Abstract
Conceptual frameworks are useful in research because they can highlight priority research domains, inform decisions about interventions, identify outcomes and factors to measure, and display how factors might relate to each other to generate and test hypotheses. Discovery, translational, and implementation research are all critical to the overall mission of genomic medicine and prevention, but they have yet to be organized into a unified conceptual framework. To fill this gap, our diverse team collaborated to develop the Genomic Medicine Integrative Research (GMIR) Framework, a simple but comprehensive tool to aid the genomics community in developing research questions, strategies, and measures and in integrating genomic medicine and prevention into clinical practice. Here we present the GMIR Framework and its development, along with examples of its use for research development, demonstrating how we applied it to select and harmonize measures for use across diverse genomic medicine implementation projects. Researchers can utilize the GMIR Framework for their own research, collaborative investigations, and clinical implementation efforts; clinicians can use it to establish and evaluate programs; and all stakeholders can use it to help allocate resources and make sure that the full complexity of etiology is included in research and program design, development, and evaluation.
Keywords: genomics, conceptual, framework, model, diversity, translational research, implementation
Introduction
As the pace of genomics research continues to accelerate, it is transitioning from bench to bedside and from a discovery-only technology to one that is demonstrating utility in clinical care. The complexity of genomic technologies presents significant and unique challenges. Unlike a chemistry panel that is interpreted in the same way regardless of context or phenotype, genomic tests can have different interpretations and different uses across various areas of medicine. For example, a primary care provider might focus on genetic disease risk, a pediatric endocrinologist might focus on genomic diagnosis, and a pain specialist might focus on pharmacogenomic results. Disease risk interpretation is also influenced by an individual’s phenotype, family health history, and ancestry, so the same test result may have different implications for clinical management in different individuals and be interpreted differently by different clinicians, individuals, communities, payers, and policymakers.
Genomic discoveries must be relevant to and meet the needs of individuals and providers from a variety of backgrounds and in a range of settings, necessitating inclusion of diverse populations in all aspects of genomics research, from discovery to translation to implementation in clinical care. To avoid undermining the scientific integrity of conclusions drawn from research studies, diversity in research is important. Researchers should ensure the collected data use standardized measures that reflect the multidimensional nature of a person’s identity, especially within the context of race, ethnicity, socioeconomic status, cultural identity, family background, and geographic ancestry.1 There also is a concerted effort to diversify other domains of genomics research: from bench to bedside, from rare disease to common disease focus, from academic to community sites, and from European ancestry to diverse ancestry populations.
Leading the way are two U.S. national research consortia funded by the NIH National Human Genome Research Institute (NHGRI). The first, the Clinical Sequencing Evidence-Generating Research (CSER) consortium (also funded by the National Institute on Minority Health and Health Disparities and by the National Cancer Institute),2 is tasked with addressing challenges related to implementation of sequencing in diverse populations. The second, the Implementing Genomics in Practice (IGNITE) network, focuses on implementation of genomic testing and technologies in diverse clinical settings and populations.3, 4 Both recognize that gaining complete insights about the relative roles of genomic variation, social context, and physical environment in human traits, health, and disease requires greater participation of individuals with ancestors from all regions of the world.
Today, the opportunity exists to support research in diverse populations that promises to offer a scientific basis for challenging the extrapolations made by the current uses of race and ethnicity.5
This new emphasis on broadening genomics research has not yet had use of a common conceptual framework to guide research into translation and implementation and to feed back into and inform discovery research. Conceptual frameworks are a way of organizing ideas.6 They can provide a structure that intuitively and clearly presents underlying factors that may inform or lead to interventions, concepts that can be measured, outcomes relevant for evaluation, and connections positing how these factors might relate to each other. Conceptual frameworks can focus on discovery research (uncovering causes and influences of specific factors on specific disease outcomes), translational research (evaluating clinical impact of interventions), and/or the emerging science of implementation research (evaluating best practices to integrate effective interventions into clinical care).
Discovery, translational, and implementation frameworks are critical to the overall mission of genomic medicine and could be but have not yet been combined into a unified conceptual framework. There are many validated frameworks in discovery and translational fields, and frameworks such as the Consolidated Framework for Implementation Research (CFIR) are emerging in implementation science.7 A unified genomics research framework would integrate discovery (e.g., whether gene-environment interactions increase disease risk), translation (e.g., the effect of gene sequencing of sick individuals on healthcare utilization and quality of life), and implementation (e.g., what types of clinical site characteristics facilitate or impede use of genomics in routine care). One could also use a unified framework to examine the wider array of factors that contribute to health disparities, including social determinants of health, to test equity-enhancing strategies and to identify best practices to sustain or scale successful interventions or programs.8
Existing frameworks, though valuable, are neither capable of representing the complexity of genomic medicine implementation research nor flexible enough to address the full spectrum of genomic discovery, translational, and implementation research needs. CSER and IGNITE members therefore worked together to build a framework to comprehensively reflect the needs of genomic medicine researchers. We developed this framework to highlight priority research domains and factors across multiple levels—including affected individuals, providers, health systems, and social environments—to consider when developing new hypotheses and research ideas. Our multi-level framework can also be used to inform the choice of interventions undertaken, select measures to evaluate the processes that influence their uptake, evaluate the effects of interventions on individuals and families, and evaluate the implications for health and social policy. This inclusive but simple framework may also be of use to the wider genomics community when developing research questions, strategies, and measures and to those actively integrating genomic medicine into clinical practice. In this paper, we describe the foundation and results of that work: The Genomic Medicine Integrative Research (GMIR) Framework. Because of the broad expertise and experience of the diverse experts and stakeholders involved in its development, GMIR may be generalizable across a variety of clinical settings (e.g., primary and specialty care, urban and rural settings) and participant characteristics (e.g., race/ethnicity, socioeconomic status, access to care).
