Abstract
Huntington disease (HD) is caused by a CAG repeat expansion in the huntingtin (HTT) gene. Although the length of this repeat is inversely correlated with age of onset (AOO), it does not fully explain the variability in AOO. We assessed the sequence downstream of the CAG repeat in HTT [reference: (CAG)n-CAA-CAG], since variants within this region have been previously described, but no study of AOO has been performed. These analyses identified a variant that results in complete loss of interrupting (LOI) adenine nucleotides in this region [(CAG)n-CAG-CAG]. Analysis of multiple HD pedigrees showed that this LOI variant is associated with dramatically earlier AOO (average of 25 years) despite the same polyglutamine length as in individuals with the interrupting penultimate CAA codon. This LOI allele is particularly frequent in persons with reduced penetrance alleles who manifest with HD and increases the likelihood of presenting clinically with HD with a CAG of 36–39 repeats. Further, we show that the LOI variant is associated with increased somatic repeat instability, highlighting this as a significant driver of this effect. These findings indicate that the number of uninterrupted CAG repeats, which is lengthened by the LOI, is the most significant contributor to AOO of HD and is more significant than polyglutamine length, which is not altered in these individuals. In addition, we identified another variant in this region, where the CAA-CAG sequence is duplicated, which was associated with later AOO. Identification of these cis-acting modifiers have potentially important implications for genetic counselling in HD-affected families.
Keywords: age of onset, CAG repeat, genetic modifier, Huntington disease, loss of interruption, polyglutamine, reduced penetrance alleles, somatic instability
Introduction
The age of clinical onset (AOO) of Huntington disease (HD [MIM: 143100]) is significantly influenced by the length of the expanded CAG repeat, which is translated into polyglutamine (see GeneReviews in Web Resources). However, this repeat polymorphism does not fully explain variability in AOO, and individuals with HD with identical expanded CAG repeat lengths frequently present with clinical symptoms at different ages.1
This variability in AOO is particularly evident in carriers of reduced penetrance (RP) alleles (36–39 CAGs), where the majority of these individuals may remain asymptomatic into old age. However, some of these individuals do present with HD at much earlier ages2, 3 than predicted from CAG repeat length. The factors influencing this variability at the same polyglutamine length remains unexplained.4
Differences in AOO between individuals with HD have been shown to be influenced by heritable factors, suggesting that other genetic modifiers play an important role in modifying disease onset.5, 6, 7 Recent studies have identified candidate modifier regions for HD onset, both at the HTT locus8 and across the genome.9, 10 Therefore, to investigate this further, we focused on examining the cis-acting HTT sequence, where the polyglutamine tract is encoded by a CAG trinucleotide repeat, interrupted by a penultimate CAA codon. This was motivated by the fact that sequence variants within this region have been previously described,11, 12, 13, 14 yet no formal analysis of AOO has been performed. To investigate the role of variants in this region on AOO, particular focus was placed on screening RP-carrying individuals. This was motivated by the fact that the pedigree in the original study that identified these variants contained numerous individuals in this repeat range.
Subjects and Methods
Study Populations
Genomic DNA from individuals with HD was obtained from the HD Biobank at the University of British Columbia (UBC) or through collaborators from HD pedigrees found to be carrying interrupting sequence variants. Further, in order to identify symptomatic individuals with HD with the canonical interrupting sequence, we also randomly screened HD-affected individuals with AOO information in the UBC HD Biobank. All samples were collected, stored, and accessed with informed consent and ethical approval from the UBC/Children’s and Women’s Health Centre of British Columbia Research Ethics Board (UBC C&W REB H06-70467 and H06-70410). AOO was determined by the clinicians treating the individuals or ascertained from their medical records. The predicted AOO for individuals with HD based on CAG repeat length was calculated according to the Langbehn et al. formula,4 and AOO ratios (i.e., predicted/observed AOO), along with related percentiles, were calculated as previously described.8
HTT CAG and CCG Repeat Sizing and Interrupting Sequence Characterization
HTT CAG and CCG repeat sizing was performed with control samples of known repeat lengths, using previously described methods, at the Centre for Molecular Medicine and Therapeutics at UBC in Vancouver, Canada.2, 15 Variants in the interrupting sequence between the HTT CAG-CCG repeat tracts were genotyped by clonal sequencing. Briefly, polymerase chain reaction (PCR) products encompassing the HTT CAG-CCG repeat tracts (HTT-CAG-3-F-EcoRI: 5′-GATCGAATTCATTGCCCCGGTGCTGAGCG-3′ and HTT-CAG-3-R-HindIII: 5′-GATCAAGCTTGCGGGCCCAAACTCACGGTC-3′) were cloned into pUC19 plasmids following restriction enzyme double digest (six units of EcoR1 and HindIII) and ligation. Vectors were subsequently transformed into DH5-α E. coli cells and positive clones were identified via colony PCR, then cultured overnight for extraction with QIAprep Spin Miniprep Kits (QIAGEN) and Sanger sequencing with the M13-R primer (5′-CAGGAAACAGCTATGAC-3′).
HTT Haplotyping and Genome-wide Array Analyses
Haplotyping employing single-nucleotide polymorphisms (SNPs) spanning the HTT locus was carried out as previously described.16 HD subjects were genotyped using the Infinium Global Screening Array v2.0 (Illumina) for 665,608 variants. Genotype data were subsequently clustered using the GenomeStudio Software (Illumina) and additional variant and sample filtering and analyses was performed using PLINK 1.9. Principal component analyses were performed on linkage disequilibrium pruned (R2 > 0.25) array data to determine the genetic ancestry of individuals using EIGENSOFT v5.0, including the 1000 Genomes Project Phase 3 samples as a reference. In order to identify tag variants for the interrupting sequence modifiers, array data from chromosome 4 were imputed using the Haplotype Reference Consortium Panel (version r1.1 2016) as previously described.17 Variants located 1 Mb up- and downstream of the HTT locus (CAG)n-CAA-CAG sequence were then extracted and SNPs displaying within genotyped cohort minor allele frequencies > 0.01 and imputation R2 > 0.5 were analyzed. We then assessed linkage disequilibrium with these SNPs and the modifier variants of interest, stratified by sub-haplotype.
Genotyping of HTT (CAG)n-CAA-CAG Interrupting Sequence Variants in General Population Controls
The frequency of the (CAG)n-CAA-CAG interrupting sequence variants were determined in a cohort of 1,657 unrelated general population control subjects, recruited as unaffected parents in an autism spectrum disorder study (a specific cohort within the Autism Speaks MSSNG Project).18 Sequence-graph-based alignment of PCR-free whole-genome sequence data from these individuals was performed with ExpansionHunter (v3.0.0-rc1),19, 20 which explicitly models the HTT CAG and CCG repeats, as well as the interrupting sequence region. This updated version of the software20 has been specifically designed to genotype repeats and interrupting sequence variants from PCR-free whole-genome sequencing data. We restricted analyses to samples with CCG repeat calls within what has been detected from traditional fragment analysis sizing (i.e., CCG repeats between 5 and 12). For each interrupting sequence in each sample, we calculated the ratio of observed reads that fully span the interruption to their expected per-haplotype number (O/E ratio), calling non-canonical variants where the absolute value of 1.0 minus the O/E ratio was greater than 0.2.
HTT CAG Somatic Expansion Ratio Calculations and Germline Instability Estimates
Electropherogram traces from fluorescently labeled CAG sizing PCR products were used to calculate an expansion ratio to measure the somatic instability of the pathogenic repeat, since similar approaches have been successfully employed to measure huntingtin CAG instability.21 For these analyses, additional LOI carriers were identified by screening additional family members in the carrier HD pedigrees, including all samples where there was information regarding subject age at the time of sample collection. Reactions were performed in triplicate and PCR products were diluted (1:60) before being run on the ABI Prism 3130xl Genetic Analyzer using manufacturer protocols (Applied Biosystems). Traces were assigned using GeneMapper Software v4.0 (Thermo Fisher Scientific) and the expansion ratio was calculated using the area under all expanded CAG repeat lengths relative to the area under the most prominent peak (i.e., diagnostic CAG repeat size for the subject).
