Abstract
Less than 1% of all microorganisms of the available environmental microbiota can be cultured with the currently available techniques. Metagenomics is a new methodology of high-throughput DNA sequencing, able to provide taxonomic and functional profiles of microbial communities without the necessity to culture microbes in the laboratory. Metagenomics opens to a ‘hypothesis-free’ approach, giving important details for future research and treatment of ocular diseases in ophthalmology, such as ocular infection and ocular surface diseases.
Keywords: ophthalmology, metagenomics, sequencing, eye, ocular surface, cornea, microbiome
Current knowledge about the eye microbiome
The ocular surface (OS) microbiome is an understudied topic, compared with other host-associated environments. While the Human Microbiome Project initially studied five main body areas—the skin, the gastrointestinal tract, the urogenital tract, the oral and the nasal mucosa1—an emerging area of research is focusing on the eye and the microbiota of the OS.2
Recent studies demonstrated that OS hosts a number of commensal microorganisms.3 Earlier culture-based surveys suggested that the OS are colonised by microbial communities dominated by Gram-positive Firmicutes, in particular, species belonging to the Staphylococcus, Streptococcus, Corynebacterium and Propionibacterium.4 A screening including approximately 1000 16S rRNA reads revealed that the diversity of healthy conjunctiva was higher than previously thought.5 Other recent studies based on traditional microbiological techniques have examined the microbiota of the OS,6–11 although a more comprehensive analysis of microbial diversity of OS has been hindered by the limitations of conventional cultivation techniques.12–14 More recent screening of OS-associated microbiome, using molecular metagenomic techniques, extended further the knowledge about OS microbial diversity.2 5 15 16 Shestopalov and colleagues estimated using real-time PCR that in 1 ng of extracted DNA, the number of bacterial genomes (ie, bacterial richness) was on average 79.8 and 729 in the conjunctiva and cornea, respectively. Significant amounts (22 over 55) were detected in the eye for the first time.17 Dong et al detected 59 distinct bacterial genera using a 16S rDNA gene pyrosequencing approach on the OS of four healthy individuals15 (figure 1). Despite the low number of individuals examined, this is one of the first studies focusing on the bacterial diversity of the OS microbiome. Healthy OS microbiome is dominated by Proteobacteria, Actinobacteria and Firmicutes. The most common taxa at the genus level were P seudomonas, Propionibacterium, Bradyrhizobium, Corynebacterium, Acinetobacter, Brevundimonas, Staphylococci, Aquabacterium, Sphingomonas, Streptococcus, Streptophyta and Methylobacterium (figure 1). This is in general agreement with the previous studies, although many false positives may derive from contamination.4
Figure 1.
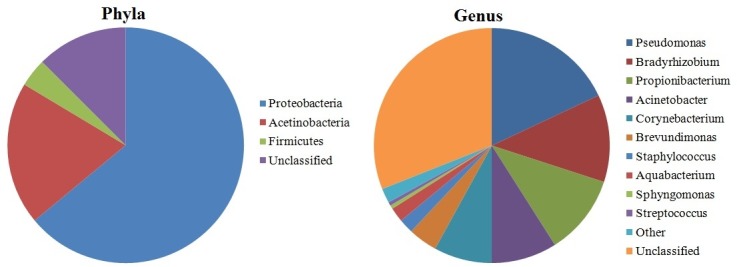
Pie charts displaying the relative proportions of taxa at the phylum level and at the genus level in healthy eye microbiome (according to Dong et al 15 [15]).
Microbial invasion into the OS compromises corneal clarity and causes inflammation in blinding conditions like keratitis, endophthalmitis and trachoma. Herpes simplex type 1, hepatitis B and C viruses can be detected using PCR in the tears of asymptomatic volunteers,18 thus suggesting that simply focusing solely on the bacterial constituents of the OS may result in an incomplete understanding of the OS microbiome. In recent time, Zhou et al 19 showed how the changes in the conjunctival microbiome occur in trachomatous disease compared with normal controls. Wen et al 20 showed how the microbiome of healthy OS is shaped by age and sex and how the ocular microbiome of house finches changed during experimentally induced mycoplasmal conjunctivitis.21
Study of microbiome in OS disease has significant potential to improve the diagnosis, treatment and management of potential blinding diseases.
