Abstract
The developmental and epileptic encephalopathies (DEEs) are heterogeneous disorders with a strong genetic contribution, but the underlying genetic etiology remains unknown in a significant proportion of individuals. To explore whether statistical support for genetic etiologies can be generated on the basis of phenotypic features, we analyzed whole-exome sequencing data and phenotypic similarities by using Human Phenotype Ontology (HPO) in 314 individuals with DEEs. We identified a de novo c.508C>T (p.Arg170Trp) variant in AP2M1 in two individuals with a phenotypic similarity that was higher than expected by chance (p = 0.003) and a phenotype related to epilepsy with myoclonic-atonic seizures. We subsequently found the same de novo variant in two individuals with neurodevelopmental disorders and generalized epilepsy in a cohort of 2,310 individuals who underwent diagnostic whole-exome sequencing. AP2M1 encodes the μ-subunit of the adaptor protein complex 2 (AP-2), which is involved in clathrin-mediated endocytosis (CME) and synaptic vesicle recycling. Modeling of protein dynamics indicated that the p.Arg170Trp variant impairs the conformational activation and thermodynamic entropy of the AP-2 complex. Functional complementation of both the μ-subunit carrying the p.Arg170Trp variant in human cells and astrocytes derived from AP-2μ conditional knockout mice revealed a significant impairment of CME of transferrin. In contrast, stability, expression levels, membrane recruitment, and localization were not impaired, suggesting a functional alteration of the AP-2 complex as the underlying disease mechanism. We establish a recurrent pathogenic variant in AP2M1 as a cause of DEEs with distinct phenotypic features, and we implicate dysfunction of the early steps of endocytosis as a disease mechanism in epilepsy.
Keywords: clathrin-mediated endocytosis, developmental and epileptic encephalopathy, synaptic transmission, computational phenotypes, Human Phenotype Ontology, neurodevelopmental disorders
Introduction
A substantial proportion of childhood epilepsies present as developmental and epileptic encephalopathies (DEEs), characterized by intractable epilepsy with associated cognitive comorbidities.1 DEEs often occur in the absence of explanatory imaging or metabolic findings, and genetic causes are increasingly implicated.2, 3, 4 Recent progress through massively parallel sequencing technologies has enabled the discovery, in the last decade, of an increasing number of associated genes,5, 6, 7 most commonly found due to pathogenic de novo variants.5 The genetic landscape of DEE is heterogeneous, and pathogenic variants in single genes often explain fewer than one percent of the diagnoses of all individuals.7 Although a clear gene-phenotype association is seen for some genetic etiologies, such as Dravet syndrome and pathogenic variants in SCN1A (MIM: 607208),8, 9 many genetic epilepsies demonstrate significant phenotypic heterogeneity and have overlapping clinical presentations associated with a wide spectrum of genetic etiologies.10, 11
The discovery of underlying genetic causes in the epilepsies and neurodevelopmental disorders has primarily advanced through the ability to process and analyze large genomic datasets.12, 13 In contrast, phenotypic data are frequently collected in non-standard formats and therefore cannot be used for a systematic analysis across larger cohorts of affected individuals.14 The Human Phenotype Ontology (HPO) has been developed as a standardized format to provide both terminology and semantics to a broad range of phenotypic features, including neurological features.15, 16 This standardized vocabulary has already been used by researchers to identify individuals with rare monogenic diseases in large cohorts17, 18 and is frequently applied in a diagnostic setting to define phenotypic overlap for variant interpretation. In addition, methods to determine phenotypic similarity have been developed that incorporate the hierarchical structure of the ontology.19, 20 Given their phenotypic complexity, the childhood epilepsies lend themselves to analysis methods that capitalize on available phenotypic information in addition to genomic data.
Here, we analyzed trio whole-exome sequencing data in 314 individuals phenotyped with HPO terms. We assessed exome sequencing data for potential de novo variants and determined, observed, and predicted phenotypic similarity in individuals with de novo variants in the same gene. Two individuals with a de novo c.508C>T (p.Arg170Trp) AP2M1 (MIM: 601024; GenBank: NM_004068.3) variant had a higher phenotypic similarity than expected by chance, and more detailed phenotyping identified a clinical phenotype consistent with epilepsy with myoclonic-atonic seizures, also known as Doose syndrome. We subsequently identified two additional individuals with the identical de novo c.508C>T (p.Arg170Trp) AP2M1 variant and comparable phenotypes in a large diagnostic cohort. Functional analysis revealed that the p.Arg170Trp AP2M1 variant encoding the μ-subunit of the clathrin adaptor complex AP-2 impairs the early stages of clathrin-mediated endocytosis (CME), thereby identifying defective CME as a disease mechanism for neurodevelopmental disorders.
Material and Methods
Participant Recruitment
Informed consent for participation in this study was obtained from the parents of all probands in agreement with the Declaration of Helsinki. All studies were completed per protocol and had local approval by institutional review boards (IRBs). For study inclusion, all probands underwent a clinical data review of medical history information, including developmental and seizure history, neurological findings, and morphological details. Available electroencephalogram (EEG) and brain imaging data were reviewed for all individuals. Epilepsy syndromes and seizure types were classified according to the International League Against Epilepsy (ILAE) classification criteria.1, 21 The initial discovery cohort includes individuals from four major cohorts: the EuroEPINOMICS-RES cohort (RES, n = 135); a cohort of individuals who have epileptic encephalopathies and were recruited through a study by the German Research Foundation (DFG; n = 109); a cohort of individuals recruited through the Genomics Research and Innovation Network (GRIN; n = 48); and a cohort of individuals recruited through the Epilepsy Genetics Research Project at the Children’s Hospital of Philadelphia (EGRP; n = 8). The confirmation cohort includes 2,310 epilepsy-affected individuals who underwent diagnostic whole-exome sequencing at Ambry Genetics.
Genetic Analysis
Trio-based whole-exome sequencing (WES) on all probands and parents was performed in a research or diagnostic context with various platforms and enrichment kits. All variants of interest were confirmed by Sanger sequencing. For the RES cohort, WES was performed at the Wellcome Trust Sanger Institute as part of the EuroEPINOMICS-RES project with the Illumina TruSeq DNA Sample Preparation Kit, the Agilent Technologies SureSelect Human All Exon 50Mb Kit, and the Illumina HiSeq2000 according to the manufacturers’ protocols and as previously described.12, 22, 23 For the DFG cohort, WES was performed at the Institute of Clinical Molecular Biology at the University of Kiel and the Cologne Center for Genomics with NimbleGen SeqCap EZ Human Exome Library v2.0, Nextera Rapid Capture Exome, Nextera Rapid Capture Expanded Exome, Agilent SureSelect Human All Exon V5, and Agilent SureSelect Human All Exon 50Mb. For the GRIN cohort, WES was performed at the Broad Institute with the Nextera Rapid Capture Exome kit. For the EGRP cohort, exome sequencing was performed in a diagnostic setting at GeneDx (n = 6) with the SureSelect Human All Exon V4 (50Mb) kit and at the Division of Genomic Diagnostics at the Children’s Hospital of Philadelphia (n = 2) with the SureSelect Clinical Research Exome kits.
For the confirmation cohort, including Individual 3 and Individual 4, trio-based diagnostic whole-exome sequencing with the Integrated DNA Technologies (IDT) xGen Exome Research Panel v1.0 was performed on the proband and unaffected parents at Ambry Genetics. Genomic DNA extraction, exome library preparation, sequencing, bioinformatics, and data analyses were performed as previously described.24, 25 Identified candidate variants were confirmed via Sanger sequencing.
All genetic data on individuals included in the initial discovery cohort were re-analyzed with a standardized pipeline. The raw data was aligned to the HS37d5 human reference genome with the Burrows Wheeler Alignment (v 0.7.12) MEM algorithm. After alignment for each sample, mate tags (MC and MQ) were added to the paired-end lines with Samblaster (v 0.1.20). Base Quality Score Recalibration (BQSR) was performed with GATK tools (v4.0.0.0) before SNP and indel calling was done with HaplotypeCaller with interval lists specific to the exome enrichment kit that was used for each sample. gVCF files were combined with PICARD tools (v2.0.1) for each trio, and then genotyping was performed with the genotypeGVCF tool implemented in GATK. Variant selection and filtration were done with GATK tools before a merged variant file (VCF) was generated with the MergeVcfs functionality of PICARD tools. Annotation of the VCF file was performed by a customized version of ANNOVAR. De novo, homozygous, heterozygous, and rare variants were extracted from the annotated file if they passed the following quality criteria: (1) read depth in the proband and parents was ≥10×; (2) genotype quality in the proband and parents should be ≥20; (3) allele frequencies were <1% in all population databases; (4) the Residual Variation Intolerance Score (RVIS) percentile was <70; and (5) the read ratio was ≥25% of the alternate allele in the proband of the trio.