Small-pool PCR data for the HTT CAG repeat in sperm from 34 male European ancestry subjects were also analyzed to assess germline CAG instability, as described by Semaka et al.,22 using CAG sizing primers identical to those used in the expansion ratio analysis.
Statistical Analyses
Statistical and bioinformatic analyses were performed in R. Significant differences between genotype groups with regards to AOO and related information were calculated using a Wilcoxon rank-sum test. Significant differences in log-transformed somatic and germline instability/expansion measures and LOI carrier status, CAG repeat length, and age were assessed using linear regression. Residuals were checked for normality with the Shapiro-Wilk test. p values < 0.05 were considered significant in all analyses.
Results
Loss of the Penultimate CAA Codons Hastens AOO in HD
Sequencing of the (CAG)n-CAA-CAG sequence in HD pedigrees identified a variant that results in the loss of the penultimate CAA codon (i.e., CAA-CAG to CAG-CAG), without changing the length of the polyglutamine tract (Figures 1 and 2), referred to as the loss of interruption (LOI) variant. This variant is also characterized by another transition that causes an uninterrupted CCG repeat in the adjacent proline codons (i.e., CCG-CCA to CCG-CCG), occurring in complete linkage disequilibrium with the CAA to CAG transition in symptomatic individuals with HD. LOI carriers therefore have identical polyglutamine tract lengths as subjects with interrupting adenine nucleotides/interrupting reference sequence, but uninterrupted CAG residues.
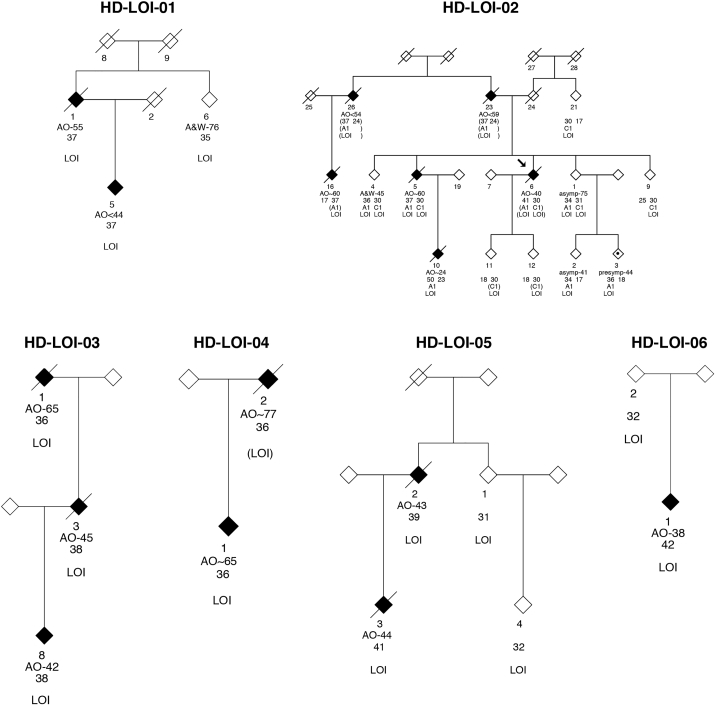
Figure 1.
Huntington Disease Loss of Interruption (LOI) Pedigrees Included in the Current Study
Affected status, individual identifier, age-of-onset information, CAG size, and LOI genotype are indicated.
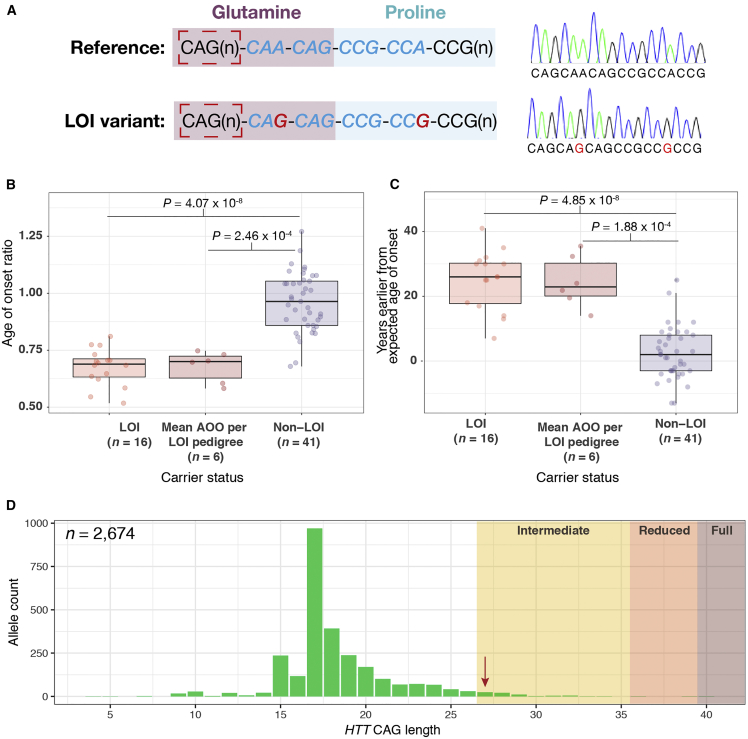
Figure 2.
The Loss of Interruption (LOI) Variant Is Associated with an Earlier Age of Onset (AOO) in Individuals with Huntington Disease (HD) and Occurs on Expanded HTT CAG Alleles
(A) The interrupting sequence between the exon one HTT CAG and CCG repeats and representative Sanger electropherograms for the reference sequence and LOI variant are shown. The interrupting sequence is depicted in blue italic font and red nucleotides show point mutations in this region that can result in the LOI variant. The dashed red box indicates the CAG repeat that is measured in diagnostic assays for HD. Nucleotides encoding the glutamine (i.e., CAG/CAA) and proline (i.e., CCG/CCA) tracts are shaded to show that the LOI variant alters the number of contiguous CAG repeats but not the number of glutamine residues in these persons.
(B) The LOI is associated with earlier AOO as determined by the AOO ratio.
(C) LOI carriers present with HD approximately 25 years earlier than predicted on average compared to current models for prediction of AOO. These calculations were performed using data from all HD-LOI subjects as well as mean values for each HD-LOI pedigree, versus HD subjects with the reference interrupting sequence.
(D) Distribution of the HTT CAG repeat lengths in the general population ascertained through genotyping from whole-genome sequencing data. The LOI allele was detected in one research participant and was found on an intermediate allele (indicated with an arrow). Intermediate, reduced penetrance and fully penetrant alleles are shaded.
In total, we identified 16 symptomatic HD-affected subjects from six pedigrees of European genetic ancestry from five countries (Australia, Canada, Italy, United States, and the Netherlands) with this LOI variant (Figures 1, 2, and S1, Table 1, mean CAG length = 39). Notably, 12 of the 16 clinically manifesting HD-LOI subjects (75%), from five of the six pedigrees, carried RP alleles (i.e., CAG 36–39). The remaining four LOI subjects (25%) were found in three of the six families and carried the LOI variant on fully penetrant HD alleles with CAG sizes of 41, 41, 42, and 50, respectively (Table 1). We did not detect the LOI variant in persons with CAG greater than 39 who are not clinically affected. However, the number of persons with fully penetrant alleles that we have examined is small (n = 25), so no definitive conclusions can be made at this stage.
Table 1.