Material and Methods
Transdisciplinary, Stakeholder-Engaged Collaboration
To ensure that the framework is relevant and useful for the diverse stakeholders interested in translational genomics research and practice, we invited individuals from key stakeholder groups—individuals receiving care, clinicians, health systems leaders, advocates, researchers, ethicists, funders, policymakers, payers, and entrepreneurs—to join the development team.9 To ensure we captured a diversity of perspectives and foci, we also included researchers with genomics, health services, economics, psychology, survey, disparities, and implementation research backgrounds, as well as diverse healthcare providers (primary care, specialist, physician, nurse, genetic counselor, trainees, and faculty) from diverse practice sites and settings (rural/urban, academic/community, inpatient/outpatient). Through virtual and in-person meetings, we developed consensus on the domains (major categories) and sub-domains in the framework.
Conceptualization
Through literature review and consensus-building discussions, our team agreed on the framework’s foundational principles and potential uses:
-
•
Built with and for the professionally, geographically, and socio-demographically diverse stakeholders focused on genomics discovery and implementation.
-
•
Recognizing the importance of and relevance of the framework for providers and individuals receiving care in diverse clinical settings, and to achieving health equity.
-
•
Including domains not easily measurable but important to consider. For example, it is difficult to measure unequal distribution of resources such as educational opportunities but it remains crucial to recognize that health is largely determined by factors outside of medical settings.
-
•
Displaying key factors relevant to genomic discovery and implementation for consideration when developing research questions, translating and interpreting findings, and generating and testing hypotheses regarding potential interactions between factors.
-
•
Identifying constructs and developing or selecting measures for these key factors.
-
•
Providing a comprehensive framework that will allow investigation of multiple domains, thus contributing to standardization of outcome measures in genomic medicine.
-
•
For single projects: prioritizing proposed measures intended for data collection.
-
•
For collaborations: harmonizing and standardizing data collection and outcomes across studies and facilitating easy integration with external efforts.
-
•
Generating hypotheses that are testable across diverse populations.
-
•
Further delineating activities for development and validation of novel metrics, instruments, and associations, and identifying gaps to inform future investigations.
Framework Development
While building new research programs to carry out the goals of the CSER consortium,1 investigators wanted to create a framework with a consistent set of standards around a system of ideas and objectives. Our team began by studying existing frameworks to identify relevant domains. These included: a genomics framework developed at one site using stakeholder engagement and focused on genetic testing for chronic disease risk;10, 11 a draft framework based on experiences in IGNITE, the first NHGRI translational genomics research network;3 and the Consolidated Framework for Implementation Research (CFIR).7 We expanded and refined these frameworks to: aid in organizing efforts to prioritize and harmonize genomics research initiatives across all sites; measure utility domains of genomic sequencing through individual and family responses to genomic testing; and evaluate potential mitigating factors, stakeholder engagement, and the interplay of diverse healthcare systems that may affect outcomes associated with the genomic applications.
To refine and finalize the main domains of the framework, we assigned each domain to an associated working group whose members conducted literature review and used their expertise to add sub-domains under the domain on which they were focused. We then reviewed this draft framework with CSER members and partners from other NHGRI networks including IGNITE, the eMERGE consortium of biorepositories returning genomic results to participants,12 the Population Architecture using Genomics and Epidemiology network (PAGE),13, 14 and other stakeholders in our transdisciplinary group. The development process involved numerous rounds of prioritization and revision to arrive at the GMIR Framework shown in Figure 1.
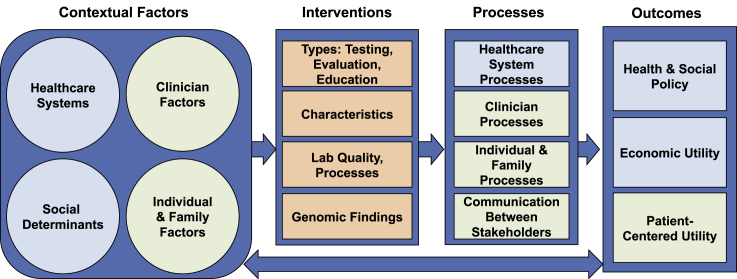
Figure 1.
The Genomic Medicine Integrative Research (GMIR) Framework
Results
The GMIR Framework
As shown in Figure 1, we organized the GMIR Framework into four overarching domains: (1) contextual factors, (2) interventions, (3) processes, and (4) outcomes, each comprising multiple sub-domains and concepts (see also Tables 1, 2, 3, and 4). Arrows in the framework illustrate general associations and known and hypothesized relationships between the major domains. On the figure’s left are contextual factors (Table 1) relevant to genomic discovery and implementation research and practice in diverse populations and settings: healthcare systems factors (infrastructure), social determinants, clinician factors, and individual factors. These sub-domains may influence each other. For example, healthcare resources such as availability of genomics specialists and testing labs may influence access to and usage of these resources by healthcare providers and individuals.
Table 1.