Phenotypic Similarity Analysis
For the 314 individuals included in the discovery cohort, 3,529 HPO terms were assigned, and there was a median of 9 terms per individual. 2,579 terms had been assigned manually by the clinicians of the EuroEPINOMICS-RES project, and 950 terms were translated from the EuroEPINOMICS phenotype database where categorical and free-text terms were added. For the final analysis, all terms per individual were merged, duplicates removed, and obsolete terms replaced by synonyms in the HPO version that was used for this study (HPO version 1.2; release format-version: 1.2; data-version: releases/2017-12-12; downloaded on 3/10/18). This resulted in a combined set of 11,146 HPO terms, including all initially assigned HPO terms and all unique ancestral terms per individual; the frequency p of each assigned or ancestral HPO term in the cohort of 314 individuals were assessed with these terms (Table S1). This allowed for the computation of the Information Content (IC) for each term, defined as −log2(p).19
Phenotypic Similarity between Two Individuals
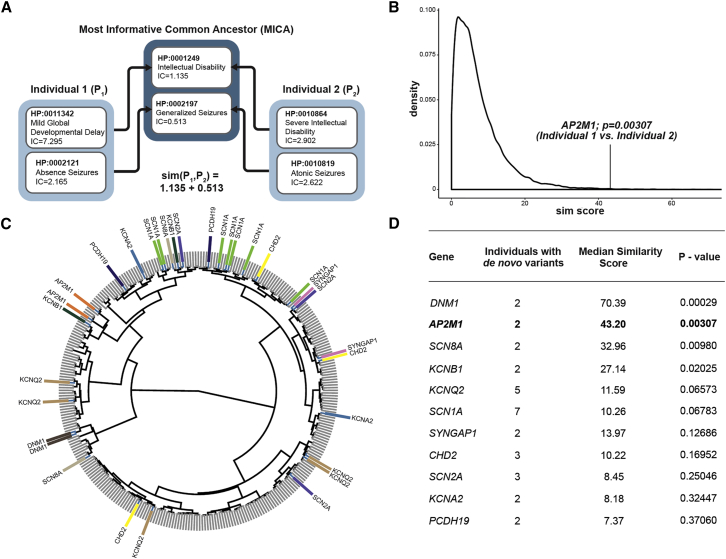
Phenotypic similarity was assessed by summing over the most informative common ancestor for all pairs of HPO terms between two individuals according to Resnik (P. Resnik, 1995, International Joint Conference on Artificial Intelligence, conference). For two individuals (P1 and P2), a matrix s holds all HPO terms in individual P1 (n terms as rows) and all HPO terms in individual P2 (m terms as columns). Each position sij represents a pair of HPO terms; the common parent term with the highest Information Content (IC) is chosen as the most informative common ancestor (MICA) as a score for sij. (Figure 1).
Figure 1.
HPO-Based Analysis Demonstrating Phenotypic Similarity in Individuals with De Novo Variants
(A) An illustration of the similarity score calculation made with the Most Informative Common Ancestor (MICA) approach.
(B) A distribution, using 100,000 permutations, of phenotypic similarities between n = 2 individuals in the cohort of 314 individuals. The vertical line indicates the observed value for two individuals with de novo variants in AP2M1. The observed value of 43.20 is in the top 0.3 percentile of the distribution, translating into an exact p value of 0.00307.
(C) A dendrogram of 314 individuals clustered by phenotypic similarity bya ward.D2 algorithm for the clustering of the similarity matrix. The gene labels refer to individuals with de novo variants in genes with two or more de novo variants in the entire cohort.
(D) Exact p values for observed versus predicted phenotypic similarity for all 11 genes shared by two or more individuals with de novo variants in the cohort of 314 individuals. P values are uncorrected and refer to the distribution of expected similarities for each number of individuals.
Two different methods, referred to as simmax and simav, generate a symmetric score for the similarity for s by summing over either the maximum or the average of all rows and columns with appropriate normalization, respectively.26 For our primary analysis, we used simmax (Equation 1).
(1) The simav method has initially been suggested for semantic similarities in the gene ontology26 and has been applied to Human Phenotype Ontology research by other authors.27
(2) The simmax method is the more conservative method for assessing semantic similarity in our hands.
Observed Versus Expected Phenotypic Similarity Scores per Gene
The expected similarity scores for genes with n de novo variants were assessed by determining with 100,000 permutations the distribution of median similarities of n individuals randomly selected from the overall cohort. Exact p values for the observed similarities for all genes with n de novo variants were determined on the basis of the distribution of similarity scores in 100,000 permutations. The custom computer code that we used to generate results for the phenotypic similarity analysis in this manuscript is publicly available.
Structural Modeling and Normal Mode Analysis
The structures of the AP-2 complex molecule were taken from the Protein Data Bank (PDB-101; accession numbers PDB: 2XA7 and 4UQI for the active and inactive state, respectively).28 Each in silico missense variant was created by the mutagenesis plugin in PyMol Molecular Graphics System Version 1.8 (Schrödinger). Wild-type (WT) and missense structures were analyzed by an ENCoM coarse-grained normal mode analysis method in order to evaluate the effect of variants on the stability of the protein. This method is based on an ENCoM entropic considerations C package29 (available at the ENCoM development website) that was compiled and used on a Ubuntu platform (Canonical Group). The calculation of the entropic difference (ΔG) between the WT AP-2 complex and missense variants was done with MATLAB software (Mathworks). For each variant, ΔG was calculated by subtracting WT from variant entropy. ΔG was normalized to the maximum absolute values, and cluster analysis was performed with MATLAB software.
Clustering of Entropy Changes for APM21 and AP2S1 Variants
To assess whether the AP2M1 p.Arg170Trp variant in the AP-2 complex results in entropy changes comparable with those of other variants known to cause disease in AP-2 subunits, we compared the effect of pathogenic variants in AP2S1 (MIM: 602242) and rare population variants in AP2M1 (Table S2). Pathogenic variants in AP2S1 cause autosomal-dominant hypocalciuric hypercalcemia (MIM: 600740), a rare genetic kidney disease. No other human diseases have been associated with variants in genes encoding the AP-2 subunit so far. ΔG between the WT AP-2 complex and missense variants was assessed with MATLAB software (Mathworks). Each variant was normalized to WT values by calculating a delta between them (ΔG).
Cell Lines and Primary Astrocytes
HeLa and HEK293T cells were obtained from ATCC. Cells were cultured with 4.5g/L glucose (Lonza) in DMEM containing 10% heat-inactivated fetal bovine serum (FBS) (GIBCO) and 1% penicillin/streptomycin (GIBCO) during experimental procedures. Cells were routinely tested for mycoplasma contamination. The conditional AP-2μ knockout (KO) mouse (AP-2lox/lox × inducible CAG-Cre) was described previously.30 Primary mouse astrocytes were prepared from 1- to 3-day-old pups. The cerebral cortices were dissected and the meninges were carefully removed in cold, sterile Hank’s balanced salt solution (HBSS). The tissue was trypsinized for 10 min at 37°C and mechanically dissociated in complete DMEM medium with 10% FBS, 1% penicillin/streptomycin, and 40 U/mL DNase I (Sigma) 10 times through a small-pore fire-polished Pasteur pipette. The cell suspension was pelleted and resuspended in fresh, complete DMEM, filtered through a 100 μm nylon membrane (BD Falcon), and plated into 10 cm2 cell culture dishes. When the cells reached confluence, the astrocytes were trypsinized, plated in poly-L-lysine-coated (Sigma) glass coverslips in 12-well plates at about 100,000 cells/cm2, and cultured for another 7–10 days. To deplete AP-2μ, culture astrocytes from floxed animals expressing a tamoxifen-inducible Cre recombinase were treated with 0.1 μM (Z)-4-hydroxytamoxifen (Sigma) the day after plating. Astrocytes derived from floxed littermates that were Cre negative were used as controls and treated with equal amounts of (Z)-4-hydroxytamoxifen. All animal experiments and procedures have been approved by the Landesamt für Gesundheit und Soziales (LaGeSo) Berlin according to §8.1 German Animal Welfare Act.
Molecular Biology
The AP-2μ p.Arg170Trp variant was first generated by overlap extension PCR with the plasmid AP2u2-mCherry (Addgene plasmid #27672) and the plasmid AP-2μ IRES mRFP in an AAV-HBA-EWB vector. siRNA-resistant AP-2μ WT and p.Arg170Trp variants were generated with the plasmid AP-2μ p.Arg170Trp-mCherry by overlap extension PCR (primers available upon request). The integrity of all cloned constructs was confirmed by DNA sequencing.
siRNA and Plasmid Transfections
HeLa cells were transfected with siRNA via jetPRIME (Polyplus) according to the manufacturer’s protocol. To achieve optimal knockdown efficiency, two rounds of silencing were performed. Thus, cells were consecutively transfected on day 1 and day 3, and the experiment was performed on day 5. For transient overexpression of proteins in knockdown cells, plasmids were transfected on day 3, together with the second round of siRNA, also with jetPRIME. For AP-2μ silencing, the siRNA we used was μ2-adaptin 5′-GUGGAUGCCUUUCGGGUCA-3′. Transfection of MISSION siRNA universal negative control #1 (Sigma) served as control siRNA. Primary astrocytes were transfected with lipofectamin 2000 (Invitrogen) at a 1:2 ratio (DNA:lipofectamin) in Opti-MEM medium (as described in the manufacturer’s instructions), and after 4 h, the medium was replaced with complete DMEM, and the astrocytes were incubated for 48 h at 37°C.
Antibodies
We performed immunoblotting and used alpha-adaptin (AP-2α) (BD transduction 610502, 1:500, mouse), mu-adaptin (AP-2μ) (BD transduction 611351, 1:500, mouse), clathrin heavy chain (clone TD.1, IgG from tissue culture supernatant, 1:500, mouse), Rab5 (BD transduction 610724, 1:250, mouse), GAPDH (Sigma G8795, 1:5000, mouse), and SNAP25 (Synaptic Systems 111011, 1:1000, mouse) as primary antibodies, and we used LI-COR 800CW and 680RD infrared as secondary antibodies. We performed immunoblot development by using a LI-COR Odyssey Fc imager, and we quantified immunoblot bands with Image Studio Lite Version 4.0 software (LI-COR). For immunostaining, we used AP-2α (Homebrew, 1:100, mouse), clathrin heavy chain (clone X22, IgG from tissue culture supernatant, 1:250, mouse), and RFP (MBL PM005, 1:500, rabbit) as primary antibodies and Alexa-568 goat anti-rabbit and Alexa 488 goat anti-mouse (1:500, Invitrogen) as secondary antibodies. We used RFP-Trap_M (Chromotek) beads for the immunoprecipitation of the Cherry-tagged AP-2μ WT and p.Arg170Trp protein variants.