Background and evolution of metagenomics
The non-culturability of microbes dates back to 1898 by Heinrich Winterberg, formalised as the ‘great plate count anomaly’ by Staley and Konopka in 1985.22 Several theories have been proposed to explain the non-culturability of microbes: (1) the cultural media used have been developed to grow microorganisms usually involved in human or animal diseases, missing a diversity of other microorganisms; (2) some bacteria may require long incubation time to form visible colonies; (3) physiological features (ie, obligate symbiosis with other species or strong quorum sensing signalling processes) may hinder the cultivation of some species; and (4) some bacteria may require a specific combination of nutritional features and aerobic requirements. The pioneering work by Woese23 in 1985 identified the 16S rRNA gene as an evolutionary chronometer for bacterial phylogeny. This gene has three unique features: (A) the ubiquity in the bacterial kingdom, (B) the structure of the gene itself, made of both variable and conserved regions (this is due to the secondary structure of the transcribed RNA, made of stretches and loops), and (C) the low (if any) amount of horizontal gene transfer. Pace et al in 1985 had the idea that 16S rRNA gene isolated from the environmental samples can directly be cloned.24 In 1991, Schmidt et al 25 successfully cloned 16S rRNA gene sequences from marine picoplankton with the use of bacteriophage lambda vector.26 In 1998, the term metagenome was introduced.27 In the last two decades metagenomic analyses have been performed on majority of the natural environments, for example, soils,28 marine picoplankton,29 hot springs,30 surface water from rivers,31 glacier ice,32 Antarctic desert soil33 and gut of ruminants.26 34 Earlier, major parts of the metagenomic studies were based on low-throughput approaches, like terminal restriction fragment-length polymorphism analysis,35 denaturing gradient gel electrophoresis36 or Sanger sequencing.37 The comparison with the sequences included in curated databases like the ones of Ribosomal Database Project II,38 Greengenes39 and SILVA40 has allowed the taxonomic classification and community profiling of environmental 16S rRNA gene sequences.
Clinical metagenomics and its potential as diagnostic tool
Classical microbiological methods are able to identify only the presumed cause of ocular infection in about 40% cases. In contrast, a metagenomic approach promises to provide important detail regarding all microbiota, allowing the identification of a greater portion of previously unidentified and the so-called ‘uncultured majority’ of microorganisms,41 whether being prokaryotes, eukaryotes or viruses. Efficient next-generation sequencing (NGS) technologies have developed greatly in the last decade, along with a reduced cost, gaining interest in the scientific community in several fields (such as medicine, biotechnology, agriculture or genetics).42 The deep impact on metagenomics given by the NGS technologies allowed the study of microbes with a much higher throughput. This enabled a ‘hypothesis-free’ approach providing all the components of the microbiome. The data produced by NGS have also proved to be particularly suitable for taxonomic and functional profiling. Thus, metagenomics opened the way to explore the remarkable genetic potential of bacteria; in some cases it was used to get hints on the metabolic requirements of yet-to-be cultured bacteria.43 More often, the application of NGS to metagenomics studies aimed to profile and compare taxonomy and function of microbial communities from different sources.23 For example, comparing human samples from a disease state to samples from healthy controls allows clinicians to get a more holistic view of the quantitative shifts of specific taxa during the course of the disease.1 44 The extraction of DNA from a human-derived sample is usually the first step of the workflow, then two possible downstream analyses can be done: (1) amplify a marker gene (usually a portion of the rRNA genes) and sequence the PCR amplification product; or (2) sequencing the extracted, fragmented DNA directly (this approach is known as ‘shotgun’ metagenomics sequencing).
