Abstract
Cargo transport along the cytoplasmic microtubular network is essential for neuronal function, and cytoplasmic dynein-1 is an established molecular motor that is critical for neurogenesis and homeostasis. We performed whole-exome sequencing, homozygosity mapping, and chromosomal microarray studies in five individuals from three independent pedigrees and identified likely-pathogenic variants in DYNC1I2 (Dynein Cytoplasmic 1 Intermediate Chain 2), encoding a component of the cytoplasmic dynein 1 complex. In a consanguineous Pakistani family with three affected individuals presenting with microcephaly, severe intellectual disability, simplification of cerebral gyration, corpus callosum hypoplasia, and dysmorphic facial features, we identified a homozygous splice donor site variant (GenBank: NM_001378.2:c.607+1G>A). We report two additional individuals who have similar neurodevelopmental deficits and craniofacial features and harbor deleterious variants; one individual bears a c.740A>G (p.Tyr247Cys) change in trans with a 374 kb deletion encompassing DYNC1I2, and an unrelated individual harbors the compound-heterozygous variants c.868C>T (p.Gln290∗) and c.740A>G (p.Tyr247Cys). Zebrafish larvae subjected to CRISPR-Cas9 gene disruption or transient suppression of dync1i2a displayed significantly altered craniofacial patterning with concomitant reduction in head size. We monitored cell death and cell cycle progression in dync1i2a zebrafish models and observed significantly increased apoptosis, likely due to prolonged mitosis caused by abnormal spindle morphology, and this finding offers initial insights into the cellular basis of microcephaly. Additionally, complementation studies in zebrafish demonstrate that p.Tyr247Cys attenuates gene function, consistent with protein structural analysis. Our genetic and functional data indicate that DYNC1I2 dysfunction probably causes an autosomal-recessive microcephaly syndrome and highlight further the critical roles of the dynein-1 complex in neurodevelopment.
Keywords: autosomal recessive, microcephaly, intellectual disability, DYNC1I2, dynein, zebrafish, apoptosis, developmental delay, dysmorphic facial features, mitotic spindle
Introduction
Intracellular cargo transport is important for eukaryotic cell function; cargoes are transported by energy-dependent motors that shuttle along the microtubule network (kinesin and dynein) or actin cytoskeleton (myosin). There are two types of motor proteins associated with the microtubule network: members of the kinesin protein family transport cargo toward the growing plus-end (anterograde), and the cytoplasmic dynein motor proteins move the cargo, such as mRNA particles, vesicles, and organelles, toward the minus-end (retrograde) of microtubules.1, 2, 3 There are two dynein transport molecular complexes that function as motors; cytoplasmic dynein-1 (also termed cytoplasmic dynein) is mainly involved in the retrograde transport of cargo within the cytoplasmic microtubule network, and cytoplasmic dynein-2 (also referred to as intraflagellar transport dynein) functions predominantly for retrograde trafficking of cargo in cilia and flagella.4, 5 The core of the cytoplasmic dynein-1 protein complex in humans is composed of a homodimer of force-generating heavy chains encoded by DYNC1H1 (MIM: 600112); these are associated with two intermediate chains encoded by DYNC1I1 (MIM: 603772) and DYNC1I2 (MIM: 603331). The heavy and intermediate chains then associate with light-intermediate chains encoded by DYNC1LI1 (MIM: 615890) and DYNC1LI2 (MIM: 611406) and three classes of light chains encoded by six genes (DYNLRB1 [MIM: 607167], DYNLRB2 [MIM: 607168], DYNLL1 [MIM: 601562], DYNLL2 [MIM: 608942], DYNLT1 [MIM: 601554], and DYNLT3 [MIM: 300302]).4, 5, 6
Although not many protein subunits in the dynein-1 complex have been implicated in human disorders, there is an emerging theme that the dynein-1 complex is particularly critical for neurodevelopment and homeostasis.7, 8 These observations can be supported by DYNC1H1-associated human disorders, which can give rise to a diverse array of autosomal-dominant neurological disorders including Charcot-Marie-Tooth (MIM: 614228),9 spinal muscular atrophy (MIM: 158600),10 intellectual disability (MIM:614563),11, 12 and lissencephaly.13 Furthermore, copy number variation (CNV) deletions encompassing DYNC1I1 at 7q21.3 have been implicated in intellectual disability, developmental delay, and craniofacial dysmorphism with or without ectrodactyly,14, 15 although the precise gene driver for the neurological features has yet to be elucidated. The involvement of the dynein-1 molecular module in neurodevelopment is exemplified further by the observation that some dynein-1 complex genes, such as DYNC1I1 and DYNC1I2, have tightly regulated expression and undergo extensive alternative splicing to enable production of isoforms that are involved in discrete cargo specificity and binding tasks in neuronal tissues.16, 17
In this study, we report three unrelated families comprising five affected individuals with an overlapping, complex neurodevelopmental disorder characterized by microcephaly, intellectual disability, cerebral malformations, and dysmorphic facial features. We performed whole-exome sequencing coupled with homozygosity mapping or copy number variant analysis to identify four different recessive alleles (splice donor site, whole gene deletion, nonsense, and missense variants) in dynein cytoplasmic 1 intermediate chain 2 (DYNC1I2); the alleles segregated with the disease in each family. Ablation of the orthologous zebrafish locus recapitulated phenotypes of the affected individuals in our cohort, and in vivo complementation studies demonstrated the loss-of-function effect of the missense mutation identified in humans. These data reinforce the importance of the cytoplasmic dynein-1 complex in neurodevelopment and specifically implicate DYNC1I2 in syndromic microcephaly.
Material and Methods
Participant Families and Ethics Approval
Family 1 (F157) was recruited and enrolled by the Institute of Basic Medical Sciences (IBMS), Khyber Medical University, Peshawar, Pakistan and was studied at the Department of Genetic Medicine and Development, University of Geneva, Switzerland. The study was approved by the Bioethics Committee of the University Hospitals of Geneva (Protocol number: CER 11-036) and by the ethical committee of the Khyber Medical University. Family 2 was studied at the Atlantic Health System, Morristown, NJ, USA, and this family was studied after an IRB approval by established institutional processes. Family 3 was evaluated in the Brain Development and Genetics Clinic at Boston Children’s Hospital and was enrolled according to protocol 05-05-076R, approved by the institutional review board of Boston Children’s Hospital. We obtained informed consent from guardians of all affected individuals who participated in this study. We obtained peripheral blood from all research participants and extracted genomic DNA according to standard protocols.
Genetic Analysis
In family 1, we performed exome sequencing of the proband (1-VI:2) by using SureSelect Human All Exon v6 reagents (Agilent Technologies) on an Illumina HiSeq4000 platform. We used an in-house custom pipeline to analyze the exome sequencing data. The pipeline is comprised of published algorithms including the Burrows-Wheeler aligner tool (BWA),18 SAMtools,18 PICARD, and the Genome Analysis Toolkit (GATK).19 We aligned sequenced reads to the GRCh37 (hg19)20 reference human genome and performed variant filtering as described.21, 22, 23 We conducted genotyping of five nuclear family members (1-IV:7, 1-V:4, 1-VI:1, 1-VI:2, and 1-VI:3) with the Illumina 720K SNP array (HumanOmniExpress Bead Chip by Illumina), and we used PLINK24 to perform homozygosity mapping. A run of homozygosity (ROH) was defined as a window of 50 consecutive homozygous SNPs; the window allowed a maximum of one mismatch in a homozygous region and was demarcated by the first heterozygous SNP at the edge of each homozygous region. We used CATCH to intersect variants from the exome sequencing data with ROH that segregated recessively in the family and with disease phenotype.25 This program also filters variants on the basis of zygosity, quality score, minor allele frequency, pathogenicity prediction, and conservation scores as described.21, 22, 23 All candidate variants were verified further by Sanger sequencing.
We carried out SNP microarray analysis in the family 2 index individual (2-II:1) by using the Affymetrix Cytoscan HD platform, according to the manufacturer’s instructions. We analyzed the data with Chromosome Analysis Suite according to the GRCh37 (hg19) assembly. We then captured exonic regions and flanking splice junctions of the exome by using the Clinical Research Exome kit (Agilent Technologies) on lymphocyte-derived genomic DNA from the family 2 index individual and both parents. Massively parallel sequencing data were generated on an Illumina system with 100 bp or greater paired-end reads. The reads were aligned to human genome build GRCh37 (UCSC hg19) and analyzed for sequence variants with a custom-developed analysis tool, as described.26 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission URL (see Web Resources).
In family 3, whole-exome sequencing and data processing were performed by the Genomics Platform at the Broad Institute of Harvard and MIT, Cambridge, MA, USA. We performed whole-exome sequencing on DNA samples (>250 ng of DNA at >2 ng/μl) by using Illumina exome capture reagent (38 Mb target). Our exome-sequencing pipeline included sample plating, library preparation (2-plexing of samples per hybridization), hybrid capture, sequencing (150 bp paired reads), sample identification quality control check, and data storage. The exome sequencing data were de-multiplexed, and sequence data from each sample were aggregated into a single Picard BAM file. Exome sequencing data were processed through a pipeline that was based on Picard and which used base quality score recalibration and local realignment at known indels. We used the BWA aligner to map reads to the human genome build 37 (hg19). Single nucleotide variants (SNVs) and insertions/deletions (indels) were called jointly across all samples with GATK HaplotypeCaller package version 3.4. Default filters were applied to SNV and indel calls according to the GATK Variant Quality Score Recalibration (VQSR) approach. The variants were annotated with the Variant Effect Predictor (VEP). Additional information is provided in the Supplemental Data.27 The variant call set was uploaded on to Seqr, and analysis was performed according to all possible inheritance patterns. Candidate variants were validated further by Sanger sequencing.