Carriers of the HTT Loss-of-Interruption (LOI) Variant Manifest with Huntington Disease (HD) Earlier than Predicted
| Pedigree | Individual Identifier | Repeat Range | Expanded CAG | Related CCG | Actual AOO | Expected AOO | Years Earlier | AOO Ratio | AOO Percentile | HTT Haplotype |
|---|---|---|---|---|---|---|---|---|---|---|
| HD-LOI-1 | 1 | RP | 37 | 10 | 55 | 85 | 30 | 0.65 | <10th | A1 LOI |
| HD-LOI-1 | 5 | RP | 37 | 10 | 44 | 85 | 41 | 0.52 | <10th | A1 LOI |
| HD-LOI-2 | 5 | RP | 37 | 7 | 60 | 85 | 25 | 0.70 | <10th | A1 LOI |
| HD-LOI-2∗ | 6 | FP | 41 | 7 | 40 | 57 | 17 | 0.70 | <10th | A1 LOI |
| HD-LOI-2∗ | 10 | FP | 50 | 7 | 24 | 31 | 7 | 0.77 | <10th | A1 LOI |
| HD-LOI-2 | 16 | RP | 37 | 7 | 60 | 85 | 25 | 0.70 | <10th | A1 LOI |
| HD-LOI-2 | 23 | RP | 37 | 7 | 59 | 85 | 26 | 0.69 | <10th | A1 LOI |
| HD-LOI-2 | 26 | RP | 37 | 7 | 54 | 85 | 31 | 0.63 | <10th | A1 LOI |
| HD-LOI-3 | 1 | RP | 36 | 10 | 65 | 95 | 30 | 0.68 | <10th | A1 LOI |
| HD-LOI-3 | 3 | RP | 38 | 10 | 45 | 77 | 32 | 0.59 | <10th | A1 LOI |
| HD-LOI-3 | 8 | RP | 38 | 10 | 42 | 77 | 35 | 0.55 | <10th | A1 LOI |
| HD-LOI-4 | 1 | RP | 36 | 10 | 77 | 95 | 18 | 0.81 | <15th | C1 LOI |
| HD-LOI-4 | 2 | RP | 36 | 10 | 65 | 95 | 30 | 0.68 | <10th | C1 LOI |
| HD-LOI-5 | 2 | RP | 39 | 10 | 43 | 69 | 26 | 0.62 | <10th | C1 LOI |
| HD-LOI-5∗ | 3 | FP | 41 | 10 | 44 | 57 | 13 | 0.77 | <10th | C1 LOI |
| HD-LOI-6∗ | 1 | FP | 42 | 10 | 38 | 52 | 14 | 0.73 | <10th | C1 LOI |
The variant occurs on both fully penetrant (indicated with asterisk) and reduced penetrance alleles in these individuals. Abbreviations: AOO, age of onset; FP, full penetrance; LOI, loss of interruption; RP, reduced penetrance.
LOI carriers (n = 16) presented with HD with an average of 25 years earlier than model predictions,4 which is significantly different from HD-affected subjects with the reference interrupting sequence [i.e., (CAG)n-CAA-CAG; n = 41, p = 4.85 × 10−8; Figure 2]. This effect was particularly apparent in RP-LOI (36–39 CAG) carriers (average of 29.1 years earlier than predicted AOO based on CAG size), compared to the fully penetrant (≥40 CAG) HD-LOI allele carriers (average of 12.8 years earlier than predicted AOO based on CAG size). Strikingly, 15 of the 16 HD-LOI subjects presented with an extremely early AOO based on their CAG repeat length (<10th percentile of predicted AOO for CAG repeat length)4 and displayed a significantly lower AOO ratio (i.e., predicted/observed AOO) compared to reference interrupting sequence subjects (p = 4.07 × 10−8).
We screened all RP-carrying individuals in the UBC HD Biobank with CAG repeat lengths in the RP range (i.e., CAG 36–39, n = 95, Table 2), revealing that 33.3% of clinically manifesting RPs (n = 36) carried the LOI variant in this range compared to only 5.1% of the asymptomatic RPs (n = 59) carrying the LOI variant in this range (p = 7.6 × 10−4). Only a minority of the cohort of asymptomatic RP carriers were expected to have presented with HD when last assessed (i.e., 10.2%), which is to be expected, since the majority of RPs do not present with HD in their lifetime. However, the statistically significant enrichment of LOI carriers in symptomatic RPs indicates that the LOI variant has a profound effect on AOO in RP-carrying individuals. At time of sampling, the three asymptomatic LOI RP carriers would not have been expected to present with signs of HD yet, regardless of the presence of the LOI variant (CAG 36 RPs: 44 and 45 years old, CAG 39 RP: 28 years old).
Table 2.
Systematic Screening of All Carriers of Reduced Penetrance (RP) Alleles in the UBC HD Biobank (n = 95) Reveals that the LOI Variant Is Enriched in Individuals Who Manifest with Huntington Disease (HD) in This Repeat Range
|
Clinically Manifest |
No Signs of Symptoms of HD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RP CAG Size | Expected AOO | LOI Carriers | Non-LOI Carriers | Total Screened | LOI Carrier Frequency | AOO Range | LOI Carriers | Non-LOI Carriers | Total Screened | LOI Carrier Frequency | Age Range |
| CAG 36 | 95 | 3 | 0 | 3 | 100% | 65–77 | 2 | 10 | 12 | 16.7% | 9–85 |
| CAG 37 | 85 | 6 | 0 | 6 | 100% | 44–60 | 0 | 2 | 2 | 0.0% | 28–73 |
| CAG 38 | 77 | 2 | 6 | 8 | 25.0% | 42–45 | 0 | 16 | 16 | 0.0% | 19–85 |
| CAG 39 | 69 | 1 | 18 | 19 | 5.3% | 43 | 1a | 28 | 29 | 3.4% | 22–79 |
| All RPs | NA | 12 | 24 | 36 | 33.3% | 42–77 | 3 | 56 | 59 | 5.1% | 9–85 |
The LOI variant is particularly relevant for individuals carrying RP alleles with fewer than 39 CAG repeats. Abbreviations: AOO, age of onset; LOI, loss of interruption; NA, not applicable; RP, reduced penetrance.
Carrier of an LOI allele where only the penultimate CAA is changed to CAG and CCA codon is unaltered: i.e., CAG(39)-CAG-CAG-CCG-CCA-CCG(10).
Remarkably, when assessing RPs that presented with HD extremely early in life (i.e., <10th percentile of AOO ratio, n = 13), 84.6% were LOI carriers. The influence of the LOI was more pronounced in the RP-carrying individuals in the 36–38 CAG range, where 64.7% of individuals manifesting with HD carried the LOI, compared to only 6.7% of non-manifesting individuals in this repeat class (p = 3.9 × 10−5). Among unrelated symptomatic RP allele pedigrees in the CAG 36–38 range, 45.5% (n = 5) carried the LOI.
In our RP allele screening, we detected one asymptomatic RP-carrying individual where only the penultimate CAA is changed to CAG and the CCA codon is unaltered (Table 2). This was the only observation of this sub-allele in the entire study, representing 31 LOI alleles, and therefore the effect on AOO of this extremely rare allele still needs further characterization.
Loss of the Penultimate CAA Codon Is Rare in the General Population
Analysis of allele samples that passed quality control in the general population cohort (n = 1,337) revealed that the LOI variant is rare in unaffected individuals (minor allele frequency = 0.04%, i.e., 1 in 2,674 alleles), with only one general population LOI variant detected, occurring on an intermediate CAG allele of 27 (Figure 2). This was confirmed via Sanger sequencing. In this general population cohort, there were 69 alleles (2.5%) in the intermediate CAG range (27–35). The LOI variant was therefore present in 1 of 69 intermediate alleles (1.45%) and found exclusively on alleles in CAG ranges ≥ 27. This agrees with routine clonal sequencing of this region that has been performed by our group, where no LOI alleles have been detected in 234 unexpanded normal repeats (CAG < 27) assessed to date.
