Abstract
Haematopoietic stem cells (HSCs) maintain balanced self-renewal and differentiation, but how these functions are precisely regulated is not fully understood. N6-methyladenosine (m6A) mRNA methylation has emerged as an important mode of epitranscriptional gene expression regulation affecting many biological processes. We show that deleting the m6A methyltransferase, Mettl3, from the adult haematopoietic system led to an accumulation of HSCs in the bone marrow and marked reduction of reconstitution potential due to a blockage of HSC differentiation. Interestingly, deleting Mettl3 from myeloid cells using Lysm-cre did not impact myeloid cell number or function. m6A sequencing revealed 2,073 genes with significant m6A modification in HSCs. Myc was identified as a direct target of m6A in HSCs. Mettl3-deficient HSCs failed to up-regulate MYC expression upon stimulation to differentiate and enforced expression of Myc rescued differentiation defects of Mettl3-deficient HSCs. Our results revealed a key role of m6A in governing HSC differentiation.
Introduction
HSCs give rise to all blood cells, and are characterized by their ability for life-long self-renewal and multilineage differentiation. HSC function is regulated by complex cell-intrinsic1 and -extrinsic pathways2, but the mechanisms that control balanced self-renewal and differentiation are poorly understood. Transcriptional, translational and epigenetic regulators have been shown to critically regulate HSC function1. Whether other molecular mechanisms also regulate HSCs is unclear.
m6A is a modified base found in eukaryotic mRNAs3. This modification of adenosine is abundant and functions as an epitranscriptomic regulator of target mRNAs through multiple mechanisms, including stability and translation efficiency4. m6A is deposited onto mRNAs by a methyltransferase complex, composed of methyl transferase-like 3 (METTL3), methyl transferase-like 14 (METTL14) and Wilms-tumor associated protein (WTAP) and other accessory proteins5. METTL3 is the essential catalytic component of the complex6–9. m6A has been shown to impact fundamental cellular processes, including DNA damage repair10, meiosis11, and circadian clock12. Transcriptome-wide mapping of m6A modification has revealed cell type-specific methylation targets, suggesting that m6A regulates cell type-specific processes13. Interestingly, m6A has been shown to impact stem and progenitor cell fate. Disruption of the m6A pathway blocks the decay of naïve embryonic stem cell program6,14. m6A also plays a critical role in embryonic neurogenesis by regulating fatal neural stem and progenitor cells15,16. The role of m6A in the haematopoietic system is emerging. Using zebrafish as a model, it has been shown that m6A is required for endothelial to haematopoietic transition and the emergence of haematopoietic stem and progenitor cells17. shRNA knockdown of Mettl3 or Mettl14 in human haematopoietic stem and progenitor cells leads to myeloid differentiation in vitro18,19. These studies show that m6A plays critical roles for HSC emergence and in vitro function, but its role in mammalian adult HSCs and haematopoiesis in vivo remained unclear.
Results
Deletion of Mettl3 disrupts haematopoiesis and leads to accumulation of HSCs
We performed quantitative real-time PCR (qPCR) analysis to assess the expression of Mettl3 in the haematopoietic system. Mettl3 transcripts were expressed at approximately 4.5-fold higher levels in CD150+CD48−Lin−Sca1+cKit+ HSCs compared with whole bone marrow cells (Supplementary Fig. 1a), suggesting that METTL3-mediated m6A may regulate the function of HSCs.
To test whether m6A regulates HSCs and haematopoiesis in vivo, we generated and validated a floxed allele of Mettl3 (Supplementary Fig. 1b), and crossed it with Mx1-cre20 to generate Mx1-cre; Mettl3fl/fl mice. We conditionally deleted Mettl3 from the adult haematopoietic cells by intraperitoneally injecting polyinosinic-polycytidylic acid (pIpC) into 6–8 week old Mx1-cre; Mettl3fl/fl mice (Supplementary Fig. 1b). Efficient deletion in HSCs was achieved by 10 days after the last pIpC injection (Supplementary Fig. 1c and d). Ten to 14 days (short term) after the last pIpC injection, complete blood count analyses revealed a significant decrease in platelet count in Mx1-cre; Mettl3fl/fl mice compared with pIpC-treated controls (Figs. 1a, b and Supplementary Fig. 2a). Recent work in the field has proposed that platelets can be directly generated from HSCs21,22. The platelet phenotype raises the possibility that m6A may regulate HSCs. The same phenotype persisted 2–3 months after the last pIpC injection (Figs. 1a, b and Supplementary Fig. 2a). By 4 months, white blood cell counts were also significantly reduced, with an altered white blood cell distribution (Figs. 1a and Supplementary Fig. 2b). These data suggest that m6 A is required for haematopoiesis.
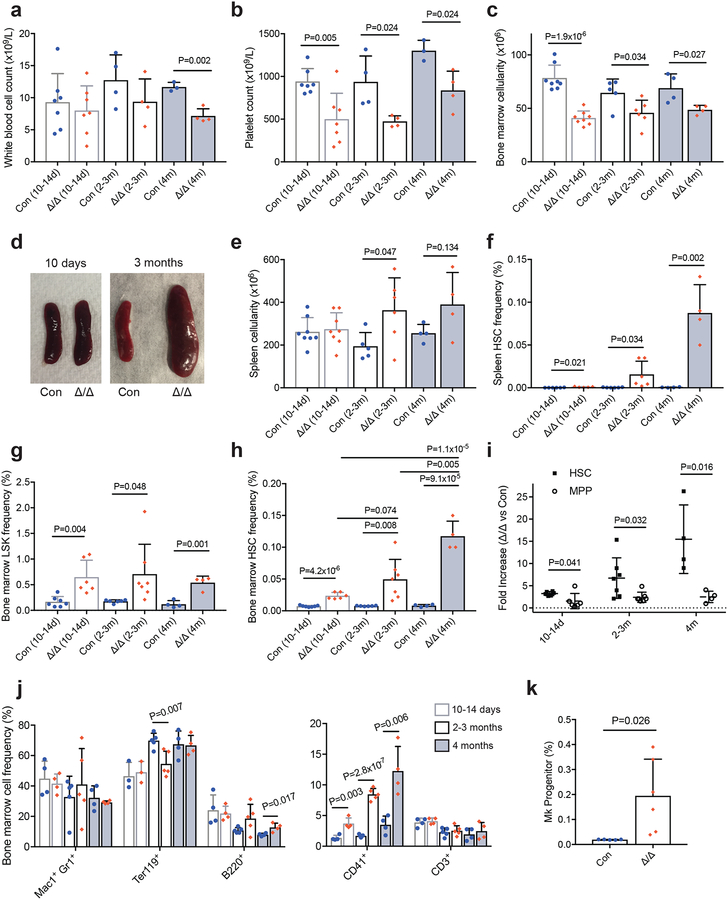
Figure 1. Loss of Mettl3 leads to accumulation of HSCs and perturbed haematopoiesis.
(a,b) White blood cell (WBC) (a) and platelet peripheral blood counts (b) from pIpC-treated control and Mx1-cre; Mettl3fl/fl mice (n=7 control (10–14d), n=7 Mx1-cre; Mettl3fl/fl (10–14d), n=4 control (2–3m), n=4 Mx1-cre; Mettl3fl/fl (2–3m), n=3 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(c) Bone marrow cellularity per hindlimb (n=28 control (10–14d), n=8 Mx1-cre; Mettl3fl/fl (10–14d), n=5 control (2–3m), n=6 Mx1-cre; Mettl3fl/fl (2–3m), n=4 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(d) Representative images of the spleens from Mx1-cre; Mettl3fl/fl and control mice 10 days and 3 months after pIpC treatment, as indicated.
(e) Spleen cellularity (n=8 control (10–14d), n=8 Mx1-cre; Mettl3fl/fl (10–14d), n=5 control (2–3m), n=6 Mx1-cre; Mettl3fl/fl (2–3m), n=4 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(f) Spleen HSC frequency (n=6 control (10–14d), n=5 Mx1-cre; Mettl3fl/fl (10–14d), n=6 control (2–3m), n=6 Mx1-cre; Mettl3fl/fl (2–3m), n=4 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(g) Frequencies of bone marrow Lin−Sca-1+c-Kit+ (LSK) progenitors (n=7 control (10–14d), n=6 Mx1-cre; Mettl3fl/fl (10–14d), n=6 control (2–3m), n=7 Mx1-cre; Mettl3fl/fl (2–3m), n=4 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(h) Frequency of bone marrow HSCs (n=7 control (10–14d), n=6 Mx1-cre; Mettl3fl/fl (10–14d), n=6 control (2–3m), n=7 Mx1-cre; Mettl3fl/fl (2–3m), n=4 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(i) Fold increase in Mx1-cre; Mettl3fl/fl bone marrow HSC or MPP frequency compared to littermate control frequencies at indicated times after pIpC treatment (n=6 (10–14d), n=7 (2–3m), n=4 (4m)).