Molecular Modeling of the DYNC1I2 Missense Variant
We mined the Protein Data Bank (PDB) for the following cryo-EM structures of dynein-dynactin complexes with different adaptors in order to predict the potential structural consequences of the p.Tyr247Cys variant (equivalent amino acid in GenBank: NP_001369.1): (A) 6F1T, structure of two dynein tail domains bound to dynactin and BICDR1; (B) 6F38, structure of two dynein tail domains bound to dynactin and HOOK3; (C) 6F3A, structure of a single dynein tail domain bound to dynactin and BICD2N; and (D) 6F1Z, structure of the roadblock-1 region of the dynein tail/dynactin/BICDR1 complex. We used UCSF Chimera software28 to model the Tyr247 and Tyr247Cys side chains and to visualize the resulting protein structures.
Zebrafish Husbandry and Embryo Maintenance
We performed all zebrafish experiments according to protocols approved by the Duke University institutional animal care and use committee (IACUC). We obtained embryos by natural mating of adult zebrafish carrying the -1.4col1a1:egfp transgenic reporter.29 Embryos were grown in egg water (0.3 g/L NaCl, 75 mg/L CaSO4, 37.5 mg/L NaHC03, 0.003% methylene blue) at 28°C until reaching respective phenotypic endpoints (time ranged from 1–3 days post-fertilization [dpf]).
CRISPR-Cas9 Genome Editing in Zebrafish Embryos
By using reciprocal BLAST, we identified in the zebrafish genome two DYNC1I2 orthologs: dync1i2a (Ensembl ID: GRCz10: ENSDART00000007756.9) and dync1i2b (Ensembl ID: GRCz10: ENSDART00000159673.2). To identify single guide (sg) RNA sites targeting either dync1i2a or dync1i2b, we used the CHOPCHOPv2 web tool.30 Using synthetic oligos as a template, we generated sgRNAs with the GeneArt precision gRNA synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. We injected a 1 nL cocktail containing each sgRNA with or without Cas9 protein (PNA Bio) into the cell of single-cell staged embryos at the following concentrations: dync1i2a, 200 pg sgRNA + 400 pg Cas9; or dync1i2b, 100 pg sgRNA + 200 pg Cas 9. To determine mosaicism at the targeted genomic locus in F0 animals, we extracted genomic DNA from individual embryos (n = 10) at 2 dpf, PCR-amplified the sgRNA targeted region, and performed heteroduplex analysis as described.31 PCR products flanking sgRNA target sites were denatured, reannealed slowly, and then migrated on a 16% polyacrylamide gel (Thermo Fisher Scientific) to separate heteroduplexes. We randomly selected a subset of PCR products corresponding to individual embryos (n = 5), then gel-purified and cloned them in TOPO-TA vector (Thermo Fisher Scientific). Individual colonies (n = 36/embryo) were sequenced with BigDye terminator 3.1 chemistry (Applied Biosystems).
Transient Suppression of dync1i2a and dync1i2b
We obtained the morpholinos (MOs) e2i2 and e5i5, targeting exons 2 and 5, respectively, to block the dync1i2a splice donor sites, as well as the MO dync1i2b e4i4, targeting exon 4 (Gene Tools). We suppressed each ortholog independently by injecting 1 nL MO into embryo batches at three different doses (3 ng, 6 ng, or 9 ng) at the one-to-four cell stage. To determine MO efficiency, we extracted total RNA from pooled embryos at 2 dpf (n = 20; controls or 9 ng MO) by using Trizol (Thermo Fisher Scientific), synthesized cDNA with the QuantiTect Reverse Transcription kit (QIAGEN), and performed RT-PCR with primers flanking the MO target region. We separated amplified PCR products on a 1% agarose gel, and we excised and purified PCR products with the QIAquick gel extraction kit (QIAGEN). We cloned purified PCR products with TOPO TA cloning vector and sequenced the resulting colonies (n = 5/sample). For all in vivo complementation assays, we used 9 ng of either dync1i2a e2i2 or e5i5 MOs.
Human DYNC1I2 ORF Constructs and Site-Directed Mutagenesis
We obtained a Gateway-compatible DYNC1I2 full-length wild-type (WT) human open reading frame (ORF) entry clone (Dharmacon; Catalog # OHS1770-202314378; GenBank: NM_001271789.1 and NP_001258718.1). This clone corresponds to the most abundant transcript in the brain. In this isoform, c.662A>G (p.Tyr221Cys) corresponds to GenBank: NM_001378.2: c.740A>G (p.Tyr247Cys) and c.790C>T (p.(Gln264∗) corresponds to GenBank: NM_001378.2: c.868C>T (p.Gln290∗). We transferred the ORF to a Gateway-compatible pCS2+ destination vector via LR-clonase II mediated recombination (Thermo Fisher Scientific). Next, we incorporated a stop codon (TAG) by site-directed mutagenesis, as described,32 and we used it as a template for the introduction of variants, c.662A>G, (p.Tyr221Cys) and c.790C>T (p.Gln264∗), identified in affected individuals, or a negative control variant, dbSNP: rs767705533, c.1468C>G (p.Pro490Ala), which corresponds to GenBank: NM_001378.2: c.1546C>G (p.Pro516Ala). There are two homozygotes in ∼120,000 individuals in gnomAD (accessed January 3rd, 2019) for this variant. Subsequent to sequence confirmation of the entire ORF, we linearized pCS2+ vectors with NotI and used them as a template for mRNA transcription with the mMessage mMachine SP6 Transcription kit (Ambion), according to the manufacturer’s instructions. For all in vivo complementation assays, we used 50 pg human DYNC1I2 mRNA.
Live, Automated Phenotyping of Zebrafish Larvae
We assessed head size and craniofacial patterning phenotypes at 3 dpf by using the Vertebrate Automated Screening Technology (VAST) Bioimager (Union Biometrica), mounted on an AXIO microscope (Zeiss) with a 10× objective lens and facilitated with an Axiocam camera (Zeiss) and ZenPro software (Zeiss). We anesthetized larvae with tricaine and acquired dorsal and ventral images (bright field and GFP, respectively), as described.33, 34, 35, 36, 37
TUNEL Assay and Whole-Mount Immunostaining
TUNEL was performed essentially as described.33, 37, 38, 39 In brief, we dechorionated embryos at 2 dpf and fixed them in 4% paraformaldehyde (PFA) overnight at 4°C. They were dehydrated in 100% methanol for 2 h at −20°C prior to rehydration and permeabilization with proteinase K for 10 min and post fixation in 4% PFA for 20 min. Embryos were treated with equilibration buffer for 1 h and incubated overnight in TdT enzyme at 4°C. Embryos were then treated with the digoxigenin provided in the ApopTag red in situ apoptosis detection kit (Millipore) for 2 h and washed three times for 10 min each with PBST.
Immunostaining was performed as described.33, 37, 38, 39, 40 Dechorionated embryos at 2 dpf (pH3); and embryos at 1 dpf or larvae at 3 dpf (acetylated α-tubulin) were fixed overnight in 4% PFA (pH3) or Dent’s solution (80% methanol, 40% DMSO; acetylated tubulin), respectively, dehydrated in methanol, and rehydrated gradually with PBST in methanol. We bleached for 10 min in 9mL PBST + 1mL H2O2 + 0.05 g KOH and washed three times for 10 min each in PBST. Embryos were permeabilized with proteinase K for 10 min, then incubated overnight in primary antibody (anti-pH3, 1:500, Santa Cruz Biotechnology, A11012 or anti-α-acetylated tubulin, 1:1,000 Sigma-Aldrich, T7451). The embryos were treated with secondary antibody (Alexa Fluor 488 goat anti-rabbit IgG, 1:500, Thermo Fisher Scientific, A11001) for 2 h and washed three times with immunofluorescence (IF) buffer (1% BSA in PBST).
Fluorescent Microscopy of Whole-Mount Zebrafish
We imaged the fluorescent signal of fixed whole-mount embryos or larvae with a Nikon AZ100 microscope equipped with a Nikon camera and controlled by Nikon NIS Elements Software. We observed dividing cells immunostained with anti-α-acetylated tubulin antibody and counterstained with DAPI at 1 dpf (Sigma, T5168, 1: 200) by using a Zeiss710 confocal microscope and Zen software.
Image Analysis
We used ImageJ (NIH) to measure the morphological features of zebrafish larvae. To assess head size on dorsal images, we drew a straight line connecting the lateral-posterior part of both eyes and outlined the remainder of the head region, excluding the eyes. To quantify craniofacial patterning defects, we measured the ceratohyal cartilage angle on ventral fluorescent images. To evaluate neurological organization on dorsal fluorescent images, we measured the area of the optic tecta and the number of axon tracts crossing the midline between the optic tecta. We counted TUNEL- or pH3-positive cells with the image-based tool for counting nuclei (ICTN) ImageJ plugin, and we normalized counts by dividing by the area of the region of interest in the forebrain (excluding the eyes). We assessed the spindle organization of dividing cells to assign mitotic stage and assess whether they were normal or abnormal according to objective criteria, as described.41
Statistical Analysis
We conducted statistical comparisons of embryo or larval batches with Prism 8 software (GraphPad) using a Student’s t test for quantitative measures (morphometric analyses) or Fisher’s exact test for qualitative data (spindle analysis).