Immunocytochemistry and Confocal Imaging
HeLa cells seeded on coverslips were fixed for 13 min with 4% paraformaldehyde (w/v, PFA) in phosphate-buffered saline solution (PBS) on ice and washed three times with PBS. Cells were permeabilized and blocked in blocking solution (PBS, 10% goat serum and 0.3% Triton X-100) for 30 min and then incubated with primary antibodies (diluted in blocking solution) for 1 h. After three washes with PBS, secondary antibodies diluted in blocking solution were incubated for 1 h, then washed three times in PBS. Coverslips were mounted in Immu-Mount (Thermo Fisher Scientific) with 1.5 mg/mL DAPI (Sigma) and visualized with a Zeiss laser scanning confocal microscope LSM780. Co-localization experiments were analyzed with ImageJ software. Data are presented as mean values ± standard error of the mean (SEM) from 5 independent experiments (N). Statistical testing was performed with a paired t test.
Cell Lysates, Co-immunoprecipitation, and Membrane Fractionation Experiments
HEK293T cells were transfected with calcium phosphate to express mCherry or the mCherry-tagged versions of AP-2μ WT or p.Arg170Trp variant. Cells were washed three times in ice-cold PBS and harvested in lysis buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 2 mM MgCl2, 1 mM PMSF, 0.1% protease inhibitor cocktail, and 1% Triton X-100). Lysates were incubated on a rotating wheel at 4°C for 30 min, then centrifuged at 17,000 g for 10 min at 4°C. Protein levels were quantified with the BCA Kit (Pierce, Thermo Fisher Scientific), and equally concentrated lysates were boiled for 5 min in Laemmli sample buffer. Between 15 and 40 μg of protein was loaded onto a 10% acrylamide gel for SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and analyzed via immunoblot with LI-COR 800CW and 680RD infrared secondary antibodies. For co-immunoprecipitation experiments, cells were harvested 48 h post-transfection in lysis buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 2 mM MgCl2, 1 mM PMSF, 0.1% protease inhibitor cocktail, and 1% Triton X-100) and incubated on ice for 30 min, then centrifuged at 17,000 g for 10 min at 4°C. Proteins in supernatants were quantified with the BCA Kit (Pierce, Thermo Fisher Scientific), and 1 mg of protein was mixed with RFP-Trap beads for 1 h at 4°C on a rotating wheel. Beads were pelleted and washed four times in lysis buffer, and bound protein was eluted in 40 μl of Laemmli sample buffer. Eluates were loaded onto a 10% acrylamide gel for SDS-PAGE and then immunoblotting. In the case of membrane fractionation studies, cells at 48 h post-transfection were harvested in homogenization buffer (20 mM HEPES [pH 7.4], 10 mM KCl, 2 mM MgCl2, 1 mM PMSF, and 0.1% protease inhibitor cocktail) and homogenized with a 1 mL syringe through a 25 G needle 10–20 times. Total cell lysate was collected after centrifugation at 720 g for 5 min at 4°C. To obtain the cytosolic fraction, total cell lysate was centrifuged at 100,000 g for 1 h at 4°C, the supernatant was collected, and the protein concentration and volume were determined. The membrane pellet was washed twice in homogenization buffer and resuspended in lysis buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 2 mM MgCl2, 1 mM PMSF, 0.1% protease inhibitor cocktail, and 1% Triton X-100) by pipetting and passed through a 25 G needle in half of the volume corresponding to the cytosolic fraction with lysis buffer (20 mM HEPES [pH 7.4], 100 mM KCl, 2 mM MgCl2, 1 mM PMSF, 0.1% protease inhibitor cocktail, and 1% Triton X-100). Protein detection in both cytosolic and membrane fractions was determined with the BCA Kit (Pierce, Thermo Fisher Scientific). Equal amounts of membrane and cytosolic fractions were loaded onto a 10% acrylamide gel for SDS-PAGE and then immunoblotting. Protein levels of AP-2α, AP-2μ, and CHC in the total cell lysates were normalized to Rab5, and protein levels in the membrane fraction were normalized to SNAP25. Data are presented as mean values ± SEM from 3–8 independent experiments (N). Statistical testing was performed with a one-sample t test.
Transferrin Uptake
For HeLa cells, AP-2μ-depleted cells were rescued by the reintroduction of siRNA-resistant AP-2μ WT and p.Arg170Trp variants, both fused to mCherry. For AP-2μ KO astrocytes, the re-expression of AP-2μ WT or p.Arg170Trp variants was performed with a construct containing RFP inserted after an internal ribosomal entry site (IRES). Transferrin endocytosis assays were essentially done as described earlier.31, 32 In brief, HeLa cells seeded on coverslips were serum-starved for 1 h and treated with 25 μg ml−1 Tf-Alexa647 (Life Technologies) for 10 min at 37°C. Primary astrocytes were seeded on coverslips, serum-starved overnight, and treated with 50 μg ml−1 Tf-Alexa647 (Life Technologies) for 5 min at 37°C. Cells were washed twice with ice-cold PBS and acid washed at pH 5.3 (0.1 M Na-acetate, 0.2 M NaCl) for 1 min on ice. The coverslips were washed twice with ice-cold PBS and fixed with 4% PFA for 30 min at room temperature. Cells were washed three times with PBS, then subjected to immunocytochemistry staining as described in Immunocytochemistry. Transferrin uptake was analyzed with a Zeiss laser-scanning confocal microscope LSM780 and quantified with ImageJ software. At least ten images were taken per condition. Cells positive for mCherry (HeLa cells) or RFP (primary astrocytes) were selected manually, and regions of interest (ROIs) were drawn around them. After background subtraction (by employing the rolling circle method with a radius of 50 pixels), the integrated density for AF647 was measured. Data are presented as mean values ± SEM. N = the number of independent experiments. n = the total number of analyzed cells.
Results
HPO-Based Similarity Analysis Demonstrates Significant Phenotypic Overlap in Individuals with the Recurrent AP2M1 c.508C>T (p.Arg170Trp) De Novo Variant
We identified 11 genes with de novo variants in two or more individuals in our cohort of 314 individuals with DEEs (Figure 1). To assess whether de novo variants in the identified 11 genes are associated with a gene-phenotype relationship, we performed a semantic similarity analysis of phenotypic features across all 314 individuals, using quantitative phenotypic similarity based on the Information Content of HPO terms (Figure 1). We calculated the median phenotypic similarity among all individuals with de novo variants in the same gene and compared the observed value per gene to the expected phenotypic similarity scores for groups of the same size, computed by permutation analysis (Figure S1). On the basis of this distribution, we determined whether individuals with de novo variants in the same gene had a higher phenotypic similarity than expected by chance (Figure 1). AP2M1 was the only gene in this group not previously associated with DEEs and was identified in two individuals with an identical c.508C>T (p.Arg170Trp) de novo variant (GRCh37 chr3: g.183898715C>T). Both individuals with the identical AP2M1 c.508C>T (p.Arg170Trp) de novo variant had a significant phenotypic similarity (p = 0.003), suggesting a significant gene-phenotype relationship and a narrow phenotypic spectrum. As a next step, we queried a diagnostic cohort of 2,310 epilepsy-affected individuals who underwent diagnostic whole-exome sequencing. We identified two additional individuals with the same AP2M1 de novo variant, resulting in a total of four individuals with the recurrent AP2M1 GenBank: NM_004068.3 c.508C>T (p.Arg170Trp) de novo variant.
Detailed Phenotypic Review of Individuals with the Recurrent AP2M1 c.508C>T (p.Arg170Trp) De Novo Variant Suggests a Recognizable Phenotype
We manually reviewed the phenotypes of the four individuals with the recurrent AP2M1 GenBank: NM_004068.3 c.508C>T (p.Arg170Trp) de novo variant in detail. All affected individuals presented with global developmental delay apparent in the first six months of life and had seizure onset between 21 months and 4 years. Two out of four individuals had received a diagnosis of autism spectrum disorder, and all affected individuals were female. Three out of four individuals had atonic seizures and generalized epileptiform discharges on EEG (Table 1, see Supplemental Note). Accordingly, AP2M1-related epilepsies share a recognizable phenotype reminiscent of epilepsy with myoclonic-atonic seizures, demonstrating that the phenotypic similarity assessed by HPO for the first two individuals (Individuals 1 and 2) was recapitulated in the second two individuals (Individuals 3 and 4), and together the key clinical features conform to a reasonably recognizable electroclinical syndrome. Within our first cohort, 64/314 individuals had a diagnosis of epilepsy with myoclonic-atonic seizures, also referred to as myoclonic-astatic epilepsy (MAE) or Doose syndrome, suggesting that up to 3% of individuals with MAE may have de novo variants in AP2M1 (point estimate 3.125%; 95% CI 2.3–14.0). Of note, both individuals with AP2M1 de novo variants in the first cohort had been independently phenotyped at two different centers prior to sequencing, excluding the possibility that assigned HPO terms were influenced by knowledge of the underlying genetic etiology or phenotyping bias within a single contributing center.
Table 1.
Phenotypic Details of Individuals with the Recurrent AP2M1 c.508C > T (p.Arg170Trp) Variant
|
Proband ID |
||||
|---|---|---|---|---|
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | |
| Age and sex | 7 years, f | 15 years, f | 4 years, f | 8 years, f |
| Development | globally delayed | globally delayed | globally delayed | globally delayed |
| Age at seizure onset | 1 year, 9 months | 1 year, 3 months | 3 years | 4 years |
| Seizure types | atypical absence; myoclonic atonic; absence with eyelid myoclonia |
atonic; atypical absence; absence with eyelid myoclonia |
atonic; bilateral tonic-clonic |
focal impaired-awareness |
| Seizure outcome | drug-responsive | drug-resistant | drug-resistant | partially responsive |
| Intellectual disability (severity) | moderate | moderate | severe | severe |
| Autism spectrum disorder | absent | absent (aggressive and self-harming behaviors) | present | present |
| Ataxia | truncal and gait ataxia | absent | gait ataxia only | truncal and gait ataxia |
| Other exam findings | hypotonia | hypotonia | hypotonia; chorea and myoclonus; a prominent maxilla and a thin upper lip |
hypotonia; tremor; long, thin hands and feet |
| MRI findings | parieto-occipital white matter abnormalities | normal | normal | normal |
| EEG findings | generalized polyspike-wave discharges | 3–4 Hz generalized spike-wave discharges | generalized spike-wave discharges | multifocal epileptiform activity |
Abbreviations are as follows: MRI = magnetic resonance imaging and EEG = electroencephalogram.