(j) Frequencies of mature cell populations in the bone marrow (n=4 control (10–14d), n=4 Mx1-cre; Mettl3fl/fl (10–14d), n=5 control (2–3m), n=5 Mx1-cre; Mettl3fl/fl (2–3m), n=4 control (4m), n=4 Mx1-cre; Mettl3fl/fl (4m)).
(k) Frequency of megakaryocyte progenitors (Lineage−Sca1−cKit+CD150+CD41+) cells in the bone marrow >10 days after pIpC treatment (n=5 control, n=6 Mx1-cre; Mettl3fl/fl).
All samples are from biologically independent mice. Values are shown as individual points with mean ± s.d. P values were determined by unpaired two-sided Student’s t-test.
Deletion of Mettl3 led to a significant reduction in bone marrow cellularity (Fig. 1c), but not spleen cellularity 10–14 days after the last pIpC injection (Figs. 1d and e). However, by 2–4 months after the last pIpC injection, in addition to a significant bone marrow cellularity reduction, the spleen size and cellularity were significantly increased with a distortion of cell type distribution (Figs. 1c–e and Supplementary Fig. 2c). The spleens contained more HSCs in Mx1-cre; Mettl3fl/fl mice compared with controls (Fig. 1f). These data are suggestive of extramedullary haematopoiesis after loss of m6A.
In the bone marrow, Lin−Sca1+cKit+ (LSK) haematopoietic progenitors (Fig. 1g) and HSCs (Fig. 1h and Supplementary Fig. 2d and e) were significantly increased at all time points examined. The HSC pool uniquely expanded over time from 10–14 days to 4 months after the last pIpC injection: progressing from a 3-fold to a 17-fold increase in HSC frequency (Figs. 1h, Supplementary Fig. 2d and e). In contrast, CD150−CD48−LSK MPP frequency was not significantly increased while CD150−CD48+LSK progenitor frequency was only modestly increased (Fig. 1i and Supplementary Fig. 2f). CD150+CD48+LSK megakaryocyte-skewed multipotent progenitor frequency was significantly increased (Supplementary Fig. 2f), suggesting that there is also an effect on the megakaryocyte lineage. Thus, at the top of the haematopoietic hierarchy, loss of m6A preferentially leads to HSC accumulation.
We also examined other haematopoietic progenitors in the bone marrow. These included Lin−Sca1lowcKitlowFlt3+IL7Rα+ common lymphoid progenitors (CLPs), CD34+FcγR−Lineage−Sca1−cKit+ common myeloid progenitors (CMPs), CD34+FcγR+Lineage−Sca1−cKit+ granulocyte/macrophage progenitors (GMPs), and CD34−FcγR−Lineage−Sca1−cKit+ megakaryocytic/erythroid progenitors (MEPs). The frequencies of these haematopoietic progenitors were unchanged in Mx1-cre; Mettl3fl/fl mice compared with controls until 4 months after the last pIpC treatment, when there was a modest decrease in GMP and a modest increase in MEP frequencies (Supplementary Fig. 2g). There was a significant increase in Lin−Sca1−cKit+CD150+CD41+ megakaryocyte progenitors and CD41+ megakaryocytic cells in the bone marrow (Fig. 1j and k), suggesting that the reduction in platelet counts (Fig. 1b) is due to a differentiation defect in the megakaryocyte lineage. These data suggest that loss of Mettl3 preferentially impacts HSCs, with effects on the megakaryocyte lineage.
Deletion of Mettl3 preferentially blocks HSC differentiation
HSCs from pIpC-injected Mx1-cre; Mettl3fl/fl mice did not incorporate more BrdU (Fig. 2a), but underwent slightly more cell death (Fig. 2b), suggesting that the accumulation of HSCs may be the result of blocked differentiation rather than enhanced self-renewal. The Mettl3-deficient HSCs did not have elevated levels of reactive oxygen species (ROS) or γH2Ax staining (Supplementary Fig. 2h and i). Consistent with an HSC differentiation defect, pIpC-injected Mx1-cre; Mettl3fl/fl mice had worse survival compared to controls when challenged with a myeloablative agent, 5-fluorouracil (5FU) (Fig. 2c).
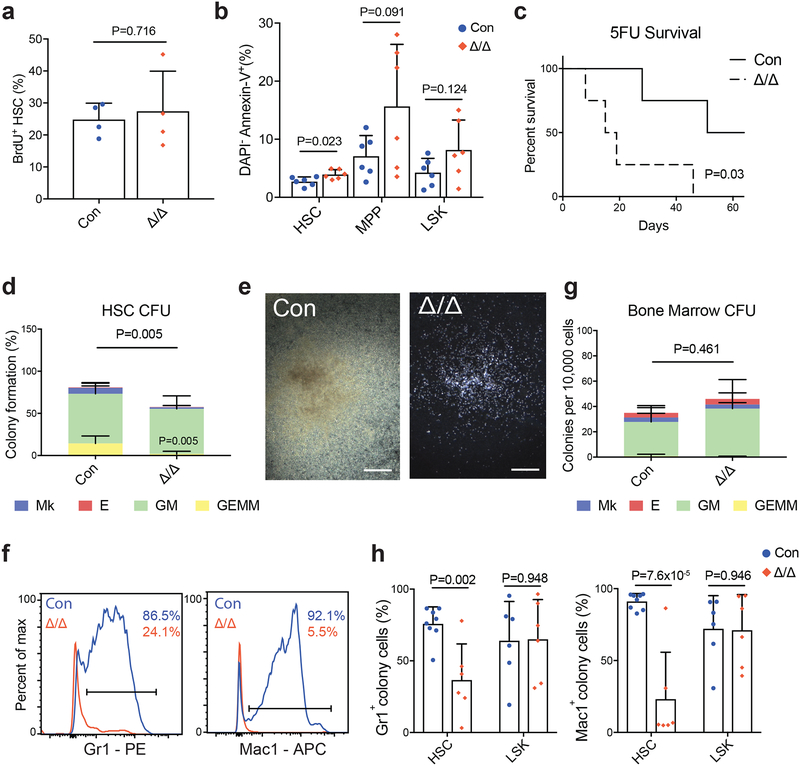
Figure 2. Mettl3 is essential for adult HSC differentiation in vitro.
(a) Frequency of BrdU+ HSCs 10–14 days after pIpC treatment (n=4 for control, n=4 for Mx1-cre; Mettl3fl/fl).
(b) Frequency of DAPI− AnnexinV+ HSC, MPP and LSK populations 10–14 days after pIpC (n=6 for control, n=6 for Mx1-cre; Mettl3fl/fl).
(c) Kaplan-Meyer survival analysis of control and Mx1-cre; Mettl3fl/fl mice treated with weekly 5-FU injections 10 days after the last dose of pIpC. P value by log-rank test (n=4 for control, n=4 for Mx1-cre; Mettl3fl/fl).
(d) Frequencies of colony formation from single sorted HSCs. Colony-forming units (CFU) scored as granulocyte, erythroid, macrophage, megakaryocyte (GEMM), granulocyte, macrophage (GM), megakaryocyte (Mk), or erythrocyte (E) colonies (n=7 for control, n=7 for Mx1-cre; Mettl3fl/fl).
(e) Representative image of CFU-GM from single HSCs from control (left) and Mettl3-deleted (right) single HSCs, genotypes confirmed by PCR. Scale bars are 400um.
(f) Representative flow cytometric histograms of mature myeloid cell surface markers Gr-1 and Mac-1 on HSC-derived colony cells from pIpC-treated control and Mx1-cre; Mettl3fl/fl mice.
(g) Numbers of colonies formed from 10,000 bone marrow cells, scored as CFU-GEMM, CFU-GM, CFU-E, or CFU-MK (n=5 for control, n=5 for Mx1-cre; Mettl3fl/fl).
(h) Quantification of expression of Gr-1 and Mac-1 cell surface markers on HSC and LSK colony cells by flow cytometry (HSC, n=8 for control, n=6 for Mx1-cre; Mettl3fl/fl; LSK, n=6 for control, n=6 for Mx1-cre; Mettl3fl/fl).
All samples are from biologically independent animals. Values are shown as individual points with mean ± s.d. P values were determined by unpaired two-sided Student’s t-test.