Results
Clinical Evaluation of Cases with Syndromic Microcephaly
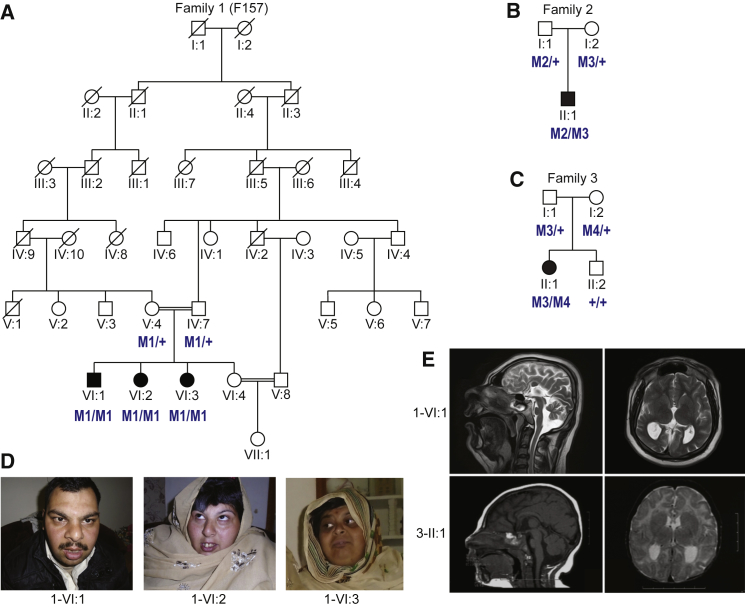
Family 1 originates from the Kohat region in the Khyber Pakhtunkhwa province of Pakistan and is a multigenerational, consanguineous pedigree with seven generations documented. Pedigree analysis indicated that the unaffected parents of the nuclear family are third cousins (Figure 1A) and have four children; three are affected (1-VI:1, 1-VI:2, and 1-VI:3) and one is a healthy individual (1-VI:4). The affected individuals exhibit similar features hallmarked by microcephaly, severe intellectual disability, short stature, and facial dysmorphisms (Table 1). In particular, all three individuals displayed upslanting palpebral features, anteverted nares, and a bulbous nose (Figure 1D). Additionally, all three cases had dysarthric speech with nasal intonation. The parents describe irritable mood with aggressive behavior in all affected children. Brain magnetic resonance imaging (MRI) of the affected male, VI:1 of family 1, at 20 years of age showed near complete agenesis of the corpus callosum, with remnants in its body portion connecting both cerebral halves, along with colpocephaly. Both cerebral cortical and brain stem hypoplasia were present, with cortical white matter hypoplasia and simplification of the cerebral cortical gyral pattern, especially in the right hemisphere. (Figure 1E).
Figure 1.
Segregation of Rare DYNC1I2 Variants in Three Pedigrees with Overlapping Neurodevelopmental Phenotypes
(A–C) Pedigrees showing segregation of rare deleterious DYNC1I2 variants. The variants, named according to GenBank: NM_001378.2, are coded as: M1, c.607+1G>A; M2, 374 kb deletion at 2q31.1; M3, c.740A>G (p.Tyr247Cys); and M4, c.868C>T (p.Gln290∗). Unfilled and shaded shapes denote healthy and affected individuals, respectively. Squares represent males, circles represent females, and double horizontal lines represent consanguineous marriage.
(D) Photographs of three affected individuals (VI:1, VI:2, and VI:3) in family 1.
(E) Representative MRI images of an affected individual (VI:1, age 20 years; T-2 weighted) from family 1 and an affected individual (II-1, age 8 months; T-1 weighted) from family 3.
Table 1.
Clinical Features of Affected Individuals with Recessive DYNC1I2 Variants
| Family ID | Family 1 (F157) | Family 2 | Family 3 | ||
|---|---|---|---|---|---|
| Variation in DYNC1I2 (GenBank:NM_001378.2and NP_001369.1) | Allele 1: c.607+1G>A, Allele 2: c.607+1G>A | Allele 1: c.740A>G (p.Tyr247Cys), Allele 2: 374 kb deletion at 2q31.1 | Allele 1: c.740A>G; (p.Tyr247Cys), Allele 2: c.868C>T; (p.Gln290∗) | ||
| Origin | Pakistani | Russian/Polish/Austrian/Ashkenazi Jewish/Irish/Swedish/German | Polish, Russian | ||
| Individual ID | 1-VI:1 | 1-VI:2 | 1-VI:3 | 2-II:1 | 3-II:1 |
| Sex | male | female | female | male | female |
| Age at last evaluation (years) | 20 | 32 | 34 | 3 | 8 months |
| Height (cm) | 156 | 152 | 145 | 90 | 61 |
| Head circumference (cm) | 47 (<1st percentile) | 48 (<1st percentile) | 48 (<1st percentile) | 41.8 (<1st percentile) | 35 (<1st percentile) |
| Intellectual disability | severe | severe | severe | yes | yes |
| Developmental delay | yes | yes | yes | yes | yes |
| Speech | delayed | delayed | delayed | delayed | delayed |
| Motor milestones | delayed | delayed | delayed | delayed | delayed |
| Behavioral problems | ADHD, aggressive | ADHD, aggressive | ADHD, aggressive | unknown | unknown |
| Facial dysmorphism | yes | yes | yes | yes | yes |
| Brain MRI | Microcephaly with simplified gyral pattern, near complete agenesis of corpus callosum with colpocephaly, and mild brain atrophy | not done | not done | Microcephaly with simplified gyral pattern, mega-cisterna magna, absence of the rostrum and genu of the corpus callosum and the septum pellucidum, and partial absence of the splenium. | Microcephaly with simplified gyral pattern, hypogenesis of the corpus callosum—small genu and splenium, reduced white matter volume, ventricle abnormality—and large trigones |
| Other symptoms | N/A | N/A | N/A | Seizures, multiple hemangiomas | Seizures, hypothyroidism, gastresophageal reflux, joint hyperextensibility, and unilateral hearing loss due to left cochlear defect |
Abbreviations are as follows: hom = homozygous; ADHD = Attention deficit hyperactivity disorder, and MRI = magnetic resonance imaging.
Individual II:1 of family 2 (Figure 1B and Table 1) was assessed at 4 months of age. He was delivered full term via normal spontaneous vaginal delivery to unaffected parents of European/Ashkenazi Jewish descent after an uncomplicated pregnancy. At the 2-month-old visit, a head ultrasound was performed because of microcephaly since birth, and it detected an absent corpus callosum. Furthermore, a brain MRI revealed the absence of the rostrum and genu of the corpus callosum and partial absence of the splenium, as well as absence of the septum pellucidum and mega cisterna magna. On a physical exam, he displayed diffuse spasticity and irregular ear helices bilaterally.
Individual II:1 of Family 3 (Figure 1C and Table 1) was born at full term via planned cesarean section to healthy parents of Eastern European ancestry. At birth, her head circumference was 31.75 cm (−2.07 SD) and her weight was 2.49 kg (−1.78 SD). She presented at 2 months of age with poor head growth and was noted then to also have a sloping forehead and prominent eyes and nose. At 4 months, her head circumference was 33 cm (−6.37 SD), her height was 59.1 cm (−0.91 SD), and her weight was 5.78 kg (−0.47 SD). Within the first year of life, she developed gastroesophageal reflux, recurrent otitis media and upper respiratory infections, global developmental delay, seizures, and hyperreflexia. Neurologic evaluation at 25 months noted a head circumference of 38 cm (−6.67 SD), hypotonia, and hyperextensible wrists and ankles. Developmentally she had a few words through sign language, was able to use a pincer grasp, was very social, and maintained eye contact but was not able to walk. Brain MRI at 3 months of age revealed a hypoplastic corpus callosum, prominent ventricles, reduced white matter volume, and simplified gyral pattern, and subsequent brain MRI at 7 months re-demonstrated the microcephaly with simplified gyral pattern and mild enlargement of the posterior horn of the ventricles (Figure 1E).
In summary, affected individuals from all three families share similar radiographic features, including profound microcephaly and overall brain hypoplasia; white matter hypoplasia ranging from severe to almost complete, in general but affecting the corpus callosum particularly; and simplification of the gyral pattern.
Genetic Analysis Identified Probable Causal Variants in DYNC1I2
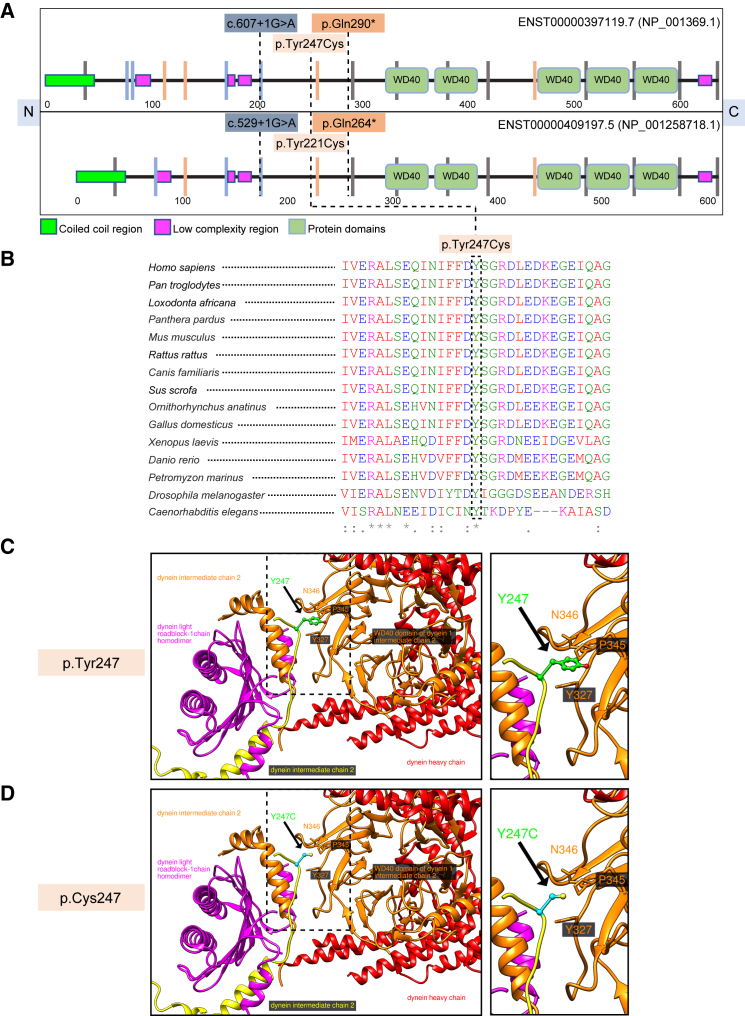
We performed exome sequencing on one affected individual (1-VI:2) of family 1 and obtained 101 million sequence reads after removing duplicates; these data covered on average 97% of the coding region with >20 reads. We used our custom analysis paradigm, which employs a minor allele frequency (MAF) threshold of <0.02 in public databases or our in-house, ethnically-matched exomes. The intersection of this dataset with a prioritization algorithm21, 22, 23 that merges outputs from in silico prediction tools yielded no pathogenic variants in the known causative genes for microcephaly, intellectual disability, or developmental delay. Next, we overlaid exome data with homozygosity mapping information constructed by genotyping five members of family 1. We identified a single-nucleotide homozygous splicing variant (GenBank: NM_001378.2:c.607+1G>A) in DYNC1I2 (Figure 2A). We confirmed the variant by segregation analysis in all members of the family via Sanger sequencing (Figure S1A). This change is at the first nucleotide of the splice donor site of a 96 bp exon present in all seven RefSeq transcripts (Table S1), and it is predicted to result in abnormal RNA processing. However, an RNA sample was unavailable for further studies. This variant was not present in our local cohort of 300 controls from the same ethnicity, nor was it present in the gnomAD database.27
Figure 2.
