Abstract
Fragile X syndrome results from a loss of the RNA-binding protein fragile X mental retardation protein (FMRP). How FMRP regulates neuronal development and function remains unclear. Here, we show that FMRP-deficient immature neurons exhibit impaired dendritic maturation, altered expression of mitochondrial genes, fragmented mitochondria, impaired mitochondrial function, and increased oxidative stress. Enhancing mitochondrial fusion partially rescued dendritic abnormalities in FMRP-deficient immature neurons. We show that FMRP deficiency leads to reduced Htt mRNA and protein levels and that HTT mediates FMRP regulation of mitochondrial fusion and dendritic maturation. Mice with hippocampal Htt knock-down and Fmr1 knockout mice showed similar behavioral deficits that could be rescued by treatment with a mitochondrial fusion compound. Our data unveil mitochondrial dysfunction as a contributor to the impaired dendritic maturation of FMRP-deficient neurons and suggest a role for interactions between FMRP and HTT in the pathogenesis of Fragile X syndrome.
Introduction
Fragile X syndrome (FXS) is the most common heritable cause of intellectual disability and the highest single-gene contributor to autism1. FXS patients exhibit extensive behavioral deficits including impaired executive functions, defective learning, heightened anxiety, and impaired social ability1, 2. FXS arises largely from mutations in the fragile X mental retardation (FMR1) gene resulting in deficiency of fragile X mental retardation protein (FMRP), a brain and neuron-enriched RNA binding protein3. How FMRP deficiency impairs brain function remains unclear. Studies of postmortem adult human FXS brains have identified several neuropathological features including immature dendritic spines of neurons4. However, whether this deficit results from impaired development or altered plasticity during adulthood remains unclear. It has been shown that neurons differentiated from FXS patient-derived induced pluripotent stem cells (iPSCs) display impaired neurite extension5. In addition, both forebrain neurons differentiated from FXS human embryonic stem cells6, 7 and hippocampal neurons isolated from neonatal Fmr1 knockout (KO) mice8, 9 exhibit deficits in synaptic plasticity and neurite extension. Furthermore, we have shown that FMRP-deficient adult hippocampal new neurons also exhibit impaired dendritic and spine maturation9. Therefore, FMRP seems to have important roles in neuronal maturation across neuronal types and developmental ages. However, despite the identification of many mRNAs regulated by FMRP3,10,11, how FMRP deficiency affects neuronal development remains unclear.
Neurons depend on mitochondria, which not only provide energy to power cellular function through oxidative phosphorylation, but also regulate cellular oxidation-reduction status, calcium levels, signal transduction, and apoptosis12. During embryonic development, mitochondria are important for neural progenitor proliferation13 and neuronal survival14. Alterations in mitochondrial morphology and function directly impact morphological development of neurons15. In adult brains, decreased mitochondrial ATP production impairs dendritic maturation of adult-born hippocampal neurons16. A large number of neurodegenerative diseases have been associated with disruptions of mitochondrial function17. However, it remains unexplored whether mitochondrial dysfunction contributes to pathogenesis of FXS. A limited number of studies have shown metabolic changes in the brain of Fmr1 KO mice, including increased rate of glucose metabolism18, elevated metabolic and oxidative stress19, increased ROS production, and abnormal nitric oxide metabolism20, 21, as well as altered energy metabolism at the systemic level22. FMRP promotes protein translation of Superoxide Dismutase, a regulator of oxidative stress23. However, whether FMRP deficiency affects mitochondrial function and its implication for neuronal development remain unknown.
In the present study, we report that FMRP has a critical role in dendritic maturation of adult new neurons, neonatal hippocampal neurons, and human neurons developed in transplanted mouse brains. We discover that FMRP-deficient immature neurons exhibited altered expression of mitochondrial genes, fragmented mitochondria, impaired mitochondrial function, and increased oxidative stress. Enhancing mitochondria fusion by either a chemical activator or exogenous expression of mitochondrial fusion genes rescued both mitochondrial morphology and dendritic maturation deficits of FMRP-deficient neurons. We discovered that FMRP deficient neurons had reduced HTT levels and acute knockdown of HTT recapitulates both mitochondrial fusion and neuronal maturation deficits seen in Fmr1 KO neurons. We used guide RNAs to target modified CRISPR/Cas9 (dCas9VP64-SAM) to selectively activate the endogenous Htt gene in neurons and show that increased Htt transcription rescued both mitochondrial fusion and dendritic maturation deficits of Fmr1 KO neurons. Finally, we show that mice with HTT knockdown in the hippocampus exhibit several behavioral deficits similar to Fmr1 mutant mice and treatment with a mitochondrial fusion compound rescued behavioral deficits of both Fmr1 KO mice and mice with hippocampal knockdown of HTT. Our data demonstrate that mitochondrial dysfunction contributes to the impaired maturation of FMRP-deficient developing neurons and present a crosstalk between FMRP and HTT in pathogenesis of human diseases.
Results
Selective deletion of FMRP from immature neurons leads to impaired transition into mature neurons.
We have shown that deletion of FMRP from NSCs in the adult DG leads to reduced neurogenesis and cognitive functions which can be rescued by restoring FMRP functions in adult NSCs either genetically or pharmacologically24-27. However, the specific role of FMRP during maturation of adult-born neurons remains unexplored. Doublecortin (DCX) is a microtubule-associated protein required for neuronal migration and is exclusively expressed in neuronal precursor cells and immature neurons. In the adult DG, neuronal precursors begin to express DCX while actively dividing, and continue to express DCX for 2 to 3 weeks as they develop into mature neurons that express neuronal nuclear antigen (NeuN, gene product of Rbfox3)28. We generated tamoxifen (TAM)-inducible conditional FMRP knock out mice (cKO;Cre;tdT or cKO) and littermate controls (Ctrl;Cre;tdT or control) by crossing Dcx promoter-driven inducible Cre (Dcx-CreERT2) mice, Cre-dependent tdTomato (tdT) reporter mice, and heterozygote female Fmr1-floxed (Fmr1loxP/+) mice (Fig. 1a and Supplementary Fig. 1a, b). We injected 6-weeks old cKO and control mice with TAM and analyzed at 3, 7, 14, 28, or 56 days after injections, corresponding to key developmental stages of adult new neurons28 (Fig. 1b). At 3 days after the last TAM injection, a majority of DCX+ cells in the DG of both cKO and control mice were tdT+ (Fig. 1e) which persisted to at least day 56. FMRP expression was detected in the tdT+DCX+ cells of the control mice but not the cKO mice (Fig. 1d, Supplementary Fig. 2), indicating that TAM injection resulted in efficient Cre-mediated recombination in DCX+ population. There is no detectable tdT expression in the DG of mice without Cre (Supplementary Fig. 3). The expression of tdT was found in the proliferating cells (Ki67+, Supplementary Fig. 1c) and neuroblasts (TBR2+, Supplementary Fig. 1d) but not in radial glia-like NSCs (RGLs, NESTIN+GFAP+) (Supplementary Fig. 1e), astrocytes (S100β+), or mature neurons (NeuN+) (Supplementary Fig. 1f). Therefore FMRP was specifically deleted in DCX+ cells.
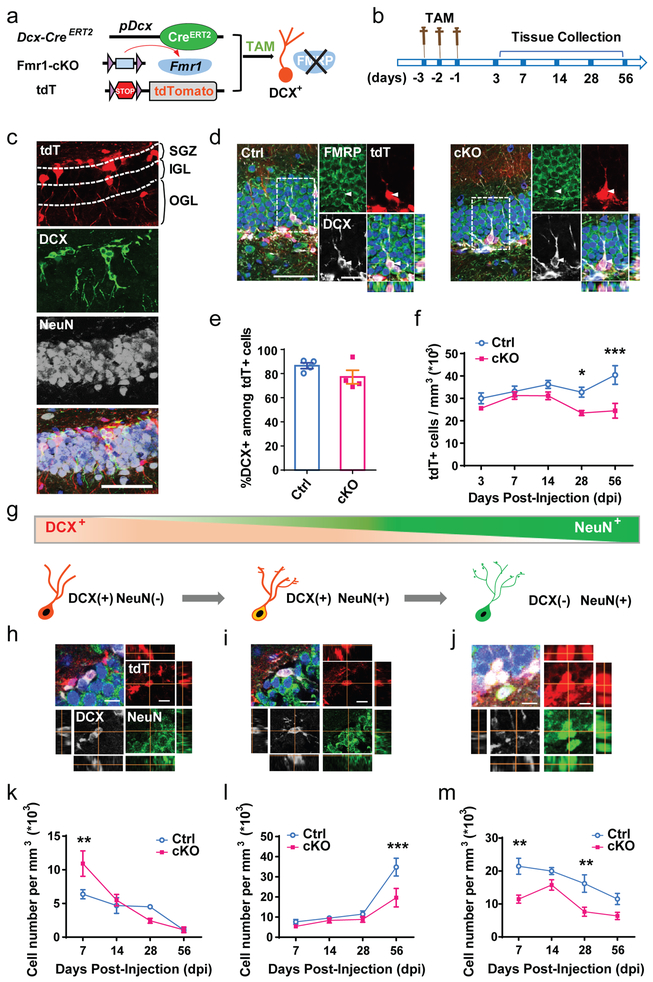
Figure 1: Specific deletion of FMRP from immature neurons leads to reduced neuronal number.
a, Schematic diagram showing generation of inducible conditional FMRP deletion in immature neurons by administration of tamoxifen. b, Experimental scheme for assessing the impact of specific deletion of FMRP from immature neurons on neuronal number in the DG. c, Representative confocal images (from 3 independently repeated experiments with similar results) used for the following analysis including new-born neuronal number and specific stages of development in the DG. The boundaries of SGZ, IGL and OGL were marked by the dashed line. Red, tdT; green, DCX; white, NeuN. Scale bars, 50 μm. d, Representative confocal images (from 3 independently repeated experiments with similar results) of FMRP (green), DCX (white) staining of brain sections from Ctrl;Cre;tdT (left) and cKO;Cre;tdT (right) mice. Scale bars, 20 μm. e, Percentage of DCX+ in total tdT+ cells in DG of cKO;Cre;tdT mice and Ctrl;Cre;tdT mice at Day 7 after tamoxifen injection (Student’s t-test, two-sided, t(6) = 1.529, P = 0.1771. Ctrl: 86.54 ± 2.393, n = 4 mice; cKO: 77.19 ± 5.629, n = 4 mice). f, Quantitative comparison of tdT+ neuronal numbers in the SGZ of cKO;Cre;tdT mice and Ctrl;Cre;tdT mice. (Two-way ANOVA with Bonferroni post hoc test, two-sided: F(4,40) = 2.617, P = 0.0493. Day 3: Ctrl: 30.01 ± 2.446, n = 5; cKO: 25.61 ± 0.7324, n = 6. Day 7: Ctrl: 33.11 ± 2.370, n = 5; cKO: 31.28 ± 1.660, n = 4. Day 14: Ctrl: 36.24 ± 1.753, n = 5; cKO: 31.14 ± 1.727, n = 5. Day 28: Ctrl: 32.76 ± 2.234, n = 6; cKO: 23.47 ± 1.140, n = 5. P = 0.0246. Day 56: Ctrl: 40.42 ± 4.207, n = 5; cKO: 24.48 ± 3.296, n = 4. P = 0.0002). g, Schematic diagram showing the cell markers across stages of maturation that were used: immature DCX-only: DCX+NeuN-; transitioning neurons: DCX+NeuN+; mature neurons: DCX-NeuN+. h-j, Representative confocal images (from 3 independently repeated experiments with similar results) of neurons for analysis of specific stages of development in the SGZ and IGL. h, DCX+NeuN-; i, DCX+NeuN+; j, DCX-NeuN+. Red, tdT; green, NeuN; white, DCX. Scale bars, 20 μm. k-m, Quantitative comparison of specific stages of development: k, DCX+NeuN- (Two-way ANOVA with Bonferroni post hoc test, two-sided, F(3,27) = 4.243, P = 0.014; Day 7: Ctrl: 6.370 ± 0.6726, n = 4; cKO: 10.91 ± 1.899, n = 4. P = 0.0088. Day 14: Ctrl: 4.693 ± 1.169, n = 5; cKO: 5.496 ± 0.8511, n = 5. Day 28: Ctrl: 4.505 ± 0.2918, n = 4; cKO: 2.426 ± 0.3657, n = 4. Day 56: Ctrl: 1.049 ± 0.3171, n = 5; cKO: 1.083 ± 0.4259, n = 4. ; l, DCX+NeuN+ (Two-way ANOVA with Bonferroni post hoc test, two-sided, F(3,27) = 3.475, P = 0.0297; Day 7: Ctrl: 7.636 ± 1.176, n = 4; cKO: 5.403 ± 0.4007, n = 4. Day 14: Ctrl: 9.523 ± 0.6073, n = 5; cKO: 8.345 ± 1.243, n = 5. Day 28: Ctrl: 11.47 ± 1.585, n = 4; cKO: 8.848 ± 1.424, n = 4. Day 56: Ctrl: 1.049 ± 0.3171, n = 5 34.82 ± 4.433, n = 5; cKO: 19.65 ± 4.579, n = 4. P = 0.0009); m, DCX-NeuN+ (F(3,27) = 1.292, P = 0.297; Day 7: Ctrl: 21.45 ± 2.428, n = 4; cKO: 11.46 ± 1.229, n = 4. P = 0.0019. Day 14: Ctrl: 19.99 ± 1.064, n = 5; cKO: 15.77 ± 1.573, n = 5. Day 28: Ctrl: 16.18 ± 2.672, n = 4; cKO: 7.635 ± 1.388, n = 4. P = 0.0087. Day 56: Ctrl: 11.51 ± 1.706, n = 5; cKO: 6.372 ± 1.110, n = 4) of tdT+ neuronal numbers in the SGZ of cKO;Cre;tdT mice and Ctrl;Cre;tdT mice. *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M.
We quantified the number of tdT+ cells in the subgranular zone (SGZ, Fig. 1c) and the internal granular layer (IGL, Supplementary Fig. 4a), where the cell bodies of immature neurons are typically located24,26. The cKO mice did not show significant change in the number of tdT+ new cells from day 3 to day 14 post TAM injection. There was no significant difference in Ki67+ dividing population between genotypes at day 3 (Supplementary Fig. 4b), suggesting that FMRP deletion did not affect proliferation and generation of DCX+ neuroblasts. However, cKO mice had a significantly lower number of tdT+ cells at day 28 and day 56 compared to control mice (Fig. 1f), without change in the overall volume of the DG (Supplementary Fig. 4c). Adult born DG neurons switch from expressing DCX to expressing NeuN during maturation28; therefore DCX and NeuN expression have been used to assess stages of new neuron development (Fig. 1g). We found that the number of DCX-only immature cells (DCX+NeuN-, Fig. 1h) increased significantly in the SGZ of cKO mice at day 7 but exhibited no significant change after day 14, compared to the control mice (Fig. 1k). The number of immature neurons that are transitioning from immature to mature stage (DCX+NeuN+, Fig. 1i) did not show significant differences between genotypes until day 56 when cKO mice exhibited significantly reduced DCX+NeuN+ cells compared to controls (Fig. 1l). On the other hand, the number of NeuN-only mature neurons (DCX-NeuN+, Fig. 1j) were substantially lower in the cKO compared with the control mice from day 7 to day 56 with statistically significant reductions at both day 7 and day 28 (Fig. 1m). These data suggest that selective deletion of FMRP from DCX-expressing immature neurons led to initially transient increase of immature neurons but significant reduction in final stage of maturation leading to reduced number of mature new neurons in the DG.