Contextual Sub-domains and Concepts
| Sub-domains | Concepts |
|---|---|
| Healthcare System Factors | |
| Medical care | access to and quality of care; EHR type |
| System features | resources and infrastructure: genomic; mix of individuals receiving care (number, payers, demographics) |
| type: in/outpatient; academic/community; rural/urban; private/public | |
| culture: openness to change; structural discrimination | |
| Social Determinants | |
| Social | networks; support; resources; influence; engagement |
| Built environment | housing; poverty; crime; walkability; exposure to pollution, toxins, pathogens; food; racism; unequal distribution of resources (policies, systems, education, economic) |
| Clinician Factors | |
| Demographics | age; race/ethnicity; sex/gender; training; specialty; experience |
| Psychosocial | psychosocial: stress; satisfaction; self-efficacy; reaction to medical uncertainty |
| stressors/resources: time; access to information; experts; referrals; perceived racism | |
| attitudes, knowledge and understanding: genetic (testing, return of results, expected utility); implicit bias | |
| Individual & Family Factors | |
| Demographics | age; race/ethnicity; sex/gender; language; literacy; numeracy; income; employment; education; insurance; self-reported ancestry |
| Bio-psychosocial | biological/clinical: family history; comorbidity; genetics, including genetic ancestry |
| psychosocial: depression; stress; anxiety; self-efficacy; stigma; racism; perceived racism; activation; cultural norms; family issues | |
| stressors/resources: family; community; occupational; financial; illness | |
| knowledge, attitudes and understanding: genetic testing, results & their disclosure; intervention; trust (in testing, results, providers); expected utility; activation | |
Table 2.
Intervention Sub-domains and Concepts
| Sub-domains | Concepts |
|---|---|
| Genomics-informed intervention components | type: of genetic & genomic testing, sequencing purpose: to evaluate/inform risk assessment, diagnosis, therapy disclosure of results: how and by whom education: of individuals receiving care, families, clinicians |
| Intervention characteristics | cost, evidence, complexity |
| Laboratory quality and processes | technical attributes, bioinformatics pipelines, variant interpretation processes |
| Genomic findings | pathogenicity, actionability, diagnostic yield |
Table 3.
Process Sub-domains and Concepts
| Sub-domains | Concepts |
|---|---|
| Healthcare System Processes | |
| Intervention adoption | participation of individuals receiving care, sites, health systems & providers; uptake adherence; satisfaction; sustained delivery; payer coverage |
| Implementation climate | implementation readiness; access to resources; knowledge & information; compatibility with workflow; leadership engagement |
| Clinician Processes | |
| Intervention-related | adoption; recommendations |
| Behaviors | cultural appropriateness; communication style |
| Individual & Family Processes | |
| Biological/clinical processes and markers | biomarkers measured in blood, urine, saliva, sweat |
| Psychological and behavioral processes | experiences; satisfaction with procedures and materials; understanding of results; health behavior changes in response to results; psychological effects of testing and results (positive and negative); impact of medical recommendations and following of medical recommendations |
| Communication | |
| Nature & quality of communication between stakeholders | individuals in care, families, providers, labs, payers |
| stakeholder engagement and input into processes | |
Table 4.
Outcome Sub-domains and Concepts
| Sub-domains | Concepts |
|---|---|
| Patient-centered utility | health status: physical and mental health; quality of life; mortality; health of individuals, families, communities, populations; perceived, self-reported or personal utility life and reproductive planning health equity |
| Economic utility | cost/value: from individuals receiving care, family, health system and societal perspectives cost effectiveness and healthcare utilization |
| Health and social policy | healthcare, educational, public health, environmental and industry policies, laws and regulations |
Contextual factors may also influence the middle boxes in the figure, i.e., the interventions that researchers and other practitioners put in place, the processes (Table 2) prompted by the interventions among clinicians, individuals, and healthcare systems, and how these stakeholders work together and communicate with each other. For example, healthcare infrastructure (contextual factors) may influence the feasibility of implementing different types of genomics-informed strategies (interventions) in specific settings. Individual-level factors such as attitudes of providers and the individuals they care for may subsequently influence the type of intervention (Table 3) researchers choose to test, for example, based on what is feasible and acceptable. Contextual factors will also influence whether and/or how testing, results disclosure, and education will proceed (interventions), as well as what providers recommend to the individuals they care for and whether individuals follow recommendations (processes). And, contextual factors may directly affect outcomes (Table 4), e.g., demographics such as race and income are directly associated with morbidity and mortality. Interventions may directly prompt processes and influence outcomes, and processes may directly affect outcomes: e.g., high-risk pathologic variants affect health and provider recommendations (processes) often impact healthcare utilization (outcomes). Finally, outcomes may loop back and impact contextual factors: e.g., health policies affect health infrastructure and severity of people’s illnesses may affect individual and provider attitudes toward genetic testing or genomic sequencing.
Concepts, Measures, Harmonization, Hypothesis Development, and Examples
CSER working groups used the finalized the GMIR framework to identify and review existing measures that mapped to the domains. We identified myriad measures and prioritized those that had been validated, particularly in diverse populations. Next, we identified gaps for which stakeholders agreed we needed to develop new scales or measures. Teams representing working group co-chairs, leads of each CSER research site, and funders then decided which measures would be harmonized (implemented by every site) and recommended (used if sites choose to measure a specific domain). We subsequently built a repository of all measures and prioritized measures and data collection time points that would be successful in the face of competing requirements, including minimizing individual burden and ensuring applicability to site-specific clinical and research contexts. We describe key measures that we agreed to harmonize below.
Underlying contextual factors we harmonized in CSER include individual’s demographics, perceived access to care, clinician and individual knowledge, attitudes, understanding of the genomic intervention that is the focus of the research project, and organizational readiness to change from the healthcare system perspective.
Interventions harmonized include evaluation of the technical (laboratory processes) attributes related to differing bioinformatics pipelines and variant interpretation processes, and the tools and resources necessary to inform case-level reporting and diagnostic yield rates.
Processes harmonized include measurement of perceived utility; provider experience with genomics; provider’s and tested individual’s understanding of, reactions to, satisfaction with, and psychosocial effects of testing, results disclosure and medical recommendations; tested individuals following of or adherence to medical recommendations; and intervention-based attributes (characteristics, adoption).
Outcomes harmonized include patient-centered and economic utility using quality of life tools and associated costs from multiple perspectives, and healthcare resource utilization. We anticipate that analyses of these outcomes will broaden the genomic medicine evidence base to inform health policies and payers that serve diverse populations.