DYNC1I2 Protein Schematic, Multiple Sequence Alignment, and Molecular Modeling of the Recurrent p.Tyr247Cys Variant
(A) Top, a schematic diagram of the DYNC1I2 (GenBank: NP_001369.1) protein sequence showing the location of three variants, c.607+1G>A, c.740A>G (p.Tyr247Cys), and c.868C>T (p.Gln290∗), found in affected individuals from three different families. Bottom, a schematic diagram of the DYNC1I2 (GenBank: NP_001258718.1) protein sequence showing the location of the same three variants, c.529+1G>A, c.662A>G (p.Tyr221Cys), and c.790C>T (p.Gln264∗), in the most abundantly expressed isoform that we used in our zebrafish experiments. The schematic was adapted from SMART. N = amino terminus; C = carboxy terminus; and vertical lines indicate intron position and phase (gray = phase 0; blue = phase 1; and orange = phase 2).
(B) A multiple-sequence alignment generated with Clustal Omega showing conservation of the missense variant. The amino acid color scheme is: red = hydrophobic or aromatic (AVFPMILWY); blue = acidic (DE); magenta = basic (RHK); and green = hydroxyl, amine, basic, and glutamine (STYHCNGQ). Asterisks (∗) indicate conserved sequence identity and (:) indicates amino acid similarity.
(C) A model of the position of GenBank: NP_0011378.2 p.Tyr247 residue (green) of DYNC1I2 in the 6F1T cryo-EM structure of the dynein-dynactin complex. The residue is located on the linker, and its side chain is pointing toward the WD40 domain (orange). The linker seems to be involved in the homodimerization and in the interactions of the WD40 domain with the light roadblock chain of dynein (magenta).
(D) Modeling of the GenBank: NP_0011378.2 p.Tyr247Cys (cyan) mutant in the 6F1T cryo-EM structure of dynein/dynactin complex. The mutation to Cys might also influence the folding process and change the structure of the DYNC1I2 protein, impairing its interactions in the complex. Panels C and D show an overview (left) and a magnified view, outlined by a dashed line (right).
Individual II:1 of family 2 underwent a two-tiered clinical genetic analysis consisting of chromosomal microarray analysis (CMA) and whole-exome sequencing. We identified a 374 kb interstitial heterozygous deletion: arr[hg19] 2q31.1(172,318,311–172,692,048)×1. This region encompasses four genes, DCAF17 (MIM: 612515), CYBRD1 (MIM: 605745), DYNC1I2, and SLC25A12 (MIM: 603667), and has not been reported previously in DECIPHER (Figure S2). We reasoned that this CNV in isolation was insufficient to explain the proband’s phenotype because confirmatory testing showed it to be paternally inherited, and each of the four genes showed low probability of being loss-of-function intolerant (pLI range from 0–0.22). We conducted whole-exome sequencing of all three members of the trio to an average coverage of 140× and 98% of the target was covered by >10 reads. We did not identify any likely-pathogenic variants in phenotype-relevant genes. However, an examination of the coding regions encompassed by the 2q31.1 interval in the proband exome uncovered a rare heterozygous change, c.740A>G (p.Tyr247Cys), inherited from the mother (MAF = 1.8×10−5 in gnomAD; heterozygous variants were present in four individuals of Ashkenazi Jewish descent), (Figures 2A and S1B). Together, the CNV analysis intersection with whole-exome sequencing data pointed to DYNC1I2 as a likely-recessive cause of the phenotype (Figure 1B and Tables 1 and S1).
We conducted genetic studies on individual II:1 of family 3 to investigate the possibilities that there were either chromosomal abnormalities or pathogenic SNVs in the exome. We evaluated the female proband by karyotype, subtelomeric fluorescence in situ hybridization (FISH), and CMA, all of which were nondiagnostic. The CMA identified a maternally inherited heterozygous loss of the 6p pericentromeric region at 6p11.2, reported as a variant of uncertain significance. Next, we carried out sequencing analysis of several genes, including ASPM (MIM: 605481), ZNF335 (MIM: 610827), and NDE1 (MIM: 609449), related to autosomal-recessive microcephaly, but resulting data were also nondiagnostic. Then, we performed trio-based whole-exome sequencing, achieving coverage of >90% of targets at 20× and a mean target coverage of ∼100×. No pathogenic variants in genes known to be implicated in microcephaly, intellectual disability, or developmental delay were found. Next, we focused on de novo and recessive variants in non-disease-associated genes, filtering for rare variants that segregated with the proband’s phenotype. Although no rare de novo variants were identified in the index individual, we found bi-allelic variants in DYNC1I2; these variants were the same rare c.740A>G (p.Tyr247Cys) change identified in family 2 and a nonsense variant, c.868C>T (p.Gln290∗) (Figure 2A), that was absent from the gnomAD database. Both SNVs were Sanger sequenced in the familial samples, then confirmed to be heterozygous in the parents and compound-heterozygous in the proband (Figures 1C and S1C and Tables 1 and S1). These data point to DYNC1I2 variation as the most likely genetic cause in a third pedigree with phenotypically overlapping affected individuals united through the data sharing platform GeneMatcher.42
Molecular Modeling of the DYNC1I2 Missense Variant
There is strict amino acid conservation of Tyr247 from humans to C. elegans even though it does not reside in an annotated protein domain (Figure 2B). To investigate the possible structural consequences of p.Tyr247Cys, we modeled the cryo-EM structures of dynein tail(s) bound to dynactin and to different adaptors (PDB: 6F1T, 6F38, 6F3A, and 6F1Z; Figures 2C and 2D).43 An evaluation of the longest DYNC1I2 isoform (GenBank: NP_001369.1) showed that Tyr247 is located on the linker after the N-terminal helix of DYNC1I2 (Figure 2C). The N-terminal helices, which form a symmetric homodimer, interact with the dynein light chain roadblock-type 1 homodimer (Figure 2C). The linker containing Tyr247 is directed toward the first WD40 domain proximal to the N terminus (Figures 2A and 2C) of its DYNC1I2 dimerization partner. Because of low structural resolution of 6F1T, 6F38, and 6F3A, the positions of the atoms, except for the Tyr247 carbon beta atom, in the Tyr247 side chain could not be determined. Also, we were limited in solving the roadblock-1 (6F1Z) domain interaction of the Tyr247 side chain position, indicating high mobility of the linker fragment. Nevertheless, the position of the carbon beta suggests that Tyr247 interacts with some residues, such as Tyr327, Pro345 and Asn346, of the WD40 domain (Figure 2C).
WD40 domains typically have structural roles in different complexes and serve as a rigid scaffold for interactions. In DYNC1I2, the WD40 domain links the catalytic heavy chain that has a scaffolding function and provides energy for the dynein movement with the light roadblock (DYNLRB) chain (Figure 2C). The WD40 domain also interacts with the heavy chain of another dynein complex and stabilizes its position, therefore facilitating a role in increasing the speed of vesicle transport.43 The Tyr247Cys variant (Figure 2D) might be problematic not only because of lost Tyr interactions, but also because it might create nonnative disulphide bonds during folding and thus change the structure of DYNC1I2 and consequently influence the homodimerization process or interactions with other chains in the dynein/dynactin complex. We speculate that altered structure might weaken or damage the interactions between the dynein intermediate and light chains and possibly impact the dynein heavy chain and other molecules in the complex indirectly.
In Vivo Modeling of DYNC1I2 Loss Results in Neuroanatomical and Craniofacial Phenotypes
To bolster the candidacy of DYNC1I2 as the cause of a microcephaly syndrome, we developed zebrafish models. We used reciprocal BLAST to identify two zebrafish orthologs, dync1i2a (80% identity, 89% similarity) and dync1i2b (82% identity, 91% similarity), versus human protein, respectively. Because we did not know a priori whether both paralogs perform redundant functions, we queried the literature and our in-house zebrafish RNA-seq dataset (WT heads at 5 dpf; GEO: GSE63191)44 for information about spatio-temporal expression and relative abundance. Previous whole-mount RNA in situ hybridization studies placed dync1i2a in a near-ubiquitous pattern in the one-cell to 3 dpf stages.45 Additionally, dync1i2a was expressed at ∼4.7-fold greater abundance than dync1i2b (29 versus 6 reads per kilobase of transcript, per million mapped reads [RPKM]) in the head transcriptome at 5 dpf. Bereft of criteria to exclude either paralog, we developed reagents to disrupt both non-syntenic loci.