The AP2M1 c.508C>T (p.Arg170Trp) Variant Affects the Thermodynamic Entropy of the AP-2 Complex
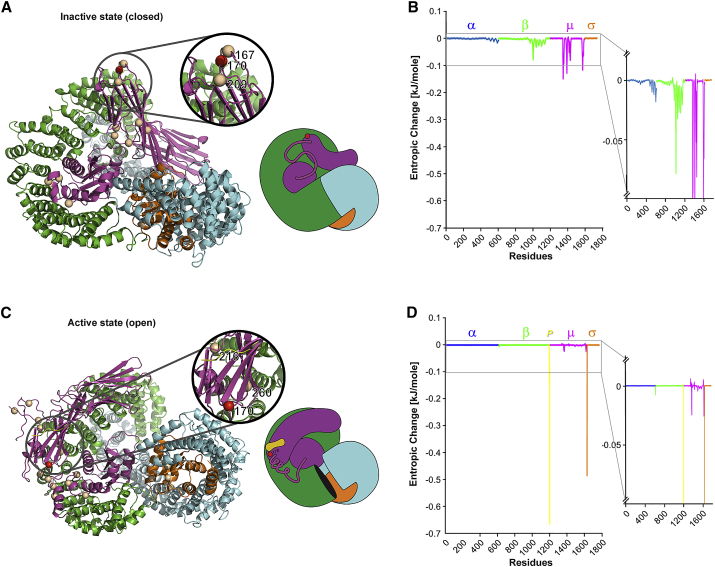
Given the important roles of AP-2 in the brain, we hypothesized that impaired AP-2 function in the presence of the p.Arg170Trp variant may underlie DEEs in humans. Arg170 is part of a basic phospholipid-binding patch within AP-2μ that, on the basis of on structural data, is postulated to stabilize the active open conformation of AP-2 and its association with cargo membrane proteins.33 To test whether the p.Arg170Trp variant may affect AP-2 activation, we assessed protein thermodynamics in the inactive (closed) and active (open) conformation of AP-2 WT or the AP-2 p.Arg170Trp variant by molecular modeling (Figure 2). We found that the p.Arg170Trp variant showed a significant increase of entropy compared with the WT. In the inactive (closed) state of the protein, the p.Arg170Trp variant caused increased entropy, mainly located in the α, β, and μ subunits. In the active (open) state of the protein, the p.Arg170Trp variant caused a significant increase in entropy compared to WT in the μ and σ subunits, as well as at the cargo protein.
Figure 2.
Effect of the AP2M1 p.Arg170Trp Variant on the Thermodynamic Entropy of the AP-2 Complex
(A) Both the entire structure and a simplified cartoon model of the AP-2 complex are shown in the inactive closed state. The color-coding is a follows: AP-2α (blue), AP-2β (green), AP-2μ (magenta), AP-2 σ (orange), and the AP-2-bound cargo peptide P (yellow). The p.Arg170Trp variant is depicted as a red sphere. Further pathogenic variants in AP2S1 (AP-2σ) and rare population variants in AP2M1 (AP-2μ) variants are shown as golden spheres.
(B) Differences in entropy (entropic change; ΔG) between the wild-type (WT) AP-2 complex and the AP-2 complex containing the AP2M1 p.Arg170Trp variant are graphically depicted as ΔG for each residue across the entire AP-2 complex for the inactive closed state. The inlay shows an enlarged y axis to emphasize the differences in ΔG for each subunit.
(C) The structure of the AP-2 complex and a simplified cartoon model including the bound cargo peptide P (yellow) are shown in the active state, and the AP2S1 and AP2M1 variants are labelled.
(D) The difference in entropy between the WT AP-2 complex and the AP-2 complex containing the AP2M1 p.Arg170Trp variant for the active open state.
In order to estimate the functional effect of the entropy changes described above, we compared the entropy changes of the AP2M1 p.Arg170Trp variant with rare population variants in AP2M1 and pathogenic variants in AP2S1, the causative genetic etiology for autosomal-dominant hypocalciuric hypercalcemia. Regarding the inactive (closed) state of the protein, cluster analysis of entropic changes indicated a clear separation between all tested rare population variants and the pathogenic variants in AP2S1. The cluster analysis showed that entropy change regarding the p.Arg170Trp variant was separated significantly from both (Figure S2). In the active (open) state of the protein, the entropy change due to the p.Arg170Trp variant clustered with the pathogenic variants in AP2S1 and was separated from the rare population variants (Figure S2). Given that only active AP-2 is capable of associating with cargo membrane proteins, these results suggest that the AP2M1 p.Arg170Trp variant might functionally impair AP-2-mediated cargo recognition and, thereby, CME.
Clathrin-Mediated Endocytosis Is Reduced in Human HeLa Cells and AP-2μ KO Mouse Astrocytes Expressing the Pathogenic AP2M1 c.508C>T (p.Arg170Trp) Variant
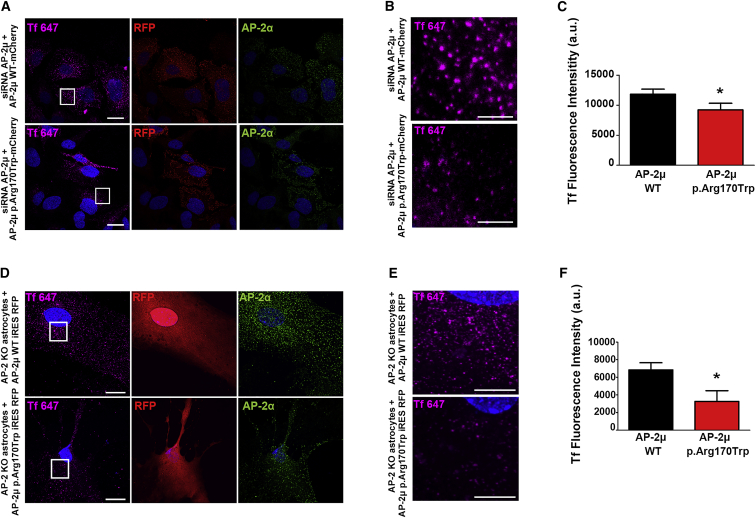
On the basis of the molecular modeling data, we hypothesized that the recurrent AP2M1 c.508C>T (p.Arg170Trp) variant may affect CME. We probed this prediction experimentally by quantitatively determining the efficacy of CME in AP-2μ-depleted human HeLa cells rescued by plasmid-based re-expression of siRNA-resistant mCherry-labeled WT or p.Arg170Trp variant AP-2μ. HeLa cells expressing the pathogenic p.Arg170Trp AP-2μ variant showed reduced levels of internalized AlexaFluor647-labeled transferrin after 10 min compared with cells expressing the WT AP-2 complex (Figures 3 and S4). A similar endocytic defect was observed in primary astrocytes derived from conditional AP-2μ knockout (KO) mice upon the re-expression of the p.Arg170Trp variant of AP-2μ (Figures 3 and S5). These results suggest that the pathogenic AP2M1 c.508C>T (p.Arg170Trp) variant is affecting AP-2-mediated cargo recognition and, thereby, CME.
Figure 3.
Defective Clathrin-Mediated Endocytosis in Cells Expressing the AP2M1 p.Arg170Trp Variant
(A) Representative images of HeLa cells depleted of endogenous AP-2μ; the cells were rescued by the re-expression of siRNA-resistant mCherry-AP-2μ wild-type (WT) or p.Arg170Trp mutant and allowed to internalize AlexaFluor647-labeled transferrin (Tf) for 10 min at 37°C. Cells were fixed and immunostained for endogenous AP-2α and RFP. RFP was labeled to amplify the signal for mCherry and to identify transfected cells. The scale bars represent 20 μm.
(B) A zoom of the marked area in (A) illustrates reduced Tf endocytosis in cells expressing the p.Arg170Trp variant. The scale bars represent 5 μm. Note that the punctate distribution of WT or Arg170Trp mutant mCherry-AP-2μ is consistent with its proper targeting to endocytic pits (see also Figure S4).
(C) A quantification of data shown in (A). The data represent mean ± SEM, N = 3 independent experiments (wherein n = 198 for AP-2μ WT and n = 172 for AP-2μ p.Arg170Trp total cells analyzed). ∗p < 0.05 from a paired two-tailed t test.
(D) Representative images of primary astrocytes from WT or AP-2μ knockout (KO) mice; the cells were rescued by re-expression of untagged AP-2μ WT or p.Arg170Trp together with soluble RFP and allowed to internalize AlexaFluor647-labeled transferrin (Tf) for 5 min at 37°C. Cells were fixed and immunostained for endogenous AP-2α and cytoplasmic RFP. RFP expressed from the same construct after an internal ribosomal entry site (IRES) was labeled to identify transfected cells. The scale bars represent 20 μm.
(E) A zoom of the marked area in (D) shows less Tf in astrocytes expressing the mutant variant of AP-2μ. The scale bars represent 10 μm.
(F) A quantification of data shown in (D). The data represent mean ± SEM, N = 5 independent experiments (wherein n = 74 for AP-2μ WT and n = 72 for AP-2μ p.Arg170Trp total cells analyzed). ∗p < 0.05 from an unpaired t test.