We are now using the GMIR framework to develop and refine working group hypotheses that we will test through research across the CSER network and in collaboration with other consortia and that will guide analytic plans across sites. Strengths from this data harmonization effort include providing an increased sample size for the domains and hypotheses under investigation and allowing comparison of the same measures across diverse clinical and population settings.
Below, we outline three examples of how investigators have used this framework to plan their research, develop and test hypotheses, and contextualize their findings and show how we modified the GMIR Framework for these examples in Figures 2, 3, and 4.
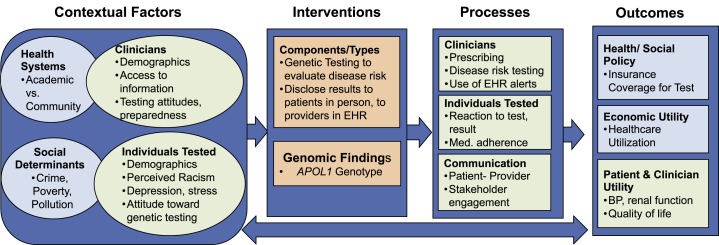
Figure 2.
GMIR Framework Adapted for Chronic Disease Risk Genetic Testing
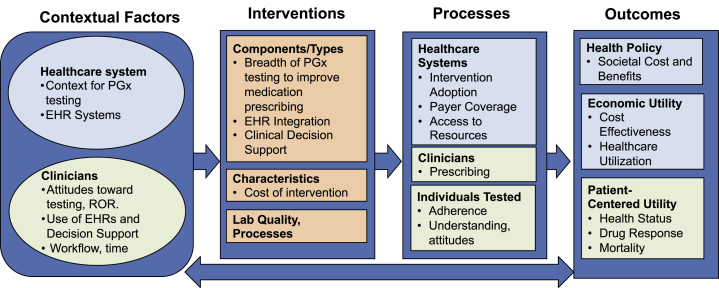
Figure 3.
GMIR Framework Adapted for Pharmacogenomic Testing
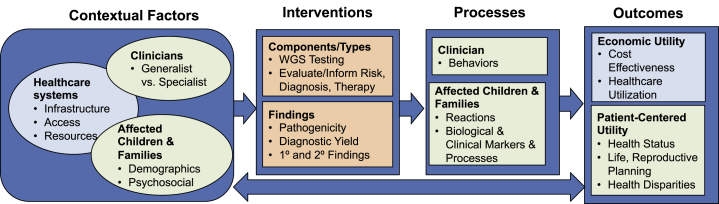
Figure 4.
GMIR Framework Adapted for WGS in Children with Rare Diseases
Genomic Chronic Disease Risk in African Ancestry Populations
The presence of two APOL1 (MIM: 603743) risk variants, nearly exclusively found in African ancestry (AA) people, is associated with a 5- to 10-fold increased risk for hypertension-related kidney failure and explains a significant proportion of black-white renal disease disparities.15 As part of the IGNITE Consortium, a NYC community-clinical-academic team developed a randomized clinical trial to test AA adults for high-risk APOL1 renal disease variants.10 Using a tailored GMIR Framework (Figure 2), the team engaged with a range of stakeholders to explore contextual factors (individual, provider, and investigator attitudes toward race, ancestry, APOL1, and kidney disease), what they would want in an intervention, and what processes and outcomes they thought would be important to assess.11, 16 We chose to assess baseline provider and individual psycho-behavioral contextual factors and selected an intervention to return APOL1 test results to providers through best practice alerts in electronic health records and to individuals receiving care through lay persons. Next, we chose to measure the effect of the intervention on processes including clinical care and on outcomes (individual’s blood pressure and quality of life and clinician’s renal testing). We generated and tested hypotheses based on the framework, such as whether providers’ experience would increase their preparedness and confidence in working with individuals who had chronic disease risk genetic testing.17 The team is also studying whether underlying and contextual factors (social determinants such as pollution and crime) modify the association between an intervention component (APOL1 test result) and an outcome (renal function).
Pharmacogenomic Testing, Addressing Organizational, Provider, and Technology Barriers, Assessing Effect on Morbidity, Cost Effectiveness, and Societal Implications
Pharmacogenomic (PGx) variants are potent determinants of drug disposition or metabolism and significantly contribute to adverse outcomes of many common therapies. The PREDICT (Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment) trial assessed the clinical and economic effects of preemptively testing a panel of pharmacogenes across a large population. The contextual factors assessed related to health systems and clinicians. The team first assessed the organizational and clinical context of the implementation to identify and address barriers, including laboratory testing quality and processes, clinician attitudes, organizational and technology infrastructure, governance mechanisms, and integration of PGx testing, returning results and clinical decision support into electronic health records and clinical workflows18, 19 (Figure 3). Next, the team measured process and outcomes (clinical and economic) related to EHR-integrated PGx results, particularly with respect to CYP2C19 (MIM: 609535)-directed antiplatelet therapy after percutaneous coronary intervention.20 They evaluated program effectiveness via measures of genetic data, provider attitudes and behaviors, clinical adoption of the program, and examination of EHR prescribing logs.21, 22 For the other key outcomes—economic and societal utility—the investigators discovered it was challenging to directly measure program costs, so the team conducted standard cost-effectiveness modeling from a societal perspective.23
Whole-Exome Sequencing (WES) in Children with Rare Diseases
As clinicians are increasingly utilizing next-generation technologies such as WES as diagnostic tools, it is important to define the clinical situations in which these technologies will demonstrate maximal clinical utility and cultural acceptability, particularly in underrepresented and underserved populations.24, 25, 26, 27 The Program in Prenatal and Pediatric Genome Sequencing Study is assessing these factors in diverse pediatric populations with rare diseases and couples in pregnancy. The GMIR Framework illustrates aspects of the child’s demographics and social environment (contextual factors), testing intervention, results provision, and clinical and personal outcomes that the team will study (Figure 4). The team used aspects of the GMIR Framework to plan the intervention. To address underlying factors of access and infrastructure, the team will offer testing at institutions that do not have genetic services and that are heavily utilized by underserved parents (contextual factors), enrolling children during routine clinical visits (intervention) and comparing testing provided by genetic and non-genetics providers (process). This study will assess the effects of disclosing primary and secondary findings: key outcomes will include economic-healthcare utilization and patient-centered utility measures, including reproductive planning and physical and emotional state.