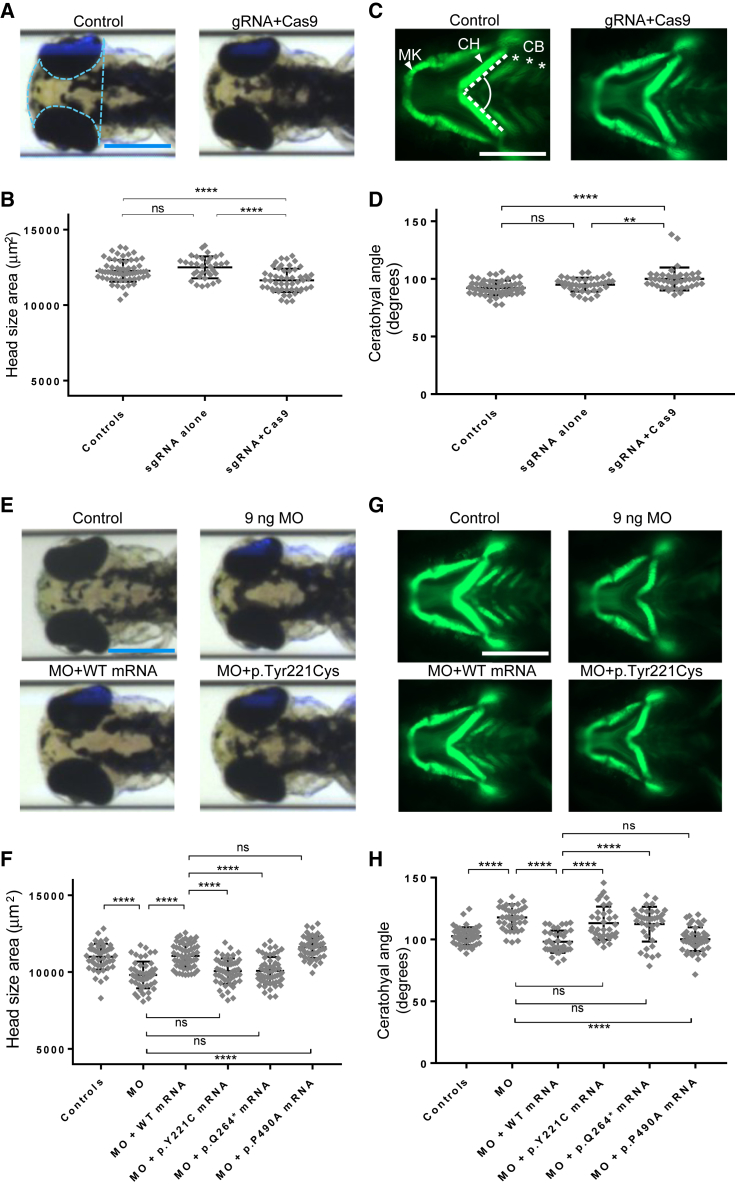
To introduce small insertions or deletions in each of the two paralogs, we performed CRISPR-Cas9 genome-editing. We generated single guide (sg)RNAs targeting the candidate principal isoforms of dync1i2a (exon 15) or dync1i2b (exons 4 and 15) (Table S2 and Figures S3A and S4A) and injected them in the presence or absence of Cas9 protein into -1.4col1a1:egfp reporter embryos at the one-cell stage. Heteroduplex analysis, cloning, and direct sequencing of DNA derived from single embryos (one control, five sgRNA + Cas9-injected embryos, with 16 clones per embryo at 1 dpf) resulted in 70%, 84%, and 50% estimated mosaicism, respectively (Figures S3A, S3B, S3C, S4A, S4B, and S4C). We have shown previously that zebrafish have robust anatomical surrogates for head size and craniofacial patterning,33, 35, 36, 37, 38, 39, 40, 46 phenotypes that we can capture simultaneously in a semi-automated fashion by using a capillary-based zebrafish imaging platform.47 We injected embryo batches with the dync1i2a or dync1i2b sgRNAs with or without Cas9, reared animals to 3 dpf, and acquired dorsal bright-field images to measure head size and ventral fluorescent images of GFP-positive cells painting the pharyngeal skeleton. In dync1i2a F0 mutants, we observed significant differences in both measures: a 7% reduction in head size area (p < 0.0001 versus sgRNA alone) and a 5° increase in ceratohyal angle (p < 0.01 versus sgRNA alone); a total of three biological replicates produced similar results (Figures 3A–3D). However, in dync1i2b F0 mutant larvae originating from either of the two sgRNAs developed, we detected no significant differences in either head area or ceratohyal angle compared to sgRNA alone or control counterparts (two replicates; Figures S4F and S4G). Together, our data intimated at a role for dync1i2a, but not dync1i2b, in the development of anterior brain and cartilaginous structures, a phenomenon that has been documented for pairs of duplicate genes in the zebrafish genome.48
Figure 3.
Zebrafish Models of dync1i2a Disruption Display Neuroanatomical and Craniofacial Patterning Defects, and GenBank: NM_001271789.1: c.662A>G (p.Tyr221Cys) Is a Loss-of-Function Variant
(A) Representative dorsal bright field images of control and F0 mutant larvae.
(B) Quantification of head size area in F0 mosaic mutants
(C) Representative ventral images of the GFP signal in the anterior region of -1.4col1a1:egfp transgenic reporters for control and F0 mutant larvae. Abbreviations are as follows: MK = Meckel’s cartilage; CH = ceratohyal cartilage; and CB = ceratobranchial arches.
(D) Quantification of the CH angle in F0 mutants
(E) Representative dorsal bright field images of control, e2i2 morphant (MO), WT, or mutant mRNA rescue larvae.
(F) Quantification of complementation assays that used head size as a phenotype readout; p.P490A is a negative control variant (dbSNP: rs767705533, 2 homozygotes in ∼120,000 individuals in gnomAD)
(G) Representative ventral images of the GFP signal in the anterior region of -1.4col1a1:egfp transgenic reporters for control, e2i2 morphant, WT, or mutant mRNA rescue larvae.
(H) Quantification of complementation assays that used the CH angle as a phenotype readout; data are concordant with the scoring of head size. All phenotyping was performed at 3 days post-fertilization.
The pale blue dotted line in panel A indicates the area measured to obtain the data in panels B and F. The angle indicated between the dashed white lines in panel C indicates the measurement used to obtain data for panels D and H. Scale bars in panels A, C, E, and G represent 300 μm; image sizing is consistent across panels. ∗∗∗∗p < 0.0001; ∗∗p < 0.01; and ns = not significant; n = 40–67/condition, and three biological replicates showed similar results for data shown in panels B, D, F, and H. Error bars represent standard deviation of the mean. Note that the amino acid codons Y221C, Q264∗, and P490A of the most abundant isoform (GenBank: NM_001271789.1) used in the zebrafish experiments correspond to Y247C, Q290∗, and P516A of the longest isoform (GenBank: NM_001378.2) of DYNC1I2.
In Vivo Complementation Studies Show that p.Tyr247Cys Is Pathogenic
Multiple lines of evidence implicate the recurrent missense variant as likely to be deleterious: (A) it was observed in two unrelated cases assessed as part of our small DYNC1I2 cohort (Figures 1B and 1C and Table 1); (B) it is a highly conserved residue present in eukaryotic taxa from mammals to nematodes (Figure 2B); and (C) it is predicted to alter the structure of DYNC1I2 pairing through WD40 domains (Figures 2C and 2D). To test experimentally this hypothesis, we performed in vivo complementation studies according to our established paradigm.32
First, we sought alternative reagents to validate our dync1i2a and dync1i2b F0 mutant data, and we obtained splice-blocking MOs targeting donor sites in either paralog (Table S2). To rule out any major contribution of dync1i2b in head size or craniofacial patterning, we showed that the exon 4-intron 4 (e4i4) MO induces a frameshifting exon deletion, eliminating a substantial proportion of WT dync1i2b mRNA (Figures S4A, S4D, and S4E). However, injection of this reagent into zebrafish embryo batches at increasing doses (3 ng, 6 ng, and 9 ng), followed by our dual-phenotype imaging and measurement paradigm, reproduced the dync1i2b F0 mutant result: we observed there was a lack of a detectable phenotype compared to controls, even at the highest dose tested and in replicate experiments (Figures S4H and S4I).
We then turned our efforts to dync1i2a, the paralog in zebrafish likely to be the functionally relevant DYNC1I2 ortholog. Recognizing that, like human DYNC1I2, the D. rerio dync1i2a locus is subject to alternative splicing, we targeted splice donor sites present in all annotated transcripts (exon 2 and exon 5 of the canonical isoform; termed e2i2 and e5i5) and showed efficient knockdown of both reagents (Figures S3A and S3D–F). We injected increasing doses of either MO using the same dose-curve testing and phenotyping paradigms, and we observed robust dose dependency of phenotype in larvae scored at 3 dpf. The highest dose of e2i2 tested (9 ng) resulted in a 10% reduction in head size (p < 0.0001 versus controls; Figures 3E, 3F, and S3G) and a 30° increase in ceratohyal angle (p < 0.0001 versus controls; Figures 3G, 3H, and S3H). Importantly, we could rescue these defects significantly by coinjection of 50 pg WT human DYNC1I2 mRNA (GenBank: NM_001271789.1; Table S1), which has been shown to display the most copious expression in human brain regions (p < 0.0001 for MO alone versus MO plus WT mRNA; both phenotypes, triplicate experiments; Figures 3E–3H). These data were recapitulated for the e5i5 MO (Figures S3I, S3J, and S5).