To directly assess the differentiation potential of Mettl3-deficient HSCs, we performed methylcellulose assays. Single HSCs from pIpC-treated Mx1-cre; Mettl3fl/fl or control mice were directly sorted into methylcellulose, which supports myelo-erythroid differentiation in vitro23,24. Significantly fewer Mettl3-deficient HSCs formed colonies compared with control HSCs, particularly multipotent granulocyte, erythrocyte, monocyte and megakaryocyte (GEMM) colonies (Fig. 2d). The vast majority of the colonies formed by Mettl3-deficient HSCs were significantly smaller compared with those formed by control HSCs (Fig. 2e and Supplementary Fig. 3a). Flow cytometric analysis on cells from these colonies showed that Mettl3-deficient HSCs failed to differentiate while control HSCs readily differentiated into Gr1+ and Mac1+ myeloid cells (Figs. 2f and h). A few colonies were formed at normal sizes by Mettl3-deficient HSCs and genotyping revealed that these colonies had escaped complete Mettl3 deletion (Supplementary Fig. 3b and c). Mettl3-deficient HSCs in the spleens also failed to differentiate (Supplementary Fig. 3d).
In contrast, when Mettl3-deficient whole bone marrow cells were plated into methylcellulose, colonies formed at similar numbers, size and types compared with controls (Fig. 2g). We further plated sorted Mettl3-deficient restricted progenitors (LSK, CD150−CD48+LSK and CD150+CD48+LSK), into methylcellulose. They formed normal numbers of colonies, similar in size and type compared with controls (Supplementary Fig. 3e). Flow cytometry analysis confirmed that these colonies contained normal numbers of differentiated cells (Fig. 2h and Supplementary Fig. 3f). Taken together, these data suggest that loss of Mettl3 preferentially leads to a differentiation block in HSCs but not in restricted progenitors in vitro.
Deletion of Mettl3 leads to cell-intrinsic HSC reconstitution and differentiation defects in vivo
To test whether loss of Mettl3 leads to HSC defects in vivo, we performed competitive reconstitution assays by transplanting 500,000 bone marrow cells from Mx1-cre; Mettl3fl/fl or control mice 10 days after the last pIpC injection along with 500,000 wild-type competing bone marrow cells into lethally irradiated recipient mice. Mettl3-deficient bone marrow cells contributed significantly lower levels of overall, myeloid, B and T lineage cells in the peripheral blood and a trend towards lower levels of HSCs in the bone marrow compared with controls (Fig. 3a and Supplementary Fig. 4a). Genotyping analysis on sorted residual donor-derived peripheral blood cells demonstrated that these residual cells escaped complete Mettl3 deletion (Supplementary Fig. 4b). To specifically test whether HSCs were functionally defective, we sorted and transplanted Mettl3-deficient and control HSCs along with wild-type competing bone marrow cells into lethally irradiated mice. While control HSCs readily reconstituted the recipient mice in all major haematopoietic lineages, Mettl3-deficient HSCs failed to give any discernable reconstitution (Fig. 3b and Supplementary Fig. 4c). These data suggest that Mettl3-deficient HSCs are defective in reconstituting recipient mice.
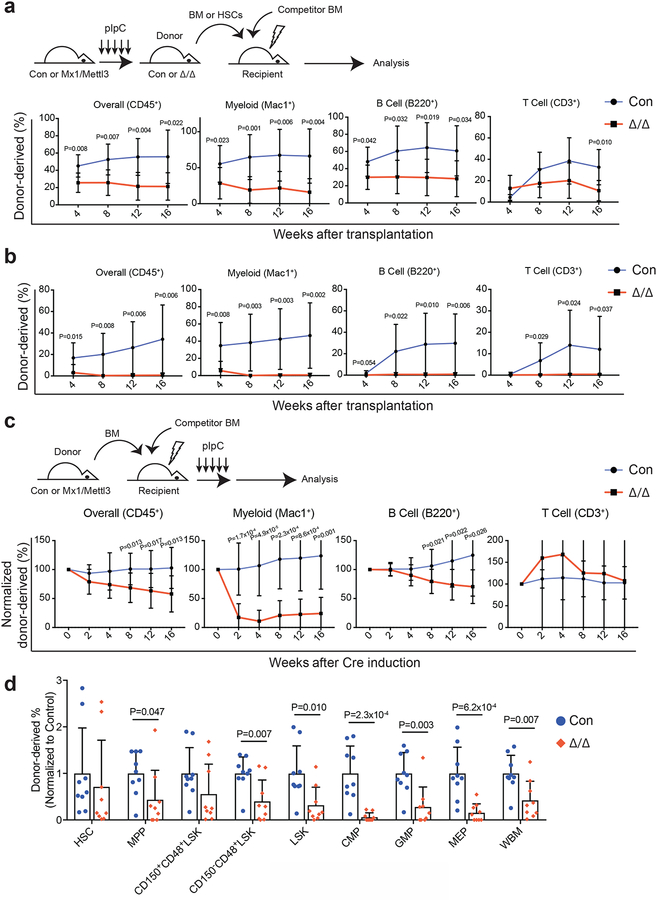
Figure 3: Loss of Mettl3 disrupts HSC differentiation in vivo.
(a) Competitive transplantation of 500,000 donor whole bone marrow cells from pIpC-treated Mx1-cre; Mettl3fl/fl or Mettl3fl/fl controls with 500,000 competitor cells. Multi-lineage chimera levels in the peripheral blood were assessed up to 16 weeks after transplantation (n=7 for control, n=8 for Mx1-cre; Mettl3fl/fl; samples were from independent recipients of two independent donor pairs from two experiments).
(b) Competitive transplantation of 50 sorted HSCs from pIpC-treated Mx1-cre; Mettl3fl/fl or controls with 300,000 competitor cells. Multi-lineage chimera levels in the peripheral blood were assessed up to 16 weeks after transplantation (n=9 for control, n=9 for Mx1-cre; Mettl3fl/fl; samples were from independent recipients of two independent donor pairs from two experiments).
(c) 500,000 donor whole bone marrow cells from untreated Mx1-cre; Mettl3fl/fl or controls with 500,000 competitor cells were transplanted into lethally irradiated recipient mice. Recipients were treated with pIpC after stable peripheral blood chimerism was established. Multi-lineage peripheral blood chimera levels shown as the percentage of the original chimera level up to 16 weeks after pIpC treatment (n=11 for control, n=12 for Mx1-cre; Mettl3fl/fl; samples were from independent recipients of three independent donor pairs from three experiments).
(d) Chimera levels of indicated bone marrow cell populations 20 weeks after pIpC treatment from (c) (n=9 for control, n=9 for Mx1-cre; Mettl3fl/fl; samples were from independent recipients of three independent donor pairs from three experiments).
Values are shown as individual points with mean ± s.d. P values were determined by unpaired two-sided Student’s t-test.
To determine whether Mettl3 is required cell-intrinsically for HSC function, we transplanted Mx1-cre; Mettl3fl/fl or control bone marrow cells, without pIpC treatment, along with wild-type competitor bone marrow cells into irradiated recipient mice. We waited at least 8 weeks after transplantation for stable bone marrow chimerism to be established. Mettl3 was then deleted by pIpC injection and donor-derived peripheral blood cells were monitored over time. Overall reconstitution by Mettl3-deficient bone marrow cells was significantly lower than controls (Fig. 3c). The reconstitution defects were mainly in the myeloid and B lineages, but not in the T cell lineages (Fig. 3c), consistent with the slow turnover kinetics of T cells in adults in vivo. After 16 weeks, the recipient mice were sacrificed and analyzed for the frequencies of donor-derived bone marrow HSCs and haematopoietic progenitors. Mettl3-deficient HSCs were present at similar levels compared with control HSCs (Fig. 3d), suggesting that HSC self-renewal is largely normal. However, these HSCs generated gradually less mature haematopoietic cells along the differentiation hierarchy (Fig. 3d). Consistent with the depletion of donor-derived myeloid cells in the peripheral blood (Fig. 3c), very little CMPs, GMPs and MEPs were derived from Mettl3-deficient HSCs (Fig. 3d). Genotyping on sorted donor-derived cells in recipient mice showed that these residual cells escaped complete Mettl3 deletion (Supplementary Fig. 4d). These data suggest that Mettl3 is required for HSC differentiation in vivo.
We also conditionally deleted Mettl3 from haematopoietic cells using Rosa26-creER25. We noncompetitively transplanted bone marrow cells from Rosa26-creER; Mettl3fl/fl mice into irradiated recipient mice and then deleted Mettl3 by administering tamoxifen. Deletion of Mettl3 led to a significantly higher frequency of HSCs in the recipient mice (Supplementary Fig. 4e), recapitulating the Mx1-cre; Mettl3fl/fl model (Fig. 1h). We also competitively transplanted Rosa26-creER; Mettl3fl/fl bone marrow cells together with recipient-type bone marrow cells and waited at least 8 weeks after transplantation for stable bone marrow chimerism before administering tamoxifen. Tamoxifen-induced deletion of Mettl3 also led to a significant reduction in reconstitution activity (Supplementary Fig. 4f), similar to Mx1-cre; Mettl3fl/fl mice (Fig. 3c).