Discussion
In the Genomic Medicine Integrative Research (GMIR) Framework, our team aimed to provide a simple, clear, and comprehensive framework that genomics researchers and other stakeholders can use to identify factors that underlie the relationship between genomics and population health, interventions that may affect translation and implementation of genomics into practice, the processes that influence implementation, and key outcomes of interest. Our process successfully demonstrates how diverse stakeholders can develop a unifying genomic framework by coming to consensus on which factors or domains should be included; how to use the framework to generate hypotheses and choose key topics of interest across multiple sites, settings, and projects; how to form subcommittees to focus on these topics; and how to choose and harmonize measures for topics of interest. Our examples show how groups with different priority research areas employed the GMIR Framework to guide their research and advance the knowledge base for diverse populations.
The GMIR Framework may be useful for other genomic medicine research groups or consortia that need to select factors, hypotheses, and measures for individual research projects. It may also help research groups and consortia set the context for large-scale, multi-site research and to generate shared knowledge. There are several strengths of this framework. It was developed collaboratively by researchers from multiple institutions and from multiple disciplines, as well as individuals, clinicians, advocates, funders, payers, and policymakers, using robust stakeholder engagement and team science methods. We carefully collated measures for domains and summarized them on a website. This list of measures will become more comprehensive over time as we and others identify additional validated measures and develop new measures. While we recognize the value of other frameworks relevant to genomic medicine such as ClinGen28 (another NHGRI-funded consortium), these prior frameworks are relatively limited in scope, while the GMIR Framework is considerably more comprehensive.
Our team worked to ensure that the GMIR Framework would be useful for groups studying genomics in diverse populations and clinical settings. Awareness of genetic testing or genomic sequencing as an option, knowledge of the testing process, implicit bias, overcoming concerns regarding testing, proximity to healthcare, and demographic factors exemplify hurdles that can compromise participation at the individual and interpersonal levels.29, 30 Cost, insurance coverage, concerns about stigmatization and discrimination based on test results, and lack of preparedness of systems to facilitate testing and disclosure of results are major barriers to delivering genetic testing equitably at a health system level. Our framework highlights health equity as a critical outcome requiring consideration and highlights key sets of contextual and intervention factors, processes, and outcomes to consider as potential contributors to disparities.
Members of several research networks are employing the GMIR Framework, including CSER,1 IGNITE,15 and eMERGE, which used elements of this framework to create a provider survey.10 Other consortia, such as IGNITE-2 (Web Resources) and All of Us,31 may use the framework to guide selection of hypotheses, measures, and outcomes for participants who have yet to be recruited. The common use of the GMER Framework across multiple consortia may facilitate studies across multiple consortia, increasing collective exploration and sample size power. Ultimately, the contextualization of research findings will help make stronger links back to the underlying healthcare, social determinant, individual, and provider factors that can guide best uses of genomic approaches.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank our partners in the CSER and the IGNITE networks (https://ignite-genomics.org/about-ignite), the Mount Sinai Genomics Stakeholder Board, and our funders: NHGRI, NCI, and NIMHD (for CSER) U01HG009610, U01HG006485, U01HG007301, U01HG007292, U01HG006487, U01HG009599, ZIAHG200387, U24 HG007307; NHGRI (for IGNITE) U01HG007278 and U01HG007282; and NCATS (for the Mount Sinai CTSA-ConduITS) UL1TR001433.