FMRP-deficient DCX immature neurons in the adult DG exhibited impaired maturation
Immature DG new neurons undergo morphological changes during maturation28. Our previous studies have shown that deletion of FMRP from NESTIN-expressing NSCs leads to reduced dendritic maturation of DG new neurons9,24. However, it is unclear whether this deficit results from impaired NSC differentiation26,29 or defective neuronal maturation. We therefore injected retrovirus expressing Cre driven by a Dcx promoter (Retro-pDcx-Cre) together with retrovirus expressing Cre-dependent GFP (Retro-Flip-GFP) into the DG of FMRP-cKO (Fmr1loxP/y) and FMRP-cON (Fmr1loxP–Neo/y) mice that express less than 10% FMRP in the absence of Cre24 (Fig. 2a-c). Retroviruses transduce only dividing cells, but Cre will be active only after the infected NSCs have differentiated into DCX+ immature neurons. At 28 days after viral injection, GFP+ neurons in cKO mice exhibited reduced FMRP expression whereas those cON mice exhibited increased expression (Supplementary Fig. 5). The DCX+ newborn neurons in cKO mice showed significant reductions in dendritic complexity, total dendritic length, number of branching points, and number of dendritic ends compared to GFP+ neurons in WT mice (Fig. 2d-h). On the other hand, restoration of FMRP selectively in DCX+ new neurons in the cON mice rescued dendritic morphology deficits (Fig. 2e-h). Therefore, FMRP expression in DCX+ newborn neurons is crucial for dendritic maturation.
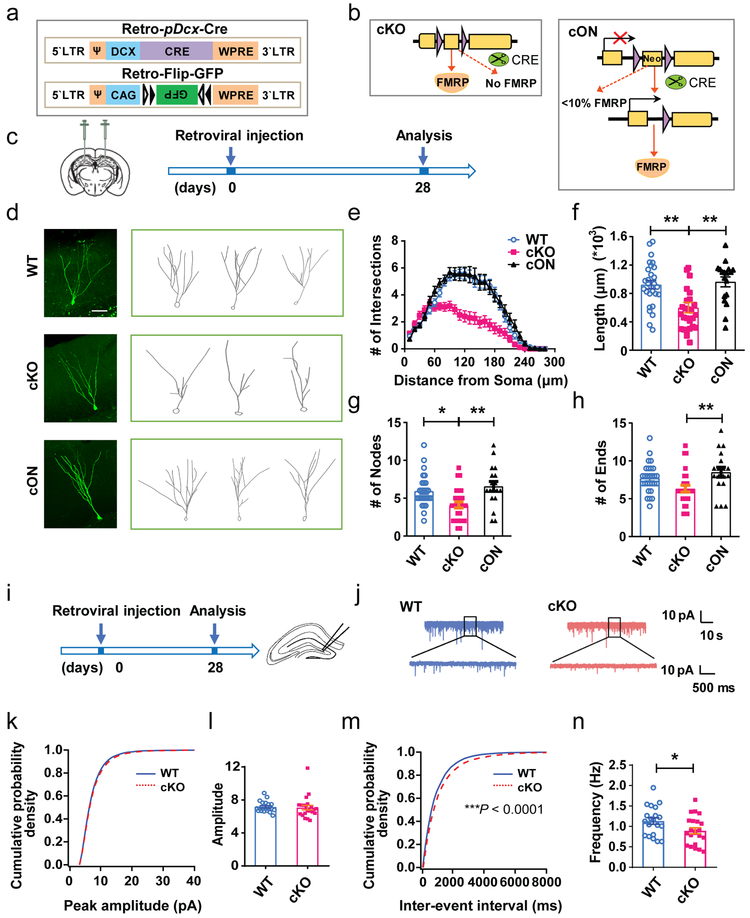
Figure 2: FMRP-deficient DCX+ immature neurons in the adult DG exhibited impaired maturation.
a, Schematic illustration of dual retroviral vectors expressing Cre driven by DCX promoter (Retro-DCX-Cre) and expressing Cre-dependent GFP (Retro-Flip-GFP) to infect DCX+ cells. b, Schematic diagram showing selective deletion or restoration of FMRP using FMRP-cKO (left) and FMRP-cON (right) mice combined with Cre virus–mediated recombination. c, Experimental scheme for assessing the impact of specific deletion of FMRP from immature neurons on dendritic maturation. d, Representative confocal images (from 3 independently repeated experiments with similar results) and Neurolucida software-created traces of GFP+ neurons in WT (top), cKO (middle) and cON (bottom) mice at 28 d post-viral injection. Scale bar, 50 μm. e-f, Dendritic complexity analysis of GFP+ neurons in WT and cKO and cON mice. e, Scholl analysis (Multi-ANOVA, F(2,72) = 16.024, P < 0.0001. WT vs. cKO: P < 0.0001, WT vs. cON: P = 0.772, cKO vs. cON: P < 0.0001. WT: n = 28 neurons in 5 mice, cKO: n = 27 neurons in 4 mice, cON: n = 18 neurons in 3 mice); f, Dendritic length (one-way ANOVA with Bonferroni post hoc test: F(2,70) = 8.842, P = 0.0004; Bonferroni post hoc test, WT vs. cKO: P = 0.0018; WT vs. cON: P > 0.9999; cKO vs. cON: P = 0.0019. WT: 0.9240 ± 0.05876, n = 28 neurons in 5 mice, cKO: 0.6010 ± 0.07261, n = 27 neurons in 4 mice, cON: 0.9645 ± 0.06987, n = 18 neurons in 3 mice). g, Dendritic nodes (one-way ANOVA with Bonferroni post hoc test: F (2, 70) = 7.437, P = 0.0012; Bonferroni post hoc test, WT vs. cKO: P = 0.0123, WT vs. cON: P> 0.9999, cKO vs. cON: P = 0.0022. WT: 5.929 ± 0.3948, n = 28 neurons in 5 mice, cKO: 4.111 ± 0.4077, n = 27 neurons in 4 mice, cON: 6.556 ± 0.6428, n = 18 neurons in 3 mice). h, Dendritic ends (one-way ANOVA with two-sided Bonferroni post hoc test: F (2, 70) = 5.169, P = 0.0081; Bonferroni post hoc test, WT vs. cKO: P = 0.0902, WT vs. cON: P = 0.7935; cKO vs. cON: P = 0.0089. WT: 7.714 ± 0.3668, n = 28 neurons in 5 mice, cKO: 6.333 ± 0.4498, n = 27, n = 27 neurons in 4 mice, cON: 8.500 ± 0.6530, n = 18 neurons in 3 mice). i, Experimental scheme for assessing the impact of specific deletion of FMRP from immature neurons on synaptic transmission of DG newborn neurons. j, Representative traces of mEPSCs recorded from GFP+ DG neurons in acute hippocampal slices derived from WT and cKO mice 4 weeks after retroviral injection. (7 independently repeated experiments with similar results). k-l, Cumulative probability distribution (WT, n = 20 cells from 7 mice; cKO, n = 22 cells from 8 mice. P = 0.084, nonparametric Wilcoxon Rank test, two-sided) and average amplitude (Student’s t-test, two-sided, t(40) = 0.2630, P = 0.7936. WT: 7.154 ± 0.1483; cKO: 7.068 ± 0.2818) of mEPSC. m-n, Cumulative probability distribution (WT, n = 20 cells; cKO, n = 22 cells. P < 0.0001, nonparametric Wilcoxon Rank test, two-sided) and average frequency (Student’s t-test, two-sided, t(40) = 2.191, P = 0.0344. WT: 1.132 ± 0.08012; cKO: 0.8934 ± 0.07382) of mEPSC. *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M
To assess the functional impact of FMRP deletion in immature neurons on synaptic transmission of DG newborn neurons, we injected retrovirus expressing neuronal Synapsin promoter driven Cre into the DG of adult cKO mice. At 4 to 5 weeks after viral injection, we recorded evoked action potential (AP) and pharmacologically isolated spontaneous miniature excitatory postsynaptic currents (mEPSCs) of GFP+ neurons (Fig. 2i, j). We found no significant differences in the intrinsic properties, action potential firing, resting membrane potential (RMP), membrane capacitance, input resistance, and membrane time constant of GFP+ newborn DG cells between cKO and control mice (Supplementary Fig. 6a-g). In addition, we found no significant difference in the amplitude (Fig. 2k, l), rise time (Supplementary Fig. 6h) and time constant of decay (Supplementary Fig. 6i) of mEPSCs between GFP+ cells from cKO and WT mice (Fig. 2j). However the frequency of mEPSCs recorded from GFP+ DG cells in cKO mice was significantly decreased, as compared with those in WT mice (Fig. 2m, n). These results suggest that FMRP deficiency in immature neurons impairs their ability to receive excitatory transmission which may result from dendritic maturation deficits. Therefore, FMRP is essential for proper neuronal maturation.
FMRP-deficient developing neurons have altered expression of metabolic genes
To investigate the molecular mechanism underlying FMRP-regulated maturation of newborn neurons, we crossed Fmr1 KO mice with Dcx-DsRed transgenic mice in which DCX+ cells express DsRed30. We isolated DsRed+ cells from the DG of 6-week-old Fmr1-/y;Dcx-DsRed and control Fmr1+/y;Dcx-DsRed littermates using fluorescence activated cell sorting (FACS; Fig. 3a, b). Consistent with our previous observation30, FACS-isolated DsRed+ cells expressed high levels of Dcx and DsRed mRNA, but not Nestin or Rbfox3 mRNA (Supplementary Fig. 7). Transcriptomic analysis of DsRed+ cells of two pairs of KO and WT samples yielded 519 differentially expressed (DE) genes (FDR-adjusted P < 0.05) (Supplementary Table 1a, b). These 519 genes were enriched in pathways involved in metabolism, organelle organization, synaptic transmission, wounding, intracellular transport and cell development (Fig. 3c-d, Supplementary Table 2). Among them, the metabolic pathways containing 146 (21.9 %) DE genes ranked second among all pathways (Fig. 3d, Supplementary Table 3a).
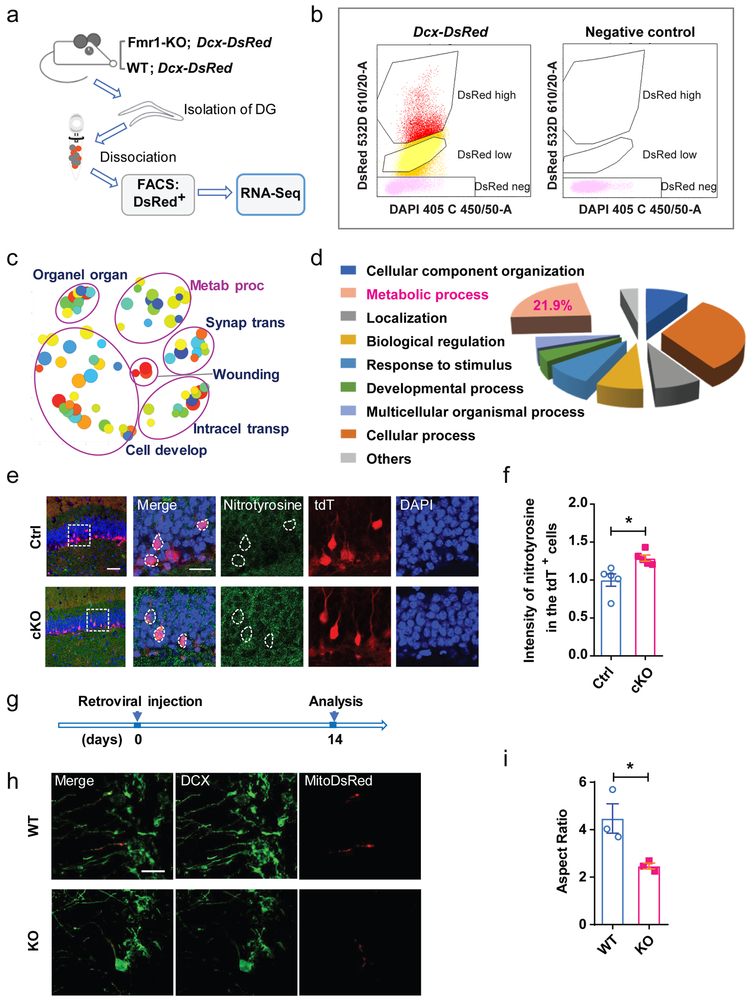
Figure 3: Metabolic process related genes changed in FMR1-deficient developing neurons.
a, Experimental workflow of FACS-seq showing dissection of adult DG, direct cell isolation using FACS, RNA-seq, and bioinformatics analysis. b, Example of sorting gates used to separate for DsRed+ and DsRed− single cells dissociated from DG tissue. For each sorting, gates were drawn based on the profile of a WT mouse that did not express DsRed. (3 independently repeated experiments with similar results). c, REVIGO scatterplot for GO terms cluster representatives for changed genes in Fmr1 KO Dcx-DsRed cells. Abbreviation, full name: Organ organ, Organelle organization; Metab proc, Metabolic process; Synap trans, Synaptic transmission; Intracel transp, Intracellular transport; Cell develop, Cell development. d, A GO biological process was used to categorize differentially expressed genes in Fmr1 KO Dcx-DsRed cells by PATHER. e, Sample confocal images of nitrotyrosine (green) staining of brain sections from Ctrl;Cre;tdT (left) and cKO;Cre;tdT (right) mice (day 28 after TAM injection). 3 independently repeated experiments with similar results. Scale bars, 50 μm. f, The expression level of nitrotyrosine in the tdT+ cells of the DG from cKO; Cre; tdTomato mice (Student’s t-test, two-sided, t(8) = 3.191, P = 0.0128. Ctrl: 1.000 ± 0.08076, n = 5 mice; cKO: 1.288 ± 0.04050, n = 5 mice). g, Experimental scheme for assessing the mitochondrial morphology in the DCX+ immature neurons of DG from Fmr1 KO and WT mice. h, Representative confocal images (from 3 independently repeated experiments with similar results) of mitochondria in the DCX+ immature neurons in the DG of Fmr1 KO and WT mice: green, DCX; Red, mitoDsRed. Scale bar, 50 μm. i, Quantitative comparison of mitochondrial aspect ratio of the DCX+ immature neurons of DG from Fmr1 KO and WT mice (Student’s t-test, two-sided, t(4) = 3.146, P = 0.0337. WT: 4.472 ± 0.6183, n = 3 mice; KO: 2.468 ± 0.1255, n = 3 mice). *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M. .
Neuronal metabolism is highly dependent on mitochondrial functions12. We confirmed that several DE genes involved in mitochondrial functions had altered levels in hippocampal neurons isolated from Fmr1 KO mice (Supplementary Fig 8). The impact of FMRP deficiency on mitochondria has not been determined. We therefore assessed the levels of nitrotyrosine, a marker for oxidative stress. We found that the tdT+ new adult DG neurons in cKO;Cre;tdT mice exhibited increased nitrotyrosine level compared with those in the control mice (Fig. 3e, f). To visualize mitochondria in DG newborn neurons, we injected retrovirus carrying mitochondria-targeted DsRed (Retro-pCAG-mitoDsRed)16 into the DG of 6-week-old Fmr1 KO and WT mice and assessed the mitochondrial morphology at 14 days after viral injection (Fig. 3g). The mitochondria in the proximal dendrites of DCX+ immature neurons of the KO mice were significantly shorter (reduced aspect ratio) compared with those in WT mice (Fig. 3h, i), suggesting that the mitochondria were more fragmented in FMRP-deficient immature neurons. Therefore, deletion of FMRP from DCX-expressing immature neurons significantly impacts metabolic processes which may result from mitochondrial dysfunction.
FMRP-deficient DCX+ immature neurons have impaired mitochondrial fusion
To determine whether FMRP deficiency affects mitochondrial morphology in developing neurons and identify the developmental time points of this effect, we transfected primary hippocampal neurons8,9 with vectors expressing mitochondrial targeting GFP (mitoGFP) and an shRNA against Fmr1 (shFmr1)9 at 4, 6, and 9 days after plating (DIV) and analyzed on DIV 15 (Supplementary Fig. 9). We found that shFmr1-transfected neurons had more fragmented mitochondria compared to controls (shNC) at all time points, with neurons transfected at DIV 4 exhibiting the most profound changes, suggesting that the impact of FMRP knockdown on mitochondrial shape occurred early during neuronal maturation. We therefore focused on the early time point (DIV4) for the remaining studies.
Next, we assessed mitochondria in neurons isolated from Fmr1 KO mice transfected with mitoDsRed and cytosolic GFP. All mitoDsRed+ cells were DCX+ (Supplementary Fig. 10a). We found no significant difference in either mitochondrial density or coverage area between KO and WT neurons (Supplementary Fig. 10b, c). However, mitochondria in Fmr1 KO neurons exhibited significantly reduced aspect ratio compared with those in WT neurons (Fig. 4a, b), similar to our observation in the adult DG (Fig. 3). We then assessed the function of mitochondria by examining the mitochondrial membrane potential (MMP) and KO neurons exhibited significantly reduced MMP compared to WT neurons (Fig. 4c,d). Recently, discrepancies between findings in mouse and human have raised extensive discussion1. To evaluate the development of human FXS neurons, we transplanted neural progenitors (NPCs) differentiated from control and FXS patient derived iPSCs5, 31 into the cortex of immune-deficient mouse brains and analyzed at 4 months post-transplantation. We found that FXS human neurons developed in transplanted mouse brains also exhibited impaired dendritic maturation and increased oxidative stress (Supplementary Fig. 11). Hence, our data suggest that FMRP deficiency impairs mitochondrial and metabolic functions in both mouse and human neurons.