There is a wealth of studies on the human microbiome, spanning several physiological conditions related to age, disease, race and many more variables. Starting from the first ‘inocula’, that is, transmission from mothers to children.45 A significant role has been attributed to the birth delivery method, with marked differences in microbiota between children born through vaginal delivery versus caesarean section.46 Different studies have debated determining the ‘core’ bacterial taxa of gut and skin.1 47 The latest analysis suggests that instead of the core taxa, homeostatic communities are defined by the presence of a core microbial gene set that encodes essential metabolic pathways.12 48
In addition, several studies have highlighted the variability of the microbiome according to body sites, race and ethnicity as outlined by Gupta et al.49 The genetic asset of the host itself influences the microbiome; this is well studied in metabolic dysfunctions such as obesity, diabetes and inflammatory bowel disease.50 Two simultaneous projects: the European project, MetaHIT (Metagenomics of the Human Intestinal Tract—www.metahit.eu), and the American Human Microbiome Project,51 use metagenomics to facilitate the study of human intestinal microbiome. The use of drugs has also an influence on the microbiome. Bioinformatic tools are being developed and updated almost monthly for the analysis of the data52 53 and in developing integrated databases, for example, https://portal.hmpdacc.org/. They are, however, over-represented by strong biases towards samples from stool, and oral and vaginal microbiomes.
Selecting the test
The two methods (shotgun and marker-based metagenomics) can be used in different instances: the marker-based approach is used to get the taxonomic profiles of the community under study, whereas shotgun approach gives wider information on function and an extended phylogenetic breadth.54 For both methods, there are pros and cons: marker-based studies are well suited for analysis of large number of samples, that is, multiple patients, longitudinal studies, and so on, and are cheaper; however, there are well-known amplification biases and the amount of information is limited to the taxonomy.55 On the other hand, shotgun metagenomics is usually more expensive. It may miss low-abundant species and when host-associated metagenomes are studied, most of the reads derive from the host genome, especially when studying sites with low bacterial biomass. It offers, however, increased resolution, enabling the possibility to discover new microbial genes and genomes as well as a more specific taxonomic and functional classification of sequences (in some cases). Importantly, shotgun metagenomics allows the simultaneous study of viruses, bacteriophages, archaea and eukaryotes.56 Sample collection and storage methods are critical for most metagenomic studies: they are often arbitrary and rely on the common practices developed in single laboratories or even by single researchers.57 However, in some cases, such as the study of the human faecal microbiome, there are well-established standard procedures.58
A standardised protocol for sample collection, handling and storage for metagenomic studies in ophthalmology is still under development (data not shown). In addition, as all low biomass samples, corneal surfaces are particularly vulnerable to external contaminations, which could also derive from the reagent kits,59 therefore, a proper experimental design should include a number of blank controls and the use of ultrapure reagents to minimise this risk. Several significant efforts to unravel bacterial identity with a resolution as high as the level of strain have already been published.60 The integration of the metaomics (collective name that stands for metagenomics, metatranscriptomics, metaproteomics, and so on) with information such as clinical history, dietary information and genetic background of the patient may be useful in the implementation of mechanistic models explaining the microbiome structure and function.61 Biomarker discovery needs a high number of replicates; one pipeline developed for this task is LEfSe62 which relies on the linear discriminant analysis of effect size. It detects consistent abundance patterns among features (that can be either taxa or coding genes) in a multidimensional data set such as a species-per-sample metagenomic matrix. It is highly scalable and it has proved to achieve a discrete performance in reducing the false-positive detection, although as explicitly admitted by the developer, the false-negative rate is slightly higher. Other pipelines are also available for biomarker discoveries,63 however, a benchmark among them is beyond the scope of this review. Last, but not least, the complex tasks described above require high computational power and specific expertise in the field of biostatistics and informatics.64
From bench to bedside: clinical applications
Clinical applications in OS
