gliomas.
Importance of the Study
Gliomas form a heterogeneous group of malignant brain tumors. The choice of appropriate tailored therapies is crucial for patients’ outcome but requires precise identification of glioma subtype. The diagnostic and prognostic stratification of brain gliomas highly benefits from the identification of molecular markers such as codeletion of chromosome arms 1p and 19q, recently recognized as a favorable predictive factor. The in vivo detection of cystathionine in 1p/19q codeleted gliomas by MRS opens up the possibility to investigate cancer-specific metabolic pathways noninvasively and to monitor cancer treatments in patients harboring these tumors.
Diffuse lower-grade gliomas (LGGs), which include World Health Organization (WHO) grades II and III astrocytomas and oligodendrogliomas, form a biologically heterogeneous group.1 Histology alone is often inadequate to provide a precise diagnosis needed for tailored therapy. Major improvements in the diagnostic and prognostic stratification of brain gliomas have been enabled by combining histopathological evaluation with molecular markers. Mutations in isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2)2,3 and complete codeletion of chromosome arms 1p and 19q4–6 have been recognized as favorable prognostic molecular markers. As these markers have represented a major breakthrough in the diagnosis of brain tumors, they have been integrated into the 2016 WHO classification of gliomas.7
IDH mutations can be found in both astrocytomas and oligodendrogliomas and are characterized by a specific cellular metabolism, causing the accumulation of 2-hydroxyglutarate (2HG) in tumor cells.2,3 Conversely, 1p/19q codeletion is linked specifically to the oligodendroglial histologic subtype and, despite its first description in 1994,4 the biological effects are still unclear. While metabolomic analyses in gliomas have led to variable results to date,9–12 two reports have surprisingly found normal levels for most metabolites involved in intermediary metabolism.13,14 Among the exceptional changes observed, one report identified alterations of cystathionine, an immediate precursor of cysteine and thus glutathione, with some evidence of altered antioxidant response in IDH-mutated tumors.14 In IDH-mutated 1p/19q codeleted gliomas, higher expression of cystathionine-β-synthase (CBS), the first enzyme of the transsulfuration pathway, was associated with better prognosis in this subset of gliomas,14 highlighting this pathway’s contribution to glutathione generation.15
In this study, we investigated the effect of 1p/19q codeletion on cancer cell metabolism by combining 1H (proton) magnetic resonance spectroscopy (MRS) measurements in human brain gliomas in vivo with the analysis of a series of standard amino acids by liquid chromatography–mass spectroscopy (LC-MS) in human glioma biopsies. MRS offers a unique opportunity to measure noninvasively a large number of brain metabolites and potentially enables in vivo detection and monitoring of molecules accumulating in certain brain tumors.16–19 The detection of cystathionine in vivo by MRS, herein reported for the first time, opens up the possibility of an accurate and noninvasive identification of glioma subtypes with potential monitoring of specific treatment responses.
Materials and Methods
Human Subjects
Sixty-five subjects (35 males; median age: 45 y, range: 22–76 y) with a suspected or diagnosed LGG were recruited at the Hospital Pitié-Salpêtrière in Paris and Hospital Foch in Suresnes over a 4-year period (2014–2017). The entire cohort includes subjects scanned with MRI and subjects from whom brain tissues were obtained and used for tissue and quantitative polymerase chain reaction (qPCR) analyses. Integrated diagnoses according to the WHO 2016 classification7 are reported in Table 1.
Table 1.
Clinical and histomolecular features of the cohort
| Histological Diagnosis | # Subj | WT | IDH1 | IDH2 | 1p/19q Codel | MRS | Tissue Analysis | qPCR |
|---|---|---|---|---|---|---|---|---|
| Oligodendroglioma grade II | 12 | 0 | 10 | 2 | 12 | 6 | 7 | 7 |
| Anaplastic oligodendroglioma grade III | 17 | 0 | 11 | 6 | 17 | 9 | 12 | 10 |
| Diffuse astrocytoma grade II | 8 | 0 | 7 | 1 | 0 | 4 | 8 | 7 |
| Anaplastic astrocytoma grade III | 14 | 0 | 14 | 0 | 0 | 9 | 7 | 9 |
| Glioblastoma | 13 | 11 | 1 | 1 | 0 | 2 | 12 | 0 |
| Ganglioglioma | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Total | 65 | 12 | 43 | 10 | 29 | 31 | 47 | 33 |
Human Subjects for MRI/MRS
Thirty-one subjects (17 males; median age: 44 y, range: 22–68 y) underwent the MRI/MRS examination. Additional inclusion criteria were: age >18 years, Karnofsky performance status score >60, and ability to provide written informed consent prior to inclusion in the study. The study was approved by the local ethical committee (CPP-Paris 6) in accordance with Declaration of Helsinki principles.
Of these subjects, 12 were included prospectively for suspected diagnosis of LGG estimated from T2-weighted fluid attenuation inversion recovery (FLAIR) images, and scheduled to receive subtotal tumor resection. These subjects underwent the MRI/MRS at a median interval of 1 day before the surgery. Genotyping assay for IDH identified 10 IDH mutants (5 with 1p/19q codeletion) and 2 IDH wild-type (WT) cases. Tissue and qPCR analyses were also performed for these subjects.
The remaining 19 subjects had a past tissue diagnosis of an IDH1-mutated glioma (10 with 1p/19q codeletion) and were scanned post surgery (no brain tumor tissue was available for further analysis).