Mettl3 is not required for myelopoiesis
To directly test whether the observed predominant myeloid lineage reconstitution defect (Fig. 3c) was a consequence of loss of Mettl3 in myeloid cells, we conditionally deleted Mettl3 from myeloid cells by generating Lysm-cre; Mettl3fl/fl mice. Consistent with previous reports26, Lysm-cre efficiently recombined in bone marrow myeloid cells (Supplementary Fig. 4g). Six-8 week old Lysm-cre; Mettl3fl/fl mice had normal peripheral blood counts (Figs. 4a and b, and Supplementary Fig. 4h), normal bone marrow and spleen cellularity (Fig. 4c and Supplementary Fig. 4i), and normal frequencies of myeloid cells in the bone marrow and spleens (Fig. 4d).
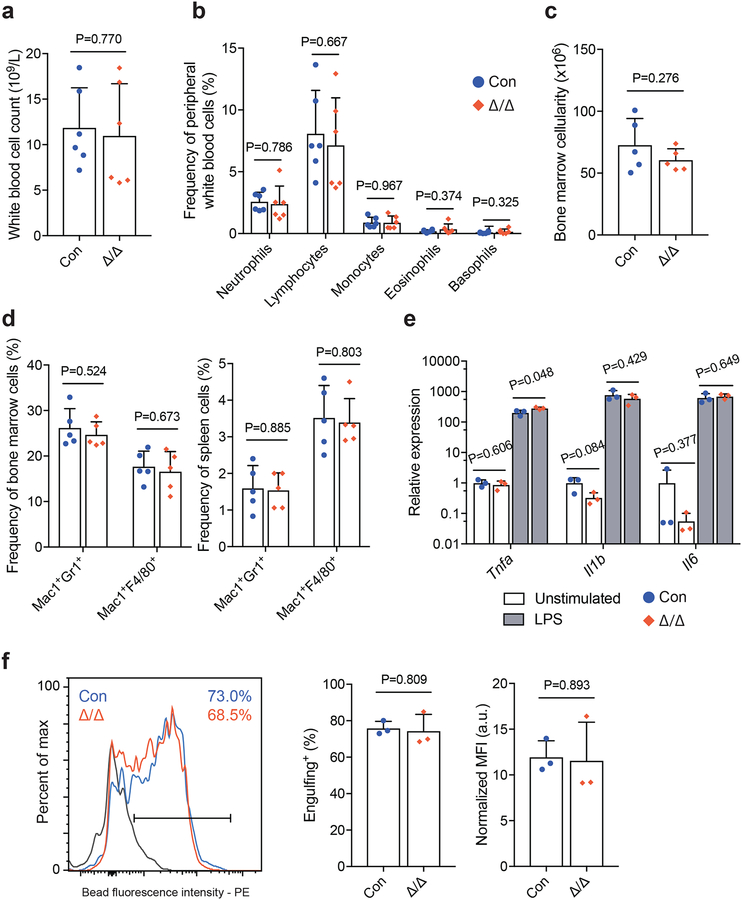
Figure 4. Loss of Mettl3 has no impact on myeloid cell maintenance or function.
(a) White blood cell counts in LysM-cre; Mettl3fl/fl mice (n=6 for control, n=6 for LysM-cre; Mettl3fl/fl).
(b) Frequencies of differential white blood cell counts in LysM-cre; Mettl3fl/fl mice (n=6 for control, n=6 for LysM-cre; Mettl3fl/fl).
(c) Bone marrow cellularity (n=5 for control, n=5 for LysM-cre; Mettl3fl/fl).
(d) Frequencies of bone marrow (left) and spleen (right) neutrophils (Mac1+Gr1+) and macrophages (Mac1+F4/80+) by flow cytometry (n=5 for control, n=5 for LysM-cre; Mettl3fl/fl).
(e) qPCR analysis of cytokine expression by unstimulated and LPS-stimulated bone marrow macrophages (n=3 for control, n=3 for LysM-cre; Mettl3fl/fl).
(f) Representative histogram (left) and quantification (right) of bone marrow macrophage phagocytosis of fluorescent-labeled beads (n=3 for control, n=3 for LysM-cre; Mettl3fl/fl). Shaded area denotes background staining (4°C incubation).
All samples were from biologically independent animals. Values are shown as individual points with mean ± s.d. P values were determined by unpaired two-sided Student’s t-test.
Mettl3-deficient bone marrow myeloid cells from Lysm-cre; Mettl3fl/fl mice formed macrophages with normal morphology (Supplementary Fig. 4j). When challenged with lipopolysaccharide (LPS), these macrophages showed normal up-regulation of inflammatory cytokines, Tnfa, Il1b and Il6 (Fig. 4e). Mettl3-deficient bone marrow-derived macrophages also displayed robust engulfing activity, similar to controls (Fig. 4f). These data suggest that Mettl3 is not required for the maintenance or function of mature myeloid cells and the observed reconstitution deficiencies (Fig. 3c) are a consequence of HSC differentiation defects. Furthermore, these data provide conclusive evidence that there is a stage-specific dependence on Mettl3 and m6A during haematopoietic differentiation in vivo.
Identification of m6A targets in HSCs
To identify direct m6A targets in rare HSCs, we developed an m6A-tagged mRNA immunoprecipitation sequencing (meRIP-seq) method to analyze the m6A methylome of 2,000 freshly sorted wild-type HSCs from 6–8 week old mice (Supplementary Figs. 5a and b). 2,073 transcripts (FDR<0.05 and fold>2) were significantly enriched in the m6A antibody-bound fraction compared to the non-bound fraction and were defined as high-confidence m6A targets (Figs. 5a and b, and Supplementary Table 1). This represented only ~9% of the genes expressed in the input mRNA fraction, suggesting that m6A preferentially marks a subset of genes in HSCs. We also performed meRIP-seq on Mettl3-deficient HSCs from Mx1-cre; Mettl3fl/fl mice 10 days after pIpC administration. The enrichment of 995 transcripts (~48% of the high-confidence m6A targets) was significantly reduced (p<0.05) in Mettl3-deficient HSCs, suggesting that the m6A modification of these targets depended acutely on METTL3 (Supplementary Figs. 5c and d). The lack of complete elimination of m6A may be due to residue METTL3 protein, slow turnover of some target mRNAs or antibody detection of cap region m6Am, which unlike m6A, is known not to be regulated by Mettl3 but is also detected by the anti-m6A antibodies used5.
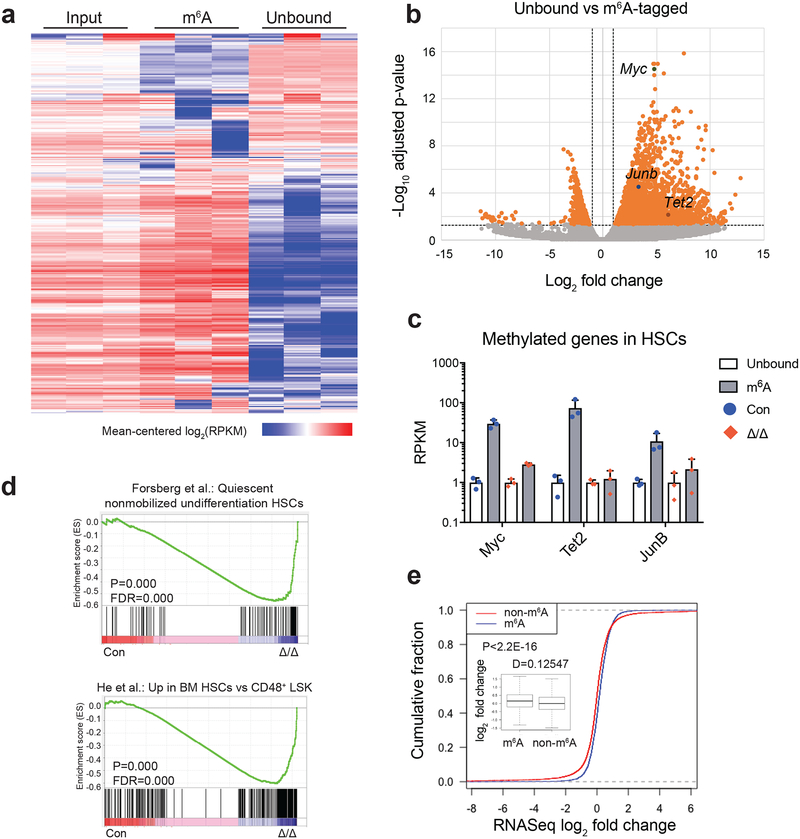
Figure 5. Identification of m6A methylation targets in HSCs.