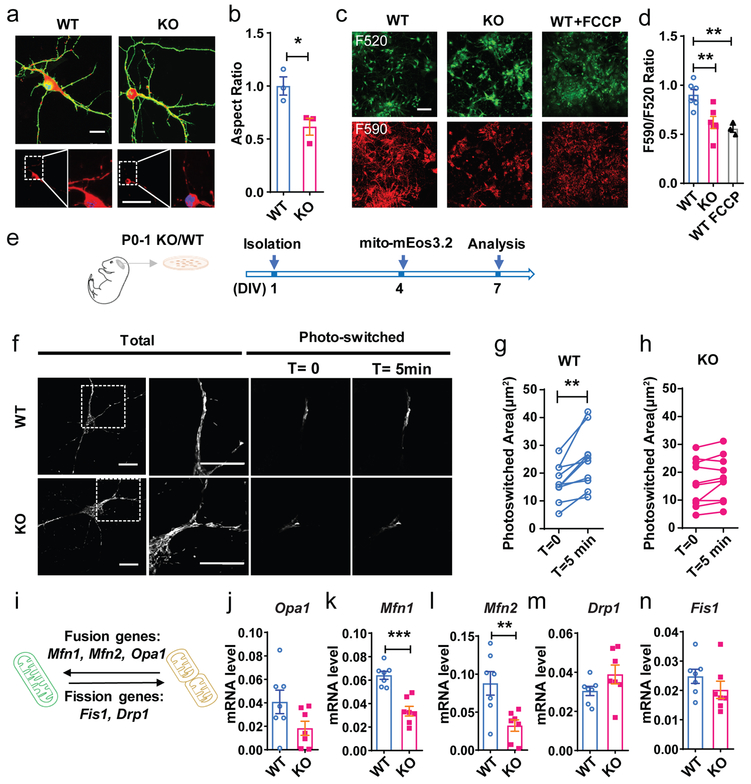
Figure 4: Impaired mitochondrial fusion in FMRP-deficient DCX+ immature neurons.
a, Representative confocal images (from 3 independently repeated experiments with similar results) and 3D reconstruction of mitochondria in the Fmr1 KO and WT hippocampal neurons. Scale bar, 5 μm. b, Mitochondrial aspect ratio (Normalized to WT. Student’s t-test, two-sided, t(4) = 3.268, P = 0.0308. WT: 1.000 ± 0.08617, n = 3 mice; KO: 0.6158 ± 0.07997, n = 3 mice). c, Representative images (from 3 independently repeated experiments with similar results) of JC-10 dye-stained Fmr1 KO and WT neurons, and WT neurons with FCCP treatment at DIV 7. Scale bar, 50 μm. d, Quantification of the ratio of red (F590) and green (F520) fluorescence in Fmr1 KO and WT neurons (Student’s t-test, two-sided, WT vs. KO: t(10) = 3.474, P = 0.006; WT vs. WT+FCCP: t(7) = 4.24, P = 0.0038. e, Experimental scheme for assessing the mitochondrial dynamics of DIV 7 primary hippocampal neurons from P0-1 Fmr1 KO pups or their WT littermates, transfected with mitochondrial targeted photoconvertible mEOS3.2. f. Sample images (from 2 independently repeated experiments with similar results) of live imaging experiments showing total mitochondria before photo-switching (Total) and photo-switched mitochondria at T=0 and T=5min in WT and KO neurons. Scale bar, 10 μm. Scale bar, 10 μm. g, h, Quantitative analysis showing photo-converted mitochondria exhibited significantly more spread in WT (g) neurons but not in KO (h) neurons (Student’s t-test, two-sided, WT: t(11) = 4.053, P = 0.0019; T =0: 17.08 ± 1.761, T =5 min: 23.92 ± 2.750, n = 12 cells, KO: t(9) = 2.121, P = 0.0542; T =0: 16.15 ± 2.624, T =5 min: 17.87 ± 2.543, n = 10 cells from 3 KO and 3 WT littermates). i, Schematic illustration of mitochondrial fusion and fission genes in regulation of mitochondrial dynamics. j-n, Quantification analyses of mRNA levels of mitochondrial fusion and fission genes in WT and Fmr1KO primary hippocampal cells. Gapdh was used as the internal control for quantitative PCR analysis. (Student’s t-test, two-sided. Opa1, WT = 0.04076 ± 0.01003, n = 7 mice; KO = 0.01826 ± 0.006013, n = 7 mice, t(12) = 1.924, P = 0.0784. Mfn1, WT = 0.06412 ± 0.003417, n = 7 mice; KO = 0.03351 ± 0.004111, n = 7 mice, t(12) = 5.727, P < 0.0001; Mfn2, WT = 0.08783 ± 0.01551, n = 7 mice; KO = 0.03231 ± 0.007524, n = 7 mice, t(12) = 3.220, P = 0.0072; Drp1, WT = 0.03046 ± 0.002363, n = 7 mice; KO = 0.03891 ± 0.004793, n = 7 mice, t(12) = 1.582, P = 0.1397; Fis1, WT = 0.02480 ± 0.002401, n = 7 mice; KO = 0.02020 ± 0.002939, n = 7 mice, t(12) = 1.213, P = 0.2483). *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M.
Enhancing mitochondrial fusion rescues FMRP-deficient immature neurons
Mitochondria undergo frequent morphological changes through fission and fusion17. Since the overall area of mitochondria coverage did not change (Supplementary Fig. 10), the reduced aspect ratio in FMRP-deficient neurons might be a result of either increased fission or reduced fusion. We therefore used a photo-switching assay to assess mitochondrial fission and fusion dynamics (Fig. 4e-g). We transfected Fmr1 KO and WT neurons with mito-mEos3.2. mEOS3.2 is a green fluorescent protein but can be converted to a red fluorescent protein with 405 nm light. Nearly all mitochondria in mito-mEos3.2-transfected neurons were green (Fig. 4e) We then selectively converted a fraction of them to red and used live imaging to monitor mitochondrial dynamics in a fragment of dendrite. Any green mitochondria that was not originally photo-switched but turned red must either be a part of the original mitochondria or it must have fused with the photo switched mitochondria. We compared the area of photo-switched mitochondria between time T=0 and time T=5 min. Within the 5-minute period, a dynamic mitochondrial network would usually spread out or connect with neighbor mitochondria, In WT neurons, mitochondria spread out and increased the area of photo-switched mitochondria by about 40% within the 5 min period (Fig. 4g). However in KO neurons this increase was only ~10% (Fig. 4h). These data support our hypothesis that Fmr1 KO neurons exhibited reduced mitochondrial fusion.
A number of proteins are involved in mitochondrial fusion and fission17. Mitofusin (MFN) 1 and MFN2 mediate the fusion of mitochondrial outer membranes while Optic Atrophy 1 (OPA1) acts in inner membrane. Dynamin-related protein 1 (DRP1) and fission 1 protein (FIS1) mediates mitochondrial fission (Fig. 4i). Mfn2, Mfn1 and Drp1 mRNA were downregulated in KO dsRed+ adult immature neurons (Supplementary Table 1, Supplementary Fig. 10d). In addition, MFN1 and MFN2 were significantly lower in KO hippocampal neurons (DIV7) at both mRNA and protein levels, whereas Opa1, Fis1, and Drp1 mRNAs exhibited no significant change although OPA1 and DRP1 protein levels were reduced, compared to WT neurons (Fig. 4i-n; Supplementary Fig. 12, Supplementary Fig. 23). These results suggest that FMRP is important for maintaining the levels of mitochondrial fusion factors in developing neurons.
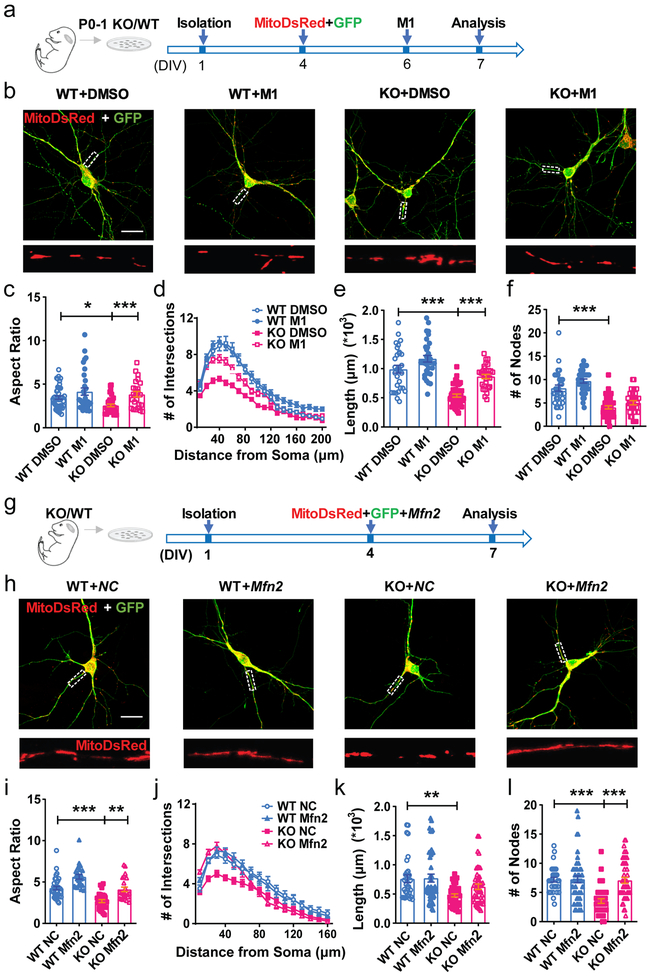
To test the hypothesis that mitochondrial fusion deficits in FMRP-deleted immature neurons contribute to impaired maturation, we treated Fmr1 KO and WT neurons with M1, a cell-permeable phenylhydrazone that promotes mitochondrial fusion and restores mitochondrial tubular network in neuronal cells32 (Fig. 5a). M1-treatment restored aspect ratio of mitochondria in the KO neurons to the levels of the WT neurons (Fig. 5b, c). In addition, M1 treatment partially rescued dendritic maturation deficits of KO neurons, without significant effect on WT neurons (Fig. 5d-f) or significant impact on protein levels of MFN1 and MFN2 (Supplementary Figs 13-14, Supplementary Fig 24). Furthermore, exogenous MFN2 rescued both fragmented mitochondrial (Fig. 5g-i) and dendritic (Fig. 5j-l) morphogenesis in Fmr1 KO neurons, without a significant effect on WT neurons. Exogenous MFN1 exhibited similar effect as MFN2 (Supplementary Fig. 15). Together, these data demonstrate that of mitochondrial fusion significantly ameliorated impaired dendritic maturation induced by FMRP deficiency.
Figure 5: Impaired mitochondrial fusion in FMRP-deficient DCX+ immature neurons.
a, Experimental scheme for assessing the effect of mitochondrial fusion promoting compound M1 on the dendritic maturation of KO neurons. b, Sample confocal images of mitochondria in the Fmr1 KO and WT hippocampal neurons treated with DMSO or M1 (10μM). Enlarged view of the boxed area is shown at the bottom of each image. 3 independently repeated experiments with similar results. Scale bars, 20 μm. c, Quantification of mitochondrial aspect ratio in mitoDsRed expressing Fmr1 KO or WT neurons treated with DMSO or M1 (10μM) (Two-way ANOVA with two-sided Bonferroni post analysis for multiple comparisons: treatment × genotype: F(1, 128) =1.073, P = 0.3022. WT:DMSO vs. KO:DMSO, P = 0.0358; KO:DMSO vs. KO:M1, P = 0.0009. WT:DMSO = 3.449 ± 0.1885, n = 45; KO:DMSO = 2.561 ± 0.1544, n = 36; WT:M1 = 4.143 ± 0.4195, n = 31; KO:M1 = 3.842 ± 0.3202, n = 24 cells from 3 independent experiments). d, Sholl analysis of dendritic complexity of Fmr1 KO or WT neurons treated with DMSO or M1 (10μM) (Multi-ANOVA, WT DMSO vs. KO DMSO: F(1,92) = 42.671, P < 0.0001; WT DMSO vs. WT M1: F(1,81) = 8.045, P = 0.006; KO DMSO vs. KO M1: F(1,92) = 45.808, P < 0.0001. WT:DMSO, n = 30; KO:DMSO, n = 43; WT:M1, n = 32; KO:M1, n = 30 cells from 3 independent experiments). e, Quantification of total dendritic length in mitoDsRed expressing Fmr1 KO or WT neurons treated with DMSO or M1 (10μM) (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F(1,129) = 66.55, P < 0.0001. WT:DMSO = 0.9901 ± 0.06578, n = 30; KO:DMSO = 0.5379 ± 0.02668, n = 43; WT:M1 = 1.170 ± 0.05598, n = 32; KO:M1 = 0.8647 ± 0.03644, n = 30 cells from 3 independent experiments). f, Dendritic nodes (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (1, 131) = 0.3239. WT DMSO vs. KO DMSO: P < 0.0001. WT:DMSO = 8.167 ± 0.6610, n = 30; KO:DMSO = 4.047 ± 0.3344, n = 43; WT:M1 = 9.688 ± 0.4409, n = 32; KO:M1 = 5.033 ± 0.4510, n = 30 cells from 3 independent experiments); g, Experimental scheme for assessing the effect of restoration of MFN1 and MFN2 on the dendritic maturation of KO neurons. h, Sample confocal images of mitochondria in the Fmr1 KO and WT hippocampal neurons transfected with Mfn2 or NC. Enlarged view of the boxed area is shown at the bottom of each image. 3 independently repeated experiments with similar results. Scale bars, 20 μm. i, Quantification of mitochondrial aspect ratio in Fmr1 KO or WT neurons transfected with Mfn2 or NC (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (1, 108) = 0.009781. WT:NC vs. KO:NC, P < 0.0001; WT:NC vs. WT:Mfn2, P = 0.0009; KO:NC vs. KO:Mfn2, P = 0.0013. WT:NC = 4.271 ± 0.2085, n = 36; KO:NC = 2.702 ± 0.1834, n = 28; WT:Mfn2 = 5.635 ± 0.2736, n = 24; KO:Mfn2 = 4.112 ± 0.2697, n = 24 cells from 3 independent experiments). j, Sholl analysis of dendritic complexity of Fmr1 KO or WT neurons transfected with Mfn2 or NC (Multi-ANOVA, WT NC vs. KO NC: F(1,61) = 10.436, P = 0.002; KO NC vs. KO Mfn2: F(1,74) = 4.141, P = 0.045. WT:NC, n = 32; KO:NC, n = 30; WT:Mfn2, n = 45; KO:Mfn2, n = 45 cells from 3 independent experiments). k, Quantification of dendritic length in Fmr1 KO or WT neurons transfected with Mfn2 or NC (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (1, 158) = 1.560. WT:NC vs. KO:NC, P = 0.0041. WT:NC = 0.7613 ± 0.06684, n = 32; KO:NC = 0.4768 ± 0.02672, n = 40; WT:Mfn2 = 0.7729 ± 0.06793, n = 45; KO:Mfn2 = 0.6257 ± 0.04641, n = 45 cells from 3 independent experiments). l, Dendritic nodes (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (1, 158) = 12.02, P = 0.0007. WT NC vs. KO NC: P < 0.0001; KO NC vs. KO Mfn2: P < 0.0001 WT:NC = 7.188 ± 0.3951, n = 32; KO:NC = 3.525 ± 0.3971, n = 40; WT:Mfn2 = 7.267 ± 0.5907, n = 45; KO:Mfn2 = 7.089 ± 0.4993, n = 45 cells from 3 independent experiments). *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M.