Human Subjects for Tissue and qPCR Analyses
Forty-seven brain tumor tissue samples from 47 individuals were analyzed (26 males; median age: 45 y, range: 22–76 y). Of these, 33 snap-frozen samples were used also for gene expression analysis by qPCR (15 males; median age: 41 y, range: 25–58 y).
Magnetic Resonance Acquisition
In vitro MRS
Cystathionine phantom ([cystathionine] = 1 mM, pH 7.2) was prepared using phosphate buffer with 4,4,-dimethyl-4-silapentane-1-sulfonic acid added for chemical shift referencing and measured at physiological temperature. MRS acquisition was performed using a 3 T whole-body Siemens Prismafit system equipped with a 32-channel receive-only Siemens head coil. MR spectra were measured, as in the in vivo acquisitions, using a single-voxel Mescher–Garwood point resolved spectroscopy (MEGA-PRESS)20 sequence: repetition time (TR) = 2 s, echo time (TE) = 68 ms, number of averages = 512, volume of interest (VOI) = 3 cm3 (see In vivo MRI/MRS section).
In vivo MRI/MRS
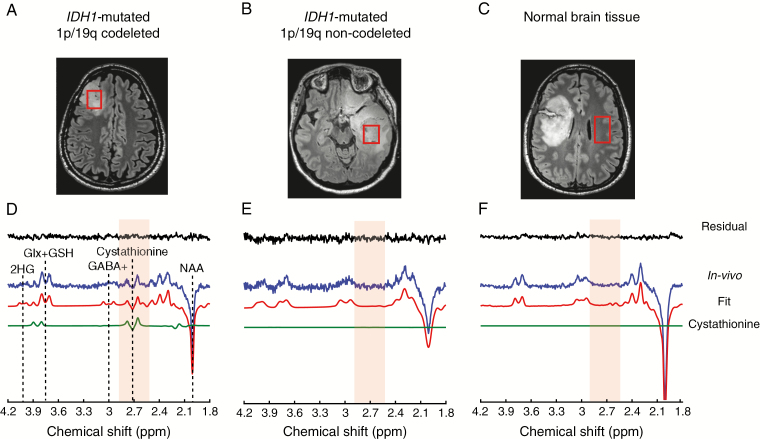
Acquisitions were performed at CENIR using a 3 T whole-body system (Magnetom Verio, Siemens) equipped with a 32-channel receive-only head coil. Three-dimensional FLAIR images (field of view = 255 × 255 × 144 mm3, resolution: 1.0 × 1.0 × 1.1 mm3, TR/TE = 5000/399 ms, scan time = 5.02 min) were acquired to position the spectroscopic VOI in the glioma (hyper-intense region in the images) (Figure 1A, B). The VOI size was adapted to the tumor size with 6 cm3 as a minimum size. MR spectra were acquired with a single-voxel spectral editing MEGA-PRESS20 sequence (TR = 2 s, TE = 68 ms) enabling detection of 2HG and cystathionine, using previously described procedures and parameters.19 The editing pulse was applied at 1.9 ppm for the edit-on condition and at 7.5 ppm for the edit-off condition, in an interleaved fashion (128 pairs of scans, scan time = 8.5 min). The final spectra were obtained by subtracting the spectra acquired at the edit-on and edit-off conditions. For 8 subjects, the MEGA-PRESS acquisition was performed also in the contralateral region outside the visible lesions (Figure 1C).
Fig. 1.
In vivo detection of cystathionine. The location and size of the VOIs are shown on FLAIR images in (A) an IDH1-mutated 1p/19q codeleted glioma, (B) an IDH1-mutated non-codeleted glioma, and (C) normal brain tissue. (D, E, F) In vivo spectra (blue) are shown together with LCModel fits (red), the cystathionine contribution (green), and residuals (black). The cystathionine pattern is visible at 2.7 ppm (light orange shading) in the IDH1-mutated 1p/19q codeletion glioma (D), while it is not detectable in the IDH1-mutated non-codeleted glioma (E), and normal tissue (F). Spectra are scaled with respect to the water signal. No line broadening was applied. NAA: N-acetylaspartate; GABA+: GABA + macromolecules; Glx + GSH: glutamate + glutamine + glutathione.
The acquired spectra were processed in Matlab (MathWorks Inc., Natick, MA) and analyzed using LCModel v6.3-0G21 (Stephen Provencher, Inc., Oakville, ON, Canada) as previously described19 with cystathionine added to the basis set and macromolecular spectrum removed. The reported concentrations are semi-quantitative.
Tissue Analysis
Automated immunohistochemical analysis of IDH1 R132H and mutational status of IDH1 and IDH2 were determined as previously described.22 All cases in this series scoring negative for IDH1 R132H immunostaining were analyzed for IDH1 and IDH2. The presence of 1p/19q codeletion was assessed by copy number variation analysis from next-generation sequencing targeted gene capture.