To test the pathogenicity of the p.Tyr247Cys change, we compared the efficiency of variant DYNC1I2 mRNA to rescue the morphant phenotype versus that of WT for both head size and craniofacial patterning in the context of the most abundant isoform, GenBank: NM_001271789.1. We used the most common variant from gnomAD, p.Pro490Ala, as a negative control (dbSNP: rs767705533, 2 homozygotes in ∼120,000 individuals), mRNA bearing p.Gln264∗ as a positive control, and p.Tyr221Cys as the test variant. Tyr221 is equivalent to the GenBank: NM_001378.2:p.(Tyr247) in the isoform (GenBank: NM_001271789.1) that we used in these experiments. We found that both of the case-associated SNVs scored as pathogenic (significantly worse than WT mRNA, but no different from MO alone). However, the negative control variant scored consistently as benign (not significantly different from WT mRNA rescue; Figures 3E–3H; three replicate experiments with similar results). Importantly, equivalent doses of each DYNC1I2 mRNA injected in the absence of a MO did not produce any appreciable head-size or mandibular phenotypes that differed from WT mRNA (Figures S6A and S6B). In conclusion, our variant testing data support the notion that p.Tyr221Cys impairs protein function, producing a lack of rescue similar to the nonsense p.Gln264∗ change.
Mechanistic Underpinnings of Microcephaly in dync1i2a Zebrafish Models
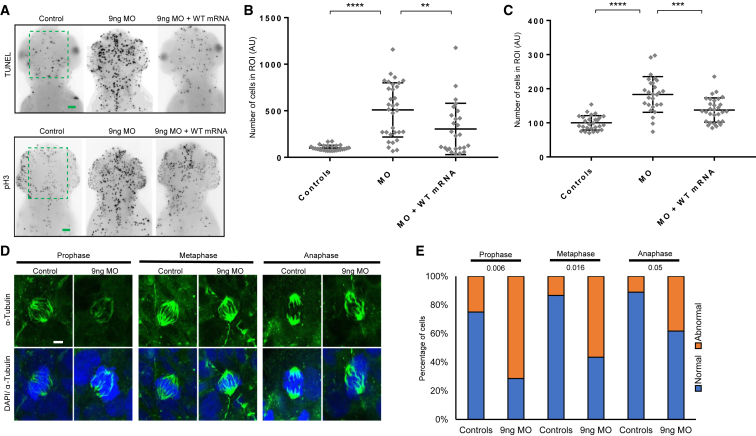
To probe further the neurological phenotype induced by loss of DYNC1I2, we performed acetylated α-tubulin immunostaining to mark the axon tracts of neurons, imaged the dorsal aspect of the zebrafish head at 3 dpf, and measured features relevant to humans in our cohort. Individuals in each pedigree (1-VI:1, 2-II-1, and 3-II-1) showed hypogenesis or agenesis of the corpus callosum (Figure 1E and Table 1), so we first measured the number of commissural axon tracts connecting the zebrafish optic tecta in controls, dync1i2a MO-injected animals, and animals injected with MO plus WT DYNC1I2 mRNA, as described.46 We observed a modest but significant reduction in the number of axon tracts crossing the dorsal midline in MO-injected batches (p < 0.0001 versus controls), and this defect could be rescued significantly by the presence of WT mRNA (p < 0.0001 versus MO alone; replicate batches; Figures S7A and S7B). Additionally, we measured the size of the optic tecta, neuron-rich sensory processing centers in the zebrafish brain whose size correlates with overall head size.39, 44, 49 Consistent with our bright-field dorsal head-size measurements, we observed a significant reduction in the area of the optic tecta in dync1i2a morphants (p < 0.01 versus controls), but the size of these structures was restored significantly when MO was coinjected with WT DYNC1I2 mRNA (p < 0.01 versus MO alone; Figures S7A and S7C). Overall, we observed marked disorganization of axonal patterning in the zebrafish head.
We wondered whether apoptosis could account for the reduction in head size of dync1i2a zebrafish models, as had been shown previously for other models of microcephaly.33, 37, 38, 39, 50, 51 To test this, we performed TUNEL staining in embryos at 2 dpf and quantified TUNEL-positive cells within a normalized area of the dorsal forebrain between the eyes. In dync1i2a morphants, we observed a significant increase in cell death that could be rescued with co-injection of WT human DYNC1I2 mRNA (p < 0.0001, MO versus control; p < 0.01, MO versus MO+WT mRNA; Figures 4A and 4B). We hypothesized that the increased cell death in the context of dync1i2a loss might be relevant to altered cell cycle progression. As a first test, we monitored cells undergoing G2/M transition with phospho-histone-H3 (pH3) immunostaining. Similar to the TUNEL result, we observed a significant increase in pH3-stained cells in morphants, and this increase was rescuable by human WT mRNA, indicating a specific defect in cell cycle progression (p < 0.001, MO versus control; p < 0.001, MO versus MO+WT mRNA; repeated with similar results; Figures 4A and 4C). To explore further the pH3 immunostaining result, we evaluated the organization of mitotic spindles during cell division in dync1i2a morphants versus controls. We fixed embryos at 1 dpf, performed whole-mount immunostaining with anti α-tubulin antibody, counterstained nuclei with DAPI, and imaged mitotic cells on the spinal cord by using confocal microscopy. We scored dividing cells on the basis of spindle orientation to categorize mitotic stage (prophase, metaphase, or anaphase), and we made a qualitative assignment of normal or abnormal spindle appearance, as described.41 We observed a significantly greater number of cells with abnormal spindle morphology in morphants versus controls in prophase and metaphase, but nominal differences in anaphase cells (n = 33–42 cells/condition; Figures 4D and 4E). Together, these data suggest that the microcephaly phenotype of dync1i2a morphants is a result of augmented cell death, probably due to altered cell cycle progression that could be linked to spindle morphology during the early stages of mitosis.
Figure 4.
Apoptosis, Altered Cell Cycle Progression, and the Effect on Mitotic Spindle Organization in dync1i2a Morphants
(A) Representative dorsal inverted fluorescent images showing TUNEL-positive cells (top) and phospho-histone H3 (pH3) immunostained cells (bottom) in controls, e2i2 morphants (MO), and WT mRNA rescue animals. The green dashed box indicates the region of interest (ROI) quantified in panels B and C. The scale bars represent 30 μm, and there is consistent image sizing across panels.
(B) Quantification of TUNEL-positive cells as a readout of apoptosis; n = 30–35/condition; repeated twice with similar results.
(C) Quantification of pH3-positive cells as an indicator of cell cycle progression (G2/M transition); n = 28–32/condition; repeated twice with similar results. In panels B and C, experiments were performed on larvae at 2 days post-fertilization (dpf). ∗∗∗∗p < 0.0001; ∗∗∗p < 0.0001; and ∗∗p < 0.01; error bars represent standard deviation of the mean.
(D) Representative confocal microscope images of fluorescently stained dividing cells scored at three different stages of the cell cycle on the spinal cord of wholemount embryos at 1 dpf. α-tubulin marks mitotic spindles and DAPI indicates nuclei. The scale bar represents 2 μm.
(E) Qualitative scoring of the embryos shown in panel D for the appearance of normal versus abnormal mitotic spindle organization. Stages of mitosis were defined as follows: prophase = chromosomes are condensed, centrosomes are at opposite poles, and spindle fibers begin to emerge; metaphase = chromosomes are lined up at the metaphase plate, and each chromatid is attached to a spindle fiber; and Anaphase = chromatids are pulled to opposite poles, and the cell is elongated. If a cell displayed misalignment of poles and/or uneven distribution of spindle fibers to chromosomes it was scored as abnormal. P values showing statistical differences between controls versus morphants are shown at the top of the chart. n = 33–42 embryos/condition; repeated.
Discussion
Here, we expand the repertoire of human phenotypes caused by variants in the cytoplasmic dynein-1 complex. We report five affected individuals, from three unrelated families, who harbor different recessive DYNC1I2 variants, all of which are likely to impair protein function (Figures 1A–C and2A). All individuals in our cohort present similar clinical features, predominantly microcephaly, developmental delay, facial dysmorphism, and similar brain MRI findings, which include hypoplasia of the white matter, most notably hypogenesis or almost complete agenesis of the corpus callosum (Table 1B), and simplification of cerebral cortical gyration. Additionally, we note the identification of a shared missense variant, p.Tyr247Cys, in two unrelated affected individuals with eastern European ancestry. This allele might come from a common ancestral origin, or it could reside in a mutational hotspot. Haplotype analysis in both pedigrees would be required to clarify these two possibilities. Notably, and different from family 1, both individuals harboring p.Tyr247Cys present with seizures, but given that individual II:1 of family 2 has a heterozygous deletion at 2q31.1, it is not possible to discern whether the seizures could be linked to the p.Tyr247Cys missense allele or to other sites in trans within the genome. We anticipate that the inclusion of the DYNC1I2 locus in clinical testing would identify larger cohorts of pathogenic DYNC1I2 variants, and these cohorts will enable genotype-phenotype correlation studies.
We support the candidacy of DYNC1I2 variants in autosomal-recessive human disease with dync1i2a CRISPR-Cas9 F0 or morphant zebrafish larvae. First, we show physiological relevance for loss of gene function in phenotypes relevant to our human cohort; these phenotypes include microcephaly, craniofacial patterning defects, and intertectal axon deficits (Figures 3 and S7). Furthermore, we used in vivo complementation testing to estimate, within the limits of our assay detection, that p.Tyr247Cys, tested in the context of the most abundant transcript (GenBank: NM_001271789.1), has a loss of protein function indistinguishable from that of the nonsense variant identified by this study, p.Gln290∗.