Reduced HTT levels in FMRP-deficient developing neurons contribute to mitochondrial dysfunction and neuronal maturation deficits
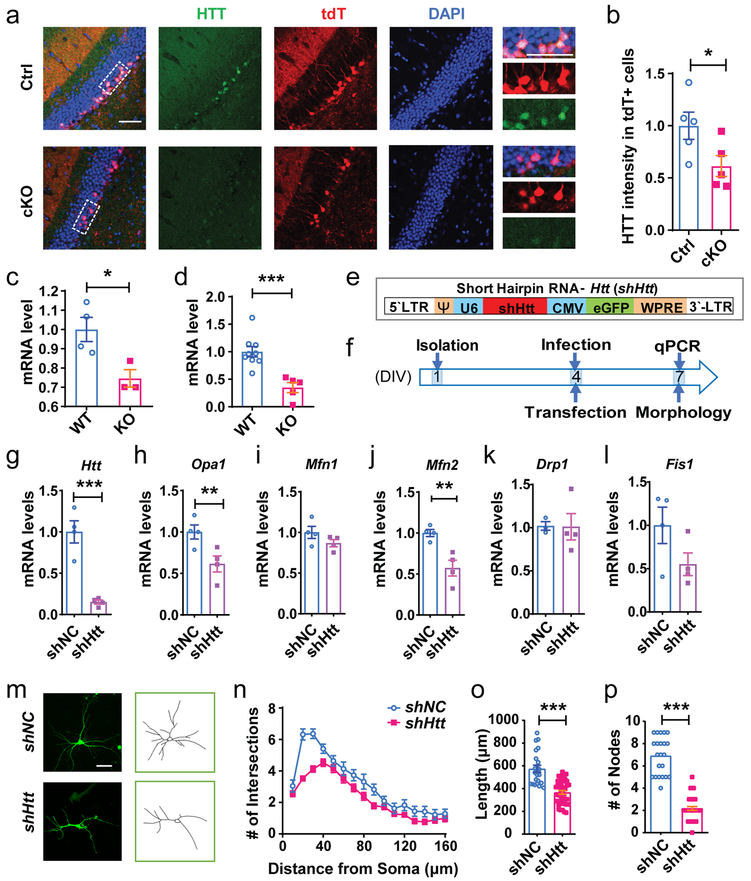
We next investigated how FMRP regulates mitochondrial fusion. There were 40 common FMRP targets identified in mouse forebrain3, human HEK293 cells11 and mouse hippocampus10 (Supplementary Table 4). However, none of these 40 FMRP targets was among the 519 DE genes we found in Fmr1 KO immature neurons (Fig. 3), suggesting that these 519 DE genes might be indirectly regulated by FMRP. We then used BioGRID database (Supplementary Table 5) to identify genes with known one-degree physical interactions with the 519 DE genes and discovered 18 of them were among the 40 FMRP targets (Supplementary Fig. 16, Supplementary Table 6). We also used BioGRID to search for genes that had first degree interaction with the 7 mitochondria-related genes (defined by PATHER, Supplementary Table 3b) among the 519 DE genes. Surprisingly, only Htt was found to be among the 40 FMRP targets, suggesting that Htt is the most likely FMRP target involved in FMRP regulation of mitochondria in immature neurons (Supplementary Fig. 16). Other than genome-wide studies3,10,11, a direct interaction between FMRP and Htt mRNA has not been validated. We performed RNA binding protein immunoprecipitation and identification of co-precipitated RNA (RNA-IP) using an FMRP antibody coupled with real-time PCR analysis. The FMRP RNA-IP sample was highly enriched with Htt but not Mfn1, Mfn2, or Fis1 mRNAs compared to Gapdh control mRNAs (Supplementary Fig. 17a). We next assessed the levels of HTT protein in immature neurons and found that the tdT+ neurons in the DG of cKO;Cre;tdT mice exhibited significantly reduced HTT immunostaining intensity compared with those in control mice (Fig. 6a, b). In addition, Htt mRNA levels were significantly lower in Fmr1 KO DG tissue of 3-week-old mice (Fig. 6c) with a high proportion of DCX+ cells24. Furthermore, Htt mRNA levels were significantly lower in Fmr1 KO primary hippocampal neurons, compared with controls (Fig. 6d). To determine how FMRP deficiency affects Htt mRNA levels, we determined the stability of Htt mRNA in KO and WT hippocampal neurons treated with transcriptional inhibitor actinomycin D. We found that Htt mRNA had a reduced half-life (T1/2) in Fmr1 KO neurons (3.86 hours) compared to WT neurons (4.18 hours; Supplementary Fig. 17b). Therefore, FMRP directly binds Htt mRNA and maintains the stability of Htt mRNAs, and FMRP deficiency leads to reduced Htt mRNA and HTT protein levels.
Figure 6: FMRP-deficient immature neurons exhibited reduced HTT expression and down-regulation of HTT led to impaired dendritic maturation.
a, Representative confocal images (from 3 independently repeated experiments with similar results) of HTT expression in the DCX+ immature neurons in the DG of cKO;Cre;tdT and Ctrl;Cre;tdT mice. Scale bars, 50 μm. b, The expression level of HTT in the tdT+ cells of the DG from cKO; Cre; tdT and Ctrl;Cre;tdT mice (Student’s t-test, two-sided, t(8) = 2.348, P = 0.0468. Ctrl: 1.000 ± 0.1304, n = 5 mice; cKO: 0.6139 ± 0.1002, n = 5 mice). c, Quantification of mRNA levels of Htt in 3-week-old WT and Fmr1 KO DG tissues. Gapdh was used as the internal control for quantitative PCR analysis. (Student’s t-test, two-sided, t(5) = 3.056, P = 0.0282. WT: 1.000 ± 0.06252, n = 4 mice; KO: 0.7457 ± 0.04554, n = 3 mice). d, Quantification of mRNA levels of Htt in DIV 7 WT and Fmr1 KO primary hippocampal cells. Gapdh was used as the internal control for quantitative PCR analysis. (Student’s t-test, two-sided, t(12) = 4.530, P = 0.0007. WT: 1.000 ± 1.000 ± 0.09355, n = 9; KO: 0.3492 ± 0.09103, n = 5 independent experiments). e, Schematic illustration of retroviral vectors expressing shNC and expressing shHtt. f, Experimental scheme for assessing the effect of down-regulation of Htt on mitochondrial fusion and fission genes and dendritic maturation of hippocampal neurons. g-l, Quantification of mRNA levels of Htt, mitochondrial fusion and fission genes in primary hippocampal cells infected by lentivirus-shHtt or lentivirus-shNC (above) or transfected with shHtt/shNC (below). (Student’s t-test, two-sided. Htt, shNC = 1.000 ± 0.1345, n = 4; shHtt = 0.1493 ± 0.02515, n = 4 biologically independent samples, t(6) = 6.291, P = 0.0008; Opa1, shNC = 1.000 ± 0.08564, n = 4; shHtt = 0.6141 ± 0.09681, n = 4, t(6) = 2.985, P = 0.0245; Mfn1, shNC = 1.000 ± 0.07478, n = 4; shHtt = 0.8673 ± 0.04440, n = 4, t(6) = 1.526, P = 0.1778; Mfn2, shNC = 1.000 ± 0.04351, n = 4; shHtt = 0.5720 ± 0.09657, n = 4, t(6) = 4.041, P = 0.0068; Drp1, shNC = 1.000 ± 0.04058, n = 4; shHtt = 1.011 ± 0.1528, n = 4, t(6) = 0.06726, P = 0.9486; Fis1, shNC = 1.000 ± 0.2099, n = 4; shHtt = 0.5505 ± 0.1316, n = 4, t(6) = 1.814, P = 0.1196). m, Examples of confocal images and Neurolucida software-created traces of DIV7 primary hippocampal neurons transfected with shHtt or shNC at DIV4. 3 independently repeated experiments with similar results. Scale bar, 20 μm. m, Sholl analysis of dendritic complexity of hippocampal neurons transfected with shHtt or shNC (Multi-ANOVA, shNC vs. shHtt: F(1,61) = 27.712, P < 0.0001. shNC, n = 22; shHtt, n = 40 cells from 3 independent experiments). o, Quantification of dendritic length of hippocampal neurons transfected with shHtt or shNC (Student’s t-test, t(60) = 6.454, P < 0.0001. shNC, 574.4 ± 31.77, n = 22; shHtt, 366.6 ± 16.34, n = 40 cells). p, Quantification of dendritic nodes of hippocampal neurons transfected with shHtt or shNC (Student’s t-test, two-sided. t(60) = 13.57, P < 0.0001. shNC, 6.909 ± 0.3598, n = 22; shHtt, 2.175 ± 0.1675, n = 40 cells). *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M.
Although pathological HTT signaling is known to induce mitochondrial changes including loss of membrane potential and abnormal mitochondrial dynamics33, nearly all functional studies of HTT have been performed in the context of CAG repeats-containing HTT gene. The impact of HTT deficiency on either neuronal or mitochondria functions remains largely unexplored. We infected hippocampal neurons with lentivirus expressing shRNA against (shHtt) and GFP. We found that shHtt infected neurons showed significant reduction in both mRNA and protein levels of HTT (Fig. 6e, f; Supplementary Fig. 18; Supplementary Fig. 25), as well as mRNA and protein levels of MFN2 and OPA1 but not Fis1 and Drp1 mRNA levels compared with shNC infected neurons, (Fig. 6g-l; Supplementary Fig. 18). In addition, hippocampal neurons with HTT knockdown (shHtt) exhibited significantly reduced dendritic complexity, dendritic length, and number of dendritic nodes (Fig. 6m-p). These results indicate that HTT deficiency in immature neurons leads to reduced mitochondrial fusion gene expression and impaired dendritic maturation, which phenocopies the deficits observed in FMRP-deficient developing neurons.
CRISPR/dCas9-targeted activation of endogenous Htt transcription rescues mitochondrial and dendritic maturation deficits of FMRP-deficient neurons
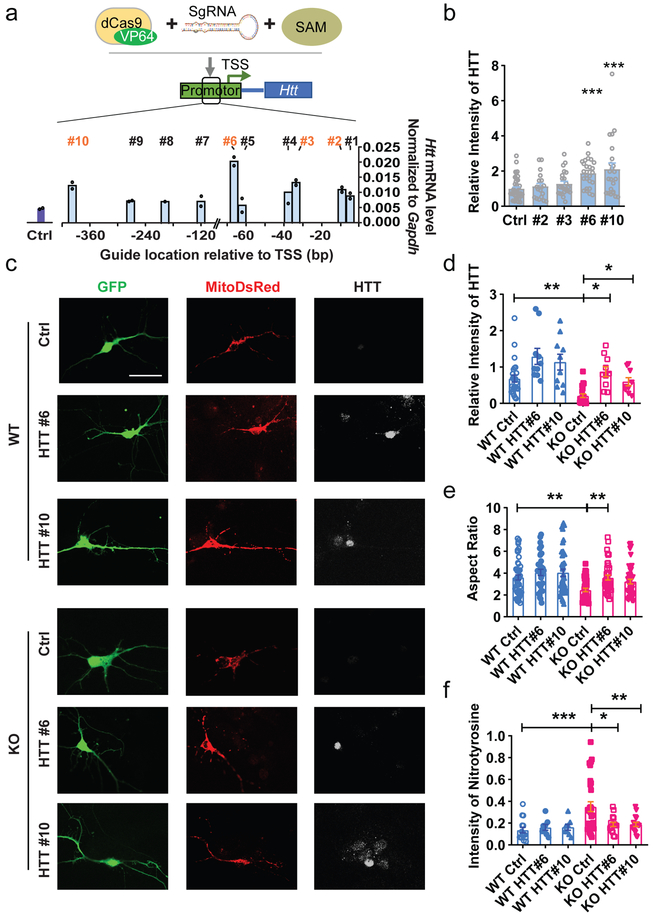
To examine whether reduced Htt is responsible for mitochondrial deficits seen in FMRP-deficient neurons, we sought to test whether increasing the expression levels of HTT may rescue mitochondrial fusion and dendritic maturation deficits of FMRP-deficient neurons. Because HTT protein is large (∼348 kDa) with 9429 nucleotide protein coding sequence for human and 9663 for mouse HTT, it is at the limit of the maximal capacity of lentiviral packaging. In addition, we were also concerned that overexpression of full length HTT may lead to off-target effects. We therefore designed a guide RNA (sgRNA)-guided sequence-specific transcription activation system that utilized a fusion protein of inactive Cas9 (dCas9) and transcriptional activation domain VP64 (dCas9–VP64) together with a synergistic activation mediator (SAM, MS2–p65–HSF1) that enhances the effect of transcription activation34. We designed 10 sgRNAs targeting various sequences in the proximal promoter, between −400 bp and −7 bp relative to transcription start site (TSS) of the mouse Htt gene. The sgRNA candidates #2, #3, #6 and #10 exhibited the strongest effects in elevating the endogenous Htt mRNA levels in transfected Neuro2A cells (Fig. 7a). Among them, sgRNA #6 and #10 significantly enhanced the levels of endogenous HTT protein levels in transfected hippocampal neurons (Fig. 7b; Supplementary Fig 19).
Figure 7: Increasing the expression levels of HTT rescued mitochondrial fusion deficits of FMRP-deficient neurons.
a, Quantification of mRNA levels of Htt in the Neuro2A cells transfected with sgRNA (or Ctrl) with dCas9–VP64 and SAM. 10 sgRNAs were designed to target the proximal promoter region (−7 to −400 bp from the TSS) of Htt. dCas9- based transcription activators of Htt consists of three components: dCas9-VP64 fusion protein, synergistic activation mediator (SAM, MS2–p65–HSF1) and sgRNA. b, Relative intensity of HTT in the primary hippocampal neurons transfected with sgRNAs (#2, 3, 6, 10 or Ctrl) with dCas9–VP6-SAM. (One-way ANOVA with Bonferroni post hoc test: F (4, 129) = 7.564, P < 0.0001; Bonferroni post hoc test, P < 0.0001. Ctrl: 1.000 ± 0.09112, n = 44; #2: 1.135 ± 0.1668, n = 19; #3: 1.276 ± 0.1424, n = 23; #6: 1.882 ± 0.1558, n = 25; #10: 2.109 ± 0.3470, n = 23 cells from 3 biological independent samples) c, Sample confocal images of DIV 7 Fmr1 KO and WT hippocampal neurons transfected with SAM complex. 3 independently repeated experiments with similar results. Scale bars, 20 μm. d, Quantification analyses of HTT expression level in the DIV 7 Fmr1 KO and WT hippocampal neurons (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (2, 80) = 0.1846. WT Ctrl vs. KO Ctrl: P =0.004; WT Ctrl vs. WT HTT#6: P =0.041; KO Ctrl vs. KO HTT#6: P = 0.0268; KO Ctrl vs. KO HTT#10: P =0.0388. WT Ctrl: 0.6814 ± 0.09871, n = 25; WT HTT#6: 1.134 ± 0.2168, n = 10; WT HTT#10: 1.291 ± 0.2200, n = 10; KO Ctrl: 0.2118 ± 0.05165, n = 21; KO HTT#6: 0.5943 ± 0.1110, n = 9; KO HTT#10: 0.8697 ± 0.1367, n = 10 cells from 3 biological independent samples). e, Quantification of mitochondrial aspect ratio (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (2, 244) = 0.9154. WT Ctrl vs. KO Ctrl: P = 0.0016; KO Ctrl vs. KO HTT#6: P = 0.0038. WT Ctrl: 3.610 ± 0.2324, n = 50; WT HTT#6: 4.069 ± 0.2899, n = 34; WT HTT#10: 4.036 ± 0.3330, n = 40; KO Ctrl: 2.462 ± 0.1607, n = 42; KO HTT#6: 3.592 ± 0.2246, n = 40; KO HTT#10: 3.225 ± 0.2189, n = 44 cells from 3 biological independent samples). f, Quantification of the expression level of nitrotyrosine (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (2, 95) = 4.283. WT Ctrl vs. KO Ctrl: P < 0.0009; KO Ctrl vs. KO HTT#6: P = 0.0136; KO Ctrl vs. KO HTT#10: P < 0.0025. WT Ctrl: 0.1364 ± 0.02563, n = 15; WT HTT#6: 0.1606 ± 0.02687, n = 8; WT HTT#10: 0.1606 ± 0.02687, n = 8; KO Ctrl: 0.3503 ± 0.04518, n = 30; KO HTT#6: 0.1878 ± 0.01902, n = 16; KO HTT#10: 0.1878 ± 0.01902, n = 16 cells from 3 biological independent samples). *P < 0.05, **P < 0.01, ***P < 0.001. All error bars reflect the Mean ± S.E.M.