Tissue samples were homogenized in bidistilled water, and the soluble protein concentration was measured by a bicinchoninic acid assay. Concentrations were measured with liquid chromatography coupled to tandem mass spectrometry (UPLC-MSMS). For accurate quantification, a stable isotope internal standard for the same structure for each metabolite (Eurisotop) was added to sample supernatant after centrifugation of homogenized tissue. Samples were first derivatized using AccQ Tag Ultra (Waters Corporation) according to manufacturer’s recommendations. Amino acid separation was performed with an Acquity UPLC system using a CORTECS UPLC C18 column coupled to microTQS tandem mass spectrometer (Waters Corporation). Internal labeled standards were labeled on all carbons and nitrogen(s) with the following exceptions: cystathionine 3,3,4,4-D4, glutamine 2,3,3,4,4-D5, asparagine 13C4, and tryptophan indole-D5. Total glutathione was measured by UPLC-MSMS on an Acquity UPLC system using a C18 column coupled to a Xevo TQD mass spectrometer (Waters Corporation) after reduction with dithiotreitol. The internal standard for glutathione measurements was labeled 13C2,15N on the glycine residue. We measured 2HG acid by gas chromatography coupled to tandem mass spectrometry (GC-MSMS) on a GC-436 Scion-TQD device (Brüker Daltonics). Samples were processed by organic (ethylacetate) extraction and derivatized by a standard silylation protocol (N,O-bis[trimethylsilyl]trifluoroacetamide + 1% trimethylchlorosilane [BSTFA-TMCS]). The 2HG acid internal standard was labeled D3.
For each tissue sample, amino acid and total glutathione concentrations were normalized to the median of amino acid concentrations calculated from threonine, proline, methionine, isoleucine, leucine, tyrosine, phenylalanine, ornithine, lysine, arginine after exclusion of highly variable metabolites (reflecting storage condition, variable glycolysis such as aspartate, glycine, glutamate, glutamine, serine, alanine) and those metabolites sometimes not reported in the literature (valine, cystine, asparagine, tryptophan, citrate, cystathionine).23
Quantitative PCR
Total RNA was extracted using the Maxwell RSC Simply RNA Tissue Kit (Promega) following the manufacturer’s instructions. The gene expression of phosphoglycerate dehydrogenase (PHGDH, 1p12) and cystathionine gamma-lyase (CTH, 1p31.1) in the tumor tissue was analyzed using real-time qPCR analysis experiments with SYBR green detection. The reference gene was PPIA (peptidylprolyl isomerase A). One IDH WT glioma sample was used as the calibrator. Sequences of primers used for PHGDH correspond to TCAGTTCGTGGACATGGTGA (forward) and TCTTTCAGGAGGCCGACAAT (reverse), and for CTH to GACATTGAAGGCTGTGCACA (forward) and AGACACCAGGCCCATTACAA (reverse). The qPCR reactions were performed using the Light Cycler 480 system as previously described.24
The 2delta-deltaCT method was used to determine the relative expression levels. The calculation of the relative amounts of the studied transcript compared with the reference transcript was performed using the Light Cycler 480 software. The final results are expressed as a ratio of the expression levels of the studied gene and the reference as previously described.24
Public Datasets for Brain Tumor Expression
Public mRNA expression data from 2 cohorts of LGGs were explored for expression of PHGDH and CTH. Data from the POLA Network25 were retrieved from ArrayExpress (E-MTAB-3892), and LGG data from The Cancer Genome Atlas (TCGA)26 were downloaded from Broad GDAC Firehose (doi: 10.7908/C1K64H78; February 21, 2019). Status of 1p/19q codeletion in TCGA samples was assigned using GISTIC2 results by chromosome arm as found on the data portal of TCGA.
Cell Models
Human retinal pigmented epithelial 1 (RPE-1) cells targeted by clustered regularly interspaced short palindromic repeat‒associated protein-9 nuclease (CRISPR-CAS9) were produced by the TacGene gene targeting platform (Inserm and the National Museum of Natural History, Paris, France), led by Dr Jean-Paul Concordet. Targeted were exon 6 of CTH, part of the catalytic domain, and exon 7 of PHGDH, mutated in serine-deficient human patients. A plasmid encoding the 2 guide RNAs and the CAS9 protein was transfected into parental RPE-1 cells. One hundred clones were genotyped by PCR. One clone harbored the 49-megabase deletion spanning the 2 guide RNA target sites between the 2 genes (referred to as “large heterozygous deletion” or “del+/−”), thus including the 3′ half of CTH and the 5′ half of PHGDH coding regions plus the intervening sequences. A second clone harbored a 2 bp insertion in CTH and a 1 bp insertion in PHGDH (referred to as CTH+/−, PHGDH+/−). And a third clone showed 3 separate mutations: a monoallelic 1 bp insertion in CTH, a 1 bp insertion in one PHGDH allele, and a 6 bp deletion in the other PHGDH allele (referred to as “PHGDH knock-out/KO” or “PHGDH−/−”).