(a) Heat map of transcript abundance between input, m6A-tagged, and unbound fractions. A total of 5,676 genes with log2(RPKM) ≥5 in ≥4 samples and stdev[log2(RPKM)]≥0.6 are shown.
(b) Volcano plot of meRIP-seq transcript expression differences between m6A-tagged and unbound fractions in HSCs. The x-axis specifies the log2 fold-changes (FC) and the y-axis specifies the log2 false discovery rate (FDR). Dashed vertical and horizontal lines indicate the filtering criteria (log2(FC)>1 and FDR<0.05). Orange dots represent 2,621 transcripts showing statistically significant differences between bound and non-bound factions, of which 2,073 are significantly enriched in the m6A-tagged fraction.
(c) meRIP-seq data showing that transcripts of some known HSC regulators are m6A-tagged in wild-type HSCs, and this methylation is largely eliminated in Mx1-cre; Mettl3fl/fl (Mettl3Δ/Δ) HSCs 10 days after Cre induction. Data were from n=3 biologically independent samples for control and Mettl3Δ/Δ. Values are shown as individual points with mean ± s.d.
(d) Gene set enrichment analysis plots showing that Mettl3-deficient HSCs lose HSC gene signature, as determined by RNA-seq profiling. Analysis completed on gene list ranked by log10 FDR and fold change sign, with enrichment determined after 1,000 permutations. Data were from n=3 biologically independent samples for control and Mettl3Δ/Δ. The statistics was computed using GSEA, and controlled for multiple comparisons by false discovery rate.
(e) Cumulative distribution of log2 (gene expression ratios, Mettl3-deficient/control). Genes were separated as m6A and non-m6A as assessed by meRIP-seq. Insert is the box plot of the log2 fold change in expression of non-m6A and m6A targets in HSCs. Plot displays the mean, standard deviation and interquartile range. P value was determined by two-sided Kolmogorov-Smirnov test.
All sequencing data were from n=3 biologically independent animals for control and Mettl3Δ/Δ.
Gene ontology (GO) analysis of m6A-tagged mRNAs demonstrated enrichment of genes related to nucleic acid metabolism, cell cycle regulation, transcription, RNA binding, and cellular stress responses, immune system development and haematopoiesis (Supplementary Fig. 5e). Supporting feed-forward regulation, several genes encoding components of the m6A pathway, including WTAP and m6A-reader proteins5, were also targets of m6A (Supplementary Fig. 5f). Consistent with the differentiation defects observed in Mettl3-deficient HSCs (Figs. 1–4), we observed significant enrichment of genes associated with haematopoietic differentiation (Supplementary Fig. 5e). These data suggest that m6A regulates HSC differentiation by directly targeting haematopoietic differentiation pathways. Several known HSC regulators were identified as major targets of m6A, including Myc, Junb, and Tet2 (Fig. 5b). More importantly, the enrichment of these genes in the m6A-bound fraction was significantly decreased when Mettl3 was deleted from HSCs, suggesting the m6A modification of these genes is dependent on METTL3 (Fig. 5c and Supplementary Figs. 5c and d).
Deletion of Mettl3 leads to a loss of HSC identity without drastic alterations of the transcriptome
To further understand the molecular mechanisms underpinning the functional defects of Mettl3-deficient HSCs, we performed RNA-seq analysis on Mettl3-deficient HSCs using Smart-seq227. 830 genes showed significant expression differences (p<0.05) (Supplementary Table 2). After stringent analysis with multiple comparisons correction, only 7 genes showed significant expression differences (FDR<0.05) between Mettl3-deficient and control HSCs (Supplementary Fig. 6a and Supplementary Table 2). As expected, Mettl3 was one of the most significantly down-regulated genes in Mettl3-deficient HSCs (Supplementary Fig. 6a). Gene set enrichment analysis (GSEA)28 showed that Mettl3 deficiency predominantly led to a loss of expression of HSC identity-associated genes defined by previous publications29–31 (Fig. 5d and Supplementary Fig. 6b). These data suggest that METTL3-mediated m6A modestly altered HSC identity without profoundly impacting global mRNA levels.
m6A has been shown to negatively regulate target mRNA stability4. We thus assessed whether m6A targets were up-regulated in Mettl3-deficient HSCs. 95 out of 384 up-regulated genes in Mettl3-defieient HSCs (p<0.05 and fold>1.2) were m6A targets (Supplementary Table 2 and Supplementary Fig. 6c). But none of the 7 high-confidence differentially expressed genes (FDR<0.05) (Supplementary Table 1 and Supplementary Fig. 6a) were high-confidence m6A targets. Compared to non-m6A targets, transcripts of m6A targets had slightly increased abundance in Mettl3-deficient HSCs (Fig. 5e). Taken together, these data suggest that although m6A may directly control the transcript levels of some genes, it predominantly regulates target mRNAs post-transcriptionally in HSCs.
Myc is a direct target of m6A in HSCs
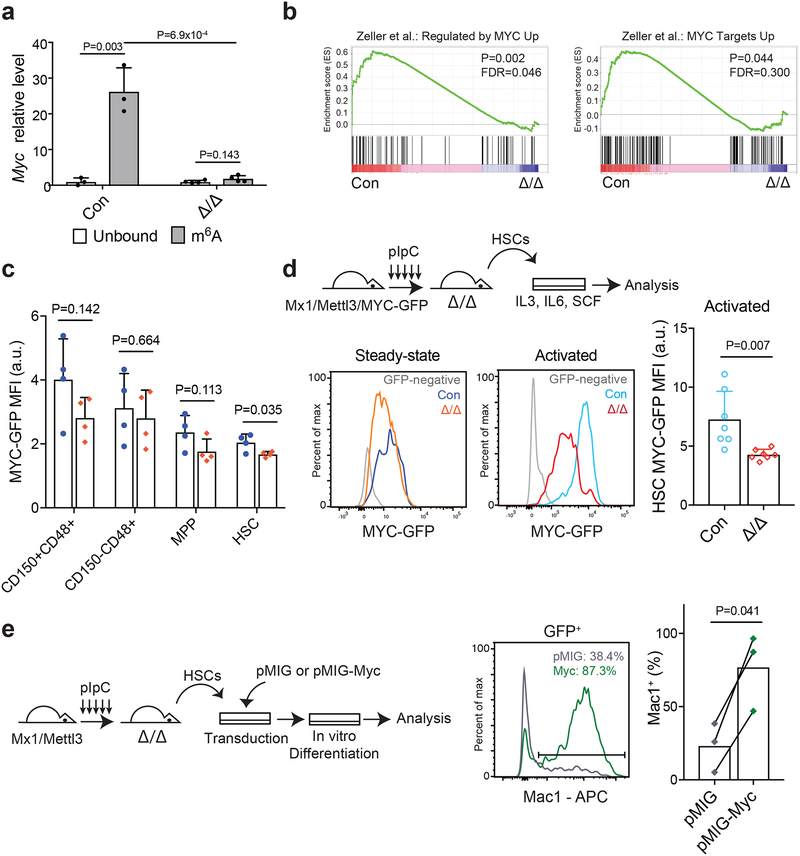
It has been shown that the MYC protein level is very low in HSCs, and is up-regulated upon differentiation through post-transcriptional mechanisms32,33. Furthermore, deletion of Myc blocks HSC differentiation while accumulated higher levels of MYC protein promotes HSC differentiation32,34. Prompted by the observations that Myc is a major determinant of HSC differentiation and the phenotypic similarity between Myc and Mettl3 conditional knockout mice (accumulation of HSCs and blockage of differentiation), we investigated whether Myc is a direct target of m6A in HSCs. Our meRIP-seq analyses revealed that Myc was one of the top candidates highly enriched in m6A antibody-bound fraction compared with non-bound fraction (~30 fold enrichment; FDR=3.96×10−15) (Figs. 5b and c), in a Mettl3-dependent manner (Fig. 5c and Supplementary Figs. 5c and d). We confirmed these meRIP-seq data using qPCR on independent samples (Fig. 6a). Interestingly, Myc transcript levels were not significantly changed in Mettl3-deficient HSCs compared with controls (Supplementary Fig. 6d). Using several previously published Myc target gene sets35,36, GSEA revealed a significant decrease in Myc target gene signatures in Mettl3-deficient HSCs (Fig. 6b and Supplementary Fig. 6e), suggesting that m6A may primarily regulate Myc mRNA translation.
Figure 6. Mettl3 regulates HSC function by targeting Myc.