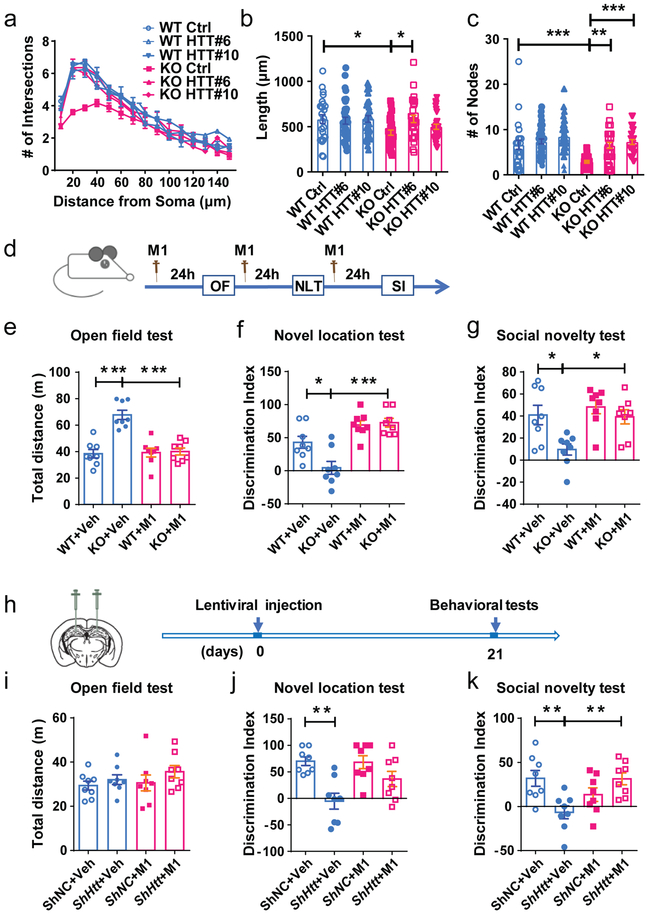
We then transfected Fmr1 KO or WT primary hippocampal neurons with either sgRNA#6 or #10 together with dCas9-V164, SAM, mitoDsRed, and GFP on DIV4 and analyzed the mitochondrial morphology and oxidative stress level of the neurons on DIV7 (Fig. 7c-f). Htt gene activation by sgRNA#6 (HTT #6) led to a significant increase in HTT protein levels in both WT and KO neurons compared to control sgRNA conditions, and more importantly, restored the HTT levels in KO neurons to the WT level (Fig. 7d). Htt gene activation increased aspect ratio of mitochondria (Fig. 7e) and reduced the nitrotyrosine level (Fig. 7f) in Fmr1 KO neurons without a significant effect on WT neurons. In addition, Htt gene activation rescued dendritic complexity of KO neurons without a significant effect on WT neurons (Fig.8a-c). Activation of HTT using sgRNA #10 showed similar effects as sgRNA#6 (Fig. 7c-f, 8a-c). Therefore, these data suggest that HTT, a target of FMRP in immature neurons, is central to FMRP regulation of mitochondrial function and neuronal maturation.
Figure 8: Increasing the expression levels of HTT rescued dendritic complexity deficits of FMRP-deficient neurons.
a, Sholl analysis of dendritic complexity (Multi-ANOVA, WT Ctrl vs. KO Ctrl: F(1,54) = 5.643, P = 0.023; KO Ctrl vs. KO HTT#6: F(1,57) = 6.616, P = 0.013; KO Ctrl vs. KO HTT#10: F(1,57) = 1.881, P = 0.176. WT Ctrl:, n = 25; WT HTT#6:, n = 35; WT HTT#10:, n = 33; KO Ctrl:, n = 30; KO HTT#6:, n = 28; KO HTT#10:, n = 28 cells from 3 biological independent samples). b, Quantification of dendritic length (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (2, 173) = 2.621. WT Ctrl vs. KO Ctrl: P = 0.0253; KO Ctrl vs. KO HTT#6: P = 0.0126. WT Ctrl: 583.5 ± 48.35, n = 25; WT HTT#6: 565.7 ± 37.64, n = 35; WT HTT#10: 585.0 ± 36.69, n = 33; KO Ctrl: 434.2 ± 28.83, n = 30; KO HTT#6: 585.7 ± 41.36, n = 28; KO HTT#10: 498.4 ± 28.12, n = 28 cells from 3 biological independent samples). c, Quantification of dendritic nodes (Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons: treatment × genotype: F (2, 173) = 2.938, P = 0.0284. WT Ctrl vs. KO Ctrl: P = 0.0007; KO Ctrl vs. KO HTT#6: P < 0.0016; KO Ctrl vs. KO HTT#10, P < 0.0001. WT Ctrl: 6.640 ± 0.9914, n = 25; WT HTT#6: 7.371 ± 0.5562, n = 35; WT HTT#10: 8.394 ± 0.6498, n = 33; KO Ctrl: 2.967 ± 0.2273, n = 30; KO HTT#6: 6.500 ± 0.6475, n = 28; KO HTT#10: 7.250 ± 0.5507, n = 28 cells from 3 biological independent samples). d, Experimental scheme for assessing cognitive functions in Fmr1 wildtype (WT) and KO mice treated with M1. e-g, M1 treatment rescued hyperactivity (e, open field activity analysis, treatment × genotype: F (1, 28) = 20.31, P = 0.0001. WT+Veh vs. KO+Veh: P < 0.0001; KO+Veh vs. KO+M1: P < 0.0001. WT+Veh: 38.35 ± 3.218; KO+Veh: 67.77 ± 3.472; WT+M1: 39.27 ± 3.315; KO+M1: 40.07 ± 2.628), spatial working memory (f, novel location test, treatment × genotype: F (1, 28) = 6.671, P = 0.0153. WT+Veh vs. KO+Veh: P = 0.0135; KO+Veh vs. KO+M1: P < 0.0001. WT+Veh: 42.99 ± 9.080; KO+Veh: 4.451 ± 9.744; WT+M1: 69.35 ± 6.482; KO+M1: 72.68 ± 6.590), and interaction with a stranger mouse (g, social novelty test, treatment × genotype: F (1, 28) = 2.756, P = 0.108. WT+Veh vs. KO+Veh: P = 0.0166; KO+Veh vs. KO+M1: P = 0.0258. WT+Veh: 40.95 ± 8.859; KO+Veh: 9.601 ± 5.049; WT+M1: 48.21 ± 6.095; KO+M1: 39.30 ± 6.442). (n = 8 mice per group). h, Experimental scheme for assessing cognitive functions in WT with HTT knockdown by stereotaxic injection of lentivirus expressing shHTT or control shNC in the DG and treated with M1. The M1 treatment scheme was the same as e. i-k, HTT knockdown does not affect hyperactivity (i, open field activity analysis, treatment × genotype: F (1, 28) = 0.1991, P = 0.6589. shNC+Veh: 29.38 ± 2.008; shHtt+Veh: 32.04 ± 2.195; shNC+M1: 30.57 ± 3.581; shHtt+M1: 35.66 ± 2.799) but impaired spatial working memory (j, novel location test, treatment × genotype: F (1, 28) = 2.954, P = 0.0967. shNC+Veh vs. shHtt+Veh: P = 0.0013. shNC+Veh: 70.10 ± 8.098; shHtt+Veh: −5.372 ± 15.03; shNC+M1: 68.25 ± 12.25; shHtt+M1: 36.52 ± 14.36) and interaction with a stranger mouse (k, social novelty test, treatment × genotype: F (1, 28) = 13.39, P = 0.001. shNC+Veh vs. shHtt+Veh: P = 0.0076; shHtt+Veh vs. shHtt+M1: P = 0.0085. shNC+Veh: 31.88 ± 8.945; shHtt+Veh: −6.624 ± 7.325; shNC+M1: 13.36 ± 7.692; shHtt+M1: 31.35 ± 6.743). (n = 8 mice per group), M1 treatment rescued these deficits. Two-way ANOVA with two-sided Bonferroni post hoc analysis for multiple comparisons was used for all data analyses of behavioral tests. *P < 0.05, **P < 0.01, ***P < 0.001. Data are presented as mean ± SEM.
M1 treatment rescues behavioral deficits of Fmr1 KO mice and mice with HTT knockdown in the hippocampus
Since M1 treatment rescued both mitochondrial and neuronal morphology in Fmr1 KO neurons (Fig. 4), we next investigated whether M1 could rescue the behavioral deficits in Fmr1 KO mice (Fig. 8d). Consistent with literature35, 36,26, vehicle-treated Fmr1 KO mice exhibited hyperactivity in an open field test (Fig 8e) without significant changes in anxiety-related center field preference (Supplementary Fig. 20a), and impaired spatial working memory in a novel location test (NLT, Fig 8f, Supplementary Fig. 20c-e), impaired interaction with a novel mouse (Social Novelty, Fig. 8g; Supplementary Fig. 21a-e). M1 treatment rescues these deficits observed in Fmr1 KO mice (Fig. 8e-g).
We next acutely knocked down HTT in adult DG of WT mice using lentivirus expressing shHtt followed by behavioral assessments at 21 days later (Fig. 8h). We found that mice with HTT knocked down in the DG exhibit no change in open field activities (Fig. 8i, Supplementary Fig. 20a), but exhibited impaired spatial working memory (Fig. 8j, Supplementary Fig. 20c, f, g) and interaction with a novel mouse (Fig. 8k; Supplementary Fig. 21f-i). Intriguingly, these deficits were rescued by M1 treatment (Fig. 8j, k; Supplementary Fig. 21f-i). Therefore, behavioral deficits in mice lacking either FMRP or HTT could be rescued by a compound that promotes mitochondrial fusion, supporting the converging deficits in mitochondrial functions of these two disease genes (Supplementary Fig. 22).
Discussion
In this study we show that FMRP deficiency in immature neurons leads to impaired mitochondrial fusion and enhancing mitochondrial fusion restores neuronal maturation in Fmr1 KO neurons. Our work provides the first direct evidence that links FMRP deficiency and mitochondrial dysfunction. Our surprising discovery of HTT as a key mediator of FMRP regulation of mitochondria unveils a previously unknown crosstalk between these two human disease genes.
Interrogating cell type-specific and developmental stage-specific roles of FMRP is critical for understanding the disease mechanism and developing effective treatment for FXS. We have previously shown that deletion of FMRP in adult NSCs leads to a reduced number and impaired dendritic complexity of new hippocampal neurons9, 24. However such deficits could be a consequence of either reduced neuronal differentiation of NSCs or impaired maturation of neurons24, 26. Here we show that selective deletion of FMRP from DCX-expressing immature neurons had no significant effect on neuroblast proliferation, in contrast to its role in regulating NSC proliferation26, but led to reduced number of new mature neurons at all time points. In addition, these DG neurons had not only impaired morphological maturation but reduced frequency of mEPSC, suggesting impaired excitatory synaptic input and deficient integration into the neural circuitry37, 38. Therefore, our present study demonstrates that FMRP has distinct roles in immature neurons, independent of its roles in NSCs24, 26 and FMRP deficiency significantly affects the final stage of maturation of new neurons.
Several studies have investigated mitochondrial function in Fragile X-associated tremor/ataxia syndrome (FXTAS) which is caused by pre-mutation expansions (55–200) of the CGG repeat in the 5′ untranslated region of the FMR1 gene leading to overexpression of mutant CGG repeat containing FMR1 mRNAs. Both hippocampal neurons isolated from premutation mouse models39 and fibroblasts from patients with FXTAS40 showed abnormal mitochondrial numbers and functions. However, FXTAS patients retain FMRP protein expression at various levels and their pathology is likely due to the presence of mutant FMR1 mRNAs rather than the absence of FMRP protein. Our current study reports for the first time the contribution of mitochondrial dysfunction in FXS. Mitochondria are dynamic organelles continuously remodeled through the balance between fusion and fission events, which are mediated by both the fusion machinery including MFN1, MFN2, and OPA1 and fission machinery including FIS1 and DRP141. Mice deficient in either MFN1 or MFN2 die in mid-gestation and embryonic fibroblasts lacking MFN1 or MFN2 display fragmented mitochondria42. MFN2 is critical in many cell types43. Specific deletion of MFN2 leads to reduced mitochondrial transport, impaired respiratory chain function, and aberrant mitochondrial morphology, leading to increased oxidative stress and neuronal death44, 45,46. MFN2 knock-down in hiPSCs results in defective neurogenesis and synapse formation, while MFN2 overexpression promotes maturation of human neurons47. Therefore, mitochondrial fusion is important for neuronal maturation. Our data suggest that MFNs and mitochondria might be potential treatment targets for FXS.
Abnormal expansion of a CAG repeat located in exon 1 of the human HTT gene leads to HD, a neurodegenerative disorder33. Studies investigating the toxic effects of mutant HTT gene in cell culture or animal models have revealed mitochondrial changes including a loss of membrane potential, abnormal mitochondrial dynamics, and increased oxidative stress33. The tissues and cells of HD patients have increased mitochondrial fragmentation due to decreased levels of MFN1, MFN2 and OPA1 levels and increased DRP1 and FIS1 levels48. However, most of these studies have been done in the context of CAG repeat containing HTT gene. The exact function of HTT protein itself remains unclear33. HTT is widely expressed and is thought to be involved in diverse cellular activities such as regulation of transcription, transporting materials, binding proteins and other structures, endocytosis and autophagy33. The HTT/Htt gene locus in both human and mouse is large, spanning 180 kb and consisting of 67 exons, therefore understanding the normal biological functions of non-mutant HTT protein remains a challenge, and activation and restoration of HTT protein with high efficiency has not been achieved49. To restore HTT levels in Fmr1 KO neurons without confound of exogenous HTT overexpression, we used a Htt gene-specific guide RNAs to direct a dCas9 -VP64 fusion protein together with SAM amplification system34 to the endogenous Htt gene promoter. We demonstrate that this is an effective method for studying functions of HTT or other large proteins in neurons or other cell types. Htt mRNA has been identified as a target of FMRP 3, 10,11,50; however the functional significance of FMRP regulation of Htt remains unexplored. We show that HTT levels directly impact the levels of Mfn2, a gene down regulated in FMRP deficient neurons. Both HTT and FMRP are widely expressed with potentially many interactors and targets. Both Fmr1 KO mice and mice with hippocampal HTT knockdown exhibited spatial working memory and social novelty deficits. The fact that treatment with a mitochondrial fusion promoting compound rescued these deficits further support that mitochondrial fusion might be one of the convergent mechanisms underlying the functions of FMRP and HTT. On the other hand, it is possible that FMRP and HTT may regulate certain mitochondrial functions independently of each other. In fact, we found that acute knockdown of Htt led to decreased Opa1 mRNA level but had no significant impact on Mfn1 (Fig. 6l, m), whereas Fmr1 KO neurons had reduced levels of both Mfn1 and Mfn2 but not Opa1 (Fig. 4k). HTT knockdown in the DG did not lead to hyperactivity as seen in Fmr1 KO mice, which could be due to either differential functions of these two proteins or due to restricted effects from regional knockdown of HTT. Nevertheless, our study has elucidated a novel role of HTT in FMRP regulation of mitochondrial function and dendritic maturation. Further studies on the crosstalk between these two proteins will shed light on our understanding of the pathogenesis of both HD and FXS.