Doan and Pinsky in 2016 showed how the healthy OS has a unique microbiome with viral and bacterial communities. In their study, quantitative 16S PCR resulted in 0.1 bacterial 16S rDNA/human actin copy on the OS compared with 10 16S rDNA/human actin copies of facial skin and higher bacterial diversity on the OS.65 Many ocular infections are acquired on the OS and the diagnosis of causative pathogen can be challenging.66 67 In postoperative endophthalmitis, the pathogen identification is between 50% and 70%68 69 and cultures failed in approximately 33% of cases.70 In a recent study on patients with uveitis, Doan et al found that herpes simplex virus type 1 (HSV-1), Cryptococcus neoformans and Toxoplasma gondii were associated with the disease.71 Other than Rubeola RNA virus, even Ebola RNA virus was detected in the ocular fluid after resolution of viraemia.72 A recent attempt provided a proof of concept for the use of metagenomics as diagnostic tool, and developed specific bioinformatic pipelines to differentiate pathogenic agents and antibiotic resistance genes with a higher resolution.73
Contact lens
Shin et al showed that wearing contact lenses makes ocular conjunctiva more similar to the skin microbiota.16 Lee et al 74 studied blepharitis using different sampling from eyelashes and tears showing increased Staphylococcus, Streptophyta, Corynebacterium and Enhydrobacter. Studies suggested how contact lenses could function as a medium for skin bacteria to come to OS.75 76 Zhang et al found how slight microbe variability was found between orthokeratology lens wearers with soft contact lenses wearers and in non-wearers.77
Connection between gut and eye microbiome
It is well acknowledged that gut microbiome influences the communication between the enteric nervous system and the central nervous system (known as gut-brain axis).78 Likewise, OS microbiome shows connection to the gut microbiome. Considering that the eye is the site of inflammatory diseases like uveitis, scleritis and Mooren’s corneal ulcer, it is possible that these autoimmune reactions are associated with dysbiosis in the gut.79 de Paiva et al found that ‘the severity of Sjögren Syndrome (SS) ocular and systemic disease was inversely correlated with microbial diversity.’80 SS is marked by a dysbiotic intestinal microbiome driven by low relative abundance of commensal bacteria and high relative abundance of potential pathogenic genera. This is associated with worse ocular mucosal disease in a mouse model of SS and in patients with SS. The lowest diversity of stool microbiota was found in subjects with the most severe keratoconjunctivitis sicca and combined systemic and ocular disease. Such result is in agreement with other findings in which a disease state correlates with the low diversity of the microbiome in a specific compartment, such as inflammatory bowel disease (where Clostridium difficile dominates the microbiome) or the pulmonary microbiome during cystic fibrosis.81 Animal models of experimental autoimmune uveitis have shown significant attenuation of this disease following administration of oral antibiotics that altered the intestinal microbiota.82 Further evidence strongly suggests that the homeostatic microbiome plays a protective role in preventing colonisation of pathogenic species. It was demonstrated that oral administration of antibiotics reduced the severity of uveitis in mice with experimentally induced autoimmune uveitis.83 84 The recent cases of persistent infection with Ebola virus,72 and possibly Zika virus,85 explain the urgency to develop better diagnostics for uveitis. These cases, with important public health consequences, highlight the eye’s role as a potential reservoir for infectious agents. In addition, epigenetic mechanisms may cooperate with microbiota to initiate ocular inflammation.86
Discussions and conclusion
Detection of intraocular infections relies heavily on molecular diagnostics. In ophthalmology, the most widely available molecular diagnostic panel to detect infections includes separate pathogen-directed PCRs: HSV, varicella zoster virus, cytomegalovirus and T. gondii. Not surprisingly, more than 50% of all presumed corneal infections fail to have a pathogen isolated.87 88 NGS has offered clear advantages to make a definite diagnosis compared with conventional diagnostic methods and advances in metagenomics have made the use of NGS more useful for clinical disgnostics.71 Metagenomic deep sequencing (MDS) has the potential to improve diagnostic yield; it can theoretically detect all pathogens in a clinical sample89 using an unbiased and hypothesis-free approach. Wilson et al showed that NGS protocols could be completed in less than 48 hours90 with a significant advantage compared with many of the culture-dependent assays. Improving our understanding of the composition and function of a normal ocular microbiome would be a good starting point for a targeted therapy and the development of probiotic products. Host-microbe and microbe-microbe interactions on the OS indicate the beneficial function of the microbiota and the understanding of these principles by the clinicians could possibly guide appropriate use of topical and systemic antibiotics.91
MDS could be used in patients with difficult-to-diagnose infections, as a front-line diagnostic tool. In difficult-to-culture samples, metagenomic shotgun is a promising test for the identification of microbial keratitis and undiagnosed encephalitis.92 93 Together with uveitis-related disorders, the microbiome study can potentially and dramatically change the management and treatment of these diseases.