Metabolic Analysis in Cell Models
About one million cells per condition and replicate were grown at confluence in Dulbecco’s modified Eagle’s medium/F12 and Glutamax dipeptide followed by incubation in the same culture medium with added 13C6 glucose (equimolar to natural glucose contained in the medium) for up to 4 hours. To test for the effects of CBS activation, 100 μM S-adenosylmethionine (SAM; Sigma) was supplemented during the labeling experiments. Cells were trypsinized and rinsed with NaCl 0.9% aqueous solution, and cell pellets were stored at −80°C until processing. Samples were mechanically disrupted in 50% methanol in water and the supernatant was analyzed. We added internal standards several mass units heavier than fully 13C-labeled metabolites, including U-13C3, 2,3,3-D3 U-15N serine, U-13C2, 2,2-D2, U-15N glycine, and D10 alloisoleucine (Eurisotop). In addition to the unlabeled U-12C ions, selected reaction monitoring transitions for the ions with maximum expected labeling for glycine, serine, and cystathionine (13C2, 13C3, and 13C3, respectively) were measured by LC-MSMS and GC-MSMS. Sample preparation and MS characteristics were as above, except that samples for GC-MSMS were extracted with a 1:1 mixture of ethylacetate and isopropanol. Isotopic enrichment was expressed as the isotopic ratio. The mean values of the ratios calculated from experiments run in parallel with added unlabeled glucose, which was considered the baseline, were subtracted. Total absolute concentrations were calculated as the sum of the labeled and unlabeled metabolites divided by valine levels (all other essential amino acids tested gave the same profile, data not shown).
Statistical Analysis
Differences of the in vivo data between groups were analyzed using a 2-tailed Student’s t-test and were considered statistically significant for P < 0.05.
Metabolite concentrations from tissue analysis were compared using a non-parametric Wilcoxon test. Linear regressions between metabolites were derived with the lm function in R software (http://www.R-project.org/ ; February 21, 2019).
Gene expression analyses for PCR data were performed using R and Prism software. Fold changes were compared using the Mann–Whitney U-test.
The public tumor dataset analyses were performed using R. The Tukey post hoc test was performed after a 1-way ANOVA to compare the mean mRNA expression values between samples according to codeletion and IDH mutation status. Differences were considered statistically significant for a Tukey adjusted P < 0.05.
Results
In Vivo Cystathionine Detection in Glioma
Thirty-one subjects with glioma underwent MRS examination (Table 1). High-quality spectra from VOI positioned in the glioma minimizing the cystic and necrotic areas (Figure 1A, B) or in healthy brain tissue (Figure 1C) were obtained. Signals attributed to cystathionine were visible at ∼2.7 and ∼3.9 ppm in the tumor of a subset of subjects (Figure 1D, Supplementary Figure 1), but not in healthy brain tissue (Figure 1F). Statistically significant higher cystathionine levels (P = 0.014; Figure 2) were detected in IDH-mutated 1p/19q codeleted gliomas (average [cystathionine] = 2.6 mM; range: 0.2–4.1 mM; average Cramér-Rao lower bound [CRLB] = 36%), compared with IDH-mutated non-codeleted gliomas (average [cystathionine] = 1.25 mM; range: 0–3.6 mM; average CRLB = 235%). Of the 2 IDH WT gliomas, cystathionine was measured in 1 case ([cystathionine] = 4.5 mM and CRLB = 14%). Cystathionine was not detected in the healthy tissue. No significant dependence of cystathionine levels on grade or mutational status was observed (data not shown).
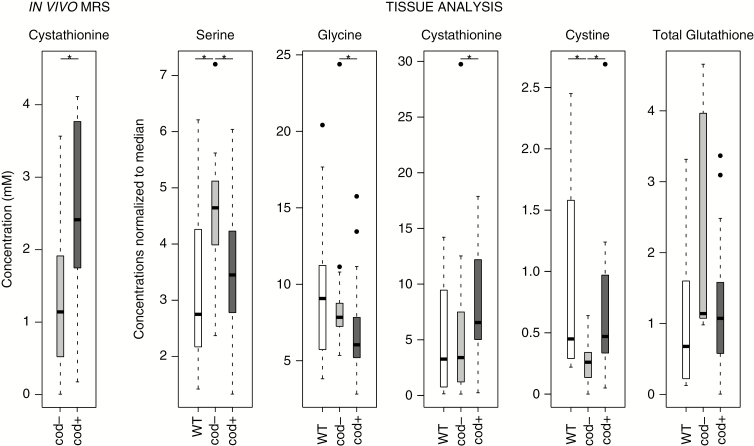
Fig. 2.
Cystathionine and metabolites involved in synthesis of glutathione. Concentrations of cystathionine from in vivo MR spectra and metabolites involved in glutathione synthesis pathway, normalized to metabolite median, from tissue analysis. *P < 0.05. Cod-: IDH1+ or IDH2+ non-codeleted; cod+: IDH1+ or IDH2 + 1p/19q codeleted.
Identification of Cystathionine
Unexpected signals observed in the edited MR spectrum in an IDH1-mutated 1p/19q codeleted glioma at ~2.7 ppm and ~3.9 ppm were identified as cystathionine based on the chemical shifts of the resonances as well as the J-couplings needed for those signals to appear in the editing scheme used (Supplementary Table 1). As shown in Supplementary Figure 2, the resonance at ~2.2 ppm is within the bandwidth of the editing pulse and J-coupled to resonances at ~2.7 and ~3.9 ppm. In vivo, the cystathionine pattern is clearly visible at ~2.7 ppm, while the multiplets at ~3.9 and ~2.2 ppm partially overlap with glutamate, glutamine, and N-acetylaspartate signals (Supplementary Figures 1 and 2). The excellent agreement between the in vivo spectrum, the phantom cystathionine spectrum, and the simulated spectrum used in the basis set affirms the correct assignment of the in vivo signal and the accuracy of the chemical shifts and J-coupling constants used to generate the basis spectra.