(a) meRIP-qPCR data showing Myc enrichment in m6A-bound fraction in control and Mx1-cre; Mettl3fl/fl HSCs 10 days after pIpC treatment (n=3 for control, n=4 for Mx1-cre; Mettl3fl/fl). Data shown were normalized to the expression of Rplp0, a target found not to be methylated in HSCs by our meRIP-Seq analysis.
(b) Gene set enrichment analysis plots, as determined by HSC RNA-seq profiling of control and Mx1-cre; Mettl3fl/fl HSCs 10 days after pIpC treatment (n=3 for control and Mettl3Δ/Δ). Mettl3-deficient HSCs significantly lose Myc target genes. Analyses were completed on gene list ranked by log10 FDR and fold-change sign. P values were determined by the GSEA algorithm after 1,000 permutations. The statistics was computed using GSEA, and controlled for multiple comparisons by false discovery rate.
(c) Quantification of the mean fluorescence intensity (MFI) of GFP expression in LSK subsets from Mx1-cre; Mettl3fl/fl; Myc-GFP mice or Myc-GFP controls at least 10 days after pIpC treatment (n=4 for Myc-GFP controls, n=4 for Mx1-cre; Mettl3fl/fl; Myc-GFP). Values were normalized to GFP-negative control cells of the same population.
(d) Schematic of experimental design (top). Representative FACS plots of MYC-GFP expression in HSCs at steady-state (left) and after cytokine activation (right). Quantification of the mean fluorescence intensity (MFI) of GFP expression in HSCs after cytokine activation (n=7 for MYC-GFP controls, n=7 for Mx1-cre; Mettl3fl/fl; MYC-GFP). Values were normalized to GFP-negative control cells.
(e) Schematic of experimental design (left). Representative FACS plot (middle) and quantification (right) of the frequency of Mac-1+ cells in virally transduced colonies from sorted HSCs from Mx1-cre; Mettl3fl/fl mice 10 days after pIpC treatment (n=3 for pMIG, n=3 for pMIG-Myc).
All samples were from biologically independent animals. Values are shown as individual points with mean ± s.d. P values were determined by paired two-sided Student’s t-test.
The Myc protein level can be directly measured by a knockin Myc-GFP translational reporter in vivo37. To assess the impact of Mettl3 deletion on the level of Myc protein in HSCs, we generated Mx1-cre; Mettl3fl/fl; Myc-GFP mice. Consistent with prior publications32,33, HSCs express low levels of MYC-GFP compared to haematopoietic progenitors at steady state (Fig. 6c). Deletion of Mettl3 led to a modest significant reduction of MYC-GFP in HSCs (Fig. 6c). Remarkably, upon activation to differentiate, Mettl3-deficient HSCs failed to up-regulate MYC-GFP while control HSCs could readily do so (Fig. 6d). Mettl3 deletion did not lead to changes in HSC Myc transcript levels under the activation condition (Supplementary Fig. 6d). Given that m6A primarily regulates target mRNA stability and translation5,38, these data suggest that m6A is required for Myc mRNA translation in HSCs, particularly upon differentiation.
We tested whether forced expression of Myc can rescue the HSC differentiation defects caused by Mettl3 deletion. We transduced Mettl3-defiecent HSCs with Myc-expressing or control retrovirus (Fig. 6e and Supplementary Fig. 6f) and assessed their differentiation potential in vitro. Forced expression of Myc significantly rescued the differentiation defect of Mettl3-deficient HSCs compared with controls (Fig. 6e). These data indicate that Myc is one of the major functional targets of m6A in HSCs.
Discussion
We demonstrate that m6A is preferentially required for adult HSC differentiation in vivo and in vitro, but not mature myeloid cell maintenance or function. These results demonstrate that m6A is not a general cellular house-keeping mechanism, but rather a modification with specific roles in HSCs. Our results are in contrast to reports that show m6A inhibits differentiation of CD34+ human haematopoietic progenitor cells in vitro18,19. Since those experiments were performed using shRNAs on cells of mixed populations of HSCs and progenitors in vitro, it is not clear what the effects on human HSCs are in vivo. Of note, we observed differences between HSCs and progenitors in their response to Mettl3 deletion (Fig. 2 and Supplementary Fig. 3), suggesting that the roles of m6A in HSCs and progenitors need to be assessed independently. Consistent with our data, Yao et al recently showed that conditional deletion of Mettl3 led to accumulation of HSCs in the bone marrow while our paper was under review39.
In contrast to its role in enabling HSC differentiation, m6A inhibits differentiation in AML leukemic cells18,19,40. This surprising difference may reflect distinct mechanisms of action for m6A in leukemic cells and HSCs, and suggests that it may be possible to develop m6A-targeted therapies against leukemia cells without damaging HSC function. More work is needed to elucidate the differences in the role of m6A between leukemic cells and HSCs.
We found that HSC self-renewal and quiescence is largely intact after Mettl3 deletion (Figs. 2a and 3d). Thus, it appears that m6A specifically regulates HSC differentiation. m6A-mediated epitranscriptional regulation in HSCs may provide a fast and flexible mechanism to control mature blood cell output, allowing the organism to adapt to ever-changing physiological demands. Mettl3-deficient HSCs failed to establish bone marrow chimera in transplantation (Supplementary Fig. 4a and c), suggesting that m6A may regulate HSC homing and/or engraftment, which will be interesting to further investigate in the future.
Methods
Mice
Mx1-cre20, Lysm-cre26, Myc-GFP37 mice were obtained from the Jackson Laboratory and maintained on C57BL/B6 background. Rosa26-creER mice25 were generated and provided by Dr. T. Ludwig at Columbia University. Mettl3fl mice were generated by electroporation of a targeting vector (obtained from the Knockout Mouse Project Repository, KOMP) into V6.5 embryonic stem (ES) cells. Correctly targeted ES cell clone, validated by PCR and southern blot analysis, was identified and injected into BDF1 blastocysts and chimeric mice were generated. Chimeric mice were bred with C57BL/6 mice to obtain germline transmission. The resulting mice were crossed with Flpe mice41 to remove the Neo cassette. These mice were backcrossed at least 6 times onto C57BL/6 background before analysis. To induce Cre expression in Mx1-cre; Mettl3fl/fl mice, 6–8wk old animals were intraperitoneally injected with 10ug polyinosinic-polycytidylic acid (pIpC, GE or InvivoGen) in PBS every other day for 5 injections. Mice were analyzed either 10–14 days, 2–3 months, or 4 months after the final injection, as indicated in each experiment. To induce Cre expression in Rosa26-CreERT2; Mettl3fl/fl mice, recipients were given 1ug tamoxifen in 50uL corn oil by gavage every other day for 5 doses. All mice were housed in specific pathogen-free, Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-approved facilities at the Columbia University Medical Center. All protocols were approved by the Institute Animal Care and Use Committee of Columbia University. The study is compliant with all relevant ethical regulations regarding animal research.
Genotyping PCR Primers
The following primers were used for genotyping:
Mettl3fl allele: OLD810: GTTGATGAAATTATCAGTACAATGGTTCTGA; OLD811: GTAAAGAACAACTCTGGTTATCGTCATCG.
Mettl3 deletion allele: OLD812: GGATGATTTCTTCTACCATTACCTCTACCC;
OLD813: CAGAAAGCCCATCCTCAGCGTG.
5FU administration
For 5-fluorouracil (5-FU; Sigma-Aldrich) treatment, Mx1-cre; Mettl3fl/fl and littermate control mice were treated with pIpC as previously described. 10 days after the last pIpC injection, mice were injected intraperitoneally with 150 mg/kg 5-FU weekly until death or experimental conclusion.
Complete Blood Counts
Peripheral blood was collected by tail vein or cardiac puncture into EDTA-coated capillary tubes. Blood samples were analyzed by Genesis (Oxford Science Inc).
Flow cytometry
Bone marrow cells were isolated by flushing the long bones or by crushing the long bones, pelvis, and vertebrae with mortar and pestle in Ca2+ free and Mg2+ free HBSS with 2% heat-inactivated bovine serum. Splenic cells were obtained by crushing the spleens between two glass slides. The splenic and bone marrow cells were passed through a 25G needle several times and filtered through 70μm nylon mesh. The following antibodies were used to perform HSC staining: lineage markers (anti-CD2 (RM2–5), anti-CD3 (17A2), anti-CD5 (53–7.3), anti-CD8a (53–6.7), anti-B220 (6B2), anti-Gr-1 (8C5), anti-Ter119), anti-Sca-1 (E13–161.7), anti-c-kit (2B8), anti-CD48 (HM48–1), anti-CD150 (TC15–12F12.2). Additionally, the following antibodies were used to perform haematopoietic progenitor staining as previously described24: lineage markers (anti-CD2 (RM2–5), anti-CD3 (17A2), anti-CD5 (53–7.3), anti-CD8a (53–6.7), anti-B220 (6B2), anti-Gr-1 (8C5), anti-Ter119), anti-Sca-1 (E13–161.7), anti-c-kit (2B8), anti-CD34 (RAM34), anti-FLT3 (A2F10), anti-CD16/32 (93), anti-IL7Rα (A7R34).