MDS may supplement or replace numerous and expensive diagnostic assays and procedures currently employed and improve patient outcomes. This approach will allow a far more comprehensive characterisation of the aetiology of infections and also complement the current diagnostic paradigm in ophthalmology. In the near future, a full genetic approach to eye infections is not far away (figure 2) where the samples will be collected, sequenced for specific targets and a specific antimicrobial used to treat the disease. With a certain clinical impact, the ophthalmologists will have precise quantification, multilocus sequence typing of single species and genetic sequence of the microbiota of ocular samples without the problems associated with tests based on conventional cultures. Hence, we foresee that metagenomics could further advance the field of ophthalmology especially in diagnosis and target-specific treatments.
Figure 2.
The process towards genetic-guided treatment in the field of ophthalmology. The figure indicates different procedures of sample collection, nucleic acid extraction, sample preparation, sequencing, bioinformatics, analysis and report writing, indication to the eye surgeon for specific drug usage for specific microorganism and metagenomic-guided eye treatment on the patient. Being highly specific and cost-effective, metagenomics could be potentially used in ophthalmology in the near future.
Footnotes
Contributors: Concept and design of the review: DB, VR, SBK, TS, LN, AF, PG, DP, AE, SF. Drafting the manuscript: DB, VR, SBK, AE, SF. Critical revision of the manuscript: DB, VR, SBK, TS, LN, AF, PG, DP, AE, SF. Supervision: SBK, SF, VR. Final approval: DB, VR, SBK, TS, LN, AF, PG, DP, AE, SF.
Competing interests: None declared.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Turnbaugh PJ, Ley RE, Hamady M, et al. . The human microbiome project. Nature 2007;449:804–10. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willcox MDP. Characterization of the normal microbiota of the ocular surface. Exp Eye Res 2013;117:99–105. 10.1016/j.exer.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 3. Lu LJ, Liu J. Human microbiota and ophthalmic disease. Yale J Biol Med 2016;89:325–30. [PMC free article] [PubMed] [Google Scholar]
- 4. Miller D, Iovieno A. The role of microbial flora on the ocular surface. Curr Opin Allergy Clin Immunol 2009;9:466–70. 10.1097/ACI.0b013e3283303e1b [DOI] [PubMed] [Google Scholar]
- 5. Graham JE, Moore JE, Jiru X, et al. . Ocular pathogen or commensal: a PCR-based study of surface bacterial flora in normal and dry eyes. Invest Ophthalmol Vis Sci 2007;48:5616–23. 10.1167/iovs.07-0588 [DOI] [PubMed] [Google Scholar]
- 6. Fernández-Rubio ME, Rebolledo-Lara L, Martinez-García M, et al. . The conjunctival bacterial pattern of diabetics undergoing cataract surgery. Eye 2010;24:825–34. 10.1038/eye.2009.218 [DOI] [PubMed] [Google Scholar]
- 7. Pramhus C, Runyan TE, Lindberg RB. Ocular flora in the severely burned patient. Arch Ophthalmol 1978;96:1421–4. 10.1001/archopht.1978.03910060175015 [DOI] [PubMed] [Google Scholar]
- 8. Chern KC, Shrestha SK, Cevallos V, et al. . Alterations in the conjunctival bacterial flora following a single dose of azithromycin in a trachoma endemic area. Br J Ophthalmol 1999;83:1332–5. 10.1136/bjo.83.12.1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hori Y, Maeda N, Sakamoto M, et al. . Bacteriologic profile of the conjunctiva in the patients with dry eye. Am J Ophthalmol 2008;146:729–34. 10.1016/j.ajo.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 10. Sankaridurg PR, Markoulli M, de la Jara PL, et al. . Lid and conjunctival micro biota during contact lens wear in children. Optom Vis Sci 2009;86:312–7. 10.1097/OPX.0b013e318199d20c [DOI] [PubMed] [Google Scholar]
- 11. Schabereiter-Gurtner C, Maca S, Rolleke S, et al. . 16S rDNA-based identification of bacteria from conjunctival swabs by PCR and DGGE fingerprinting. Invest Ophthalmol Vis Sci 2001;42:1164–71. [PubMed] [Google Scholar]
- 12. Tschöp MH, Hugenholtz P, Karp CL. Getting to the core of the gut microbiome. Nat Biotechnol 2009;27:344–6. 10.1038/nbt0409-344 [DOI] [PubMed] [Google Scholar]