Cystathionine, Glutathione, and Proteinogenic Amino Acids in Tissue
Concentrations of cystathionine, total glutathione, and proteinogenic amino acids were determined in brain tumor samples. The normalization of concentrations to the median of the biochemically most stable amino acid concentrations allowed direct comparison of our results to those from the literature23 (regression coefficient of 0.99 between the Lefauconnier et al 1976 dataset, normalized by the same amino acids, and our dataset) and reduced the standard deviations for all metabolites (data not shown).
To characterize the metabolic effects of the 1p/19q codeletion, the metabolite levels normalized to the median of pooled IDH1 and IDH2-mutated non-codeleted samples were compared with their codeleted counterparts and WT samples (Supplementary Table 2). Six metabolites were significantly altered in a univariate analysis (Wilcoxon’s test), and 4 of these metabolites are involved in the serine and glutathione biosynthesis pathways (Figure 3). Higher glycine and serine levels were observed in IDH-mutated non-codeleted samples compared with IDH-mutated codeleted samples (P = 0.023 and P = 0.005, respectively) (Figure 2). Conversely, cystathionine and cystine were significantly higher in IDH-mutated codeleted samples compared with IDH-mutated non-codeleted samples (P = 0.048 and P = 0.007, respectively). However, cystine levels might not be reliable due to tissue conservation artifacts (notably storage time-dependent binding of cysteine to proteins), and free cysteine was below quantification levels. Total glutathione shared the same trend as serine and glycine without reaching statistical significance (Figure 2).
Fig. 3.
Suggested vulnerability for 1p/19q-codeleted gliomas. Solid arrows = single metabolic step, dash arrows = multiple (not drawn) metabolic steps. PSAT: phosphoserine amino transferase; PSPH: phosphoserine phosphatase; 1p del: 1p deletion
To investigate the mechanism leading to higher levels of cystathionine in 1p/19q codeleted samples, the correlations between cystathionine and other amino acids were investigated. Six positive correlations were observed (Supplementary Table 3). Cystathionine was highly positively correlated with serine in IDH-mutated codeleted samples (r2 = 0.78; Supplementary Figure 3) and was moderately correlated with the serine-related amino acid threonine (r2 = 0.36). In IDH-mutated non-codeleted samples, cystathionine was correlated with methionine (r2 = 0.39), while in WT samples cystathionine was correlated with Krebs cycle and glutamine-related metabolites (Supplementary Table 3).
PHGDH and CTH Expression
Quantitative PCR analyses showed approximately 40–50% lower expression of both PHGDH and CTH in IDH-mutated 1p/19q codeleted gliomas compared with IDH-mutated non-codeleted gliomas (P = 0.006 and P = 0.003, respectively) (Supplementary Figure 4). These results are consistent with cis-acting transcriptional regulation accounting for most of the expression levels.
Publicly available mRNA expression datasets of LGGs provided consistent results. In the POLA dataset,25 the mean mRNA levels for PHGDH were significantly lower in IDH-mutated codeleted gliomas compared with IDH-mutated non-codeleted gliomas (P = 8.3 × 10−5; Supplementary Figure 5). In the LGG dataset of TCGA,26 the mean RNA levels for PHGDH were significantly lower in IDH-mutated codeleted gliomas compared with IDH-mutated non-codeleted gliomas (P = 3.1 × 10−7; Supplementary Figure 5).
Similarly, in the POLA dataset, lower CTH mean mRNA levels in 1p/19q codeleted gliomas compared with non-codeleted gliomas (P = 1.6 × 10−11) and IDH WT gliomas (P = 8.5 × 10−4; Supplementary Figure 5) were observed. In the LGG dataset of TCGA, significantly lower CTH mean RNA levels were observed in IDH-mutated codeleted gliomas compared with IDH-mutated non-codeleted gliomas (P < 10−13) and IDH WT gliomas (P = 3.9 × 10−7; Supplementary Figure 5).
Metabolic Flux Alterations Associated with PHGDH and CTH Hemizygosity
To help interpret the data, we produced human RPE-1 cells with heterozygous mutations in CTH and PHGDH, creating either heterozygous intra-exonic micro-rearrangements in both genes (referred to as CTH+/−, PHGDH+/−) or a large heterozygous deletion on chromosome 1 that covered the entire region between them, including half of both genes (referred to as “large heterozygous deletion” or “del+/−”). We also generated biallelic PHGDH knockout cells, and used parental cells as a reference.
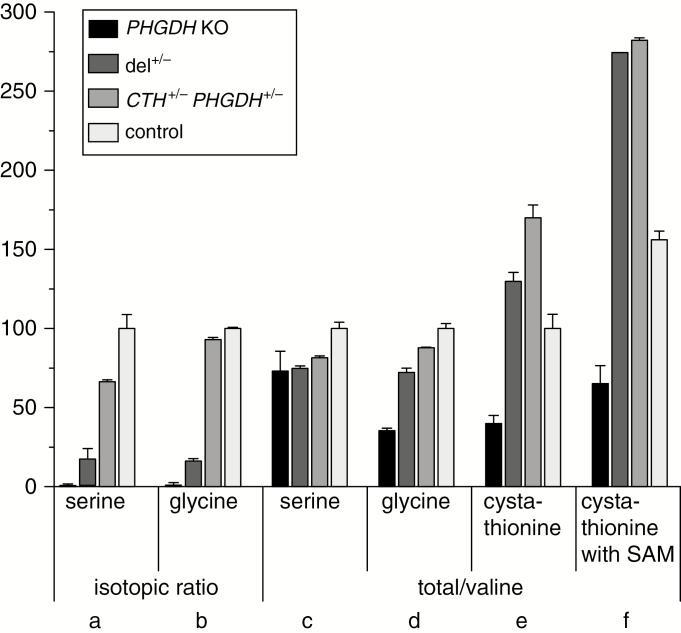
In 4 hour 13C-glucose loading experiments, label enrichment of serine and glycine was markedly reduced in PHGDH knockout cells, consistent with the expected highly reduced flux through the serine pathway (Figure 4A, B). CTH+/−, PHGDH+/−, and particularly del+/− cells showed a significant though lesser reduction of 13C enrichment, thus indicating partial deficiency in endogenous serine production (Figure 4A, B). The total intracellular pool of serine and glycine showed a less marked trend toward reduced levels, presumably reflecting compensatory mechanisms such as amino acid uptake from the supplemented culture medium (Figure 4C, D).