Cell cycle, DNA damage, cell death, reactive oxygen species analysis.
For BrdU incorporation analysis, mice intraperitoneally injected with 0.1mg/kg BrdU and maintained on 0.5mg/ml BrdU water for 5 days before the analysis. The frequency of BrdU+ cells was determined by flow cytometry using an APC BrdU Flow Kit (BD Biosciences) per manufacturer’s instructions. To measure γH2AX levels, cells were stained for SLAM markers, fixed and permeabilized (BD Biosciences BrdU Flow Kit) and stained with anti-γH2AX (Biolegend, clone 2F3), followed by flow cytometry. Annexin V staining was performed using Annexin V Apoptosis Detection Kit APC (eBioscience) per manufacturer’s instructions. ROS levels were measured by DCFDA staining (29–79-dichlorofluorescein diacetate, ThermoFisher Scientific). After antibody staining, cells were incubated with 10uM DCFDA for 15 minutes at 37°C, followed by flow-cytometry.
Long-term competitive reconstitution assay
Adult recipient mice were lethally irradiated by a Cesium 137 Irradiator (JL Shepherd and Associates) at 300 rad/min with two doses of 540 rad (total 1080 rad) delivered at least two hours apart. Cells were transplanted by retro-orbital venous sinus injection of anesthetized mice. Mice were transplanted with either 5 × 105 donor and 5 × 105 competitor bone marrow cells or 50 sorted donor HSCs and 3 × 105 competitor bone marrow cells, as indicated. Donor mice were either untreated or sacrificed10 days after pIpC treatment, as indicated. Recipient mice were temporarily maintained on antibiotic water (Baytril 0.17g/L) for 14 days after transplantation. Peripheral blood from recipient mice was analyzed by flow cytometry at indicated timepoints for at least 16 weeks after transplantation to assess the level of donor-derived blood lineages (donor chimerism), including myeloid, B, and T cells. For post-transplantation Cre-induction experiments, recipients’ peripheral blood was analyzed over at least two time points at least 6–8 weeks after transplantation to ensure stable engraftment before treatment with pIpC (10ug per mouse per dose for 5 doses) or tamoxifen (1ug per mouse per dose for 5 doses). Blood was subjected to ammonium chloride potassium red cell lysis before antibody staining. Antibodies including anti-CD45.2 (104), anti-CD45.1 (A20), anti-CD3 (17A2), anti-B220 (6B2), anti-Gr1 (8C5), and anti-Mac1 (M1/70) were used to stain cells.
Colony-forming unit assay
10,000 bone marrow cells, 100,000 spleen cells, 200 LSK cells, single MPP, single HSCs or other haematopoietic progenitors were plated in methylcellulose culture medium (3434, STEMCELL Technologies) and incubated at 37°C. Colonies were counted 9–12 days after plating on an Olympus CKX41 microscope (Olympus) and scored as derived from erythroid progenitor cells (BFU-E and CFU-E), granulocyte-macrophage progenitor cells (CFU-GM, CFU-G and CFU-M), megakaryocyte progenitor cells (CFU-MK) and multipotent granulocyte, erythroid, macrophage and megakaryocyte progenitor cells (CFU-GEMM).
Viral transduction and colony-forming assay
Retroviruses were produced by transfecting 293T cells with pMIG-Myc (Addgene plasmid #18119) or pMIG vectors along with pCL-Eco and collecting media after 48–72hrs. Virus titers were determined by transduction of NIH-3T3 fibroblasts with serial dilutions of viral media. 200 HSCs were sorted from Mx1-cre; Mettl3fl/fl mice 10 days after pIpC treatment and plated into DMEM with 15% heat-inactivated fetal bovine serum, 100ng/ml SCF, 10ng/ml IL-3, 10ng/ml IL-6 and 50μM b-mercaptoethanol. Cells were transduced with >120 MOI retrovirus particles in the presence of 8ug/mL polybrene and spun at 1400g for 1.5hrs at 20°C. Cells were then collected and plated in 1.5mL of methylcellulose culture medium and incubated at 37°C for 9–12 days. Colonies were collected in bulk and resuspended in Ca2+- and Mg2+-free HBSS with 2% heat-inactivated bovine serum before staining with antibodies. Cells were then analyzed by flow cytometry.
Macrophage Culture and phagocytosis assay
One million whole bone marrow cells were plated in 12-well plates and cultured at 37°C in Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher) supplemented with 10% Fetal Bovine Serum (LifeTech) and 30% L929 supernatant. After 6 days, non-adherent cells were removed and macrophages were re-plated in fresh medium. Stimulation and phagocytosis assays were carried out on the 7th day.
For stimulation assays, 1ug/mL lipopolysaccharides (LPS; Sigma) were added to cells for 3 hours. Stimulated and unstimulated control cells were harvested into Trizol (Invitrogen) and RNA was isolated according to manufacturer’s instructions. Samples were reverse transcribed and subject to qPCR, as previously described.
For phagocytosis assays, cells were incubated for 1 hours at 37°C or 4°C at a multiplicity of infection (MOI) of 1∶50 with Fluoresbrite® carboxylate microspheres (Polyscience), washed in PBS with 1% FBS. Non-adherent cells and excess microspheres were removed by washing with DMEM before macrophages were collected and analyzed by flow cytometry.
qRT-PCR
Cells were sorted directly into Trizol. Total RNA was extracted according to manufacturer’s instructions. Total RNA was subjected to reverse transcription using SuperScript III (Invitrogen). Quantitative real-time PCR was run using SYBR green on a CFX Connect (Biorad) system. β-actin was used to normalize the RNA content of samples. Primer sequences: β-actin: GCTCTTTTCCAGCCTTCCTT; CTTCTGCATCCTGTCAGCAA. Mettl3: AAGGAGCCGGCTAAGAAGTC; TCACTGGCTTTCATGCACTC. Myc: TCTCCCCAAGGGAAGACGAT; TTGCTCTTCTTCAGAGTCGCT Tnfa: GGAACTGGCAGAAGAGGCACTC; GCAGGAATGAGAAGAGGCTGAGAC. Il6: CCTCTGGTCTTCTGGAGTACC; ACTCCTTCTGTGACTCCAGC. Il1b: GCACTACAGGCTCCGAGATGAAC; TTGTCGTTGCTTGGTTCTCCTTGT.
MeRIP-qPCR
poly(A)+RNA was isolated from whole bone marrow cells or sorted HSCs using Dynabeads Oligo-(dT)25 magnetic beads (ThermoFisher Scientific) according to manufacturer’s instructions. 1.25 μg of anti-m6A antibody (Synaptic Systems) was pre-bound to Protein A/G magnetic beads (Millipore) in IP buffer (20-mM Tris pH 7.5, 140-mM NaCl, 1% NP-40, 2-mM EDTA) for 1 h. Sample RNA was incubated with antibody-bound Protein A/G beads for 2 hours at 4 °C. Samples were washed twice in low-salt-wash buffer (10-mM Tris pH 7.5, 5 mM EDTA), twice with high-salt-wash buffer (20-mM Tris pH 7.5, 1-M NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1-mM EDTA) and twice with RIPA buffer (20-mM Tris pH 7.5, 150-mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1-mM EDTA). All wash solutions for each sample were collected as the “unbound” fraction. RNA was eluted from the beads by incubating with 50μl 20mM N6-methyladenosine 5-monophosphate sodium salt (Sigma-Aldrich) for 1hr at 4 °C. Following ethanol precipitation, input, unbound, and m6A-bound fraction RNA was reverse-transcribed using Superscript III (Invitrogen) with random hexamers. Enrichment of m6A-containing transcripts was determined by quantitative PCR relative to Rplp0 expression. The primer sequences for Rplp0: GATGGGCAACTGTACCTGACTG and CTGGGCTCCTCTTGGAATG.
RNA-seq and meRIP-seq
For RNA-seq, libraries were prepared according to the Smart-seq2 protocol as described27 from 150 sorted HSCs from Mx-1cre; Mettl3fl/fl or control mice 10 days after pIpC treatment. Three control and four Mx-1cre; Mettl3fl/fl biological replicate samples were sequenced by Illumina HiSeq 2500 with paired-end 100-bp read length.