The AP2M1 c.508C>T (p.Arg170Trp) Variant Does Not Affect AP-2 Complex Stability, Expression, Membrane Recruitment, or Localization
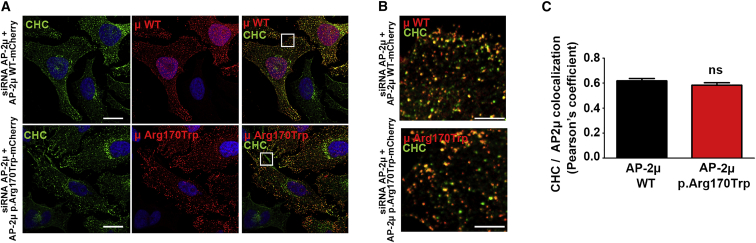
Given the functional alterations in CME mediated by the p.Arg170Trp variant, we next assessed whether the impaired CME may be a result of stability, expression levels, membrane recruitment, or localization of the p.Arg170Trp-variant-containing AP-2 complex (Figures 4 and S3). However, we found that AP-2μ WT and p.Arg170Trp variants colocalize with clathrin equally well in HeLa cells, suggesting that localization of the pathogenic AP-2μ variant in clathrin-coated pits is not altered. Likewise, the p.Arg170Trp variant of AP-2μ does not affect the expression levels, the stability, or the membrane recruitment of AP-2 complexes (Figure S3). Taken together, these results suggest that the p.Arg170Trp variant impairs AP-2 function in CME in human cells and primary brain astrocytes at an early step, probably by affecting the recognition of cargo membrane proteins (Figure S6).
Figure 4.
Intact Localization of an AP-2 Complex Carrying the p.Arg170Trp Variant
(A) Representative confocal images (maximum intensity projections) of HeLa cells depleted of endogenous AP-2μ; the cells were rescued by re-expression of siRNA-resistant mCherry-AP-2μ WT or p.Arg170Trp mutant and immunostained with clathrin heavy chain (CHC) and RFP antibodies. RFP was labeled to amplify the signal for mCherry-tagged variants and to identify transfected cells. The scale bars represent 20 μm.
(B) Merged magnified views of the boxed area in (A). The scale bars represent 5 μm.
(C) Pearson’s correlation coefficient for the co-localization of AP-2μ WT or p.Arg170Trp with clathrin heavy chain (CHC). The data represent mean ± SEM and N = 5 independent experiments. Statistical analysis was done with a paired t test.
Discussion
In our study, we identified four individuals with a homogeneous phenotype of a developmental and epileptic encephalopathy due to a recurrent AP2M1 de novo c.508C>T (p.Arg170Trp) variant. AP2M1 is highly expressed in the central nervous system and has been previously studied extensively in a functional context.30, 34 AP2M1 encodes the essential μ-subunit of the endocytic clathrin adaptor complex AP-2, which is involved in clathrin-mediated endocytosis (CME) at the plasma membrane in neurons and non-neuronal cells. CME is a major mechanism for the recycling of synaptic vesicle (SV) components at mammalian central synapses.35, 36 AP-2 plays a dual role in this process by integrating the sorting of SV protein cargo with the reformation of release-ready SVs.30 AP-2 also regulates the neuronal surface levels of GABA and glutamate receptors and thereby contributes to long-term plastic changes in neurotransmission and to excitatory/inhibitory balance.37, 38 Heterozygous mutant mice with a targeted disruption of the AP2M1 gene do not have an apparent phenotype. Homozygous mutant mice, however, die before day 3.5 postcoitus, indicating that the μ2 subunit of the AP-2 complex is critical for early embryonic development.39
AP2M1 is highly intolerant to variation in the general population and has a probability of intolerance to loss of function (pLI) score of 0.99 and a missense Z score of 5.82 in the ExAC database,40 as well as an RVIS ExAC score of −0.31 (30th percentile).41 Only a single loss-of-function variant in AP2M1 is observed in the ExAC database, compared to more than 19 expected loss-of-function variants. The AP2M1 c.508C>T (p.Arg170Trp) variant is absent from all population databases, including ExAC and gnomAD.40 Pathogenic variants in several other genes involved in neurotransmission are known to cause epilepsy and neurodevelopmental disorders, and these genes include STXBP1 (MIM: 602926), SNAP25 (MIM: 600322), STX1B (MIM: 601485), CLTC (MIM: 118955), DNM1 (MIM: 602377), and PPP3CA (MIM: 114105).5, 10, 42, 43, 44, 45 Within this group of genetic etiologies, AP2M1 stands out because the AP-2 complex mediates endocytic sorting of both presynaptic vesicle proteins and postsynaptic ion channels. AP2M1-related dysfunction may therefore provide a link between disorders of synaptic function and ion channelopathies, the two major groups of genetic etiologies identified in human epilepsy.
We present several lines of experimental evidence that support the view that the encoded p.Arg170Trp variant in AP2M1 is causal, due to impaired AP-2 complex function, for the observed epilepsy phenotype in the individuals in our study. First, by using molecular modeling in silico, we found a significant impact of the p.Arg170Trp AP2M1 variant not only on the μ-subunit but also on other subunits of the AP-2 complex, suggesting that this variant affects overall AP-2 complex function both in the inactive (closed) and active (open) state. Pathogenic variants in AP2S1 cause autosomal-dominant hypocalciuric hypercalcemia, a rare genetic kidney disease. No other human disease has been associated with disease-causing variants in genes encoding AP-2 subunits so far. In the active (open) state, the type of thermodynamic changes predicted from the p.Arg170Trp AP2M1 variant are more closely related to known pathogenic variants in AP2S1 than rare population variants in AP2M1. Although the modeling data can only provide indirect predictions about the overall functional alterations of the AP-2 complex, our normal mode analysis indicates a pronounced instability of the bound cargo protein. This instability most likely relates to the fact that Arg170 is part of the N-terminal phosphatidylinositol (4,5)-bisphosphate (PIP2)-binding site of the μ2 subunit formed by residues Lys167, Arg169, Arg170, and Lys421. It was previously reported that replacing three positively charged residues (Lys167, Arg169, and Arg170) with Glu (KRR > E) had little effect on AP-2 binding to PIP2-containing membranes in vitro, whereas additional mutations in the second PIP2-binding site of the μ2 subunit resulted in a 4-fold reduction in binding to PIP2.33 The change from the charged Arg170 to a large hydrophobic side chain (Trp; of p.Arg170Trp) may therefore conceivably lead to more unstable association of AP-2 with cargo membrane proteins at PIP2-containing membranes.33, 46 In summary, our in silico modeling that used normal mode analysis suggests that the p.Arg170Trp AP2M1 variant not only affects the function of the μ-subunit, but also globally interferes with the thermodynamic stability of the AP-2 complex and, thereby, with the recognition of cargo membrane proteins.
Second, we explored the impact of the p.Arg170Trp variant both in AP-2μ-depleted human HeLa cells and primary astrocytes derived from conditional AP-2μ KO mice. Analysis in both model systems suggested that the AP-2 μ-subunit with the p.Arg170Trp variant is present at levels comparable to WT and is integrated into functional AP-2 complexes. In contrast, the uptake of the CME cargo transferrin was significantly reduced in p.Arg170Trp AP2M1-expressing HeLa cells and astrocytes. These results indicate that the effect of the p.Arg170Trp variant is due to an alteration of AP-2 complex function rather than haploinsufficiency. Based on these data, we therefore postulate that defective endocytic sorting of either one or several AP-2-dependent cargo membrane proteins in neuronal cells underlies developmental and epileptic encephalopathies, possibly due to imbalance between excitatory and inhibitory neurotransmission. It is well established that AP-2 critically regulates both excitatory and inhibitory neurotransmission by controlling the surface number of postsynaptic glutamate and GABAA receptors, as well as the endocytic sorting of the presynaptic vesicular glutamate (vGLUT) and GABA transporters (vGAT), to reform synaptic vesicles during activity-dependent neurotransmission.37 Given the various known links between the GABA-ergic system and hyperexcitability, small changes in the efficacy of endocytic sorting of presynaptic vGAT may result in reduced GABA content of inhibitory synaptic vesicles and thereby cause excitatory/inhibitory imbalance and epilepsy. Alternatively, it is possible that excitatory/inhibitory imbalance and epilepsy result from impaired clathrin-mediated or AP-2-mediated endocytosis of postsynaptic glutamate receptors or associated factors at excitatory synapses, and thereby, elevated excitatory transmission, in addition to other possibilities. Future studies, for example in p.Arg170Trp AP2M1 knock-in mice, will be needed to address these hypothetical scenarios in detail.
Within our study, statistical evidence for an involvement of AP2M1 in genetic epilepsies was generated by a phenotypic similarity analysis based on Human Phenotype Ontology. Because moderately sized cohorts are underpowered to provide statistically significant evidence for uncharacterized gene-disease relationships on a genomic level, we leveraged the existing rich phenotypic information in this cohort to evaluate the role of de novo variants in human epilepsy. We compared predicted versus observed phenotypic similarity in groups of individuals with de novo variants in shared genes. Four out of the 11 identified genes, including genes, such as DNM1, known to be associated with neurodevelopmental disorders with a clinically distinct phenotype, had an uncorrected p value of < 0.05 (Figure 1). In contrast, genes, such as SCN2A (MIM: 182390), known to be associated with a broader neurodevelopmental phenotype were not significant in this analysis. Results for SCN1A, the gene linked to Dravet syndrome, were borderline significant. Within the list of 11 genes with de novo variants in two or more trios, AP2M1 was the only previously undescribed etiology for genetic epilepsies, and the phenotypic similarity between both individuals was significant (Table 1). Given that HPO terms were assigned manually in our study, this method is prone to bias, possibly resulting both in false positive and false negative findings. For example, both individuals with pathogenic de novo variants in DNM1 were phenotyped by clinicians from the same center, a fact that makes it impossible to distinguish a phenotyping bias from a true disease association. Although the DNM1 phenotype is known to be relatively homogeneous, we cannot exclude the possibility that at least some of the observed similarity in both individuals was due to a phenotyping bias within a single contributing group. However, both individuals with AP2M1 de novo variants were independently phenotyped by clinicians at two different centers prior to sequencing. This makes it unlikely that assigned HPO terms were influenced by knowledge of the underlying genetic etiology or phenotyping bias within a single contributing center. When comparing semantic phenotypic similarity to the probability of de novo variants in each of the genes in 314 individuals, we find that the statistical evidence based on phenotypes does not correlate with the statistical evidence from genotypes (Figure S7). This suggests that phenotypic similarity, independent of the probability of observed de novo variants, may provide statistical support for the involvement of a gene in disease. Taken together, we demonstrate that semantic phenotypic similarity can be used to provide statistical support for genetic etiologies in human epilepsy and identify recognizable disease entities. Prior studies have applied related strategies in large datasets of syndromic diseases or hospital-wide cohorts.17, 18 In addition to the success of studies in heterogeneous phenotypes, we show that HPO-based approaches can also be used in cohorts with relatively homogeneous phenotypes, such as the non-lesional pediatric epilepsies, provided that phenotypic information is sufficiently deep for underlying gene-disease relationships to be identified.