Fig. 4.
Metabolic analysis of human RPE-1 cells. Cells were generated with heterozygous inactivation of CTH and PHGDH or biallelic inactivation of PHGDH and compared with the parental cell line (see legend below). From left to right, isotopic ratios (ie, relative isotopic enrichment after 4-h incubation with 13C-labeled glucose) for (A) serine and (B) glycine, and total levels of (C) serine, (D) glycine and (E) cystathionine normalized to valine in cells cultured in standard medium. (F) Cystathionine levels increased upon addition of 100 μM SAM. Error bars: replicate standard deviation. Results are expressed relative to the mean WT levels in standard culture conditions set to 100%. PHGDH KO: monoallelic inactivation of CTH and biallelic inactivation of PHGDH; del+/−: monoallelic inactivation of PHGDH and CTH by a large deletion on 1p; CTH+/− PHGDH+/−: monoallelic inactivation of CTH and PHGDH by intra-exonic mutations; control: parental cell line.
Cystathionine was not labeled in the 4 hour 13C-glucose loading experiments under standard culture conditions (data not shown). Total cystathionine levels were only moderately altered under standard culture conditions (Figure 4E), yet markedly increased upon supplementation with SAM, a CBS activator, and the effect was greater (~+100%) in both types of double heterozygous mutant cells relative to controls (~+50%; Figure 4F). Thus, cystathionine accumulation was enhanced by the combined action of 1p deletion and gain of CBS function.
SAM supplementation induced some increased cystathionine labeling with high variance (data not shown), further indicating that in our in vitro model, serine supplementation in the culture medium was largely sufficient to fuel cystathionine production. Serine biosynthesis dysfunction was more severe in cells with the large 1p deletion compared with double heterozygosity for intragenic mutations. Possibly trans-acting regulatory sequences operating between alleles27 might be involved.
Discussion
Cystathionine as a Marker of Oxidative Stress in Codeleted Gliomas
This study reports the first measurement of cystathionine in vivo. Elevated levels of cystathionine were observed in vivo in IDH-mutated, 1p/19q codeleted gliomas compared with their non-codeleted counterparts and to normal brain tissue. This finding was confirmed by metabolomic analysis performed in glioma tissue samples and was linked to a specific cancer cell metabolic flux profile associated with the hemizygous loss of the 1p chromosomal arm. In a few subjects, high cystathionine levels were observed without codeletion, yet they may have undetected intragenic mutations (or epigenetic modifications).
In vivo edited MRS has been extensively used to measure J-coupled metabolites such as γ-aminobutyric acid in the brain20,28–30 or 2HG in gliomas.17,19 The detection accuracy of 2HG for a subset of patients from this study was reported previously.19 However, no studies reported the detection of cystathionine in vivo with MRS in either diseased or normal human brain. The identification of cystathionine was confirmed by comparing in vivo spectra acquired in 1p/19q codeleted gliomas with the cystathionine spectrum measured in a phantom. Our observation of higher, detectable cystathionine levels in IDH-mutated, 1p/19q codeleted subjects in vivo agreed with tissue analysis, which additionally suggested a mechanism of pathology with possible therapeutic implications as follows.
The discovery of selective 2HG production by mutant enzymes involved in central metabolism opened new avenues to a better understanding of the mechanisms of oncogenesis and provided a biochemical marker of great clinical interest for several cancers, including gliomas.8–14,16,31 Similar to the accumulation of 2HG occurring selectively in IDH-mutated gliomas, cystathionine accumulation and other abnormal levels of amino acids in 1p/19q codeleted gliomas (Figure 2 and Supplementary Table 2) may result from partial genetic loss of 2 enzymes located on chromosome 1p: PHGDH, the rate-limiting enzyme of the pathway diverting glycolytic intermediates into the biosynthesis of serine, and CTH, one of the 2 enzymes diverting sulfur from the methionine-homocysteine cycle to cysteine (Figure 3). Of note, serine is a methyl donor to folates, which among other functions is a significant source of nicotinamide adenine dinucleotide phosphate (NADPH). On the other hand, cysteine is the sulfur-containing precursor of glutathione, the reduced form of which is regenerated by NADPH. The latter is largely provided by the pentose phosphate pathway (Figure 3), but also by oxidation of the serine-derived methyl pool and from other sources.