Loss of the Penultimate CAA Codon, Resulting in an Uninterrupted CAG Tract, Is Associated with Increased Somatic and Germline Instability
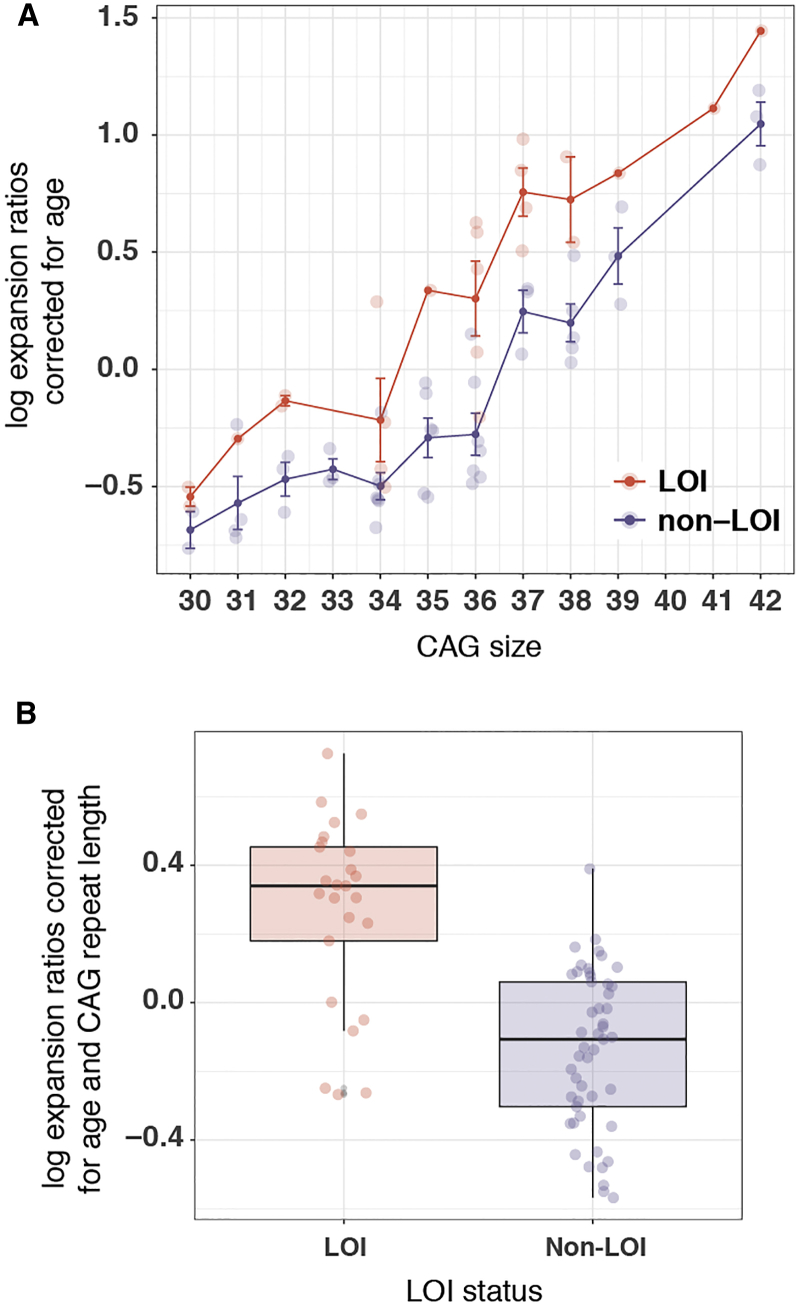
The LOI variant was associated with increased somatic instability (p = 3.5 × 10−9, Figure 3, Table S1) assessed via CAG expansion ratio in whole blood in numerous subjects (LOI carriers n = 27 versus canonical sequence subjects n = 49; CAG repeat lengths 30–42). As expected, the somatic expansion ratio was also strongly associated with increased CAG repeat size (p = 3.0 × 10−31), illustrating the accuracy of this technique. Further, older age was also associated with increased instability in these analyses (p = 2.5 × 10−3).
Figure 3.
The HTT Loss-of-Interruption (LOI) Variant Is Associated with an Increased Frequency of CAG Expansions in Whole Blood
(A) Somatic expansion ratio separated by CAG, corrected for age at sampling and stratified by LOI carrier status, shows that expansions are more frequent in 24 LOI carriers compared to 46 non-carriers. Standard error of the mean indicated where available.
(B) Expansion ratio differences after correction for age and CAG repeat length confirms that the LOI variant is associated with increased somatic instability.
These results were confirmed in the analysis of the small-pool PCR sperm data, where LOI status (p = 0.001), CAG repeat length (p = 1.6 × 10−35), and increased age (p = 0.01) were all associated with increased CAG repeat tract instability (Table S1). Of note, in both somatic and germline assays, the effect size of the LOI variant (βLOI-blood = 0.43, βLOI-germline = 0.94) on stability measures based on regression estimates was larger than that of CAG repeat length (βCAG-blood = 0.15, βCAG-germline = 0.19).
Duplication of the CAA-CAG Codons Delays AOO in HD
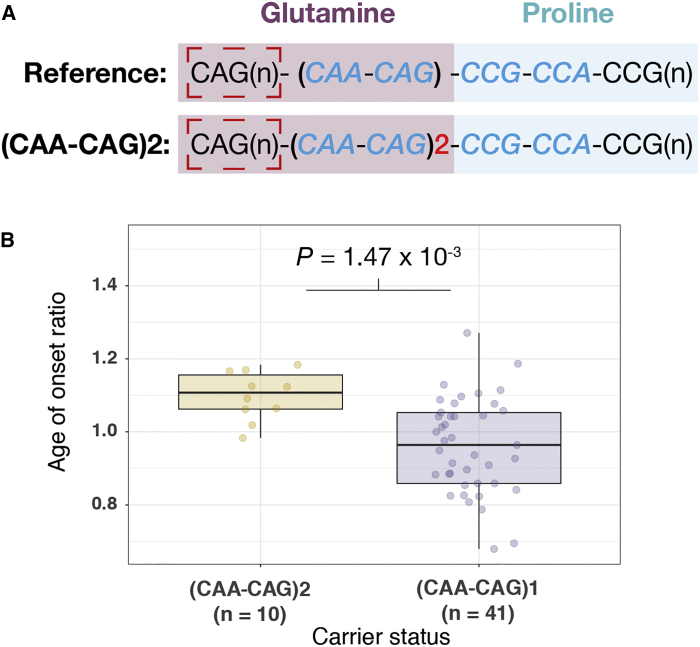
While screening samples to include as canonical HD interrupting sequence carriers for AOO analyses, we also identified a distinct variant in this region that results in a longer interrupting sequence, through the insertion of a duplicate CAA-CAG motif [i.e., (CAA-CAG)2; Figure 4]. In the three pedigrees where this variant was present, carriers (n = 10 HD-affected subjects) presented 4.2 years later than expected compared to AOO model predictions based on CAG size (AOO percentile 60th–85th, AOO ratio p = 1.47 × 10−3).4 In the general population cohort of 1,337 persons, 81 potential carriers of this variant were detected by ExpansionHunter, indicating that this variant is relatively common in these individuals, with a minor allele frequency of 2.8%.
Figure 4.
An HTT Interrupting Sequence Variant that Results in an Additional CAA-CAG Repeat Is Associated with a Later Age of Onset (AOO) in Individuals with Huntington Disease (HD)
(A) The reference HTT CAG-CCG interrupting sequence in relation to the (CAA-CAG)2 variant. Nucleotides encoding the glutamine (i.e., CAG/CAA) and proline (i.e., CCG/CCA) tracts are shaded.
(B) The (CAA-CAG)2 variant is associated with later AOO as determined by the AOO ratio in HD subjects compared to HD subjects with the reference interrupting sequence. (CAA-CAG)2 carriers present on average 4.2 years later than the majority of individuals with HD with the reference CAG repeat interruption.
Loss of the Penultimate CAA and the (CAA-CAG)2 Duplication Variants Are Each Associated with Specific Tag SNPs
The LOI variant was found to occur on subsets of two common haplotypes,16 with the A1-LOI variant occurring on two CCG7 and CCG10 configurations. Analysis of 6,554 variants that passed imputation QC at the HTT locus identified perfect tag SNPs (i.e., R2 = 1, D’ = 1) for the LOI-C1 (closest variant: rs193119731, 325 kb away) and LOI-A1-CCG10 sub-haplotypes (closest variant: rs145048189, 772 kb away, Table S2). LOI-A1-CCG7 was detected in only one pedigree and the best tags for this allele were rs73198489 and rs73200492 (R2 = 0.65, D’ = 1.00).
The duplicated (CAA-CAG)2 variant was found exclusively on a C2 HTT haplotype16 in all carriers. Numerous variants (n = 163) were in perfect linkage disequilibrium with the (CAA-CAG)2 variant, with rs10006977, closest to interrupting sequence, 289 bp away (Table S3). The frequency of rs10006977 in the Genome Aggregation Database (gnomAD)23 in non-Finnish Europeans (2.7%) agrees with the predicted frequency of the (CAA-CAG)2 variant in our general population database.