Methods
Mice
We performed all procedures involving live mice in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the protocols approved by the University of Wisconsin-Madison Animal Care and Use Committee. We generated the inducible conditional mutant mice (Fmr1loxP/y;Tg(Dcx-cre/ERT2);Gt(ROSA)26Sortm14(tdTomato)Hze, simplified as cKO;Cre;tdT) by crossing Fmr1-floxed (Fmr1loxP/+) mice with Tg(Dcx-cre/ERT2) transgenic driver (Dcx-CreERT2; MMRRC, provided by Dr. U. Mueller, Scripps Institute, San Diego, CA) mice and Gt(ROSA)26Sortm14(tdTomato)Hze reporter (tdT) mice. The Fmr1loxP/+ mice have been bred onto a C57BL/6J genetic background51. The Dcx-CreERT2 mice are 100% C57BL/6J background (stock # 032780-MU donated by Ulrich Muller). The tdT mice (C57B/L6J bacground) were from Jackson Laboratory (stock # 007914). Mice were then crossed with the Dcx-CreERT2 mice. The inducible Fmr1 conditional knockout were generated as following: Dcx-CreERT2 ; tdT male mice were crossed with heterozygote female Fmr1loxP/+ mice. The male offspring were either cKO;Cre;tdT or control (Ctrl, or Ctrl;Cre;tdT) mice to be used in experiments. We also generated the Fmr1−/Y /Dcx- DsRed mice and Fmr1+/Y /Dcx- DsRed mice. The Fmr1 KO mice bred on the C57B/L6 genetic background was as described previously29. The Dcx- DsRed mice52 (Jackson Laboratory stock #009655) have been backcrossed to a C57BL/6J background. Those male mice were crossed with heterozygote female Fmr1−/+ mice. The male offspring were used in experiments. Mice were genotyped as previously described51, 53,54. To induce recombination, mice (6 weeks old) received Tamoxifen (Sigma-Aldrich) daily for 3 days (160 mg kg–1 i.p., 40 mg ml−1 in 10% ethanol mixed with sunflower oil, Sigma-Aldrich). M1 (20 mg kg−1) or vehicle was given to 2-4-month-old mice through intraperitoneal (i.p.) injection 24 hours before each behavioral test.
Data collection timing and blinding
Data collection was carried out for a predetermined period of time, as dictated by literature or core facility-based standards, and no exclusion criteria were applied. For drug treatment, animals were randomly assigned to treatment arms with approximately equivalent numbers in each group. All cell counting and behavioral analyses were performed by experimenters who were blind to the identity and treatments of the samples.
Tissue Preparation and Immunohistochemistry
Histological analysis of mouse brains were performed as described previously with modifications29, 53. At 3, 7, 14, 28 or 56 d after the last TAM injection, mice were euthanized by intraperitoneal injection of sodium pentobarbital and then transcardially perfused with saline followed by 4% PFA. Brains were dissected out, post-fixed overnight in 4% PFA, and then equilibrated in 30% sucrose. Forty-μm brain sections were generated using a sliding microtone and stored in a −20°C freezer as floating sections in 96-well plates filled with cryoprotectant solution (glycerol, ethylene glycol, and 0.1M phosphate buffer, pH 7.4, 1:1:2 by volume). Immunohistology was performed as published previously29, 53,55. The tissue sections were pre-blocked with TBS++ (TBS containing 3% goat or donkey serum and 0.2% Triton X-100) for 1 h at room temperature, followed by incubation with primary antibodies diluted in TBS++ overnight in 4 ℃. After washing 3 times, secondary antibodies were incubated 1 h at room temperature. All sections were counterstained with a nuclear counter stain, DAPI (4’, 6-diamidine-2’-phenylindole dihydrochloride, 1:2000, Roche Applied Science, Indianapolis, IN).
The primary antibodies used were: chicken anti-GFP (Invitrogen, Carlsbad, CA, #A10262), rat anti-Ki67 (1:500, eBioscience, 14-5698, San Diego, CA, USA), rabbit anti-GFAP (1:2000, DAKO, #Z0334, Carpinteria, CA, USA), chicken anti-Nestin (1:500, Aves Labs, #NES0407, Tigard, OR, USA), rabbit anti-Doublecortin (1:500, Cell Signaling Technology, #4604S, Beverly, MA, USA), rabbit anti-cleaved caspase-3 (1:500, Cell Signaling, #9661, Danvers, MA, USA), mouse anti-NeuN (1:500, Millipore, MAB377, Billerica, MA, USA), rabbit anti-S100β (1:1000, Dako, Z0334, Carpinteria, CA, USA), mouse anti-FMRP (1:500, Millipore, MAB2160), mouse anti-Nitrotyrosine (39B6) (1:500, Santa Cruz Biotechnology, sc-32757, Texas, DA, USA), mouse anti-OPA1 (1:1000, Abcam, ab157457, Cambridge, MA) mouse anti-MFN1 (1:1000, Abcam, ab57602, Cambridge, MA), rabbit MFN2 (1:500, Proteintech, 12186-1-AP, Rosemont, IL), rabbit anti-DRP1 (1:1000, Abcam, ab184247, Cambridge, MA) and mouse anti-Huntingtin (3E10) (1:500, Santa Cruz Biotechnology, sc-47757, Texas, DA, USA).
Fluorescent secondary antibodies used by 1:500 dilution: goat anti-chicken-488 (A11039, Invitrogen), goat anti-mouse 568 (A11004, Invitrogen), goat anti-rabbit 647 (A21245, Invitrogen), donkey anti-goat 568 (A11057, Invitrogen), donkey anti-rabbit 647 (A31573, Invitrogen), goat anti-mouse 647 (A21235, Invitrogen), goat anti-rabbit 568 (A11011, Invitrogen), and donkey anti mouse 647 (A31571, Invitrogen). After staining, sections were mounted and maintained at 4°C in the dark until analysis.
In vivo cell quantification and fate mapping
Quantification and fate mapping of tdT+ cells in the DG was performed as previously described24. For quantification of tdT+ SGZ cells, 1 in 12 serial sections starting at beginning of hippocampus (relative to bregma, −1.5 mm) to the end of hippocampus (relative to bregma, −3.5 mm) were used. DG Volume and the total tdT+ cells in the subgranular zone, inner half side of granule layer, outer half side of granule layer were analyzed using unbiased stereology (StereoInvestigator, MBF Biosciences, Inc) as described elsewhere29, 55.
The signal intensity of FMRP, HTT or nitrotyrosine in tdT+ cells in the DG of each animal was quantified using Image J software as previously described27. The z-stack images (2 μm interval) were acquired using Nikon A1 confocal microscope. At least 10 individual tdT+ cells were randomly selected from DG of the brain sections in each animal and the fluorescent intensity of FMRP, HTT or nitrotyrosine was measured after subtracting background pixel intensity in the same image using Image J software (NIH). The average intensity from each animal (at least 10 cells) was count as n = 1 for statistical analysis. Samples from three to four individual animals, each from a different litter, per experimental condition were analyzed (n = 3 or 4).
The signal intensity of FMRP and GFP in GFP+ cells in the DG of retrovirus injected mice was quantified using Image J software. The z-stack images (2 μm interval) were acquired using Nikon A1 confocal microscope. A line was drawn through the soma of each GFP+ cell and the intensity of GFP and FMRP was measured along that line. The intensity of FMRP signal was then plotted alongside background FMRP signal intensity and GFP signal intensity.
In vivo fate mapping of tdT+ cells was performed as described53. Briefly, above 100 tdT+ cells in the DG were randomly selected, and their phenotypes (double labeling with NeuN, S100β, Tbr2, Nestin, GFAF, DCX or Ki67, caspase3) were determined using either a Nikon A1 laser scanning confocal microscope. Data were presented as the percentage of tdT+ cells.
Plasmids and reagents
Retro-DCX-Cre and Retro-CAG-DIO(GFP) were cloned using Retro-CAG-red fluorescent protein (RFP)56 as the backbone. Briefly, CAG and RFP coding sequence were replaced with coding sequences of DCX-Cre or DIO(GFP). Retroviral vector (Retro-SYN-GFPCre) was subcloned from RV-SYN-GTRgp (a general gift from Dr. van Praag (NIH), Dr. Suh (Cleveland Clinic), and Dr. Gage (The Salk Institute) and was described in their publication57 by adding the GFPCre fragment after syn promotor. Retro-CAG-IRES-mitoDsRed was a general gift from Dr. Lie and described in his publication16. shFmr1-mCherry and shNC-mCherry were cloned from our published retroviral vectors expressing shFmr1 and GFP9.
Synapsin-GFP and Synapsin-mitoDsRed were gifts from Dr. Chang (University of Wisconsin-Madison) and described in his publication58. All the sequences inserted into the backbones were confirmed by sequencing. Mfn1 and Mfn2 was a gift from David Chan42 (Addgene plasmid # 23212; # 23213). For in vivo tests, mitochondrial fusion promoting compound M1 (Sigma-Aldrich SML0629) was made in a stock concentration of 50 mg/ml in dimethyl sulfoxide (DMSO) and then diluted to 5 mg/ml in sunflower oil. For in vitro tests, M1 (10 μg/ml in 0.1% DMSO) was administrated 24 hours before fixation of neurons for analysis.
Lenti-shHtt and shNC were cloned using lentivirus-shNC vector as a backbone59 and the U6 or H1-shRNA cassettes were also cloned intro the backbone through HpaI/ClaI restriction sites. The efficiency and specificity of shRNA knockdown were determined by transfecting into Neuro2A using Lipofectamine 2000 (Invitrogen, #11668-027), and analyzed at 60-hr post-transfection by qPCR.
Lenti-dCAS-VP64 (Addgene #61425), lenti-MS2-P65-HSF1 (Addgene #61426) and lenti sgRNA (Addgene #61427) (SAM system) were gifts from Feng Zhang34. 10 sgRNAs targeting the proximal promoter of mouse Htt gene (HTT#1-10), between −400 bp and −7 bp relative to transcription start site (TSS), were designed in benchling based on published scoring methods 60,61 and cloned to the lentiviral sgRNA vector (Addgene #61427). The efficiency of activation of Htt were determined by transfecting the SAM plasmids into Neuro2A using Lipofectamine and analyzed at 60-hr post-transfection by qPCR.
Production of Lentivirus and Retrovirus
Lentivirus production was performed as described previously55, 62, 63. Retrovirus production was performed as described in our previous publications55, 59, 64. Briefly, lentiviral DNA was co-transfected with packaging plasmids pMDL, REV and pCMV-Vsvg into HEK293T cells using calcium phosphate method. Retroviral DNA was co-transfected with packaging plasmids pCMV-gag-pol and pCMV-Vsvg into HEK293T cells using calcium phosphate method. The viral transfer vector DNA and packaging plasmid DNA were transfected into 5X15 cm dishes of cultured HEK293T cells using the calcium phosphate method. The medium containing lentivirus was collected at 36 and 60 hours post-transfection, pooled, filtered through a 0.2-μm filter, and concentrated using an ultracentrifuge at 19 k rpm for 2 hours at 4°C using a SW27 rotor (Beckman). The virus was washed once and then resuspended in 100 μl PBS. We routinely obtained 1X109 infectious viral particles /ml for lentivirus and 1X108 infectious viral particles /ml for retrovirus.
In Vivo Retroviral Grafting and Morphological Analysis of Targeted Neurons
In vivo virus grafting was performed as described24, 59, 64. Briefly, 6-week-old male mice were anesthetized with isofluorane and placed in a stereotactic instrument (DAVID KOPF Instruments, Tujunga, CA). Microinjections were performed using custom-made injection 33-gauge needles (Hamilton, #776206, Reno, NV, USA) connected to a 10 μL syringe (Hamilton, #87930). Virus (1 μl with titer greater than 1X108/ml) was mixed and then stereotaxically injected into the dentate gyrus using the following coordinates relative to bregma, caudal: +2.0 mm; lateral: +/−1.6 mm; ventral: −2.0 mm. 4 weeks post viral grafting, mice were perfused for differential analysis. Mice were deeply anesthetized with pentobarbital and perfused with saline followed by 4% PFA. Morphological analyses of retroviral labeled new neurons were performed as described 59, 65-67. eGFP+ or mitoDsRed+ neurons were imaged on a Nikon A1 confocal or a Zeiss Apotome microscope with a 20 X objective or 60 X oil objective. Z-stacks of eGFP+ dendrites were captured at 3 μm intervals and the dendrites and the cell body of single eGFP+ neurons were analyzed by Neurolucida software with 3D module plug-in (MicroBrightField, Inc. Williston, VI, http://www.mbfbioscience.com/). The morphology of mitochondria in neurites was analyzed using ImageJ (RRID:SCR_003070) with mitochondrial morphology plug-in as previously described 68. The signal intensity of HTT or nitrotyrosine in neurons post transfection was quantified using Image J software. About 10 individual mitoDsRed+ cells each animal (3-4 animals each group) were randomly selected, the fluorescent intensity of HTT or nitrotyrosine was measured after subtracting background pixel intensity in the same image using Image J software (NIH).
Electrophysiology
Young adult male C57BL/6 wild type and transgenic mice (1-month-old) were used for stereotaxic surgery followed by patch clamp experiments. All the protocols related to animal maintenance and experiments were in accordance with the National Institute of Health guidelines and approved by the NIA Institutional Animal Care and Use Committees. Mice were anaesthetized (Avertin 0.4 mg g−1 i.p.) and stereotaxic surgery was performed to deliver two injections of 1 μl of SYN-CRE-GFP retrovirus into the right dorsal and ventral dentate gyrus (DG) using spatial coordinates relative to bregma as follows: Dorsal DG, anterior-posterior (AP) = − 2.10 mm; medial-lateral (ML) = 1.9 mm; dorso-ventral (DV) = −2.20 mm, and ventral DG, AP = 6 −3.10 mm; ML = 2.8 mm; DV = −3.20 mm.
Four to 5 weeks after retrovirus injection, mice were deeply anesthetized with isoflurane and decapitated, followed by quick transfer of brain into dissection solution comprising (mM) 110 Choline-Cl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 glucose, 1 CaCl2, 7 MgCl2, 0.6 Na+ pyruvate, 1.3 Na+ ascorbate, and 3 kynurenic acid. Horizontal slices (300 μm thick) from dorsal to mid-dorsal hippocampus were obtained and transferred to incubation solution containing (mm): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 2 CaCl2, 2 MgCl2, 20 glucose, 3 Na+ pyruvate and 1.3 Na+ ascorbate (equilibrated with 95% O2 and 5% CO2; pH 7.4, osmolarity- 310 mOsm). Slices were incubated at 34 °C for 10 min and stored at room temperature for 1 hour for recovery, followed by patch clamp recordings. Patch clamp experiments were performed in artificial cerebrospinal fluid (ACSF) containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 2 CaCl2, 1.3 MgCl2 and 20 glucose, equilibrated with 95% O2 and 5% CO2; pH 7.4, osmolarity- 305 mOsm) in a submerged-type recording chamber (flow rate 2–2.5 mL/min at 30 °C).
For recording miniature excitatory postsynaptic currents (mEPSCs), pipettes were filled with (in mM): 125 CsOH, 125 gluconic acid, 7 CsCl, 10 HEPES, 0.1 EGTA, 5 Na-ATP, 0.5 Na-GTP, 10 Na-phospocreatine (pH 7.4, adjusted with CsOH; 290 mOsm). For recording intrinsic properties, pipettes were filled with (in mM): 130 K-gluconate, 7 KCl, 0.1 EGTA, 10 HEPES, 5 Mg-ATP, 0.5 Na-GTP and 10 Na-phospocreatine (pH 7.4, adjusted with KOH). Miniature EPSCs were recorded in presence of 0.5 μM tetrodotoxin (TTX), 1 μM CGP 55845, 20 μM Picrotoxin and 20 μM Bicuculline methiodide to block sodium channels, GABAB receptors and GABAA receptors.
Adult born neurons were identified by expression of GFP under epifluorescence using differential interference contrast video microscopy and recorded either in voltage clamp or current clamp mode. Whole-cell patch-clamp recordings were filtered at 2 kHz and digitalized at 10 kHz and 50 KHz for voltage and current clamp recording, respectively using Multiclamp 700B, Digidata 1440A and pClamp 10.4 Software (Molecular Devices). Series resistance was typically within 10–30 MΩ. Miniature EPSC events were analyzed in Igor Pro (Wave Metrics Inc., Lake Oswego, OR, USA) using peak amplitude detection algorithm. Threshold was typically kept at −3 pA and adjusted with respect to the baseline noise. Artifacts were screened through visual inspection and overlapping events (event frequency of ≥100 Hz) were discarded. Raw current traces of 300s were analyzed for peak amplitude, frequency, rise time and decay time constant, which were averaged for each cell and used for further statistical analysis. A single exponential function was fitted to decay times of individual events to evaluate the decay time constant.
Input resistance was calculated by measuring the slope of the V–I curve obtained by plotting the amplitude of the steady-state voltages (V) against the corresponding step current injections (I) from −10 to 0 pA of 500-ms duration with 5 pA increment. Averaged voltage trace from 10-20 sweeps of −10 pA current injections with duration of 1000 ms was used to estimate membrane time constant and capacitance. 100 ms of the initial voltage trace was fitted with a single exponential function to deduce the membrane time constant. Membrane capacitance was calculated as a ratio of membrane time constant and input resistance, calculated from the averaged voltage trace.