IDH-mutated tumors may selectively benefit from an increased production of total glutathione to compensate for diminished NADPH. In fact, NADPH is used by IDH for 2HG production and is thus less available for oxidized glutathione recycling.32,33 Despite NADPH depletion, reduced glutathione levels are maintained in IDH-mutated tumors, suggesting alternative antioxidant pathways.14,34,35 Conversely, in codeleted tumors, both total glutathione production and NADPH regeneration could be limited by serine levels possibly because of reduced PHGDH activity due to 1p/19q codeletion. Reduced serine biosynthesis may lead to increased reliance on the CBS/CTH pathway as a critical response to increased oxidative stress, consistent with previous suggestions.19,34,35 In particular, high CBS expression was shown to confer better prognosis in IDH-mutated 1p/19q codeleted gliomas,14 in line with a previous study showing that decreased expression of CBS promotes glioma tumorigenesis in tumor xenografts.34 Correlation between higher CBS expression and survival in IDH-mutated 1p/19q codeleted gliomas was confirmed also from the POLA public dataset (data not shown). Fack et al14 reported decreased cystathionine in IDH mutant tumor xenografts compared with WT, yet in the small sample shown, cystathionine levels were roughly inversely correlated with codeletion status, which is consistent with our findings.
Cystathionine accumulation in codeleted tumors may result from partial CTH deficiency that confers increased susceptibility to metabolic overflow, as suggested by clinical findings of families segregating heterozygous CTH mutations that are associated with moderately increased plasma cystathionine.36 In this context, CBS overexpression is expected to enhance CTH overflow while also further contributing to serine depletion, and possibly accounting for the observed striking positive correlation between reduced serine and increased cystathionine in codeleted tumors (Supplementary Figures 3 and 6).
To test these hypotheses, we generated double heterozygous PHGDH and CTH deficient cells that indeed showed partial serine deficiency and accumulated cystathionine upon supplementation with a CBS activator (Figure 4). CBS overexpression is thus a promising candidate mechanism for a homeostatic response to serine deficiency induced by heterozygous loss of PHGDH and CTH. Additional experiments are required to validate the proposed mechanism in gliomas, notably in terms of endogenous serine/glycine dependence.
Therapeutic Implications
Recently, it has been shown that cystathionine accumulates selectively in human breast cancer tissues due to a breast cancer–specific metabolic adaptation that is absent in normal breast. It was speculated that this adaptation provides additional homeostatic stability to the endoplasmic reticulum and the mitochondria toward maintaining a higher apoptotic threshold that is also responsible for drug resistance.37 Nevertheless, the observed block of the CTH reaction led to an increased dependency on exogenous cysteine in order to produce glutathione, which presumably makes breast and other cancers sensitive to systemic cyst(e)inase therapy in mice.38 Our findings suggest that heterozygous loss of CTH in combination with heterozygous loss of PHGDH has similar functional effects, thus suggesting that cysteine depletion might be more effective in targeting 1p deleted gliomas. Moreover, a serine/glycine-depleted diet or PHGDH inhibition considerably reduced tumor growth in well-defined cancer models in mice, thus demonstrating a critical selective role for endogenous serine biosynthesis in sustaining tumorigenesis.39
Cystathionine and other related amino acids identified in this study, such as glycine and glutathione, can be detected noninvasively with MRS. This opens up the possibility of investigating in vivo cancer-specific metabolic pathways. Our results point to a possible selective vulnerability to serine and glutathione depletion in 1p/19q codeleted gliomas and suggest in vivo cystathionine as a candidate marker to monitor cancer treatments in patients harboring these tumors.
Funding
FB and SL acknowledge support from the program “Investissements d’avenir” ANR-11- INBS-0011—NeurATRIS: Translational Research Infrastructure for Biotherapies in Neurosciences, ANR-10-IAIHU-06—Paris Institute of Translational neuroscience, and ANR-11-INBS-0006—France Life Imaging. ALDS and MS acknowledge support from the grant INCa-DGOS-Inserm_1250 of the SiRIC CURAMUS. ALDS and MS acknowledge support from the Institut National de la Santé et de la Recherche Médicale grant: INCa-DGOSInserm_1250 (SiRIC CURAMUS). DD and MM acknowledge support from the following National Institutes of Health grants: BTRC P41 EB015894 and P30 NS076408.
Conflict of interest statement. The authors have declared that no conflicts of interest exist.
Authorship statement: Development of MRS methodology: FB, MM; Acquisition and analysis of MRS data: FB, RV, DKD, MM; Metabolomic acquisition and analysis: CP, CO; qPCR analysis: ADS, AK; Cell experiments: CP, LT, CO; Interpretation of the data: FB, CP, MS, CO, MM; Manuscript writing: all authors; Recruitment of subjects: ADS, MS; Study supervision: SL, MS, CO, MM; Design and conceptualization of the study: CO, MM
Supplementary Material
Acknowledgments
The authors would like to thank Edward J. Auerbach, PhD, for implementing MRS sequences on the Siemens platform, Marion Benazra for performing the qPCR experiments, Jamie D. Walls, PhD, and Andreas Trabesinger, PhD, for comments about the manuscript.