Discussion
The findings presented here reveal that the LOI variant, which causes loss of the penultimate CAA codon and increases the length of uninterrupted CAG repeat, is a modifier of AOO of HD and is also associated with increased somatic instability. This important finding is further supported by the fact that this variant displays familial aggregation as an autosomal-dominant trait and all carriers present with HD extremely early in life (Figure 1). This suggests that the LOI sequence variant is the major contributor toward the observed differences in predicted AOO in these individuals and families.
We have shown here that AOO of HD is significantly influenced by variants that alter the length of the uninterrupted CAG repeat, independent of polyglutamine length. This suggests that in these families, uninterrupted CAG repeat length, which is lengthened by the LOI variant, is more informative for AOO predictions than polyglutamine length alone, which would be the same for a given CAA/CAG repeat class. These findings therefore challenge the prior belief that polyglutamine length is a primary determinant of AOO and demonstrate that length of the uninterrupted CAG repeat, and not encoded polyglutamine, is the major contributor to AOO in HD. Future predictive models of AOO may therefore consider assessing the number of uninterrupted CAG residues, rather than effective polyglutamine length, in their estimates.
The LOI variant is particularly relevant for individuals with HD in the RP range, causing carriers to manifest with HD much earlier on average than predicted (29.1 years earlier than predicted AOO based on CAG size). Identifying a highly penetrant modifier variant such as the LOI variant provides an explanation for why a subset of RP individuals with clinical HD manifest with the disorder much earlier than others. In this study (Table 2), all individuals (7/7) who manifest with HD with CAG sizes of 36 and 37 had the LOI (AOO range: 44–77 years). None of the persons with CAG of 36 and 37 (10/10) who did not have the LOI (current ages 9–85 years) have manifested with signs and symptoms of HD at this time. The importance of this finding is highlighted by recent research that has shown that in the general population, RP alleles are more common than previously thought,2 suggesting extremely low penetrance in the majority of RP carriers that do not carry the LOI.
Despite being rare, the LOI variant has a large effect size compared to other HD modifiers. For example, the previously identified GWAS modifier with the largest known effect size in HD, rs146353869 in FAN1 (2% minor allele frequency), leads to a 6.1-year alteration in AOO on average at fully penetrant CAG repeat lengths.10 By comparison, the four manifest LOI carriers in our study with CAG > 39 presented with HD an average of 12.8 years earlier than reference HD-affected subjects (<10th percentile for AOO for a particular CAG allele).4 The frequency and expression of the LOI in the fully penetrant HD allele range still needs further study. However, the frequency was indirectly assessed by a study performed in the diagnostic setting, which indirectly detected the LOI variant by allelic dropout with diagnostic primers complementary to the reference interrupting sequence.13 This study described three manifest HD pedigrees that carried the LOI variant, representing 3.3% of the HD-affected families investigated. Remarkably, one of these clinically manifest HD-affected individuals carried an IA (CAG 35). A second pedigree contained RP-carrying individuals with clinical HD,13 lending further support to the variant’s impact on AOO in RP carriers. The findings presented here provide a scientific basis for one such HD-affected subject with a CAG of 35, explaining why persons with this genotype could manifest with signs and symptoms of HD. This has important implications for the diagnosis of HD in similar individuals with CAG repeat genotypes in the borderline IA and RP ranges.
In this study, all but one of the individuals carrying the LOI variant presented with HD in the earliest percentile for the corresponding polyglutamine repeat typically encoded by the reference CAG repeat and interruption. The variant was predominantly seen in RP-carrying individuals with clinical HD, comprising 75% of clinically symptomatic carriers. However, a strong effect was still observed in individuals with HD with fully penetrant alleles (manifesting with HD on average of 12.8 years earlier), indicating that the finding is potentially generalizable to the HD population. Larger population-based studies should systematically assess the impact of the LOI variant on AOO in fully penetrant HD alleles (i.e., CAG repeats greater than 39) to confirm and refine this observation. This will help elucidate whether a large proportion of individuals with fully penetrant HD alleles that have extremely early AOO carry the LOI variant, as well as whether the LOI is particularly relevant for AOO below a certain CAG repeat length threshold.
In our large cohort of population controls (n = 3,314 alleles), individuals with alleles in the normal range did not carry the LOI (n = 3,242 alleles, i.e., < 27 CAG), indicating that the variant is more prevalent at longer CAG repeat lengths, likely due to a higher likelihood of CAG expansion over generations.24, 25, 26 The data presented here have implications for relatives at risk for HD and suggest that assessment for the LOI variant would be helpful in any individual with HD who manifests much earlier than expected based on CAG length.
The LOI variant was discovered by our group through the analysis of unstable intermediate allele families.11 Over the last 20 years, through the recruitment of additional pedigrees, we now have sufficient power in the current study to show the influence of the LOI variant on AOO in HD. One limitation that should be noted is that prediction of a specific AOO for a particular person is difficult because of the broad confidence limits particularly for CAG sizes in the lower ranges, limiting accuracy. Ranges of AOO for specific CAG lengths have been previously calculated.4 Our assessments of AOO timing should be viewed in that context, reflecting earlier AOO or later AOO than might have been expected for a particular CAG, with these caveats noted.
Our analyses indicate that somatic instability, resulting in a mosaic of longer CAG repeat and polyglutamine tracts in vivo, is the most likely mechanism for modification of HD onset by the LOI variant as shown by two orthogonal methods. Somatic instability in HD has returned to the fore with recent HD-onset GWASs uncovering the importance of DNA repair genes.9, 10 Prior to these genomic studies, previous work by our group27 has shown that the composition of CAA interruptions in the CAG repeat may be responsible for phenotypic differences between HD mouse models and could be similarly mediated by differences in somatic instability. It has also been shown that the degree of somatic instability correlates with AOO in HD.28 Other research has demonstrated that somatic CAG expansion rates differ across tissues in all individuals with HD; the striatum is the most vulnerable to this phenomenon,29 with some individuals with HD exhibiting more than 1,000 CAG repeats in this brain.26, 30 Somatic instability observed in blood is less pronounced and ranges within a few CAG repeat sizes of the progenitor CAG.26
A limitation of the current study is that we have not analyzed the influence of the LOI on somatic instability in the brain. However, increased somatic mosaicism that has been observed in individuals with HD in regions of the brain that are most affected early and selectively in the disease suggests that the correlation between the LOI variant and this trait has not occurred by chance.26, 28 We hypothesize that increased somatic instability results in the expression of proteins with longer polyglutamine length, causing earlier and more severe neuronal damage. In this study with have shown that there is increased somatic mosaicism in LOI carriers and that this would be predicted to result in increased mosaicism of expanded CAG repeats in the brain.
Modification of clinical presentation by loss of glutamine-encoding CAA interruptions to pure CAG repeats has been reported in other polyglutamine disorders. For example, loss of interrupting CAA codons within the polyglutamine-encoding repeats of ATXN2 and TBP have been shown to modify the pathogenicity and onset of two spinocerebellar ataxias (SCAs), SCA2 and SCA17.31, 32, 33, 34 Our finding that loss of the reference CAA interruption hastens age of onset in HD, and that an extra CAA interruption conversely delays onset, expands the number of polyglutamine disorders where variable CAG and CAA repeat composition can result in phenotypic differences without alteration of translated polyglutamine length. Furthermore, our study suggests that rare variations in polyglutamine codon structure may be present in other polyglutamine diseases and could account for phenotypic outliers in those conditions.
We also demonstrate that LOI status and increased age are associated with a higher frequency of CAG repeat expansions in sperm. This has potential clinical implications in relation to the rate of new mutations from older fathers who have CAGs of 27–35. It also suggests that offspring of LOI fathers may be at higher risk of CAG expansion and earlier AOO of HD. We have previously shown that CAG repeat length correlates with measures of instability in sperm,22, 35 but the finding with regards to the potential influence of donor age on stability has not been previously reported.
In addition to the LOI variant, we found an interrupting sequence polymorphism that is associated with clinically meaningful, later AOO and is characterized by an extra CAA-CAG motif at the end of the polyglutamine tract (Figure 4). This lends further support to the role of somatic instability in HD as the variant may increase stability of the CAG repeat by preventing slippage during DNA replication. Based on our findings here, we predict that these individuals may have less somatic instability in the brain associated with later AOO. The data in this manuscript also raise the provocative question as to whether gene editing approaches to modifying guanine-to-adenine nucleotides in specific regions in the CAG tract might confer additional somatic stability and delay onset of HD. In vitro and in vivo studies could initially test this hypothesis.