In summary, we identify a recurrent pathogenic variant in AP2M1 as a cause of genetic epilepsies resulting in a recognizable electroclinical phenotype with features of epilepsy with myoclonic-atonic seizures or Doose syndrome. By demonstrating impairment of CME despite intact integration into functional AP-2 complexes, we establish dysfunction of the early step of CME as a disease mechanism in neurodevelopmental disorders and epilepsy.
Consortia
EuroEPINOMICS-RES Consortium Members include: Rudi Balling, Nina Barisic, Stéphanie Baulac, Hande Caglayan, Dana Craiu, Peter De Jonghe, Christel Depienne, Renzo Guerrini, Helle Hjalgrim, Dorota Hoffman-Zacharska, Johanna Jähn, Karl Martin Klein, Bobby P.C. Koeleman, Vladimir Komarek, Eric Leguern, Anna-Elina Lehesjoki, Johannes R. Lemke, Holger Lerche, Tarja Linnankivi, Carla Marini, Patrick May, Hiltrud Muhle, Deb K. Pal, Aarno Palotie, Felix Rosenow, Susanne Schubert-Bast, Kaja Selmer, Jose M. Serratosa, Sanjay Sisodiya, Ulrich Stephani, Pasquale Striano, Arvid Suls, Tiina Talvik, Sarah von Spiczak, Sarah Weckhuysen, and Federico Zara.
GRIN Consortium Members include: Paul Avillach, Anna Bartels, Sawona Biswas, Florence Bourgeois, Batsal Devkota, Tracy Glauser, Barbara Hallinan, Allison Heath, Joel Hirschhorn, Judson Kilbourn, Sek Won Kong, Ian Krantz, In-Hee Lee, Kenneth D. Mandl, Eric Marsh, Kristen Sund, Deanne Taylor, and Peter White.
Declaration of Interests
D.N.S. and S.T. are full-time employees of Ambry Genetics. Exome sequencing is one of Ambry’s commercially available diagnostic tests.
Acknowledgements
We thank the participants and their family members for taking part in the study. We would like to thank Mahgenn Cosico and Margaret O’Brien for their support in enrolling research participants and for administrative assistance. The current study was supported by the EuroEPINOMICS-Rare Epilepsy Syndrome (RES) Consortium, which provided the capacity for exome sequencing. This work was supported by intramural funds of the Children's Hospital of Philadelphia; by the German Research Foundation (HE5415/3-1 to I.H.) within the EuroEPINOMICS framework of the European Science Foundation; by the German Research Foundation (DFG; HE5415/5-1, HE5415/6-1to I.H., HA2686/13-1 [Koselleck Program] to V.H., and WE4896/3-1 to Y.G.W.); by DFG Research Unit FOR2715 (Le1030/16-1, We4896/4-1, and He5415/7-1 to I.H. and Y.G.W.); by the Luxembourg National Research Fund (INTER/DFG/17/11583046 to R.K.); by the German Society for Epileptology (to Y.G.W.); and by the Genomics Research and Innovation Network (GRIN, grinnetwork.org), a collaboration among the Children’s Hospital of Philadelphia, Boston Children’s Hospital, and the Cincinnati Children’s Hospital Medical Center. I.H. also received support through the International League Against Epilepsy. This work was also partially supported by National Institutes of Health (NIH) grant NS108874. O.S. and F.B. were supported by the Israel Science Foundation (grant No. 1010/16). C.A.E. was supported in part by a Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant, T32 NS091008-01. N.S. was in part supported from the intramural research funding program of the Faculty of Medicine Tübingen (Fortüne 2381-0-0). K.S. was supported by the Ministry of Health of the Czech Republic (AZV15–33041 and DRO 00064203). A.P. is supported by the Boston Children’s Hospital Translational Research Program.
Published: May 16, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.001.
Contributor Information
Ingo Helbig, Email: helbigi@email.chop.edu.
the EuroEPINOMICS-RES Consortium:
Rudi Balling, Nina Barisic, Stéphanie Baulac, Hande Caglayan, Dana Craiu, Peter De Jonghe, Christel Depienne, Renzo Guerrini, Helle Hjalgrim, Dorota Hoffman-Zacharska, Johanna Jähn, Karl Martin Klein, Bobby P.C. Koeleman, Vladimir Komarek, Eric Leguern, Anna-Elina Lehesjoki, Johannes R. Lemke, Holger Lerche, Tarja Linnankivi, Carla Marini, Patrick May, Hiltrud Muhle, Deb K. Pal, Aarno Palotie, Felix Rosenow, Susanne Schubert-Bast, Kaja Selmer, Jose M. Serratosa, Sanjay Sisodiya, Ulrich Stephani, Pasquale Striano, Arvid Suls, Tiina Talvik, Sarah von Spiczak, Sarah Weckhuysen, and Federico Zara
the GRIN Consortium:
Paul Avillach, Anna Bartels, Sawona Biswas, Florence Bourgeois, Batsal Devkota, Tracy Glauser, Barbara Hallinan, Allison Heath, Joel Hirschhorn, Judson Kilbourn, Sek Won Kong, Ian Krantz, In-Hee Lee, Kenneth D. Mandl, Eric Marsh, Kristen Sund, Deanne Taylor, and Peter White
Accession Numbers
The EuroEPINOMICS-RES exome-sequencing data are deposited in the European Genome-Phenome Archive, accession numbers EGAS00001000190, EGAS00001000386, and EGAS00001000048.
Web Resources
Computer Code Used for Phenotypic Similarity Analysis, https://github.com/galerp/helbig_lab
ENCoM Development, https://github.com/NRGlab/ENCoM
ExAC Database, http://exac.broadinstitute.org
gnomAD Browser, http://gnomad.broadinstitute.org/
Human Phenotype Ontology, https://hpo.jax.org/app/
Online Mendelian Inheritance in Man, http://www.omim.org
Residual Variation Intolerance Score (RVIS), http://genic-intolerance.org
Supplemental Data
References
- 1.Scheffer I.E., Berkovic S., Capovilla G., Connolly M.B., French J., Guilhoto L., Hirsch E., Jain S., Mathern G.W., Moshé S.L. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scheffer, I.E., Berkovic, S., Capovilla, G., Connolly, M.B., French, J., Guilhoto, L., Hirsch, E., Jain, S., Mathern, G.W., Moshe, S.L., et al. (2017). ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 58, 512-521. [DOI] [PMC free article] [PubMed]
- 2.McTague A., Howell K.B., Cross J.H., Kurian M.A., Scheffer I.E. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15:304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]; McTague, A., Howell, K.B., Cross, J.H., Kurian, M.A., and Scheffer, I.E. (2016). The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 15, 304-316. [DOI] [PubMed]
- 3.Helbig I., Tayoun A.A. Understanding genotypes and phenotypes in epileptic encephalopathies. Mol. Syndromol. 2016;7:172–181. doi: 10.1159/000448530. [DOI] [PMC free article] [PubMed] [Google Scholar]; Helbig, I., and Tayoun, A.A. (2016). Understanding genotypes and phenotypes in epileptic encephalopathies. Mol. Syndromol. 7, 172-181. [DOI] [PMC free article] [PubMed]
- 4.Helbig I., Heinzen E.L., Mefford H.C., International League Against Epilepsy Genetics Commission Genetic literacy series: Primer part 2-Paradigm shifts in epilepsy genetics. Epilepsia. 2018;59:1138–1147. doi: 10.1111/epi.14193. [DOI] [PubMed] [Google Scholar]; Helbig, I., Heinzen, E.L., and Mefford, H.C.; International League Against Epilepsy Genetics Commission (2018). Genetic literacy series: Primer part 2-Paradigm shifts in epilepsy genetics. Epilepsia 59, 1138-1147. [DOI] [PubMed]
- 5.Heyne H.O., Singh T., Stamberger H., Abou Jamra R., Caglayan H., Craiu D., De Jonghe P., Guerrini R., Helbig K.L., Koeleman B.P.C., EuroEPINOMICS RES Consortium De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018;50:1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]; Heyne, H.O., Singh, T., Stamberger, H., Abou Jamra, R., Caglayan, H., Craiu, D., De Jonghe, P., Guerrini, R., Helbig, K.L., Koeleman, B.P.C., et al.; EuroEPINOMICS RES Consortium (2018). De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 50, 1048-1053. [DOI] [PubMed]
- 6.Helbig K.L., Farwell Hagman K.D., Shinde D.N., Mroske C., Powis Z., Li S., Tang S., Helbig I. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet. Med. 2016;18:898–905. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]; Helbig, K.L., Farwell Hagman, K.D., Shinde, D.N., Mroske, C., Powis, Z., Li, S., Tang, S., and Helbig, I. (2016). Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet. Med. 18, 898-905. [DOI] [PubMed]
- 7.Lindy A.S., Stosser M.B., Butler E., Downtain-Pickersgill C., Shanmugham A., Retterer K., Brandt T., Richard G., McKnight D.A. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia. 2018;59:1062–1071. doi: 10.1111/epi.14074. [DOI] [PubMed] [Google Scholar]; Lindy, A.S., Stosser, M.B., Butler, E., Downtain-Pickersgill, C., Shanmugham, A., Retterer, K., Brandt, T., Richard, G., and McKnight, D.A. (2018). Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 59, 1062-1071. [DOI] [PubMed]
- 8.Djémié T., Weckhuysen S., von Spiczak S., Carvill G.L., Jaehn J., Anttonen A.K., Brilstra E., Caglayan H.S., de Kovel C.G., Depienne C., EuroEPINOMICS-RES Dravet working group Pitfalls in genetic testing: The story of missed SCN1A mutations. Mol. Genet. Genomic Med. 2016;4:457–464. doi: 10.1002/mgg3.217. [DOI] [PMC free article] [PubMed] [Google Scholar]; Djemie, T., Weckhuysen, S., von Spiczak, S., Carvill, G.L., Jaehn, J., Anttonen, A.K., Brilstra, E., Caglayan, H.S., de Kovel, C.G., Depienne, C., et al.; EuroEPINOMICS-RES Dravet working group (2016). Pitfalls in genetic testing: The story of missed SCN1A mutations. Mol. Genet. Genomic Med. 4, 457-464. [DOI] [PMC free article] [PubMed]