Published: May 16, 2019
Web Resources
CSER Harmonized Measures, https://cser-consortium.org/cser-research-materials (under CSER Phase 2 Harmonized Measures)
IGNITE, https://grants.nih.gov/grants/guide/rfa-files/RFA-HG-17-008.html
OMIM, http://www.omim.org/
References
- 1.Bonham V.L., Callier S.L., Royal C.D. Will precision medicine move us beyond race? N. Engl. J. Med. 2016;374:2003–2005. doi: 10.1056/NEJMp1511294. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bonham, V.L., Callier, S.L., and Royal, C.D. (2016). Will precision medicine move us beyond race? N. Engl. J. Med. 374, 2003-2005. [DOI] [PMC free article] [PubMed]
- 2.Amendola L.M., Berg J.S., Horowitz C.R., Angelo F., Bensen J.T., Biesecker B.B., Biesecker L.G., Cooper G.M., East K., Filipski K., CSER consortium The Clinical Sequencing Evidence-Generating Research Consortium: Integrating Genomic Sequencing in Diverse and Medically Underserved Populations. Am. J. Hum. Genet. 2018;103:319–327. doi: 10.1016/j.ajhg.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Amendola, L.M., Berg, J.S., Horowitz, C.R., Angelo, F., Bensen, J.T., Biesecker, B.B., Biesecker, L.G., Cooper, G.M., East, K., Filipski, K., et al.; CSER consortium (2018). The Clinical Sequencing Evidence-Generating Research Consortium: Integrating Genomic Sequencing in Diverse and Medically Underserved Populations. Am. J. Hum. Genet. 103, 319-327. [DOI] [PMC free article] [PubMed]
- 3.Weitzel K.W., Alexander M., Bernhardt B.A., Calman N., Carey D.J., Cavallari L.H., Field J.R., Hauser D., Junkins H.A., Levin P.A., IGNITE Network The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genomics. 2016;9:1. doi: 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Weitzel, K.W., Alexander, M., Bernhardt, B.A., Calman, N., Carey, D.J., Cavallari, L.H., Field, J.R., Hauser, D., Junkins, H.A., Levin, P.A., et al.; IGNITE Network (2016). The IGNITE network: a model for genomic medicine implementation and research. BMC Med. Genomics 9, 1. 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed]
- 4.Orlando L.A., Sperber N.R., Voils C., Nichols M., Myers R.A., Wu R.R., Rakhra-Burris T., Levy K.D., Levy M., Pollin T.I. Developing a common framework for evaluating the implementation of genomic medicine interventions in clinical care: the IGNITE Network’s Common Measures Working Group. Genet. Med. 2018;20:655–663. doi: 10.1038/gim.2017.144. [DOI] [PMC free article] [PubMed] [Google Scholar]; Orlando, L.A., Sperber, N.R., Voils, C., Nichols, M., Myers, R.A., Wu, R.R., Rakhra-Burris, T., Levy, K.D., Levy, M., Pollin, T.I., et al. (2018). Developing a common framework for evaluating the implementation of genomic medicine interventions in clinical care: the IGNITE Network’s Common Measures Working Group. Genet. Med. 20, 655-663. [DOI] [PMC free article] [PubMed]
- 5.Bonham V.L., Green E.D., Pérez-Stable E.J. Examining how race, ethnicity, and ancestry data are used in biomedical research. JAMA. 2018;320:1533–1534. doi: 10.1001/jama.2018.13609. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bonham, V.L., Green, E.D., and Perez-Stable, E.J. (2018). Examining how race, ethnicity, and ancestry data are used in biomedical research. JAMA 320, 1533-1534. [DOI] [PMC free article] [PubMed]
- 6.Jaccard J., Jacoby J. Guilford; New York: 2010. Theory Construction and Model-Building Skills: A Practical Guide for Social Scientists. [Google Scholar]; Jaccard, J., and Jacoby, J. (2010). Theory Construction and Model-Building Skills: A Practical Guide for Social Scientists (New York: Guilford).
- 7.Damschroder L.J., Aron D.C., Keith R.E., Kirsh S.R., Alexander J.A., Lowery J.C. Fostering implementation of health services research findings into practice: a consolidated framework for advancing implementation science. Implement. Sci. 2009;4:50. doi: 10.1186/1748-5908-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]; Damschroder, L.J., Aron, D.C., Keith, R.E., Kirsh, S.R., Alexander, J.A., and Lowery, J.C. (2009). Fostering implementation of health services research findings into practice: a consolidated framework for advancing implementation science. Implement. Sci. 4, 50. [DOI] [PMC free article] [PubMed]
- 8.Chinman M., Woodward E.N., Curran G.M., Hausmann L.R.M. Harnessing Implementation Science to Increase the Impact of Health Equity Research. Med. Care. 2017;55(Suppl 9 Suppl 2):S16–S23. doi: 10.1097/MLR.0000000000000769. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chinman, M., Woodward, E.N., Curran, G.M., and Hausmann, L.R.M. (2017). Harnessing Implementation Science to Increase the Impact of Health Equity Research. Med. Care 55 (Suppl 9 Suppl 2), S16-S23. [DOI] [PMC free article] [PubMed]
- 9.Horowitz C.R., Shameer K., Gabrilove J., Atreja A., Shepard P., Goytia C.N., Smith G.W., Dudley J., Manning R., Bickell N.A., Galvez M.P. Accelerators: Sparking innovation and transdisciplinary team science in disparities research. Int. J. Environ. Res. Public Health. 2017;14:225. doi: 10.3390/ijerph14030225. [DOI] [PMC free article] [PubMed] [Google Scholar]; Horowitz, C.R., Shameer, K., Gabrilove, J., Atreja, A., Shepard, P., Goytia, C.N., Smith, G.W., Dudley, J., Manning, R., Bickell, N.A., and Galvez, M.P. (2017). Accelerators: Sparking innovation and transdisciplinary team science in disparities research. Int. J. Environ. Res. Public Health 14, 225. [DOI] [PMC free article] [PubMed]