The LOI variant remains laborious to genotype with conventional clonal sequencing methods, making the development of alternative genotyping approaches with similar efficacy as PCR/electropherogram-based fragment sizing of paramount importance for diagnostic settings. This study has identified a number of tag variants for LOI sub-haplotypes, but since they do not capture all LOI alleles, analysis of the actual interrupting sequence is preferred. Our analysis of whole-genome sequencing information indicates that genotyping the interrupting sequence is feasible and that more targeted approaches, using similar technologies, should be explored for clinical genetic testing applications.
In conclusion, we have described a modifier of HD clinical onset that has a larger impact than all previously identified modifier variants, most dramatically observed in the RP range. This provides support for the role of somatic repeat instability and DNA repair in modifying HD AOO. The relevance of somatic mosaicism in HD was first documented 25 years ago,25, 26 with the greatest instability observed in the brain and particularly those regions most pertinent to the pathogenesis of HD. Our study therefore provides an explanation as to why a proportion of RP carriers present with HD signs and symptoms early in their lifetime. This may have implications for clinical practice and may provide important information for individuals that present with RP alleles. This LOI is present at high frequency in symptomatic RPs and therefore provides an explanation as to marked variability of AOO of RP alleles. These findings are also likely to be generalizable to HD-affected persons in the fully penetrant range.
Acknowledgments
This work was supported by a Canadian Institutes of Health Research Foundation grant awarded to M.R.H. S.W.S. is the GlaxoSmithKline-CIHR Chair in Genome Sciences. We would like to thank the Centre for Molecular Medicine and Therapeutics and BC Children’s Hospital Research Institute, as well as The Centre for Applied Genomics at the Hospital for Sick Children and the University of Toronto McLaughlin Centre for support. Additionally, we would like to acknowledge Léal Makaroff for his assistance with developing and validating assays for somatic instability. We also wish to thank all the individuals with HD and their families worldwide who have chosen to participate in research, including those from the Centre for Huntington Disease and the HD BioBank at UBC; Leiden University Medical Center, the Netherlands; the Children’s Hospital in Westmead, Sydney, Australia; as well those collected through Lega Italiana Ricerca Huntington (LIRH) Foundation in Rome, Italy. Without the support of HD-affected families, none of this research would be possible.
Published: May 16, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.007.
Web Resources
GeneReviews, Caron, N.S., Wright, G.E.B., and Hayden, M.R. (2018). Huntington Disease. https://www.ncbi.nlm.nih.gov/books/NBK1305/
gnomAD Browser, https://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
Declaration of Interests
M.A.E. and E.D. are employees of Illumina, Inc. The remaining authors have no potential conflicts of interest to declare.
Supplemental Data
References
- 1.Keum J.W., Shin A., Gillis T., Mysore J.S., Abu Elneel K., Lucente D., Hadzi T., Holmans P., Jones L., Orth M. The HTT CAG-Expansion Mutation Determines Age at Death but Not Disease Duration in Huntington Disease. Am. J. Hum. Genet. 2016;98:287–298. doi: 10.1016/j.ajhg.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; Keum, J.W., Shin, A., Gillis, T., Mysore, J.S., Abu Elneel, K., Lucente, D., Hadzi, T., Holmans, P., Jones, L., Orth, M., et al. (2016). The HTT CAG-Expansion Mutation Determines Age at Death but Not Disease Duration in Huntington Disease. Am. J. Hum. Genet. 98, 287-298. [DOI] [PMC free article] [PubMed]
- 2.Kay C., Collins J.A., Miedzybrodzka Z., Madore S.J., Gordon E.S., Gerry N., Davidson M., Slama R.A., Hayden M.R. Huntington disease reduced penetrance alleles occur at high frequency in the general population. Neurology. 2016;87:282–288. doi: 10.1212/WNL.0000000000002858. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kay, C., Collins, J.A., Miedzybrodzka, Z., Madore, S.J., Gordon, E.S., Gerry, N., Davidson, M., Slama, R.A., and Hayden, M.R. (2016). Huntington disease reduced penetrance alleles occur at high frequency in the general population. Neurology 87, 282-288. [DOI] [PMC free article] [PubMed]
- 3.Maat-Kievit A., Losekoot M., Van Den Boer-Van Den Berg H., Van Ommen G.J., Niermeijer M., Breuning M., Tibben A. New problems in testing for Huntington’s disease: the issue of intermediate and reduced penetrance alleles. J. Med. Genet. 2001;38:E12. doi: 10.1136/jmg.38.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]; Maat-Kievit, A., Losekoot, M., Van Den Boer-Van Den Berg, H., Van Ommen, G.J., Niermeijer, M., Breuning, M., and Tibben, A. (2001). New problems in testing for Huntington’s disease: the issue of intermediate and reduced penetrance alleles. J. Med. Genet. 38, E12. [DOI] [PMC free article] [PubMed]
- 4.Langbehn D.R., Brinkman R.R., Falush D., Paulsen J.S., Hayden M.R., International Huntington’s Disease Collaborative Group A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin. Genet. 2004;65:267–277. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]; Langbehn, D.R., Brinkman, R.R., Falush, D., Paulsen, J.S., and Hayden, M.R.; International Huntington’s Disease Collaborative Group (2004). A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin. Genet. 65, 267-277. [DOI] [PubMed]
- 5.Wexler N.S., Lorimer J., Porter J., Gomez F., Moskowitz C., Shackell E., Marder K., Penchaszadeh G., Roberts S.A., Gayán J., U.S.-Venezuela Collaborative Research Project Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc. Natl. Acad. Sci. USA. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wexler, N.S., Lorimer, J., Porter, J., Gomez, F., Moskowitz, C., Shackell, E., Marder, K., Penchaszadeh, G., Roberts, S.A., Gayan, J., et al.; U.S.-Venezuela Collaborative Research Project (2004). Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc. Natl. Acad. Sci. USA 101, 3498-3503. [DOI] [PMC free article] [PubMed]
- 6.Gusella J.F., MacDonald M.E. Huntington’s disease: the case for genetic modifiers. Genome Med. 2009;1:80. doi: 10.1186/gm80. [DOI] [PMC free article] [PubMed] [Google Scholar]; Gusella, J.F., and MacDonald, M.E. (2009). Huntington’s disease: the case for genetic modifiers. Genome Med. 1, 80. [DOI] [PMC free article] [PubMed]
- 7.Rosenblatt A., Brinkman R.R., Liang K.Y., Almqvist E.W., Margolis R.L., Huang C.Y., Sherr M., Franz M.L., Abbott M.H., Hayden M.R., Ross C.A. Familial influence on age of onset among siblings with Huntington disease. Am. J. Med. Genet. 2001;105:399–403. [PubMed] [Google Scholar]; Rosenblatt, A., Brinkman, R.R., Liang, K.Y., Almqvist, E.W., Margolis, R.L., Huang, C.Y., Sherr, M., Franz, M.L., Abbott, M.H., Hayden, M.R., and Ross, C.A. (2001). Familial influence on age of onset among siblings with Huntington disease. Am. J. Med. Genet. 105, 399-403. [PubMed]
- 8.Bečanović K., Nørremølle A., Neal S.J., Kay C., Collins J.A., Arenillas D., Lilja T., Gaudenzi G., Manoharan S., Doty C.N., REGISTRY Investigators of the European Huntington’s Disease Network A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci. 2015;18:807–816. doi: 10.1038/nn.4014. [DOI] [PubMed] [Google Scholar]; Bečanović, K., Norremolle, A., Neal, S.J., Kay, C., Collins, J.A., Arenillas, D., Lilja, T., Gaudenzi, G., Manoharan, S., Doty, C.N., et al.; REGISTRY Investigators of the European Huntington’s Disease Network (2015). A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci. 18, 807-816. [DOI] [PubMed]