- 13. Zegans ME, Van Gelder RN. Considerations in understanding the ocular surface microbiome. Am J Ophthalmol 2014;158:420–2. 10.1016/j.ajo.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perkins RE, Kundsin RB, Pratt MV, et al. . Bacteriology of normal and infected conjunctiva. J Clin Microbiol 1975;1:147–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dong Q, Brulc JM, Iovieno A, et al. . Diversity of Bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci 2011;52:5408–13. 10.1167/iovs.10-6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin H, Price K, Albert L, et al. . Changes in the eye microbiota associated with contact lens wearing. MBio 2016;7:e00198–16. 10.1128/mBio.00198-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shestopalov VI, Antonopoulos DA, Milleret D. Metagenomic analysis of bacterial community at the human conjunctiva. Invest Ophthalmol Vis Sci 2010;51. [Google Scholar]
- 18. Kaufman HE, Azcuy AM, Varnell ED, et al. . HSV-1 DNA in tears and saliva of normal adults. Invest Ophthalmol Vis Sci 2005;46:241–7. 10.1167/iovs.04-0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Y, Holland MJ, Makalo P, et al. . The conjunctival microbiome in health and trachomatous disease: a case control study. Genome Med 2014;6 10.1186/s13073-014-0099-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wen X, Miao L, Deng Y, et al. . The influence of age and sex on ocular surface microbiota in healthy adults. Invest Ophthalmol Vis Sci 2017;58:6030–7. 10.1167/iovs.17-22957 [DOI] [PubMed] [Google Scholar]
- 21. Thomason CA, Leon A, Kirkpatrick LT, et al. . Eye of the finch: characterization of the ocular microbiome of house finches in relation to mycoplasmal conjunctivitis. Environ Microbiol 2017;19:1439–49. 10.1111/1462-2920.13625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol 1985;39:321–46. 10.1146/annurev.mi.39.100185.001541 [DOI] [PubMed] [Google Scholar]
- 23. Kim D, Hofstaedter CE, Zhao C, et al. . Optimizing methods and dodging pitfalls in microbiome research. Microbiome 2017;5 10.1186/s40168-017-0267-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pace NR, Stahl DA, Lane DJ, et al. . Analyzing natural microbial populations by rRNA sequences. ASM News 1985;51:4–12. [Google Scholar]
- 25. Schmidt TM, DeLong EF, Pace NR. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 1991;173:4371–8. 10.1128/jb.173.14.4371-4378.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar S, Krishnani KK, Bhushan B, et al. . Metagenomics: retrospect and prospects in high throughput age. Biotechnol Res Int 2015;121735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Handelsman J, Rondon MR, Brady SF, et al. . Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 1998;5:R245–9. 10.1016/S1074-5521(98)90108-9 [DOI] [PubMed] [Google Scholar]
- 28. Coolon JD, Jones KL, Todd TC, et al. . Long-term nitrogen amendment alters the diversity and assemblage of soil bacterial communities in tallgrass prairie. PLoS One 2013;8:e67884 10.1371/journal.pone.0067884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stein JL, Marsh TL, Wu KY, et al. . Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol 1996;178:591–9. 10.1128/jb.178.3.591-599.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hedlund BP, Dodsworth JA, Cole JK, et al. . An integrated study reveals diverse methanogens, Thaumarchaeota, and yet-uncultivated archaeal lineages in Armenian hot springs. Antonie Van Leeuwenhoek 2013;104:71–82. 10.1007/s10482-013-9927-z [DOI] [PubMed] [Google Scholar]
- 31. Wu C, Sun B. Identification of novel esterase from metagenomic library of Yangtze River. J Microbiol Biotechnol 2009;19:187–93. [DOI] [PubMed] [Google Scholar]