- 9.Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]; Claes, L., Del-Favero, J., Ceulemans, B., Lagae, L., Van Broeckhoven, C., and De Jonghe, P. (2001). De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 68, 1327-1332. [DOI] [PMC free article] [PubMed]
- 10.Stamberger H., Nikanorova M., Willemsen M.H., Accorsi P., Angriman M., Baier H., Benkel-Herrenbrueck I., Benoit V., Budetta M., Caliebe A. STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–962. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]; Stamberger, H., Nikanorova, M., Willemsen, M.H., Accorsi, P., Angriman, M., Baier, H., Benkel-Herrenbrueck, I., Benoit, V., Budetta, M., Caliebe, A., et al. (2016). STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology 86, 954-962. [DOI] [PubMed]
- 11.Wolff M., Johannesen K.M., Hedrich U.B.S., Masnada S., Rubboli G., Gardella E., Lesca G., Ville D., Milh M., Villard L. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140:1316–1336. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]; Wolff, M., Johannesen, K.M., Hedrich, U.B.S., Masnada, S., Rubboli, G., Gardella, E., Lesca, G., Ville, D., Milh, M., Villard, L., et al. (2017). Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain 140, 1316-1336. [DOI] [PubMed]
- 12.EuroEPINOMICS-RES Consortium. Electronic address: euroepinomics-RES@ua.ac.be. Epilepsy Phenome/Genome Project. Epi4K Consortium. EuroEPINOMICS-RES Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2017;100:179. doi: 10.1016/j.ajhg.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; EuroEPINOMICS-RES Consortium. Electronic address: euroepinomics-RES@ua.ac.be; Epilepsy Phenome/Genome Project; Epi4K Consortium; EuroEPINOMICS-RES Consortium (2017). De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 100, 179. [DOI] [PMC free article] [PubMed]
- 13.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]; Allen, A.S., Berkovic, S.F., Cossette, P., Delanty, N., Dlugos, D., Eichler, E.E., Epstein, M.P., Glauser, T., Goldstein, D.B., Han, Y., et al.; Epi4K Consortium; Epilepsy Phenome/Genome Project (2013). De novo mutations in epileptic encephalopathies. Nature 501, 217-221. [DOI] [PMC free article] [PubMed]
- 14.Helbig I., Lindhout D. Advancing the phenome alongside the genome in epilepsy studies. Neurology. 2017;89:14–15. doi: 10.1212/WNL.0000000000004053. [DOI] [PubMed] [Google Scholar]; Helbig, I., and Lindhout, D. (2017). Advancing the phenome alongside the genome in epilepsy studies. Neurology 89, 14-15. [DOI] [PubMed]
- 15.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robinson, P.N., Kohler, S., Bauer, S., Seelow, D., Horn, D., and Mundlos, S. (2008). The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83, 610-615. [DOI] [PMC free article] [PubMed]
- 16.Köhler S., Vasilevsky N.A., Engelstad M., Foster E., McMurry J., Aymé S., Baynam G., Bello S.M., Boerkoel C.F., Boycott K.M. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017;45(D1):D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kohler, S., Vasilevsky, N.A., Engelstad, M., Foster, E., McMurry, J., Ayme, S., Baynam, G., Bello, S.M., Boerkoel, C.F., Boycott, K.M., et al. (2017). The Human Phenotype Ontology in 2017. Nucleic Acids Res. 45 (D1), D865-D876. [DOI] [PMC free article] [PubMed]
- 17.Bastarache L., Hughey J.J., Hebbring S., Marlo J., Zhao W., Ho W.T., Van Driest S.L., McGregor T.L., Mosley J.D., Wells Q.S. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science. 2018;359:1233–1239. doi: 10.1126/science.aal4043. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bastarache, L., Hughey, J.J., Hebbring, S., Marlo, J., Zhao, W., Ho, W.T., Van Driest, S.L., McGregor, T.L., Mosley, J.D., Wells, Q.S., et al. (2018). Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science 359, 1233-1239. [DOI] [PMC free article] [PubMed]
- 18.Akawi N., McRae J., Ansari M., Balasubramanian M., Blyth M., Brady A.F., Clayton S., Cole T., Deshpande C., Fitzgerald T.W., DDD study Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet. 2015;47:1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]; Akawi, N., McRae, J., Ansari, M., Balasubramanian, M., Blyth, M., Brady, A.F., Clayton, S., Cole, T., Deshpande, C., Fitzgerald, T.W., et al.; DDD study (2015). Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet. 47, 1363-1369. [DOI] [PMC free article] [PubMed]
- 19.Köhler S., Schulz M.H., Krawitz P., Bauer S., Dölken S., Ott C.E., Mundlos C., Horn D., Mundlos S., Robinson P.N. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kohler, S., Schulz, M.H., Krawitz, P., Bauer, S., Dolken, S., Ott, C.E., Mundlos, C., Horn, D., Mundlos, S., and Robinson, P.N. (2009). Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 85, 457-464. [DOI] [PMC free article] [PubMed]
- 20.Pesquita C., Faria D., Falcão A.O., Lord P., Couto F.M. Semantic similarity in biomedical ontologies. PLoS Comput. Biol. 2009;5:e1000443. doi: 10.1371/journal.pcbi.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pesquita, C., Faria, D., Falcao, A.O., Lord, P., and Couto, F.M. (2009). Semantic similarity in biomedical ontologies. PLoS Comput. Biol. 5, e1000443. [DOI] [PMC free article] [PubMed]
- 21.Fisher R.S., Cross J.H., French J.A., Higurashi N., Hirsch E., Jansen F.E., Lagae L., Moshé S.L., Peltola J., Roulet Perez E. Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58:522–530. doi: 10.1111/epi.13670. [DOI] [PubMed] [Google Scholar]; Fisher, R.S., Cross, J.H., French, J.A., Higurashi, N., Hirsch, E., Jansen, F.E., Lagae, L., Moshe, S.L., Peltola, J., Roulet Perez, E., et al. (2017). Operational classification of seizure types by the International League Against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 58, 522-530. [DOI] [PubMed]
- 22.Suls A., Jaehn J.A., Kecskés A., Weber Y., Weckhuysen S., Craiu D.C., Siekierska A., Djémié T., Afrikanova T., Gormley P., EuroEPINOMICS RES Consortium De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; Suls, A., Jaehn, J.A., Kecskes, A., Weber, Y., Weckhuysen, S., Craiu, D.C., Siekierska, A., Djemie, T., Afrikanova, T., Gormley, P., et al.; EuroEPINOMICS RES Consortium (2013). De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 93, 967-975. [DOI] [PMC free article] [PubMed]
- 23.Vögtle F.N., Brändl B., Larson A., Pendziwiat M., Friederich M.W., White S.M., Basinger A., Kücükköse C., Muhle H., Jähn J.A. Mutations in PMPCB encoding the catalytic subunit of the mitochondrial presequence protease cause neurodegeneration in early childhood. Am. J. Hum. Genet. 2018;102:557–573. doi: 10.1016/j.ajhg.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vogtle, F.N., Brandl, B., Larson, A., Pendziwiat, M., Friederich, M.W., White, S.M., Basinger, A., Kucukkose, C., Muhle, H., Jahn, J.A., et al. (2018). Mutations in PMPCB encoding the catalytic subunit of the mitochondrial presequence protease cause neurodegeneration in early childhood. Am. J. Hum. Genet. 102, 557-573. [DOI] [PMC free article] [PubMed]
- 24.Farwell Hagman K.D., Shinde D.N., Mroske C., Smith E., Radtke K., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Alcaraz W.A. Candidate-gene criteria for clinical reporting: Diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet. Med. 2017;19:224–235. doi: 10.1038/gim.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]; Farwell Hagman, K.D., Shinde, D.N., Mroske, C., Smith, E., Radtke, K., Shahmirzadi, L., El-Khechen, D., Powis, Z., Chao, E.C., Alcaraz, W.A., et al. (2017). Candidate-gene criteria for clinical reporting: Diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet. Med. 19, 224-235. [DOI] [PMC free article] [PubMed]
- 25.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]; Farwell, K.D., Shahmirzadi, L., El-Khechen, D., Powis, Z., Chao, E.C., Tippin Davis, B., Baxter, R.M., Zeng, W., Mroske, C., Parra, M.C., et al. (2015). Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 17, 578-586. [DOI] [PubMed]
- 26.Schlicker A., Domingues F.S., Rahnenführer J., Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schlicker, A., Domingues, F.S., Rahnenfuhrer, J., and Lengauer, T. (2006). A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics 7, 302. [DOI] [PMC free article] [PubMed]
- 27.Deng Y., Gao L., Wang B., Guo X. HPOSim: An R package for phenotypic similarity measure and enrichment analysis based on the human phenotype ontology. PLoS ONE. 2015;10:e0115692. doi: 10.1371/journal.pone.0115692. [DOI] [PMC free article] [PubMed] [Google Scholar]; Deng, Y., Gao, L., Wang, B., and Guo, X. (2015). HPOSim: An R package for phenotypic similarity measure and enrichment analysis based on the human phenotype ontology. PLoS ONE 10, e0115692. [DOI] [PMC free article] [PubMed]
- 28.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]; Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. (2000). The Protein Data Bank. Nucleic Acids Res. 28, 235-242. [DOI] [PMC free article] [PubMed]