- 10.Horowitz C.R., Abul-Husn N.S., Ellis S., Ramos M.A., Negron R., Suprun M., Zinberg R.E., Sabin T., Hauser D., Calman N. Determining the effects and challenges of incorporating genetic testing into primary care management of hypertensive patients with African ancestry. Contemp. Clin. Trials. 2016;47:101–108. doi: 10.1016/j.cct.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Horowitz, C.R., Abul-Husn, N.S., Ellis, S., Ramos, M.A., Negron, R., Suprun, M., Zinberg, R.E., Sabin, T., Hauser, D., Calman, N., et al. (2016). Determining the effects and challenges of incorporating genetic testing into primary care management of hypertensive patients with African ancestry. Contemp. Clin. Trials 47, 101-108. [DOI] [PMC free article] [PubMed]
- 11.Kaplan B., Caddle-Steele C., Chisholm G., Esmond W.A., Ferryman K., Gertner M., Goytia C., Hauser D., Richardson L.D., Robinson M., Horowitz C.R. A culture of understanding: Reflections and suggestions from a genomics research community board. Prog. Community Health Partnersh. 2017;11:161–165. doi: 10.1353/cpr.2017.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kaplan, B., Caddle-Steele, C., Chisholm, G., Esmond, W.A., Ferryman, K., Gertner, M., Goytia, C., Hauser, D., Richardson, L.D., Robinson, M., and Horowitz, C.R. (2017). A culture of understanding: Reflections and suggestions from a genomics research community board. Prog. Community Health Partnersh. 11, 161-165. [DOI] [PMC free article] [PubMed]
- 12.Gottesman O., Kuivaniemi H., Tromp G., Faucett W.A., Li R., Manolio T.A., Sanderson S.C., Kannry J., Zinberg R., Basford M.A., eMERGE Network The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gottesman, O., Kuivaniemi, H., Tromp, G., Faucett, W.A., Li, R., Manolio, T.A., Sanderson, S.C., Kannry, J., Zinberg, R., Basford, M.A., et al.; eMERGE Network (2013). The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 15, 761-771. [DOI] [PMC free article] [PubMed]
- 13.Matise T.C., Ambite J.L., Buyske S., Carlson C.S., Cole S.A., Crawford D.C., Haiman C.A., Heiss G., Kooperberg C., Marchand L.L., PAGE Study The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am. J. Epidemiol. 2011;174:849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]; Matise, T.C., Ambite, J.L., Buyske, S., Carlson, C.S., Cole, S.A., Crawford, D.C., Haiman, C.A., Heiss, G., Kooperberg, C., Marchand, L.L., et al.; PAGE Study (2011). The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am. J. Epidemiol. 174, 849-859. [DOI] [PMC free article] [PubMed]
- 14.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L. Genetic diversity turns a new PAGE in our understanding of complex traits. bioRxiv. 2017 [Google Scholar]; Wojcik, G.L., Graff, M., Nishimura, K.K., Tao, R., Haessler, J., Gignoux, C.R., Highland, H.M., Patel, Y.M., Sorokin, E.P., Avery, C.L., et al. (2017). Genetic diversity turns a new PAGE in our understanding of complex traits. bioRxiv. 10.1101/188094.
- 15.Parsa A., Kao W.H., Xie D., Astor B.C., Li M., Hsu C.Y., Feldman H.I., Parekh R.S., Kusek J.W., Greene T.H., AASK Study Investigators. CRIC Study Investigators APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]; Parsa, A., Kao, W.H., Xie, D., Astor, B.C., Li, M., Hsu, C.Y., Feldman, H.I., Parekh, R.S., Kusek, J.W., Greene, T.H., et al.; AASK Study Investigators; CRIC Study Investigators (2013). APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 369, 2183-2196. [DOI] [PMC free article] [PubMed]
- 16.Horowitz C.R., Ferryman K., Negron R., Sabin T., Rodriguez M., Zinberg R.F., Böttinger E., Robinson M. Race, genomics and chronic disease: What patients with African ancestry have to say. J. Health Care Poor Underserved. 2017;28:248–260. doi: 10.1353/hpu.2017.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Horowitz, C.R., Ferryman, K., Negron, R., Sabin, T., Rodriguez, M., Zinberg, R.F., Bottinger, E., and Robinson, M. (2017). Race, genomics and chronic disease: What patients with African ancestry have to say. J. Health Care Poor Underserved 28, 248-260. [DOI] [PMC free article] [PubMed]
- 17.Hauser D., Obeng A.O., Fei K., Ramos M.A., Horowitz C.R. Views Of Primary Care Providers On Testing Patients For Genetic Risks For Common Chronic Diseases. Health Aff. (Millwood) 2018;37:793–800. doi: 10.1377/hlthaff.2017.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hauser, D., Obeng, A.O., Fei, K., Ramos, M.A., and Horowitz, C.R. (2018). Views Of Primary Care Providers On Testing Patients For Genetic Risks For Common Chronic Diseases. Health Aff. (Millwood) 37, 793-800. [DOI] [PMC free article] [PubMed]
- 18.Peterson J.F., Bowton E., Field J.R., Beller M., Mitchell J., Schildcrout J., Gregg W., Johnson K., Jirjis J.N., Roden D.M. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet. Med. 2013;15:833–841. doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peterson, J.F., Bowton, E., Field, J.R., Beller, M., Mitchell, J., Schildcrout, J., Gregg, W., Johnson, K., Jirjis, J.N., Roden, D.M., et al. (2013). Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet. Med. 15, 833-841. [DOI] [PMC free article] [PubMed]
- 19.Pulley J.M., Denny J.C., Peterson J.F., Bernard G.R., Vnencak-Jones C.L., Ramirez A.H., Delaney J.T., Bowton E., Brothers K., Johnson K. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pulley, J.M., Denny, J.C., Peterson, J.F., Bernard, G.R., Vnencak-Jones, C.L., Ramirez, A.H., Delaney, J.T., Bowton, E., Brothers, K., Johnson, K., et al. (2012). Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther. 92, 87-95. [DOI] [PMC free article] [PubMed]
- 20.Peterson J.F., Field J.R., Unertl K.M., Schildcrout J.S., Johnson D.C., Shi Y., Danciu I., Cleator J.H., Pulley J.M., McPherson J.A. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin. Pharmacol. Ther. 2016;100:67–74. doi: 10.1002/cpt.331. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peterson, J.F., Field, J.R., Unertl, K.M., Schildcrout, J.S., Johnson, D.C., Shi, Y., Danciu, I., Cleator, J.H., Pulley, J.M., McPherson, J.A., et al. (2016). Physician response to implementation of genotype-tailored antiplatelet therapy. Clin. Pharmacol. Ther. 100, 67-74. [DOI] [PMC free article] [PubMed]