- 32. Simon C, Herath J, Rockstroh S, et al. . Rapid identification of genes encoding DNA polymerases by function-based screening of metagenomic libraries derived from glacial ice. Appl Environ Microbiol 2009;75:2964–8. 10.1128/AEM.02644-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heath C, Hu XP, Cary SC, et al. . Identification of a novel alkaliphilic esterase active at low temperatures by screening a metagenomic library from Antarctic desert soil. Appl Environ Microbiol 2009;75:4657–9. 10.1128/AEM.02597-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gong X, Gruninger RJ, Qi M, et al. . Cloning and identification of novel hydrolase genes from a dairy cow rumen metagenomic library and characterization of a cellulase gene. BMC Res Notes 2012;5 10.1186/1756-0500-5-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 2006;103:626–31. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 1993;59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sogin ML, Morrison HG, Huber JA, et al. . Microbial diversity in the deep sea and the underexplored "rare biosphere". Proc Natl Acad Sci U S A 2006;103:12115–20. 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cole JR, Chai B, Marsh TL, et al. . The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 2003;31:442–3. 10.1093/nar/gkg039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeSantis TZ, Hugenholtz P, Larsen N, et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Research 2004;32:1363–71. 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol 2003;57:369–94. 10.1146/annurev.micro.57.030502.090759 [DOI] [PubMed] [Google Scholar]
- 42. Schlaberg R, Chiu CY, Miller S, et al. . Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 2017;141:776–86. 10.5858/arpa.2016-0539-RA [DOI] [PubMed] [Google Scholar]
- 43. Jeraldo P, Hernandez A, Nielsen HB, et al. . Capturing One of the Human Gut Microbiome’s Most Wanted: Reconstructing the Genome of a Novel Butyrate-Producing, Clostridial Scavenger from Metagenomic Sequence Data. Front Microbiol 2016;7 10.3389/fmicb.2016.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qin J, Li R, Raes J, et al. . A human gut microbial gene Catalogue established by metagenomic sequencing. Nature 2010;464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Asnicar F, Manara S, Zolfo M, et al. . Stuying vertical microbiome transmission from mothers to infants by Strain-Level metagenomic Profilingm: systems 2017. 2, 2017: e00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dominguez-Bello MG, Costello EK, Contreras M, et al. . Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010;107:11971–5. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zaura E, Keijser BJF, Huse SM, et al. . Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol 2009;9 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kurokawa K, Itoh T, Kuwahara T, et al. . Comparative Metagenomics revealed commonly enriched gene sets in human gut Microbiomes. DNA Res 2007;14:169–81. 10.1093/dnares/dsm018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gupta VK, Paul S, Dutta C. Geography, ethnicity or Subsistence-Specific variations in human microbiome composition and diversity. Front Microbiol 2017;8 10.3389/fmicb.2017.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med Overseas Ed 2016;375:2369–79. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 51. Methé BA, Human Microbiome Project Consortium . A framework for human microbiome research. Nature 2012;486:215–21. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oulas A, Pavloudi C, Polymenakou P, et al. . Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform Biol Insights 2015;9:75–88. 10.4137/BBI.S12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep 2016;6 10.1038/srep19233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jovel J, Patterson J, Wang W, et al. . Characterization of the gut microbiome using 16S or shotgun Metagenomics. Front Microbiol 2016;7 10.3389/fmicb.2016.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Soergel DAW, Dey N, Knight R, et al. . Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. Isme J 2012;6:1440–4. 10.1038/ismej.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Norman JM, Handley SA, Virgin HW. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology 2014;146:1459–69. 10.1053/j.gastro.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Quince C, Walker AW, Simpson JT, et al. . Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 2017;35:833–44. 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- 58. Costea PI, Zeller G, Sunagawa S, et al. . Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol 2017;35:1069–76. [DOI] [PubMed] [Google Scholar]