FACS Purification of DCX+ cells population from adult brains.
DG tissue were isolated from 6-7 weeks old Fmr1−/y; Dcx-DsRed mice and their WT littermates as described before69 with modifications. Two pairs of WT and KO littermate mice were used and each pair was processed on the same day to minimize variabilities. Briefly, DG were dissected using forceps and 27 gauge needle (BD, #305109) and place in Hank’s balanced salt solution (HBSS; Invitrogen, # 14025-126) on ice. Tissue was spin down and digested using MACS Neural Tissue Dissociation kit (Miltenyi Biotech, # 130-090-753, San Diego, CA, USA). After dissociation with a fire-polished glass pipette, cells were filtered through a 40-μm cell strainer (BD Falcon, #252350, CA) and washed with HBSS, the single-cell suspension from each sample was collected in Neurobasal medium containing B27 (Invitrogen, # 17504-044), 20 ng/ml basic fibroblast growth factor (FGF-2; PeproTech, #K1606, Rocky Hill, NJ, USA), 20 ng/ml epidermal growth factor (EGF, PeproTech, #A2306), 1% Antibiotic-Antimycotic, and 2 mM L-glutamine. To assess cell viability, 4’,6-diamidino-2-phenylindole (DAPI; 1:2000; Roche Applied Science, Indianapolis, IN) was added to the cells before sorting. All cell populations were isolated into single cells using a Becton Dickinson FACS Aria II contained in a Biosafety Carbinet using 20 psi pressure and 100-μm nozzle aperture. 10,000 total alive or Dcx-DsRed+ alive Cells were collected directly in Trizol. Gates were set manually by using control samples (without Dcx-DsRed).
RNA Isolation, qPCR, and RNA-seq analyses of FACS-isolated cells
Total RNA from the sorted cell was isolated using the Direct-zol™ RNA MiniPrep Kit (Zymo Research Corporation. Irvine, CA, USA). For verifying the Dcx+ or Nestin+ cell population enrichment, the isolated RNA was reverse transcript by MessageBOOSTER™ Whole Transcriptome cDNA Synthesis Kit (MBWT80510, Illumina) for qPCR. Quality, size, and concentration of the RNA during library preparation were analyzed using Bioanalyser 2100 (RNA Pico Kit, Agilent). For RNA-seq, Strand-specific, poly(A) selected cDNA libraries were generated using Nugen Ovation® Ultralow Library Systems (Illumina) according to the manufacturer’s protocol. Library validation and normalization were performed using RT-PCR and Quant-iT PicoGreen (Invitrogen). Cluster generation and high-throughput sequencing were performed on a HiSeq 2500 (Illumina), using the paired-end 100 bp protocol. Reads were aligned to the mouse genome GRCm38 with annotation from Gencode (gencode.vM15.primary_assembly.annotation.gtf)using STAR (v2.5.3a) with options: --runMode alignReads --clip3pAdapterSeq AGATCGGAAG --outSAMunmapped None --outFilterMultimapNmax 5 --outFilterMultimapScoreRange 1 --outFilterMismatchNoverLmax 0.06 --alignIntronMin 20 --alignIntronMax 1000000 -alignSJDBoverhangMin 1 --outFilterIntronMotifs RemoveNoncanonicalUnannotated --outSAMtype BAM SortedByCoordinate --limitBAMsortRAM 5000000000 --quantMode GeneCounts _ --outSAMattributes All --outFilterType BySJout --outFilterScoreMin 10 --outSAMattrRGline ID:foo --alignEndsType EndToEnd. Reads mapped to the “+” strand of annotated genes were counted by STAR using the above option “--quantMode GeneCounts”.
Bioinformatics analysis
EBSeq (v1.18.0) was used to test for differential expression in two WT and KO samples and differentially expressed genes were defined using a cutoff of PPEE<0.05. Gene ontology analysis was performed with the PANTHER functional classification70. KEGG pathway enrichment analysis was performed using the WebGESTALT (Web-Based Gene Set Analysis Toolkit) with Benjamini and Hochberg multiple test adjustment71. Proteins with known physical interactions with the products of DE genes were extracted from the BioGRID database72, release 3.4.141,
Real-Time PCR
RT-PCR and real-time PCR were performed using standard methods as described 59, 64, 73. The first-strand cDNA was generated by reverse transcription with random primers using Transcriptor First Strand cDNA Synthesis Kit (Roche, #04896866001). Standard RT-PCR was performed using GoTaq DNA polymerase (Promega, #M3005). To quantify the mRNA levels using real-time PCR, aliquots of first-strand cDNA were amplified with gene-specific primers and universal SYBR Green PCR supermix (Bio-Rad, #172-5124) using a Step-1 Real-Time PCR System (Applied Biosystems). The PCR reactions contained 20-40 ng of cDNA (except the cDNA for the IP, for which 5% of the cDNA was used for each gene examined), and 300 nM of forward and reverse primers in a final reaction volume of 20 μl. The sequences of primers used for RT-PCR reactions are as the following:
| GAPDH Forward: | AATGGGAAGCTTGTCATCAACG |
| GAPDH Reverse: | GAAGACACCAGTAGACTCCACGACATA |
| DsRed Forward | AAGGTGTACGTGAAGCACCC |
| DsRed Reverse | CCCATGGTCTTCTTCTGCAT |
| NeuroD Forward | TTA AAT TAA GGC GCA TGA AGG CC |
| NeuroD Reverse | GGA CTG GTA GGA GTA GGG ATG |
| GFAP Forward | CCA AGC CAA ACA CGA AGC TAA |
| GFAP reverse | CAT TTG CCG CTC TAG GGA CTC |
| DCX Forward | TGACAACATTAACCTGCCTCA |
| DCX Reverse | TTGGGGTTGACATTCTTGGTG |
| Nestin Forward | AGGCTTCTCTTGGCTTTCC |
| Nestin Reverse | GAGTCTCAAGGGTATTAGGCAAG |
| NeuN Forward | CGACCTGCGGCAAATGTTC |
| NeuN Reverse | CGGTCAGCATCTGAGCTAGTTTC |
| Opa1 Forward | TGGAAAATGGTTCGAGAGTCAG |
| Opa1 Reverse | CATTCCGTCTCTAGGTTAAAGCG |
| Mfn1 Forward | CCTACTGCTCCTTCTAACCCA |
| Mfn1 Reverse | AGGGACGCCAATCCTGTGA |
| Mfn2 Forward | CTGGGGACCGGATCTTCTTC |
| Mfn2 Reverse | CTGCCTCTCGAAATTCTGAAACT |
| Fis1 Forward | AGGCTCTAAAGTATGTGCGAGG |
| Fis1 Reverse | GGCCTTATCAATCAGGCGTTC |
| Drp1 Forward | CAGGAATTGTTACGGTTCCCTAA |
| Drp1 Reverse | CCTGAATTAACTTGTCCCGTGA |
| Htt Forward | CTCCATGCTGTTACGACTCATC |
| Htt Reverse | AGGGAGGAAGGAGCCAAAATC |
| Mtfr1 Forward | CTGGGCTGGATTAAGTGCCTG |
| Mtfr1 Reverse | TGGCTGGTAAGCTGAAATTGAA |
| Cox6a1 Forward | TCAACGTGTTCCTCAAGTCGC |
| Cox6a1 Reverse | AGGGTATGGTTACCGTCTCCC |
| Idh3b Forward | TGGAGAGGTCTCGGAACATCT |
| Idh3b Reverse | AGCCTTGAACACTTCCTTGAC |
| Mrm2 Forward | GAAGCTGGTAGGCGTTCCC |
| Mrm2 Reverse | CCTTTGCGGCCTTCACAAAC |
| Atp5g2 Forward | CAGTGGAGTTGAAGCGACCA |
| Atp5g2 Reverse | TGTCGATGTCCCTTGAAATGG |
| Acot9 Forward | ATACGCTTGCCTGGTTCCAA |
| Acot9 Reverse | CTCCAGACTGTGGATACGCC |
Transfection and in vitro analysis of primary hippocampal neurons
Hippocampal neurons were isolated and transfected as described previously59. Briefly, hippocampal neurons from KO or WT P0-1 neonate mice were grown as dispersed mixed cell cultures. The hippocampal neurons were transfected with Synapsin-GFP or Synapsin-mitoDsRed using the calcium phosphate precipitation on DIV 4 as they were undergoing dendritic and axonal morphogenesis during this time. At 72-hours after transfection (DIV 7), transfected neurons were fixed with 4% paraformaldehyde, washed with Dulbecco’s phosphate-buffered saline, and coverslipped in DAVCO-PVA. Dendritic morphological analysis was carried out with an Olympus BX51 microscope with 20X lens, a motorized stage, and digital camera. Dendritic traces were performed in real-time using Neurolucida software (MicroBrightField). Approximately 30 neurons were analyzed for each group in each experiment. For mitochondrial morphology analysis, mitoDsRed-positive neurons were imaged using an AxioImagerZ2 ApoTome confocal microscope (Plan-APOCHOROMAT, 20×, numerical aperture = 0.8; Zeiss) or A1RSi confocal microscope system (Nikon) with a 60 × oil objective. The aspect ratio of mitochondria in neurites was analyzed using ImageJ (RRID: SCR_003070) with mitochondrial morphology plug-in as previously described74.
Photoswitched live imaging of mitochondria
The fluorescent protein mEos3.2 75 was cloned into an expression vector containing an EF1 alpha promoter and a mitochondrial import sequence from subunit VIII of human cytochrome c oxidase (pEF mito-mEos3.2/pJDV0021). The primary hippocampal neurons were transfected with pEF mito-mEos3.2 using the calcium phosphate precipitation on DIV 4. At 72-hours after transfection (DIV 7), transfected Neurons were imaged with an Olympus FV1000 spectral based BX61WI (water immersion) upright laser scanning confocal microscope. Three images were acquired. One image before photoswitching, the second image immediately after photoswitching a small area of dendritic mitochondria (T=0), and a third image five minutes later (T=5min). The photoactivated (red) channel was thresholded to mitochondrial structures and total “red” area was calculated. Data is presented as total photoswitched area at T=0 paired with total photoswitched area at T=5min. Wild type (WT) mitochondria will move and fuse to other nearby mitochondria spreading the red signal over a large are, whereas mitochondria deficient in fusion will not spread the red (photoswitched) signal to nearby mitochondria.
Measurement of mitochondrial membrane potential
The mitochondrial membrane potential in neurons on DIV 7 was determined by JC-10 Mitochondrial Membrane Potential Assay Kit (Abcam), following the manufacturer’s protocol. Briefly, neurons were stained with JC-10 solution for 30 min at 37°C. Neurons incubated with mitochondrial uncoupler FCCP (Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, 10μM, Sigma-Aldrich) for 30 min were used as a negative control. After adding buffer B, neurons were imaged using an A1RSi confocal microscope system (Nikon) with a 20 × objective. The mitochondrial membrane potential was assessed by the quantifying the ratio of the intensity of red fluorescence (emission wavelength 590 nm) to green fluorescence (emission wavelength 520 nm).
Analysis of HTT protein level in primary hippocampal neurons
DIV7 WT hippocampus neurons were infected with Lv-shHTT or shNC and the cells were fixed using 4%PFA 48 hours post-infection. DIV7 Fmr1 KO or WT hippocampus neurons were fixed using 4%PFA and analyzed. The signal intensity of HTT in GFP+ cells in the infected group or DCX+ cells in untreated group was quantified using Image J software. The images were acquired using Nikon A1 confocal microscope with a 20x objective. At least 30 individual GFP+ or DCX+ cells were randomly selected from each group and the fluorescent intensity of HTT was measured after subtracting background pixel intensity in the same image using Image J software (NIH).
RNA Immunoprecipitation
DG of 3-week-old WT mice were harvested and homogenized in 1ml of homogenization buffer (25mM Tris [pH 7.0], 25mM Tris [pH 8.0], 100mM KCl, 12mM MgCl2, 10% IGEPAL, 1mM DTT, Protease inhibitors, 1x RNase inhibitor, 200 units/mL, 100μg/mL Cycloheximide, 1mg/mL Heparin) with 2X complete protease inhibitors (Boehringer-Mannheim). Nuclei and debris were pelleted at 14,000 X g for 15 min. The resulting supernatant was pre-cleared for 1 h with 100 μl recombinant protein G agarose (Invitrogen) (washed with lysis buffer first). An aliquot of pre-cleared input was saved for RNA extraction (20 μl). A monoclonal antibody against Fmr1 (7F1-1-C, DSHB) was incubated with supernatant at 4ºC for 4 h and before add Dynabeads (lifetech). The antibody/FMRP conjugation was rotated at 4ºC overnight. After third wash with the High salt buffer (25mM Tris [pH 7.0], 25mM Tris [pH 8.0], 300mM KCl, 12mM MgCl2, 1mM DTT, 100μg/mL Cycloheximide), the immunoprecipitations was re-suspended into Trizol (Invitrogen) for RNA isolation. RNA from IP and input was used and all Real-time PCR reactions were carried out in duplicate for each sample on each amplicon. The relative expression levels of genes in specific Fmr1-immunoprecipitations compared with non-specific immunoprecipitations (mouse IgG only) was calculated.
mRNA Stability Assay
DIV7 Fmr1 KO or WT hippocampus neurons were treated with 10 μg/ml of actinomycin D (Sigma-Aldrich) to inhibit gene transcription as described elsewhere 73 and neurons were collected at various time intervals for RNA isolation and real-time PCR analysis. Htt mRNA levels were normalized to Gapdh. RNA decay kinetics and half-life were analyzed using a published method 76. Briefly, we used exponential function M =M0e−ƛt (M: amount of mRNA at t time, M0: amount of mRNA t t=). ƛ= (ln)/T1/2 (T1/2 is the half-life of the mRNA).
In Vivo Lentiviral Grafting followed by behavioral tests.
Lentivirus (Lenti-shHtt or Lenti-shNC) stereotaxic injection was performed using similar method as described for retroviral injection (see above). Briefly, 1 μl virus was stereotaxically injected into the dentate gyrus of adult mice using the following coordinates relative to bregma, caudal: +2.0 mm; lateral: +/−1.6 mm; ventral: −2.0 mm. All mice were given at least 21 days to recover before subjected to behavioral testing started with open field test to evaluate locomotor activity and general anxiety.
Open field activity test
This test was performed as described26, 27. Activity was recorded at 10-min intervals for total 30-min by means of a computer operated tracking system (Omnitech Electronics Inc, USA). Total distance moved, and distance travelled in the Centre and periphery of arena were measured.
Novel location test
The experimental procedure was developed by the core facility of Waisman Center at UW-Madison as previously described 26, 27. Briefly, mice were handled for approximately 5 min a day for a maximum of 5 days prior to the experiment. All procedures were conducted during the light cycle of the animal between 9 a.m. and 6 p.m. Before the trial session, mice were brought into testing room and were allowed to acclimate for at least 30 minutes. Testing consisted of five 6-min trials, with a 3-min intertrial interval between each trial. During the intertrial interval, the mouse was placed in a holding cage, which remained inside the testing room. In the first trial (Pre-Exposure), each mouse was placed individually into the center of the otherwise empty open arena (38.5cm Long×38.5cm wide, and 25.5cm high walls) for 6 min. For the next three trials (Sample Trials 1-3), two identical objects were placed equidistantly from the arena wall in the corners against the wall with the colored decal. Tape objects to the floor of the arena. Then, each mouse was placed individually into the center of the arena, and was allowed to explore for 6 minutes. At the end of the trial, the mouse was removed and returned to the home cages for 3 minutes. In the last trial (Test), one of the objects was moved to a novel location, and the mouse was allowed to explore the objects for 6 minutes, and the total time spent exploring each object was measured. During the test phase, exploration time was defined as any investigative behavior (i.e., head orientation, climbing on, sniffing occurring within < 1.0 cm) or other motivated direct contact occurring with each object. To control for possible odor cues, objects were cleaned with 70% ethanol solution at the end of each trial and the floor of the arena wiped down to eliminate possible scent/trail markers. During the test phase, two objects were wiped down prior to testing so that the objects would all have the same odor. Based on a study77, the discrimination index was calculated as the percentage of time spent investigating the object in the new location minus the percentage of time spent investigating the object in the old location: discrimination index = (Novel Location exploration time/total exploration time × 100) – (Old Location exploration time/total exploration time × 100). A higher discrimination index is considered to reflect greater memory retention for the novel location object. All experiments were videotaped and scored by scientists who were blinded to experimental conditions to ensure accuracy.