For meRIP-seq, poly(A)+RNA was isolated from 3,000 sorted HSCs from C57BL/6J mice or Mx-1cre; Mettl3fl/fl mice 10 days after pIpC injection using Dynabeads Oligo-(dT)25 magnetic beads (ThermoFisher Scientific) according to manufacturer’s instructions. 1.25 μg of anti-m6A antibody (Synaptic Systems) or rabbit IgG (Jackson ImmunoResearch) was pre-bound to Protein A/G magnetic beads (Millipore) in IP buffer (20-mM Tris pH 7.5, 140-mM NaCl, 1% NP-40, 2-mM EDTA) for 1 h. Sample RNA was incubated with antibody-bound Protein A/G beads for 2 hours at 4 °C. Samples were washed twice in low-salt-wash buffer (10-mM Tris pH 7.5, 5 mM EDTA), twice with high-salt-wash buffer (20-mM Tris pH 7.5, 1-M NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1-mM EDTA) and twice with RIPA buffer (20-mM Tris pH 7.5, 150-mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1-mM EDTA). All wash solutions for each sample were collected as the “unbound” fraction. RNA was eluted from the beads by incubating with 50μl 20mM N6-methyladenosine 5-monophosphate sodium salt (Sigma) for 1hr at 4 °C. Following ethanol precipitation, RNA from input, m6A-antibody unbound, and m6A-antibody eluted, and IgG eluted fractions was reverse transcribed, amplified, tagmented and index according to the Smart-seq2 protocol27. Three biological replicate samples were sequenced by Illumina HiSeq 2500 or NextSeq 500 with paired-end 100-bp or 150-bp read length.
Analysis of RNA-seq and meRIP-seq data was performed using the Quantas pipeline (https://zhanglab.c2b2.columbia.edu/index.php/Quantas)42 with minor modifications. In brief, raw reads were mapped to the mouse genome and an exon-junction database using OLego43. Due to the low input during library preparation, potential PCR duplicates were removed using Picard (http://broadinstitute.github.io/picard). Exonic and junction reads were then counted to estimate transcript abundance (read per kilobase per million reads, or RPKM) and perform differential expression analysis using edgeR44.
Cumulative distribution analysis of RNA-seq log2 fold changes in expression between Mettl3-depleted and control HSCs was performed in R using the ecdf function. Groups were defined as m6A (meRIP-seq FDR<0.05, FC>2) or non-m6A (the rest of the genes), excluding genes with RNA-seq RPKM <1 for both Mettl3-depleted and control HSCs. Significance of difference between the cumulative distribution curves was determined using Kolmogorov–Smirnov test.
Statistics and Reproducibility
All experiments were repeated on biological replicates independently with similar results. The exact sample sizes for each experiment were indicated in figure legends. All statistics comparing two groups used two-sided Student t-test. The Kaplan-Meier estimation and two-sided log-rank test were used to compare survival difference between mouse groups. Statistical analyses were performed with Microsoft Excel or GraphPad Prism7. Statistical source data were included in Supplementary Table 3.
Reporting Summary
Further information regarding research design is available in the Nature Research Reporting Summary associated with this article.
Data Availability
Sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE123527. Source data have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgements
L.D. was supported by the Rita Allen Foundation and the National Heart, Lung and Blood Institute (R01HL132074). H.L. was supported by the Columbia Medical Scientist Training Program and the National Heart, Lung and Blood Institute (F30HL142196–01). The work by S.B., Y.Q. and C.Z. is in part supported by grants from the National Institute of Health (R01NS89676, R01GM124486, and R03HG009528). S.B. is supported by a Columbia Precision Medicine Research Fellowship. J.H.H. is supported by a generous gift from Ilana and Pascal Mantoux, and research grants from the: European Research Council, Kimmel Stem Cell Research Center, Flight Attendant Medical Research Council (FAMRI), Israel Science Foundation (ISF), Israel Cancer Research Fund (ICRF), New York Stem Cell Foundation (NYSCF). J.H.H. is a New York Stem Cell Foundation (NYSCF)–Robertson Investigator. We thank M. Kissner at the Columbia Stem Cell Initiative, S. Ho at the Columbia Center for Translational Immunology and A. Figueroa at the Department of Microbiology and Immunology for flow cytometry assistance. We thank I. Ivanov at the Department of Microbiology and Immunology for sharing the Lysm-cre mice and H. Snoeck for sharing the Mx1-cre mice. We thank C. Schindler, S. Ghosh and A. Lepelley at the Department of Microbiology and Immunology for helping macrophage assays. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696.
Footnotes
Declaration of Interests
The authors declare no competing financial interests.
References
- 1.Rossi L et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell stem cell 11, 302–317, doi: 10.1016/j.stem.2012.08.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison SJ & Scadden DT The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334, doi: 10.1038/nature12984 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desrosiers R, Friderici K & Rottman F Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America 71, 3971–3975 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer KD & Jaffrey SR The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15, 313–326, doi: 10.1038/nrm3785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer KD & Jaffrey SR Rethinking m(6)A Readers, Writers, and Erasers. Annual review of cell and developmental biology 33, 319–342, doi: 10.1146/annurev-cellbio-100616-060758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geula S et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006, doi: 10.1126/science.1261417 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Sledz P & Jinek M Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, doi: 10.7554/eLife.18434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, Doxtader KA & Nam Y Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Molecular cell 63, 306–317, doi: 10.1016/j.molcel.2016.05.041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578, doi: 10.1038/nature18298 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Xiang Y et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576, doi: 10.1038/nature21671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarwala SD, Blitzblau HG, Hochwagen A & Fink GR RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS genetics 8, e1002732, doi: 10.1371/journal.pgen.1002732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fustin JM et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806, doi: 10.1016/j.cell.2013.10.026 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Chen T et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell stem cell 16, 289–301, doi: 10.1016/j.stem.2015.01.016 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Batista PJ et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell stem cell 15, 707–719, doi: 10.1016/j.stem.2014.09.019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon KJ et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 171, 877–889 e817, doi: 10.1016/j.cell.2017.09.003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nature neuroscience 21, 195–206, doi: 10.1038/s41593-017-0057-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276, doi: 10.1038/nature23883 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Vu LP et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nature medicine 23, 1369–1376, doi: 10.1038/nm.4416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng H et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell stem cell, doi: 10.1016/j.stem.2017.11.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn R, Schwenk F, Aguet M & Rajewsky K Inducible gene targeting in mice. Science 269, 1427–1429 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216, doi: 10.1038/nature25168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrelha J et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 554, 106–111, doi: 10.1038/nature25455 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Ding L, Saunders TL, Enikolopov G & Morrison SJ Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462, doi: 10.1038/nature10783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding L & Morrison SJ Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235, doi: 10.1038/nature11885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Luca C et al. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. The Journal of clinical investigation 115, 3484–3493, doi: 10.1172/JCI24059 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausen BE, Burkhardt C, Reith W, Renkawitz R & Forster I Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic research 8, 265–277 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nature protocols 9, 171–181, doi: 10.1038/nprot.2014.006 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forsberg EC et al. Molecular signatures of quiescent, mobilized and leukemia-initiating hematopoietic stem cells. PloS one 5, e8785, doi: 10.1371/journal.pone.0008785 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He S, Kim I, Lim MS & Morrison SJ Sox17 expression confers self-renewal potential and fetal stem cell characteristics upon adult hematopoietic progenitors. Genes & development 25, 1613–1627, doi: 10.1101/gad.2052911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanova NB et al. A stem cell molecular signature. Science 298, 601–604, doi: 10.1126/science.1073823 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Reavie L et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat Immunol 11, 207–215, doi: 10.1038/ni.1839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehninger A et al. Posttranscriptional regulation of c-Myc expression in adult murine HSCs during homeostasis and interferon-alpha-induced stress response. Blood 123, 3909–3913, doi: 10.1182/blood-2013-10-531038 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Wilson A et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes & development 18, 2747–2763, doi: 10.1101/gad.313104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA & Dang CV An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4, R69, doi: 10.1186/gb-2003-4-10-r69 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740, doi: 10.1093/bioinformatics/btr260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang CY, Bredemeyer AL, Walker LM, Bassing CH & Sleckman BP Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. European journal of immunology 38, 342–349, doi: 10.1002/eji.200737972 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Yue Y, Liu J & He C RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes & development 29, 1343–1355, doi: 10.1101/gad.262766.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao QJ et al. Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res 28, 952–954, doi: 10.1038/s41422-018-0062-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbieri I et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131, doi: 10.1038/nature24678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez CI et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature genetics 25, 139–140, doi: 10.1038/75973 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Yan Q et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proceedings of the National Academy of Sciences of the United States of America 112, 3445–3450, doi: 10.1073/pnas.1502849112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Anczukow O, Krainer AR, Zhang MQ & Zhang C OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic acids research 41, 5149–5163, doi: 10.1093/nar/gkt216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE123527. Source data have been provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.