- 29.Frappier V., Chartier M., Najmanovich R.J. ENCoM server: Exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 2015;43(W1):W395-400. doi: 10.1093/nar/gkv343. [DOI] [PMC free article] [PubMed] [Google Scholar]; Frappier, V., Chartier, M., and Najmanovich, R.J. (2015). ENCoM server: Exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 43 (W1), W395-400. [DOI] [PMC free article] [PubMed]
- 30.Kononenko N.L., Puchkov D., Classen G.A., Walter A.M., Pechstein A., Sawade L., Kaempf N., Trimbuch T., Lorenz D., Rosenmund C. Clathrin/AP-2 mediate synaptic vesicle reformation from endosome-like vacuoles but are not essential for membrane retrieval at central synapses. Neuron. 2014;82:981–988. doi: 10.1016/j.neuron.2014.05.007. [DOI] [PubMed] [Google Scholar]; Kononenko, N.L., Puchkov, D., Classen, G.A., Walter, A.M., Pechstein, A., Sawade, L., Kaempf, N., Trimbuch, T., Lorenz, D., Rosenmund, C., et al. (2014). Clathrin/AP-2 mediate synaptic vesicle reformation from endosome-like vacuoles but are not essential for membrane retrieval at central synapses. Neuron 82, 981-988. [DOI] [PubMed]
- 31.Posor Y., Eichhorn-Gruenig M., Puchkov D., Schöneberg J., Ullrich A., Lampe A., Müller R., Zarbakhsh S., Gulluni F., Hirsch E. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]; Posor, Y., Eichhorn-Gruenig, M., Puchkov, D., Schoneberg, J., Ullrich, A., Lampe, A., Muller, R., Zarbakhsh, S., Gulluni, F., Hirsch, E., et al. (2013). Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 499, 233-237. [DOI] [PubMed]
- 32.Schöneberg J., Lehmann M., Ullrich A., Posor Y., Lo W.T., Lichtner G., Schmoranzer J., Haucke V., Noé F. Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 2017;8:15873. doi: 10.1038/ncomms15873. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schoneberg, J., Lehmann, M., Ullrich, A., Posor, Y., Lo, W.T., Lichtner, G., Schmoranzer, J., Haucke, V., and Noe, F. (2017). Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 8, 15873. [DOI] [PMC free article] [PubMed]
- 33.Jackson L.P., Kelly B.T., McCoy A.J., Gaffry T., James L.C., Collins B.M., Höning S., Evans P.R., Owen D.J. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jackson, L.P., Kelly, B.T., McCoy, A.J., Gaffry, T., James, L.C., Collins, B.M., Honing, S., Evans, P.R., and Owen, D.J. (2010). A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141, 1220-1229. [DOI] [PMC free article] [PubMed]
- 34.Kononenko N.L., Claßen G.A., Kuijpers M., Puchkov D., Maritzen T., Tempes A., Malik A.R., Skalecka A., Bera S., Jaworski J., Haucke V. Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat. Commun. 2017;8:14819. doi: 10.1038/ncomms14819. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kononenko, N.L., Claßen, G.A., Kuijpers, M., Puchkov, D., Maritzen, T., Tempes, A., Malik, A.R., Skalecka, A., Bera, S., Jaworski, J., and Haucke, V. (2017). Retrograde transport of TrkB-containing autophagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat. Commun. 8, 14819. [DOI] [PMC free article] [PubMed]
- 35.Dittman J., Ryan T.A. Molecular circuitry of endocytosis at nerve terminals. Annu. Rev. Cell Dev. Biol. 2009;25:133–160. doi: 10.1146/annurev.cellbio.042308.113302. [DOI] [PubMed] [Google Scholar]; Dittman, J., and Ryan, T.A. (2009). Molecular circuitry of endocytosis at nerve terminals. Annu. Rev. Cell Dev. Biol. 25, 133-160. [DOI] [PubMed]
- 36.Saheki Y., De Camilli P. Synaptic vesicle endocytosis. Cold Spring Harb. Perspect. Biol. 2012;4:a005645. doi: 10.1101/cshperspect.a005645. [DOI] [PMC free article] [PubMed] [Google Scholar]; Saheki, Y., and De Camilli, P. (2012). Synaptic vesicle endocytosis. Cold Spring Harb. Perspect. Biol. 4, a005645. [DOI] [PMC free article] [PubMed]
- 37.Kittler J.T., Chen G., Honing S., Bogdanov Y., McAinsh K., Arancibia-Carcamo I.L., Jovanovic J.N., Pangalos M.N., Haucke V., Yan Z., Moss S.J. Phospho-dependent binding of the clathrin AP2 adaptor complex to GABAA receptors regulates the efficacy of inhibitory synaptic transmission. Proc. Natl. Acad. Sci. USA. 2005;102:14871–14876. doi: 10.1073/pnas.0506653102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kittler, J.T., Chen, G., Honing, S., Bogdanov, Y., McAinsh, K., Arancibia-Carcamo, I.L., Jovanovic, J.N., Pangalos, M.N., Haucke, V., Yan, Z., and Moss, S.J. (2005). Phospho-dependent binding of the clathrin AP2 adaptor complex to GABAA receptors regulates the efficacy of inhibitory synaptic transmission. Proc. Natl. Acad. Sci. USA 102, 14871-14876. [DOI] [PMC free article] [PubMed]
- 38.Froemke R.C. Plasticity of cortical excitatory-inhibitory balance. Annu. Rev. Neurosci. 2015;38:195–219. doi: 10.1146/annurev-neuro-071714-034002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Froemke, R.C. (2015). Plasticity of cortical excitatory-inhibitory balance. Annu. Rev. Neurosci. 38, 195-219. [DOI] [PMC free article] [PubMed]
- 39.Mitsunari T., Nakatsu F., Shioda N., Love P.E., Grinberg A., Bonifacino J.S., Ohno H. Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 2005;25:9318–9323. doi: 10.1128/MCB.25.21.9318-9323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mitsunari, T., Nakatsu, F., Shioda, N., Love, P.E., Grinberg, A., Bonifacino, J.S., and Ohno, H. (2005). Clathrin adaptor AP-2 is essential for early embryonal development. Mol. Cell. Biol. 25, 9318-9323. [DOI] [PMC free article] [PubMed]
- 40.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T., O’Donnell-Luria, A.H., Ware, J.S., Hill, A.J., Cummings, B.B., et al.; Exome Aggregation Consortium (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285-291. [DOI] [PMC free article] [PubMed]
- 41.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]; Petrovski, S., Wang, Q., Heinzen, E.L., Allen, A.S., and Goldstein, D.B. (2013). Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9, e1003709. [DOI] [PMC free article] [PubMed]
- 42.Schubert J., Siekierska A., Langlois M., May P., Huneau C., Becker F., Muhle H., Suls A., Lemke J.R., de Kovel C.G., EuroEPINOMICS RES Consortium Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat. Genet. 2014;46:1327–1332. doi: 10.1038/ng.3130. [DOI] [PubMed] [Google Scholar]; Schubert, J., Siekierska, A., Langlois, M., May, P., Huneau, C., Becker, F., Muhle, H., Suls, A., Lemke, J.R., de Kovel, C.G., et al.; EuroEPINOMICS RES Consortium (2014). Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat. Genet. 46, 1327-1332. [DOI] [PubMed]
- 43.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D., Nassif C., Diallo O., Monlong J., Cadieux-Dion M., Deciphering Developmental Disorders Study High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am. J. Hum. Genet. 2017;101:664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hamdan, F.F., Myers, C.T., Cossette, P., Lemay, P., Spiegelman, D., Laporte, A.D., Nassif, C., Diallo, O., Monlong, J., Cadieux-Dion, M., et al.; Deciphering Developmental Disorders Study (2017). High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am. J. Hum. Genet. 101, 664-685. [DOI] [PMC free article] [PubMed]
- 44.von Spiczak S., Helbig K.L., Shinde D.N., Huether R., Pendziwiat M., Lourenço C., Nunes M.E., Sarco D.P., Kaplan R.A., Dlugos D.J., Epi4K Consortium. EuroEPINOMICS-RES NLES Working Group DNM1 encephalopathy: A new disease of vesicle fission. Neurology. 2017;89:385–394. doi: 10.1212/WNL.0000000000004152. [DOI] [PMC free article] [PubMed] [Google Scholar]; von Spiczak, S., Helbig, K.L., Shinde, D.N., Huether, R., Pendziwiat, M., Lourenço, C., Nunes, M.E., Sarco, D.P., Kaplan, R.A., Dlugos, D.J., et al.; Epi4K Consortium; EuroEPINOMICS-RES NLES Working Group (2017). DNM1 encephalopathy: A new disease of vesicle fission. Neurology 89, 385-394. [DOI] [PMC free article] [PubMed]
- 45.Myers C.T., Stong N., Mountier E.I., Helbig K.L., Freytag S., Sullivan J.E., Ben Zeev B., Nissenkorn A., Tzadok M., Heimer G. De novo mutations in PPP3CA cause severe neurodevelopmental disease with seizures. Am. J. Hum. Genet. 2017;101:516–524. doi: 10.1016/j.ajhg.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; Myers, C.T., Stong, N., Mountier, E.I., Helbig, K.L., Freytag, S., Sullivan, J.E., Ben Zeev, B., Nissenkorn, A., Tzadok, M., Heimer, G., et al. (2017). De novo mutations in PPP3CA cause severe neurodevelopmental disease with seizures. Am. J. Hum. Genet. 101, 516-524. [DOI] [PMC free article] [PubMed]
- 46.Kadlecova Z., Spielman S.J., Loerke D., Mohanakrishnan A., Reed D.K., Schmid S.L. Regulation of clathrin-mediated endocytosis by hierarchical allosteric activation of AP2. J. Cell Biol. 2017;216:167–179. doi: 10.1083/jcb.201608071. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kadlecova, Z., Spielman, S.J., Loerke, D., Mohanakrishnan, A., Reed, D.K., and Schmid, S.L. (2017). Regulation of clathrin-mediated endocytosis by hierarchical allosteric activation of AP2. J. Cell Biol. 216, 167-179. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.