Three-chamber sociability tests
Three-chamber social behavior test was carried out as previously described 37, 78. The social behavior testing apparatus was a 3-chambered box with dimensions of each chamber being 40 cm in length, 22 cm in width, and 23 cm in height. Dividing walls were made of clear Plexiglas containing small circular doors (7 cm in diameter) allowing access into each chamber. The procedure involved 3 phases: habituation, tests of social interaction, and social novelty recognition. Mouse was first placed in the middle compartment and allowed to explore all 3 chambers for 10 min. After habituation, an unfamiliar mouse (stranger 1, age and sex matched to the test mouse) was placed into one side of the compartments (constrained by a round wire cage that permitted nose contact between the bars but prevented extensive physical contact; stimulus mice were habituated to small wire cages for 5–10 min each for 2 days before testing), whereas the wire cagethe in the opposite side compartment contained a motionless toy. For evaluation of the social interaction, the test mouse was returned to the apparatus for 10 min. The time of exploration (approach, sniffing, and rearing within 2 cm from the round cage) of the stranger 1 and the motionless toy by the test mouse was recorded and scored offline by a trained tester blind to genotype. For the social novelty recognition test, one side compartment of the apparatus contained the familiar mouse (from the previous social interaction phase), the other side contained a novel unfamiliar mouse (stranger 2). Active exploration of the familiar and the unfamiliar mouse during the 10-min period was recorded.
Human cell culture, neural differentiation and transplantation
Human FXS iPSCs (FX11-7) were published previously5. GM1 (GM00498-4) iPSCs were generated from fibroblasts from an apparently healthy 3 year old male obtained from Coriell (GM00498) using the same Yamanaka method as described for FXS iPSCs5. The iPSCs were cultured as described using similar method as described31. Briefly, iPSCs were cultured on MEF feeder layers (WiCell) with a daily change of hESC medium of DMEM/F12 (Thermo Fisher Scientific), 20% knockout serum replacement (KSR, Thermo Fisher Scientific), 0.1 mM 2-mercaptoethanol (Sigma), 1x L-Glutamine (Thermo Fisher Scientific), 6 ng/ml FGF-2 (Waisman Biomanufacturing). Cells were passaged using 6 U/ml of dispase (Thermo Fisher Scientific) in hESC medium, washed and replated at a dilution of 1:5 to 1:10. G-banding was performed by WiCell Cytogenetics Services (Madison, WI), as described previously31.
Neural induction was carried out using a dual SMAD method79 with modifications. In brief, 4 days after hPSCs were passaged onto MEFs, neural differentiation were induced by switching hESC medium to neural induction medium (NIM) of DMEM/F12:Neurobasal 1:1, 1x N2, 1x L-Glutamine, 1x Anti-Anti (GIBCO), 10 μM SB432542 (Selleck), 100 nM LDN193189 (Selleck), and 5 uM XAV-939 (Selleck). Cells were cultured in NIM for 9 days with a daily medium change. Cells were then lifted neural progenitor cell (NPC) medium (Neurobasal medium, 1x GlutaMAX (Thermo Fisher Scientific), 1x N2, 0.5x B27 without vitamin A (Thermo Fisher Scientific), 1x Anti-Anti). Cells were further cultured for 22 days with daily change of NPC medium. Most cells at this point expressed markers of forebrain NPCs, NESTIN and PAX6.
FXS and control NPCs were infected by lentivirus-H-shNC-GFP for 3-4 days before transplantation. About 2×105 human FXS or control NPCs in 2 μl artificial cerebrospinal fluid (aCSF) containing B27, 20 ng/ml BDNF were transplanted into developing mouse brain as previously described 80. Briefly, neonatal SCID mice within 36 hr of birth were cryoanesthetized with ice. Approximately 2 μl/side of cell suspension was slowly injected into bilateral cortex with a glass micropipette. During the injection, the glass pipette was slowly withdrawn so that some of the cells were placed in the cortex. 4 months post-transplantation, mice were perfused transcardially with normal saline, followed by freshly made ice-cold 4% paraformaldehyde prepared in PBS. Morphological analyses of developing neurons were performed as described.
Statistical Analysis
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications {Smrt, 2007 #125}{Guo, 2011 #10}{Gao, 2015 #37}{Guo, 2015 #34}{Li, 2018 #111}. Data distribution was assumed to be normal but this was not formally tested. Statistical analysis was performed using ANOVA and Student’s t test, unless specified, with the aid of SPSS version 22 and GraphPad software. Two tailed and unpaired t-test was used to compare two conditions. Two-way ANOVA with Tukey’s post hoc analysis was used for analyzing multiple groups. One-Way ANOVA with Bonferroni post hoc test used for comparison among for in vivo dendritic analysis of different genotypes (GraphPad software 6). Scholl analysis was carried out using multivariate analysis of variance (MANOVA) using SPSS statistical software. All data were shown as mean with standard error of mean (mean ± SEM). Probabilities of p < 0.05 were considered as significant.
Reporting Summary
Further information on research design is available in the “Life Sciences Reporting Summary” linked to this article.
Code availability
Transcriptome data for this project are available on the Gene Expression Omnibus (accession number GSE117111). We have used only published software and freely accessible software for data analyses. Further details can be requested from the corresponding author.
Data Availability Statement
Source data associated with Figs.3 can be accessed through GEO: GSE117111. All data are reported in the main text and supplementary materials, stored at the University of Wisconsin-Madison and are available from the corresponding author upon request.
Supplementary Material
Acknowledgement
We thank Y. Xing, S. Malone, H. Zhao, E. Berndt, Y. Zhao, J. Le, Y. Sun, J. Hoang, and R. Spitzer for technical assistance, Drs. Q. Bu, A. Wang, Q. Chang, D. Joshi, and W. Qiu for help with mitochondrial analysis, K. Knobel, J. Pinnow, H. and Mitchel at the Waisman IDD Model Core, UW Carbone Cancer Center Flow Cytometry lab for help with cell isolation, and S. Splinter-BonDurant and the UW-Madison Biotechnology Center for next generation sequencing services. We also thank Dr. U. Mueller (Scripps Institute, San Diego, CA) for Tg(Dcx -CreERT2) mice and Dr. D. Lie (Friedrich-Alexander University, Erlangen, Germany) and C. Chang for viral vectors expressing mitochondrial markers. This work was supported by grants from the National Institutes of Health (R01MH078972, R56MH113146, R01NS105200 and R01MH116582 to X.Z., P30HD03352, U54HD090256 to the Waisman Center, MH061876 and NS097362 to E.R.C., F32NS098604 to J.D.V.), UW Vilas Trust (Kellett Mid-Career Award) and UW-Madison and Wisconsin Alumni Research Foundation (to X.Z.), Jenni and Kyle Professorship (to X.Z.), John Merck Fund (to X.Z, and A.B)., and in part by the National Institute on Aging, Intramural Research Program (to H.v.P.).
Footnotes
Accession Codes:
Transcriptome data for this project are available on the Gene Expression Omnibus (accession number GSE117111)
Competing interests
The authors declare no competing interests.
References
- 1.Berry-Kravis EM, et al. Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat Rev Drug Discov (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagerman RJ & Polussa J Treatment of the psychiatric problems associated with fragile X syndrome. Current opinion in psychiatry 28, 107–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irwin SA, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. American journal of medical genetics 98, 161–167 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Doers ME, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev 23, 1777–1787 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telias M, Kuznitsov-Yanovsky L, Segal M & Ben-Yosef D Functional Deficiencies in Fragile X Neurons Derived from Human Embryonic Stem Cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 15295–15306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telias M, Segal M & Ben-Yosef D Immature Responses to GABA in Fragile X Neurons Derived from Human Embryonic Stem Cells. Front Cell Neurosci 10, 121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contractor A, Klyachko VA & Portera-Cailliau C Altered Neuronal and Circuit Excitability in Fragile X Syndrome. Neuron 87, 699–715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo W, et al. Fragile X Proteins FMRP and FXR2P Control Synaptic GluA1 Expression and Neuronal Maturation via Distinct Mechanisms. Cell reports 11, 1651–1666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurin T, et al. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic acids research (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ascano M Jr., et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devine MJ & Kittler JT Mitochondria at the neuronal presynapse in health and disease. Nature reviews. Neuroscience 19, 63–80 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. The Journal of cell biology 186, 805–816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishihara N, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nature cell biology 11, 958–966 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Okamoto K, Hayashi Y & Sheng M The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Steib K, Schaffner I, Jagasia R, Ebert B & Lie DC Mitochondria modify exercise-induced development of stem cell-derived neurons in the adult brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 6624–6633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schon Eric A. & Przedborski S Mitochondria: The Next (Neurode)Generation. Neuron 70, 1033–1053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin M, Kang J & Smith CB Increased rates of cerebral glucose metabolism in a mouse model of fragile X mental retardation. Proceedings of the National Academy of Sciences 99, 15758 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidovic L, et al. A metabolomic and systems biology perspective on the brain of the fragile X syndrome mouse model. Genome research 21, 2190–2202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lima-Cabello E, et al. An Abnormal Nitric Oxide Metabolism Contributes to Brain Oxidative Stress in the Mouse Model for the Fragile X Syndrome, a Possible Role in Intellectual Disability. 2016, 8548910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.el Bekay R, et al. Enhanced markers of oxidative stress, altered antioxidants and NADPH-oxidase activation in brains from Fragile X mental retardation 1-deficient mice, a pathological model for Fragile X syndrome. The European journal of neuroscience 26, 3169–3180 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Lumaban JG & Nelson DL The Fragile X proteins Fmrp and Fxr2p cooperate to regulate glucose metabolism in mice. Human Molecular Genetics 24, 2175–2184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bechara EG, et al. A novel function for fragile X mental retardation protein in translational activation. PLoS biology 7, e16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo W, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nature medicine 17, 559–565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo W, et al. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Human Molecular Genetics 21, 681–691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Science translational medicine 8, 336ra361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. Reducing histone acetylation rescues cognitive deficits in a mouse model of Fragile X syndrome. Nature communications 9, 2494 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kempermann G, Song H & Gage FH Neurogenesis in the Adult Hippocampus. Cold Spring Harbor perspectives in biology 7, a018812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y, et al. Fragile × mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet 6, e1000898 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, et al. Integrative Single-Cell Transcriptomics Reveals Molecular Networks Defining Neuronal Maturation During Postnatal Neurogenesis. Cerebral cortex (New York, N.Y. : 1991) 27, 2064–2077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, et al. Establishment of Reporter Lines for Detecting Fragile X Mental Retardation (FMR1) Gene Reactivation in Human Neural Cells. Stem cells (Dayton, Ohio) 35, 158–169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, et al. A small molecule promotes mitochondrial fusion in mammalian cells. Angewandte Chemie (International ed. in English) 51, 9302–9305 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Carmo C, Naia L, Lopes C & Rego AC Mitochondrial Dysfunction in Huntington’s Disease. Advances in experimental medicine and biology 1049, 59–83 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidhu H, Dansie LE, Hickmott PW, Ethell DW & Ethell IM Genetic removal of matrix metalloproteinase 9 rescues the symptoms of fragile X syndrome in a mouse model. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 9867–9879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolan BM, et al. Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc Natl Acad Sci U S A 110, 5671–5676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu B, et al. GRK5 Regulates Social Behavior Via Suppression of mTORC1 Signaling in Medial Prefrontal Cortex Cerebral cortex 28, 421–432 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Pratt KG, Zimmerman EC, Cook DG & Sullivan JM Presenilin 1 regulates homeostatic synaptic scaling through Akt signaling. Nat Neurosci 14, 1112–1114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan Eitan S, et al. Early mitochondrial abnormalities in hippocampal neurons cultured from Fmr1 pre‐mutation mouse model. Journal of Neurochemistry 123, 613–621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross-Inta C, et al. Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochemical Journal 429, 545–552 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westermann B Mitochondrial fusion and fission in cell life and death. Nature reviews. Molecular cell biology 11, 872–884 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of cell biology 160, 189–200 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filadi R, Pendin D & Pizzo P Mitofusin 2: from functions to disease. Cell death & disease 9, 330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pham AH, Meng S, Chu QN & Chan DC Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum Mol Genet 21, 4817–4826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berthet A, Margolis EB, Zhang J, Hsieh I & Zhang J Loss of mitochondrial fission depletes axonal mitochondria in midbrain dopamine neurons. 34, 14304–14317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang S, et al. Mfn2 ablation causes an oxidative stress response and eventual neuronal death in the hippocampus and cortex. 13, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang D, Yan S, Yu Q, Chen D & Yan SS Mfn2 is Required for Mitochondrial Development and Synapse Formation in Human Induced Pluripotent Stem Cells/hiPSC Derived Cortical Neurons. Scientific reports 6, 31462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirendeb U, et al. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum Mol Genet 20, 1438–1455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochaba J, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proceedings of the National Academy of Sciences 111, 16889–16894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Culver BP, et al. Huntington’s Disease Protein Huntingtin Associates with its own mRNA. Journal of Huntington’s disease 5, 39–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 51.Mientjes EJ, et al. The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiology of disease 21, 549–555 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Qiu R, Tsark W & Lu Q Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res 85, 3567–3573 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Lagace DC, et al. Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J Neurosci 27, 12623–12629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soriano P Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics 21, 70–71 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Smrt RD, et al. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiology of disease 27, 77–89 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao C, Teng EM, Summers RG Jr., Ming GL & Gage FH Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience 26, 3–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vivar C, et al. Monosynaptic inputs to new neurons in the dentate gyrus. Nature communications 3, 1107–1107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bu Q, Wang A & Hamzah H CREB Signaling Is Involved in Rett Syndrome Pathogenesis. 37, 3671–3685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smrt RD, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28, 1060–1070 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barkho BZ, et al. Endogenous matrix metalloproteinase (MMP)-3 and MMP-9 promote the differentiation and migration of adult neural progenitor cells in response to chemokines. Stem Cells 26, 3139–3149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, et al. Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. The Journal of biological chemistry 283, 27644–27652 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu C, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell stem cell 6, 433–444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y, et al. Inhibition of miR-15a Promotes BDNF Expression and Rescues Dendritic Maturation Deficits in MeCP2-Deficient Neurons. Stem cells (Dayton, Ohio) 33, 1618–1629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo W, et al. Fragile X Proteins FMRP and FXR2P Control Synaptic GluA1 Expression and Neuronal Maturation via Distinct Mechanisms. Cell Rep 11, 1651–1666 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giresi PG, Kim J, McDaniell RM, Iyer VR & Lieb JD FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome research 17, 877–885 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 284, 13843–13855 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo W, Patzlaff NE, Jobe EM & Zhao X Isolation of multipotent neural stem or progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nature protocols 7, 2005–2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome research 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang B, Kirov S & Snoddy J WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic acids research 33, W741–748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic acids research 34, D535–539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo W, et al. RNA-binding protein FXR2 regulates adult hippocampal neurogenesis by reducing Noggin expression. Neuron 70, 924–938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dagda RK, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry 284, 13843–13855 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang M, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9, 727–729 (2012). [DOI] [PubMed] [Google Scholar]
- 76.Beckel-Mitchener AC, Miera A, Keller R & Perrone-Bizzozero NI Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J Biol Chem 277, 27996–28002 (2002). [DOI] [PubMed] [Google Scholar]
- 77.Contestabile A, et al. Lithium rescues synaptic plasticity and memory in Down syndrome mice. J Clin Invest 123, 348–361 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gantois I, et al. Metformin ameliorates core deficits in a mouse model of fragile X syndrome. Nature medicine 23, 674–677 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guillaume DJ, Johnson MA, Li XJ & Zhang SC Human embryonic stem cell-derived neural precursors develop into neurons and integrate into the host brain. J Neurosci Res 84, 1165–1176 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data associated with Figs.3 can be accessed through GEO: GSE117111. All data are reported in the main text and supplementary materials, stored at the University of Wisconsin-Madison and are available from the corresponding author upon request.