References
- 1. Picca A, Berzero G, Sanson M. Current therapeutic approaches to diffuse grade II and III gliomas. Ther Adv Neurol Disord. 2018;11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Parsons DW, Jones S, Zhang X, et al. . An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dang L, White DW, Gross S, et al. . Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reifenberger J, Liu L, James CD, Wechsler W. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145(5):16. [PMC free article] [PubMed] [Google Scholar]
- 5. Idbaih A, Marie Y, Pierron G, et al. . Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol. 2005;58(3):483–487. [DOI] [PubMed] [Google Scholar]
- 6. Ostrom QT, Bauchet L, Davis FG, et al. . The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 8. Guo C, Pirozzi CJ, Lopez GY, Yan H. Isocitrate dehydrogenase mutations in gliomas: mechanisms, biomarkers and therapeutic target. Curr Opin Neurol. 2011;24(6):648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seltzer MJ, Bennett BD, Joshi AD, et al. . Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70(22):8981–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reitman ZJ, Jin G, Karoly ED, et al. . Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci. 2011;108(8):3270–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Metallo CM, Gameiro PA, Bell EL, et al. . Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481(7381):380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grassian AR, Parker SJ, Davidson SM, et al. . IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74(12):3317–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tateishi K, Wakimoto H, Iafrate AJ, et al. . Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell. 2015;28(6):773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fack F, Tardito S, Hochart G, et al. . Altered metabolic landscape in IDH-mutant gliomas affects phospholipid, energy, and oxidative stress pathways. EMBO Mol Med. 2017;9(12):1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McBean GJ. The transsulfuration pathway: a source of cysteine for glutathione in astrocytes. Amino Acids. 2012;42(1):199–205. [DOI] [PubMed] [Google Scholar]
- 16. Andronesi OC, Arrillaga-Romany IC, Ly KI, et al. . Pharmacodynamics of mutant-IDH1 inhibitors in glioma patients probed by in vivo 3D MRS imaging of 2-hydroxyglutarate. Nat Commun. 2018;9(1):1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andronesi OC, Kim GS, Gerstner E, et al. . Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi C, Ganji SK, DeBerardinis RJ, et al. . 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Branzoli F, Di Stefano AL, Capelle L, et al. . Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro Oncol. 2018;20(7):907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11(6):266–272. [DOI] [PubMed] [Google Scholar]
- 21. Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6): 672–679. [DOI] [PubMed] [Google Scholar]
- 22. Sanson M, Marie Y, Paris S, et al. . Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 2009;27(25):4150–4154. [DOI] [PubMed] [Google Scholar]
- 23. Lefauconnier JM, Portemer C, Chatagner F. Free amino acids and related substances in human glial tumours and in fetal brain: comparison with normal adult brain. Brain Res. 1976;117(1):105–113. [DOI] [PubMed] [Google Scholar]
- 24. Laffaire J, Di Stefano AL, Chinot O, et al. . An ANOCEF genomic and transcriptomic microarray study of the response to irinotecan and bevacizumab in recurrent glioblastomas. Biomed Res Int. 2014;2014:282815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamoun A, Idbaih A, Dehais C, et al. ; POLA network Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat Commun. 2016;7:11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. The Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koeman JM, Russell RC, Tan MH, et al. . Somatic pairing of chromosome 19 in renal oncocytoma is associated with deregulated EGLN2-mediated [corrected] oxygen-sensing response. PLoS Genet. 2008;4(9):e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terpstra M, Ugurbil K, Gruetter R. Direct in vivo measurement of human cerebral GABA concentration using MEGA-editing at 7 Tesla. Magn Reson Med. 2002;47(5):1009–1012. [DOI] [PubMed] [Google Scholar]
- 29. Marjańska M, Lehéricy S, Valabrègue R, et al. . Brain dynamic neurochemical changes in dystonic patients: a magnetic resonance spectroscopy study: MRS Study in Dystonic Patients After TMS. Mov Disord. 2013; 28(2):201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bogner W, Gagoski B, Hess AT, et al. . 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi C, Raisanen JM, Ganji SK, et al. . Prospective longitudinal analysis of 2-hydroxyglutarate magnetic resonance spectroscopy identifies broad clinical utility for the management of patients with IDH-mutant glioma. J Clin Oncol. 2016;34(33):4030–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bleeker FE, Atai NA, Lamba S, et al. . The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119(4):487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Molenaar RJ, Botman D, Smits MA, et al. . Radioprotection of IDH1-mutated cancer cells by the IDH1-mutant inhibitor AGI-5198. Cancer Res. 2015;75(22):4790–4802. [DOI] [PubMed] [Google Scholar]
- 34. Takano N, Sarfraz Y, Gilkes DM, et al. . Decreased expression of cystathionine β-synthase promotes glioma tumorigenesis. Mol Cancer Res. 2014;12(10):1398–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harris IS, Treloar AE, Inoue S, et al. . Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27(2):211–222. [DOI] [PubMed] [Google Scholar]
- 36. Kraus JP, Hasek J, Kozich V, et al. . Cystathionine gamma-lyase: clinical, metabolic, genetic, and structural studies. Mol Genet Metab. 2009;97(4):250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sen S, Kawahara B, Mahata SK, et al. . Cystathionine: a novel oncometabolite in human breast cancer. Arch Biochem Biophys. 2016;604:95–102. [DOI] [PubMed] [Google Scholar]
- 38. Cramer SL, Saha A, Liu J, et al. . Systemic depletion of L-cyst(e)ine with cyst(e)inase increases reactive oxygen species and suppresses tumor growth. Nat Med. 2017;23(1):120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maddocks ODK, Athineos D, Cheung EC, et al. . Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544(7650):372–376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.