- 21.Peterson J.F., Field J.R., Shi Y., Schildcrout J.S., Denny J.C., McGregor T.L., Van Driest S.L., Pulley J.M., Lubin I.M., Laposata M. Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J. 2016;16:393–398. doi: 10.1038/tpj.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peterson, J.F., Field, J.R., Shi, Y., Schildcrout, J.S., Denny, J.C., McGregor, T.L., Van Driest, S.L., Pulley, J.M., Lubin, I.M., Laposata, M., et al. (2016). Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J. 16, 393-398. [DOI] [PMC free article] [PubMed]
- 22.Unertl K.M., Jaffa H., Field J.R., Price L., Peterson J.F. Clinician Perspectives on Using Pharmacogenomics in Clinical Practice. Per. Med. 2015;12:339–347. doi: 10.2217/pme.15.10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Unertl, K.M., Jaffa, H., Field, J.R., Price, L., and Peterson, J.F. (2015). Clinician Perspectives on Using Pharmacogenomics in Clinical Practice. Per. Med. 12, 339-347. [DOI] [PMC free article] [PubMed]
- 23.Graves J.A., Zhou Z., Garbett S., Peterson J.F. Economic Dimensions of Personalized and Precision Medicine National Bureau of Economic Research, Inc. 2018. The Value of Pharmacogenomic Information, NBER Chapters.https://ideas.repec.org/h/nbr/nberch/13989.html [Google Scholar]; Graves, J.A., Zhou, Z., Garbett, S., and Peterson, J.F. (2018). The Value of Pharmacogenomic Information, NBER Chapters. In: Economic Dimensions of Personalized and Precision Medicine National Bureau of Economic Research, Inc. https://ideas.repec.org/h/nbr/nberch/13989.html
- 24.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]; Retterer, K., Juusola, J., Cho, M.T., Vitazka, P., Millan, F., Gibellini, F., Vertino-Bell, A., Smaoui, N., Neidich, J., Monaghan, K.G., et al. (2016). Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 18, 696-704. [DOI] [PubMed]
- 25.Stark Z., Tan T.Y., Chong B., Brett G.R., Yap P., Walsh M., Yeung A., Peters H., Mordaunt D., Cowie S., Melbourne Genomics Health Alliance A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 2016;18:1090–1096. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]; Stark, Z., Tan, T.Y., Chong, B., Brett, G.R., Yap, P., Walsh, M., Yeung, A., Peters, H., Mordaunt, D., Cowie, S., et al.; Melbourne Genomics Health Alliance (2016). A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 18, 1090-1096. [DOI] [PubMed]
- 26.Clark M.M., Stark Z., Farnaes L., Tan T.Y., White S.M., Dimmock D., Kingsmore S.F. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 2018;3:16. doi: 10.1038/s41525-018-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Clark, M.M., Stark, Z., Farnaes, L., Tan, T.Y., White, S.M., Dimmock, D., and Kingsmore, S.F. (2018). Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med 3, 16. [DOI] [PMC free article] [PubMed]
- 27.Yu J.H., Crouch J., Jamal S.M., Tabor H.K., Bamshad M.J. Attitudes of African Americans toward return of results from exome and whole genome sequencing. Am. J. Med. Genet. A. 2013;161A:1064–1072. doi: 10.1002/ajmg.a.35914. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yu, J.H., Crouch, J., Jamal, S.M., Tabor, H.K., and Bamshad, M.J. (2013). Attitudes of African Americans toward return of results from exome and whole genome sequencing. Am. J. Med. Genet. A. 161A, 1064-1072. [DOI] [PMC free article] [PubMed]
- 28.Williams J.L., Chung W.K., Fedotov A., Kiryluk K., Weng C., Connolly J.J., Harr M., Hakonarson H., Leppig K.A., Larson E.B. Harmonizing Outcomes for Genomic Medicine: Comparison of eMERGE Outcomes to ClinGen Outcome/Intervention Pairs. Healthcare (Basel) 2018;6:83. doi: 10.3390/healthcare6030083. [DOI] [PMC free article] [PubMed] [Google Scholar]; Williams, J.L., Chung, W.K., Fedotov, A., Kiryluk, K., Weng, C., Connolly, J.J., Harr, M., Hakonarson, H., Leppig, K.A., Larson, E.B., et al. (2018). Harmonizing Outcomes for Genomic Medicine: Comparison of eMERGE Outcomes to ClinGen Outcome/Intervention Pairs. Healthcare (Basel) 6, 83. [DOI] [PMC free article] [PubMed]
- 29.Scherr C.L., Vasquez E., Quinn G.P., Vadaparampil S.T. Genetic counseling for hereditary breast and ovarian cancer among Puerto Rican women living in the United States. Rev. Recent Clin. Trials. 2014;9:245–253. doi: 10.2174/1574887110666150127110314. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scherr, C.L., Vasquez, E., Quinn, G.P., and Vadaparampil, S.T. (2014). Genetic counseling for hereditary breast and ovarian cancer among Puerto Rican women living in the United States. Rev. Recent Clin. Trials 9, 245-253. [DOI] [PMC free article] [PubMed]
- 30.Peterson E.B., Chou W.S., Gaysynsky A., Krakow M., Elrick A., Khoury M.J., Kaphingst K.A. Communication of cancer-related genetic and genomic information: A landscape analysis of reviews. Transl. Behav. Med. 2018;8:59–70. doi: 10.1093/tbm/ibx063. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peterson, E.B., Chou, W.S., Gaysynsky, A., Krakow, M., Elrick, A., Khoury, M.J., and Kaphingst, K.A. (2018). Communication of cancer-related genetic and genomic information: A landscape analysis of reviews. Transl. Behav. Med. 8, 59-70. [DOI] [PMC free article] [PubMed]
- 31.Collins F.S., Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]; Collins, F.S., and Varmus, H. (2015). A new initiative on precision medicine. N. Engl. J. Med. 372, 793-795. [DOI] [PMC free article] [PubMed]