- 9.Hensman Moss D.J., Pardiñas A.F., Langbehn D., Lo K., Leavitt B.R., Roos R., Durr A., Mead S., Holmans P., Jones L., Tabrizi S.J., TRACK-HD investigators. REGISTRY investigators Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 2017;16:701–711. doi: 10.1016/S1474-4422(17)30161-8. [DOI] [PubMed] [Google Scholar]; Moss, D.J.H., Pardiñas, A.F., Langbehn, D., Lo, K., Leavitt, B.R., Roos, R., Durr, A., Mead, S., Holmans, P., Jones, L., and Tabrizi, S.J.; TRACK-HD investigators; REGISTRY investigators (2017). Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 16, 701-711. [DOI] [PubMed]
- 10.Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell. 2015;162:516–526. doi: 10.1016/j.cell.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium (2015). Identification of Genetic Factors that Modify Clinical Onset of Huntington’s Disease. Cell 162, 516-526. [DOI] [PMC free article] [PubMed]
- 11.Goldberg Y.P., McMurray C.T., Zeisler J., Almqvist E., Sillence D., Richards F., Gacy A.M., Buchanan J., Telenius H., Hayden M.R. Increased instability of intermediate alleles in families with sporadic Huntington disease compared to similar sized intermediate alleles in the general population. Hum. Mol. Genet. 1995;4:1911–1918. doi: 10.1093/hmg/4.10.1911. [DOI] [PubMed] [Google Scholar]; Goldberg, Y.P., McMurray, C.T., Zeisler, J., Almqvist, E., Sillence, D., Richards, F., Gacy, A.M., Buchanan, J., Telenius, H., and Hayden, M.R. (1995). Increased instability of intermediate alleles in families with sporadic Huntington disease compared to similar sized intermediate alleles in the general population. Hum. Mol. Genet. 4, 1911-1918. [DOI] [PubMed]
- 12.Pêcheux C., Mouret J.F., Dürr A., Agid Y., Feingold J., Brice A., Dodé C., Kaplan J.C. Sequence analysis of the CCG polymorphic region adjacent to the CAG triplet repeat of the HD gene in normal and HD chromosomes. J. Med. Genet. 1995;32:399–400. doi: 10.1136/jmg.32.5.399. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pecheux, C., Mouret, J.F., Durr, A., Agid, Y., Feingold, J., Brice, A., Dode, C., and Kaplan, J.C. (1995). Sequence analysis of the CCG polymorphic region adjacent to the CAG triplet repeat of the HD gene in normal and HD chromosomes. J. Med. Genet. 32, 399-400. [DOI] [PMC free article] [PubMed]
- 13.Williams L.C., Hegde M.R., Nagappan R., Faull R.L., Giles J., Winship I., Snow K., Love D.R. Null alleles at the Huntington disease locus: implications for diagnostics and CAG repeat instability. Genet. Test. 2000;4:55–60. doi: 10.1089/109065700316480. [DOI] [PubMed] [Google Scholar]; Williams, L.C., Hegde, M.R., Nagappan, R., Faull, R.L., Giles, J., Winship, I., Snow, K., and Love, D.R. (2000). Null alleles at the Huntington disease locus: implications for diagnostics and CAG repeat instability. Genet. Test. 4, 55-60. [DOI] [PubMed]
- 14.Yu S., Fimmel A., Fung D., Trent R.J. Polymorphisms in the CAG repeat--a source of error in Huntington disease DNA testing. Clin. Genet. 2000;58:469–472. doi: 10.1034/j.1399-0004.2000.580607.x. [DOI] [PubMed] [Google Scholar]; Yu, S., Fimmel, A., Fung, D., and Trent, R.J. (2000). Polymorphisms in the CAG repeat--a source of error in Huntington disease DNA testing. Clin. Genet. 58, 469-472. [DOI] [PubMed]
- 15.Semaka A., Kay C., Doty C.N., Collins J.A., Tam N., Hayden M.R. High frequency of intermediate alleles on Huntington disease-associated haplotypes in British Columbia’s general population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2013;162B:864–871. doi: 10.1002/ajmg.b.32193. [DOI] [PubMed] [Google Scholar]; Semaka, A., Kay, C., Doty, C.N., Collins, J.A., Tam, N., and Hayden, M.R. (2013). High frequency of intermediate alleles on Huntington disease-associated haplotypes in British Columbia’s general population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 162B, 864-871. [DOI] [PubMed]
- 16.Kay C., Collins J.A., Skotte N.H., Southwell A.L., Warby S.C., Caron N.S., Doty C.N., Nguyen B., Griguoli A., Ross C.J. Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol. Ther. 2015;23:1759–1771. doi: 10.1038/mt.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kay, C., Collins, J.A., Skotte, N.H., Southwell, A.L., Warby, S.C., Caron, N.S., Doty, C.N., Nguyen, B., Griguoli, A., Ross, C.J., et al. (2015). Huntingtin Haplotypes Provide Prioritized Target Panels for Allele-specific Silencing in Huntington Disease Patients of European Ancestry. Mol. Ther. 23, 1759-1771. [DOI] [PMC free article] [PubMed]
- 17.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]; Das, S., Forer, L., Schonherr, S., Sidore, C., Locke, A.E., Kwong, A., Vrieze, S.I., Chew, E.Y., Levy, S., McGue, M., et al. (2016). Next-generation genotype imputation service and methods. Nat. Genet. 48, 1284-1287. [DOI] [PMC free article] [PubMed]
- 18.C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]; C Yuen, R.K., Merico, D., Bookman, M., L Howe, J., Thiruvahindrapuram, B., Patel, R.V., Whitney, J., Deflaux, N., Bingham, J., Wang, Z., et al. (2017). Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 20, 602-611. [DOI] [PMC free article] [PubMed]
- 19.Dolzhenko E., van Vugt J.J.F.A., Shaw R.J., Bekritsky M.A., van Blitterswijk M., Narzisi G., Ajay S.S., Rajan V., Lajoie B.R., Johnson N.H., US–Venezuela Collaborative Research Group Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dolzhenko, E., van Vugt, J.J.F.A., Shaw, R.J., Bekritsky, M.A., van Blitterswijk, M., Narzisi, G., Ajay, S.S., Rajan, V., Lajoie, B.R., Johnson, N.H., et al.; US-Venezuela Collaborative Research Group (2017). Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 27, 1895-1903. [DOI] [PMC free article] [PubMed]
- 20.Dolzhenko E., Deshpande V., Schlesinger F., Krusche P., Petrovski R., Chen S., Emig-Agius D., Gross A., Narzisi G., Bowman B. ExpansionHunter: A sequence-graph based tool to analyze variation in short tandem repeat regions. bioRxiv. 2019 doi: 10.1093/bioinformatics/btz431. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dolzhenko, E., Deshpande, V., Schlesinger, F., Krusche, P., Petrovski, R., Chen, S., Emig-Agius, D., Gross, A., Narzisi, G., Bowman, B., et al. (2019). ExpansionHunter: A sequence-graph based tool to analyze variation in short tandem repeat regions. bioRxiv 10.1101/572545. [DOI] [PMC free article] [PubMed]
- 21.Pinto R.M., Dragileva E., Kirby A., Lloret A., Lopez E., St Claire J., Panigrahi G.B., Hou C., Holloway K., Gillis T. Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet. 2013;9:e1003930. doi: 10.1371/journal.pgen.1003930. [DOI] [PMC free article] [PubMed] [Google Scholar]; Pinto, R.M., Dragileva, E., Kirby, A., Lloret, A., Lopez, E., St Claire, J., Panigrahi, G.B., Hou, C., Holloway, K., Gillis, T., et al. (2013). Mismatch repair genes Mlh1 and Mlh3 modify CAG instability in Huntington’s disease mice: genome-wide and candidate approaches. PLoS Genet. 9, e1003930. [DOI] [PMC free article] [PubMed]
- 22.Semaka A., Kay C., Doty C., Collins J.A., Bijlsma E.K., Richards F., Goldberg Y.P., Hayden M.R. CAG size-specific risk estimates for intermediate allele repeat instability in Huntington disease. J. Med. Genet. 2013;50:696–703. doi: 10.1136/jmedgenet-2013-101796. [DOI] [PubMed] [Google Scholar]; Semaka, A., Kay, C., Doty, C., Collins, J.A., Bijlsma, E.K., Richards, F., Goldberg, Y.P., and Hayden, M.R. (2013). CAG size-specific risk estimates for intermediate allele repeat instability in Huntington disease. J. Med. Genet. 50, 696-703. [DOI] [PubMed]
- 23.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv. 2019 [Google Scholar]; Karczewski, K.J., Francioli, L.C., Tiao, G., Cummings, B.B., Alfoldi, J., Wang, Q., Collins, R.L., Laricchia, K.M., Ganna, A., Birnbaum, D.P., et al. (2019). Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 10.1101/531210.