The current study has expanded the list of dynein-1 complex protein subunits implicated in neurodevelopmental disorders. Individuals with variants affecting the function of the dynein-1 complex show overlapping clinical features, suggesting a similar pathomechanism. For example, mutations in the dynein-1 heavy chain, DYNC1H1, have been associated with neuronal migration defects, intellectual disability, microcephaly in some affected individuals, and brain malformations similar to those observed in the DYNC1I2 patients reported here.8, 11 Prior work has shown that to interact with cargoes the intermediate chains need to bind with the dynactin complex.4, 16, 52 The dynein-dynactin complex plays a vital role in synapse stabilization and during the mitotic cell division.53, 54 Variants in DCTN1 (MIM: 601143), which encodes the subunit of dynactin, cause Perry syndrome (MIM: 168605) and distal hereditary motor neuropathy (MIM: 607647).55, 56, 57 NDE1 is another protein that, by interacting with the cytoplasmic dynein, regulates cytoskeletal dynamics during mitosis and that also has a role in neuronal migration.58, 59 Pathogenic variants in NDE1 in humans and Nde1-knockout mice resulted in severe microcephaly with lissencephaly phenotypes (MIM: 614019). Additionally, Nde1 mutant mice show abnormal mitotic spindles.60, 61
Consistent with functional overlap between microtubule-based motors, we also observe an overlap in pathomechanism for dysfunction of the kinesin and dynein-1 complexes. For example, abnormal spindle morphology, caused by pathogenic variants in the kinesin component STARD9 (MIM: 614642), has been shown to cause microcephaly, severe ID, and facial dysmorphism.62 Consistently, we observe significantly altered organization of mitotic spindles in zebrafish dync1i2a morphants versus controls, particularly during the earlier stages of mitosis, such as prophase and anaphase (Figures 4D and 4E). These observations, coupled to the augmented pH3 immunostaining data and enrichment of TUNEL-positive cells in dync1i2a morphants lead us to speculate that neural progenitors undergo mitotic delay, which then results in apoptosis, compatible with the phenotype of smaller head size, as observed in other studies.63, 64, 65, 66 The notion of mitotic delay underlying the microcephaly phenotype is supported by previous studies, such as the Magoh mouse model of microcephaly, wherein prolonged mitosis alters cell fate and promotes apoptosis.67 Further work on in vivo or in vitro DYNC1I2 models will be required to understand the functional link between abnormal spindle organization and cell cycle defects.
In summary, we add DYNC1I2 to the expansive list of genes required for proper development and maintenance of the vertebrate brain. Our combined genetic and functional studies have highlighted the importance of the dynein-1 complex in cell cycle regulation via mitotic spindle organization in neurodevelopment.
Declaration of Interests
F.M. is an employee of GeneDx, Inc., a wholly owned subsidiary of OPKO Health, Inc. N.K. is a shareholder in Rescindo Therapeutics. The other authors declare no competing interests.
Acknowledgments
We thank the Swiss Government Excellence Scholarships program, which provided M.A. the opportunity to work at the University of Geneva Medical School in Switzerland. F.U. was funded by an International Research Support Initiative Program fellowship from the Higher Education Commission of Pakistan. This project was partially supported by European Research Council (ERC) grant 219968 to S.E.A. We are thankful to all the members of the families reported in this study and to the Vital-IT platform for the computational support. We thank Mr. K. Khan for critical reading of the manuscript. Sequencing and analysis of family 2 was provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung, and Blood Institute grant UM1 HG008900 to D. MacArthur and H. Rehm. C.A.W. was supported by the National Institute of Neurological Disorders and Stroke (NINDS) (R01 NS035129); E.E.D. was supported by the National Institute of Mental Health (NIMH) (R01 MH106826). N.K. is a Distinguished Jean and George W. Brumley Professor. C.A.W. is an Investigator of the Howard Hughes Medical Institute. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the National Institutes of Health (NIH) grant P41-GM103311. We are thankful to all the members of the families reported in this study.
Published: May 9, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.04.002.
Contributor Information
Erica E. Davis, Email: erica.davis@duke.edu.
Stylianos E. Antonarakis, Email: stylianos.antonarakis@unige.ch.
Web Resources
ChopChop Software, http://chopchop.cbu.uib.no
ClinVar, GeneDx Submissions, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
Clustal Omega, https://www.ebi.ac.uk/Tools/msa/clustalo/
DECIPHER, https://decipher.sanger.ac.uk
Ensembl, https://www.ensembl.org
ExAC Browser, http://exac.broadinstitute.org/
Exome Variant Server, http://evs.gs.washington.edu/EVS/
Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/
GeneMatcher, https://genematcher.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
ImageJ, https://imagej.nih.gov
Institute for Systems Biology, http://db.systemsbiology.net/gestalt/cgi-pub/genomeMapBlocks.pl
NCBI BLAST, https://blast.ncbi.nlm.nih.gov
OMIM, http://www.omim.org/
Protein Data Bank (PDB), www.rcsb.org
Simple Modular Architecture Research Tool (SMART), http://smart.embl-heidelberg.de/
UCSC Genome Browser, http://genome.ucsc.edu
Variant Effect Predictor (VEP), https://www.ensembl.org/vep
Supplemental Data
References
- 1.Akhmanova A., Hammer J.A., 3rd Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu M.M., Holzbaur E.L. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24:564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfister K.K. Distinct functional roles of cytoplasmic dynein defined by the intermediate chain isoforms. Exp. Cell Res. 2015;334:54–60. doi: 10.1016/j.yexcr.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuta A., Deng W., Morsi El-Kadi A., Banks G.T., Hafezparast M., Pfister K.K., Fisher E.M. Mouse cytoplasmic dynein intermediate chains: Identification of new isoforms, alternative splicing and tissue distribution of transcripts. PLoS ONE. 2010;5:e11682. doi: 10.1371/journal.pone.0011682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts A.J. Emerging mechanisms of dynein transport in the cytoplasm versus the cilium. Biochem. Soc. Trans. 2018;46:967–982. doi: 10.1042/BST20170568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfister K.K., Shah P.R., Hummerich H., Russ A., Cotton J., Annuar A.A., King S.M., Fisher E.M. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maday S., Twelvetrees A.E., Moughamian A.J., Holzbaur E.L. Axonal transport: Cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laquerriere A., Maillard C., Cavallin M., Chapon F., Marguet F., Molin A., Sigaudy S., Blouet M., Benoist G., Fernandez C. Neuropathological hallmarks of brain malformations in extreme phenotypes related to DYNC1H1 mutations. J. Neuropathol. Exp. Neurol. 2017;76:195–205. doi: 10.1093/jnen/nlw124. [DOI] [PubMed] [Google Scholar]
- 9.Weedon M.N., Hastings R., Caswell R., Xie W., Paszkiewicz K., Antoniadi T., Williams M., King C., Greenhalgh L., Newbury-Ecob R., Ellard S. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2011;89:308–312. doi: 10.1016/j.ajhg.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harms M.B., Ori-McKenney K.M., Scoto M., Tuck E.P., Bell S., Ma D., Masi S., Allred P., Al-Lozi M., Reilly M.M. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology. 2012;78:1714–1720. doi: 10.1212/WNL.0b013e3182556c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirier K., Lebrun N., Broix L., Tian G., Saillour Y., Boscheron C., Parrini E., Valence S., Pierre B.S., Oger M. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat. Genet. 2013;45:639–647. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willemsen M.H., Vissers L.E., Willemsen M.A., van Bon B.W., Kroes T., de Ligt J., de Vries B.B., Schoots J., Lugtenberg D., Hamel B.C. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J. Med. Genet. 2012;49:179–183. doi: 10.1136/jmedgenet-2011-100542. [DOI] [PubMed] [Google Scholar]
- 13.Di Donato N., Timms A.E., Aldinger K.A., Mirzaa G.M., Bennett J.T., Collins S., Olds C., Mei D., Chiari S., Carvill G., University of Washington Center for Mendelian Genomics Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet. Med. 2018;20:1354–1364. doi: 10.1038/gim.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos-Zaldívar H.M., Martínez-Irías D.G., Espinoza-Moreno N.A., Napky-Rajo J.S., Bueso-Aguilar T.A., Reyes-Perdomo K.G., Montes-Gambarelli J.A., Euceda I.M., Ponce-Barahona A.F., Gámez-Fernández C.A. A novel description of a syndrome consisting of 7q21.3 deletion including DYNC1I1 with preserved DLX5/6 without ectrodactyly: A case report. J. Med. Case Reports. 2016;10:156. doi: 10.1186/s13256-016-0921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delgado S., Velinov M. 7q21.3 Deletion involving enhancer sequences within the gene DYNC1I1 presents with intellectual disability and split hand-split foot malformation with decreased penetrance. Mol. Cytogenet. 2015;8:37. doi: 10.1186/s13039-015-0139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kardon J.R., Vale R.D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers K.R., Lo K.W., Lye R.J., Kogoy J.M., Soura V., Hafezparast M., Pfister K.K. Intermediate chain subunit as a probe for cytoplasmic dynein function: Biochemical analyses and live cell imaging in PC12 cells. J. Neurosci. Res. 2007;85:2640–2647. doi: 10.1002/jnr.21213. [DOI] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makrythanasis P., Nelis M., Santoni F.A., Guipponi M., Vannier A., Béna F., Gimelli S., Stathaki E., Temtamy S., Mégarbané A. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum. Mutat. 2014;35:1203–1210. doi: 10.1002/humu.22617. [DOI] [PubMed] [Google Scholar]
- 22.Ansar M., Riazuddin S., Sarwar M.T., Makrythanasis P., Paracha S.A., Iqbal Z., Khan J., Assir M.Z., Hussain M., Razzaq A. Biallelic variants in LINGO1 are associated with autosomal recessive intellectual disability, microcephaly, speech and motor delay. Genet. Med. 2018;20:778–784. doi: 10.1038/gim.2017.113. [DOI] [PubMed] [Google Scholar]