- 24.Semaka A., Collins J.A., Hayden M.R. Unstable familial transmissions of Huntington disease alleles with 27-35 CAG repeats (intermediate alleles) Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:314–320. doi: 10.1002/ajmg.b.30970. [DOI] [PubMed] [Google Scholar]; Semaka, A., Collins, J.A., and Hayden, M.R. (2010). Unstable familial transmissions of Huntington disease alleles with 27-35 CAG repeats (intermediate alleles). Am. J. Med. Genet. B. Neuropsychiatr. Genet. 153B, 314-320. [DOI] [PubMed]
- 25.Telenius H., Almqvist E., Kremer B., Spence N., Squitieri F., Nichol K., Grandell U., Starr E., Benjamin C., Castaldo I. Somatic mosaicism in sperm is associated with intergenerational (CAG)n changes in Huntington disease. Hum. Mol. Genet. 1995;4:189–195. doi: 10.1093/hmg/4.2.189. [DOI] [PubMed] [Google Scholar]; Telenius, H., Almqvist, E., Kremer, B., Spence, N., Squitieri, F., Nichol, K., Grandell, U., Starr, E., Benjamin, C., Castaldo, I., et al. (1995). Somatic mosaicism in sperm is associated with intergenerational (CAG)n changes in Huntington disease. Hum. Mol. Genet. 4, 189-195. [DOI] [PubMed]
- 26.Telenius H., Kremer B., Goldberg Y.P., Theilmann J., Andrew S.E., Zeisler J., Adam S., Greenberg C., Ives E.J., Clarke L.A. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]; Telenius, H., Kremer, B., Goldberg, Y.P., Theilmann, J., Andrew, S.E., Zeisler, J., Adam, S., Greenberg, C., Ives, E.J., Clarke, L.A., et al. (1994). Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 6, 409-414. [DOI] [PubMed]
- 27.Pouladi M.A., Stanek L.M., Xie Y., Franciosi S., Southwell A.L., Deng Y., Butland S., Zhang W., Cheng S.H., Shihabuddin L.S., Hayden M.R. Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum. Mol. Genet. 2012;21:2219–2232. doi: 10.1093/hmg/dds037. [DOI] [PubMed] [Google Scholar]; Pouladi, M.A., Stanek, L.M., Xie, Y., Franciosi, S., Southwell, A.L., Deng, Y., Butland, S., Zhang, W., Cheng, S.H., Shihabuddin, L.S., and Hayden, M.R. (2012). Marked differences in neurochemistry and aggregates despite similar behavioural and neuropathological features of Huntington disease in the full-length BACHD and YAC128 mice. Hum. Mol. Genet. 21, 2219-2232. [DOI] [PubMed]
- 28.Swami M., Hendricks A.E., Gillis T., Massood T., Mysore J., Myers R.H., Wheeler V.C. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]; Swami, M., Hendricks, A.E., Gillis, T., Massood, T., Mysore, J., Myers, R.H., and Wheeler, V.C. (2009). Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 18, 3039-3047. [DOI] [PMC free article] [PubMed]
- 29.Shelbourne P.F., Keller-McGandy C., Bi W.L., Yoon S.R., Dubeau L., Veitch N.J., Vonsattel J.P., Wexler N.S., Arnheim N., Augood S.J., US-Venezuela Collaborative Research Group Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]; Shelbourne, P.F., Keller-McGandy, C., Bi, W.L., Yoon, S.R., Dubeau, L., Veitch, N.J., Vonsattel, J.P., Wexler, N.S., Arnheim, N., and Augood, S.J.; US-Venezuela Collaborative Research Group (2007). Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 16, 1133-1142. [DOI] [PubMed]
- 30.Kennedy L., Evans E., Chen C.M., Craven L., Detloff P.J., Ennis M., Shelbourne P.F. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]; Kennedy, L., Evans, E., Chen, C.M., Craven, L., Detloff, P.J., Ennis, M., and Shelbourne, P.F. (2003). Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 12, 3359-3367. [DOI] [PubMed]
- 31.Costanzi-Porrini S., Tessarolo D., Abbruzzese C., Liguori M., Ashizawa T., Giacanelli M. An interrupted 34-CAG repeat SCA-2 allele in patients with sporadic spinocerebellar ataxia. Neurology. 2000;54:491–493. doi: 10.1212/wnl.54.2.491. [DOI] [PubMed] [Google Scholar]; Costanzi-Porrini, S., Tessarolo, D., Abbruzzese, C., Liguori, M., Ashizawa, T., and Giacanelli, M. (2000). An interrupted 34-CAG repeat SCA-2 allele in patients with sporadic spinocerebellar ataxia. Neurology 54, 491-493. [DOI] [PubMed]
- 32.Charles P., Camuzat A., Benammar N., Sellal F., Destée A., Bonnet A.M., Lesage S., Le Ber I., Stevanin G., Dürr A., Brice A., French Parkinson’s Disease Genetic Study Group Are interrupted SCA2 CAG repeat expansions responsible for parkinsonism? Neurology. 2007;69:1970–1975. doi: 10.1212/01.wnl.0000269323.21969.db. [DOI] [PubMed] [Google Scholar]; Charles, P., Camuzat, A., Benammar, N., Sellal, F., Destee, A., Bonnet, A.M., Lesage, S., Le Ber, I., Stevanin, G., Durr, A., and Brice, A.; French Parkinson’s Disease Genetic Study Group (2007). Are interrupted SCA2 CAG repeat expansions responsible for parkinsonism? Neurology 69, 1970-1975. [DOI] [PubMed]
- 33.Fujigasaki H., Martin J.J., De Deyn P.P., Camuzat A., Deffond D., Stevanin G., Dermaut B., Van Broeckhoven C., Dürr A., Brice A. CAG repeat expansion in the TATA box-binding protein gene causes autosomal dominant cerebellar ataxia. Brain. 2001;124:1939–1947. doi: 10.1093/brain/124.10.1939. [DOI] [PubMed] [Google Scholar]; Fujigasaki, H., Martin, J.J., De Deyn, P.P., Camuzat, A., Deffond, D., Stevanin, G., Dermaut, B., Van Broeckhoven, C., Durr, A., and Brice, A. (2001). CAG repeat expansion in the TATA box-binding protein gene causes autosomal dominant cerebellar ataxia. Brain 124, 1939-1947. [DOI] [PubMed]
- 34.Nakamura K., Jeong S.Y., Uchihara T., Anno M., Nagashima K., Nagashima T., Ikeda S., Tsuji S., Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]; Nakamura, K., Jeong, S.Y., Uchihara, T., Anno, M., Nagashima, K., Nagashima, T., Ikeda, S., Tsuji, S., and Kanazawa, I. (2001). SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 10, 1441-1448. [DOI] [PubMed]
- 35.Chong S.S., Almqvist E., Telenius H., LaTray L., Nichol K., Bourdelat-Parks B., Goldberg Y.P., Haddad B.R., Richards F., Sillence D. Contribution of DNA sequence and CAG size to mutation frequencies of intermediate alleles for Huntington disease: evidence from single sperm analyses. Hum. Mol. Genet. 1997;6:301–309. doi: 10.1093/hmg/6.2.301. [DOI] [PubMed] [Google Scholar]; Chong, S.S., Almqvist, E., Telenius, H., LaTray, L., Nichol, K., Bourdelat-Parks, B., Goldberg, Y.P., Haddad, B.R., Richards, F., Sillence, D., et al. (1997). Contribution of DNA sequence and CAG size to mutation frequencies of intermediate alleles for Huntington disease: evidence from single sperm analyses. Hum. Mol. Genet. 6, 301-309. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.