- 23.Ansar M., Chung H., Waryah Y.M., Makrythanasis P., Falconnet E., Rao A.R., Guipponi M., Narsani A.K., Fingerhut R., Santoni F.A. Visual impairment and progressive phthisis bulbi caused by recessive pathogenic variant in MARK3. Human Molecular Genetics. 2018;27:2703–2711. doi: 10.1093/hmg/ddy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santoni F.A., Makrythanasis P., Antonarakis S.E. CATCHing putative causative variants in consanguineous families. BMC Bioinformatics. 2015;16:310. doi: 10.1186/s12859-015-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 27.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Kague E., Gallagher M., Burke S., Parsons M., Franz-Odendaal T., Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw398. W272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X., Xu Y., Yu S., Lu L., Ding M., Cheng J., Song G., Gao X., Yao L., Fan D. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 2014;4:6420. doi: 10.1038/srep06420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niederriter A.R., Davis E.E., Golzio C., Oh E.C., Tsai I.C., Katsanis N. In vivo modeling of the morbid human genome using Danio rerio. J. Vis. Exp. 2013;78:e50338. doi: 10.3791/50338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frosk P., Arts H.H., Philippe J., Gunn C.S., Brown E.L., Chodirker B., Simard L., Majewski J., Fahiminiya S., Russell C., FORGE Canada Consortium. Canadian Rare Diseases: Models & Mechanisms Network A truncating mutation in CEP55 is the likely cause of MARCH, a novel syndrome affecting neuronal mitosis. J. Med. Genet. 2017;54:490–501. doi: 10.1136/jmedgenet-2016-104296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutson M.R., Keyte A.L., Hernández-Morales M., Gibbs E., Kupchinsky Z.A., Argyridis I., Erwin K.N., Pegram K., Kneifel M., Rosenberg P.B. Temperature-activated ion channels in neural crest cells confer maternal fever-associated birth defects. Sci. Signal. 2017;10:eaal4055. doi: 10.1126/scisignal.aal4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isrie M., Breuss M., Tian G., Hansen A.H., Cristofoli F., Morandell J., Kupchinsky Z.A., Sifrim A., Rodriguez-Rodriguez C.M., Dapena E.P. Mutations in either TUBB or MAPRE2 cause circumferential skin creases Kunze type. Am. J. Hum. Genet. 2015;97:790–800. doi: 10.1016/j.ajhg.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw N.D., Brand H., Kupchinsky Z.A., Bengani H., Plummer L., Jones T.I., Erdin S., Williamson K.A., Rainger J., Stortchevoi A. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat. Genet. 2017;49:238–248. doi: 10.1038/ng.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stankiewicz P., Khan T.N., Szafranski P., Slattery L., Streff H., Vetrini F., Bernstein J.A., Brown C.W., Rosenfeld J.A., Rednam S., Deciphering Developmental Disorders Study Haploinsufficiency of the chromatin remodeler BPTF causes syndromic developmental and speech delay, postnatal microcephaly, and dysmorphic features. Am. J. Hum. Genet. 2017;101:503–515. doi: 10.1016/j.ajhg.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks S.S., Wall A.L., Golzio C., Reid D.W., Kondyles A., Willer J.R., Botti C., Nicchitta C.V., Katsanis N., Davis E.E. A novel ribosomopathy caused by dysfunction of RPL10 disrupts neurodevelopment and causes X-linked microcephaly in humans. Genetics. 2014;198:723–733. doi: 10.1534/genetics.114.168211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan T.N., Khan K., Sadeghpour A., Reynolds H., Perilla Y., McDonald M.T., Gallentine W.B., Baig S.M., Davis E.E., Katsanis N., Task Force for Neonatal Genomics Mutations in NCAPG2 cause a severe neurodevelopmental syndrome that expands the phenotypic spectrum of condensinopathies. Am. J. Hum. Genet. 2019;104:94–111. doi: 10.1016/j.ajhg.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guissart C., Latypova X., Rollier P., Khan T.N., Stamberger H., McWalter K., Cho M.T., Kjaergaard S., Weckhuysen S., Lesca G. Dual molecular effects of dominant RORA mutations cause two variants of syndromic intellectual disability with either autism or cerebellar ataxia. Am. J. Hum. Genet. 2018;102:744–759. doi: 10.1016/j.ajhg.2018.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao L., Chen J., Wu X., Jia S., Meng A. Zebrafish cdc6 hypomorphic mutation causes Meier-Gorlin syndrome-like phenotype. Hum. Mol. Genet. 2017;26:4168–4180. doi: 10.1093/hmg/ddx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urnavicius L., Lau C.K., Elshenawy M.M., Morales-Rios E., Motz C., Yildiz A., Carter A.P. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature. 2018;554:202–206. doi: 10.1038/nature25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borck G., Hög F., Dentici M.L., Tan P.L., Sowada N., Medeira A., Gueneau L., Thiele H., Kousi M., Lepri F. BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome Res. 2015;25:155–166. doi: 10.1101/gr.176925.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thisse B., Thisse C. Fast release clones: A high throughput expression analysis. ZFIN direct data submission. 2004 http://zfin.org [Google Scholar]
- 46.Ta-Shma A., Khan T.N., Vivante A., Willer J.R., Matak P., Jalas C., Pode-Shakked B., Salem Y., Anikster Y., Hildebrandt F. Mutations in TMEM260 cause a pediatric neurodevelopmental, cardiac, and renal syndrome. Am. J. Hum. Genet. 2017;100:666–675. doi: 10.1016/j.ajhg.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulak R. Tools for automating the imaging of zebrafish larvae. Methods. 2016;96:118–126. doi: 10.1016/j.ymeth.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 48.Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Küry S., Besnard T., Ebstein F., Khan T.N., Gambin T., Douglas J., Bacino C.A., Craigen W.J., Sanders S.J., Lehmann A. De novo disruption of the proteasome regulatory subunit PSMD12 causes a syndromic neurodevelopmental disorder. Am. J. Hum. Genet. 2017;100:352–363. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carvalho C.M., Vasanth S., Shinawi M., Russell C., Ramocki M.B., Brown C.W., Graakjaer J., Skytte A.B., Vianna-Morgante A.M., Krepischi A.C. Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. Am. J. Hum. Genet. 2014;95:565–578. doi: 10.1016/j.ajhg.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Golzio C., Willer J., Talkowski M.E., Oh E.C., Taniguchi Y., Jacquemont S., Reymond A., Sun M., Sawa A., Gusella J.F. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroer T.A. Dynactin. Annu. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 53.Hammesfahr B., Kollmar M. Evolution of the eukaryotic dynactin complex, the activator of cytoplasmic dynein. BMC Evol. Biol. 2012;12:95. doi: 10.1186/1471-2148-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eaton B.A., Fetter R.D., Davis G.W. Dynactin is necessary for synapse stabilization. Neuron. 2002;34:729–741. doi: 10.1016/s0896-6273(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 55.Newsway V., Fish M., Rohrer J.D., Majounie E., Williams N., Hack M., Warren J.D., Morris H.R. Perry syndrome due to the DCTN1 G71R mutation: A distinctive levodopa responsive disorder with behavioral syndrome, vertical gaze palsy, and respiratory failure. Mov. Disord. 2010;25:767–770. doi: 10.1002/mds.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farrer M.J., Hulihan M.M., Kachergus J.M., Dächsel J.C., Stoessl A.J., Grantier L.L., Calne S., Calne D.B., Lechevalier B., Chapon F. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puls I., Jonnakuty C., LaMonte B.H., Holzbaur E.L., Tokito M., Mann E., Floeter M.K., Bidus K., Drayna D., Oh S.J. Mutant dynactin in motor neuron disease. Nat. Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 58.Wynshaw-Boris A. Lissencephaly and LIS1: Insights into the molecular mechanisms of neuronal migration and development. Clin. Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 59.Wynshaw-Boris A., Pramparo T., Youn Y.H., Hirotsune S. Lissencephaly: Mechanistic insights from animal models and potential therapeutic strategies. Semin. Cell Dev. Biol. 2010;21:823–830. doi: 10.1016/j.semcdb.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alkuraya F.S., Cai X., Emery C., Mochida G.H., Al-Dosari M.S., Felie J.M., Hill R.S., Barry B.J., Partlow J.N., Gascon G.G. Human mutations in NDE1 cause extreme microcephaly with lissencephaly [corrected] Am. J. Hum. Genet. 2011;88:536–547. doi: 10.1016/j.ajhg.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Y., Walsh C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 62.Okamoto N., Tsuchiya Y., Miya F., Tsunoda T., Yamashita K., Boroevich K.A., Kato M., Saitoh S., Yamasaki M., Kanemura Y. A novel genetic syndrome with STARD9 mutation and abnormal spindle morphology. Am. J. Med. Genet. A. 2017;173:2690–2696. doi: 10.1002/ajmg.a.38391. [DOI] [PubMed] [Google Scholar]
- 63.Tessadori F., Giltay J.C., Hurst J.A., Massink M.P., Duran K., Vos H.R., van Es R.M., Scott R.H., van Gassen K.L.I., Bakkers J., van Haaften G., Deciphering Developmental Disorders Study Germline mutations affecting the histone H4 core cause a developmental syndrome by altering DNA damage response and cell cycle control. Nat. Genet. 2017;49:1642–1646. doi: 10.1038/ng.3956. [DOI] [PubMed] [Google Scholar]
- 64.Li H., Bielas S.L., Zaki M.S., Ismail S., Farfara D., Um K., Rosti R.O., Scott E.C., Tu S., Chi N.C. Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am. J. Hum. Genet. 2016;99:501–510. doi: 10.1016/j.ajhg.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poulton J.S., Cuningham J.C., Peifer M. Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J. Cell Biol. 2017;216:1255–1265. doi: 10.1083/jcb.201607022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doobin D.J., Dantas T.J., Vallee R.B. Microcephaly as a cell cycle disease. Cell Cycle. 2017;16:247–248. doi: 10.1080/15384101.2016.1252591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pilaz L.J., McMahon J.J., Miller E.E., Lennox A.L., Suzuki A., Salmon E., Silver D.L. Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron. 2016;89:83–99. doi: 10.1016/j.neuron.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.