- 59. Salter SJ, Cox MJ, Turek EM, et al. . Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014;12 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rinke C, Schwientek P, Sczyrba A, et al. . Insights into the phylogeny and coding potential of microbial dark matter. Nature 2013;499:431–7. 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- 61. Brown J, de Vos WM, DiStefano PS, et al. . Translating the human microbiome. Nat Biotechnol 2013;31:304–8. 10.1038/nbt.2543 [DOI] [PubMed] [Google Scholar]
- 62. Wang X, Su X, Cui X, et al. . MetaBoot: a machine learning framework of taxonomical biomarker discovery for different microbial communities based on metagenomic data. PeerJ 2015;3:e993 10.7717/peerj.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Alshawaqfeh M, Bashaireh A, Serpedin E, et al. . Reliable biomarker discovery from metagenomic data via RegLRSD algorithm. BMC Bioinformatics 2017;18 10.1186/s12859-017-1738-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morgan XC, Huttenhower C. Meta'omic analytic techniques for studying the intestinal microbiome. Gastroenterology 2014;146:1437–48. 10.1053/j.gastro.2014.01.049 [DOI] [PubMed] [Google Scholar]
- 65. Doan T, Pinsky BA. Current and future molecular diagnostics for ocular infectious diseases. Curr Opin Ophthalmol 2016;27:561–7. 10.1097/ICU.0000000000000311 [DOI] [PubMed] [Google Scholar]
- 66. Tananuvat N, Salakthuantee K, Vanittanakom N, et al. . Prospective comparison between conventional microbial work-up vs PCR in the diagnosis of fungal keratitis. Eye 2012;26:1337–43. 10.1038/eye.2012.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Taravati P, Lam D, Van Gelder RN. Role of molecular diagnostics in ocular microbiology. Curr Ophthalmol Rep 2013;1:181–9. 10.1007/s40135-013-0025-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Endophthalmitis Vitrectomy Study Collaboration Results of the endophthalmitis vitrectomy study. A randomized trial of immediate vitrectomy and of intravenous antibiotics for the treatment of postoperative bacterial endophthalmitis. Arch Ophthalmol 1995;113:1479–96. [PubMed] [Google Scholar]
- 69. Gower EW, Keay LJ, Stare DE, et al. . Characteristics of endophthalmitis after cataract surgery in the United States Medicare population. Ophthalmology 2015;122:1625–32. 10.1016/j.ophtha.2015.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee AY, Akileswaran L, Tibbetts MD, et al. . Identification of torque Teno virus in culture-negative endophthalmitis by representational deep DNA sequencing. Ophthalmology 2015;122:524–30. 10.1016/j.ophtha.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doan T, Wilson MR, Crawford ED, et al. . Illuminating uveitis: metagenomic deep sequencing identifies common and rare pathogens. Genome Med 2016;8 10.1186/s13073-016-0344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Varkey JB, Shantha JG, Crozier I, et al. . Persistence of Ebola virus in ocular fluid during convalescence. N Engl J Med Overseas Ed 2015;372:2423–7. 10.1056/NEJMoa1500306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kirstahler P, Bjerrum SS, Friis-Møller A, et al. . Genomics-based identification of microorganisms in human ocular body fluid. Sci Rep 2018;8 10.1038/s41598-018-22416-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee SH, Oh DH, Jung JY, et al. . Comparative ocular microbial communities in humans with and without blepharitis. Invest Ophthalmol Vis Sci 2012;53:5585–93. 10.1167/iovs.12-9922 [DOI] [PubMed] [Google Scholar]
- 75. Chen K-J, Hou C-H, Sun M-H, et al. . Endophthalmitis caused by Acinetobacter baumannii: report of two cases. J Clin Microbiol 2008;46:1148–50. 10.1128/JCM.01604-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lai C-C, Cheng A, Liu W-L, et al. . Infections caused by unusual Methylobacterium species. J Clin Microbiol 2011;49:3329–31. 10.1128/JCM.01241-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang H, Zhao F, Hutchinson DS, et al. . Conjunctival microbiome changes associated with soft contact lens and Orthokeratology lens wearing. Invest Ophthalmol Vis Sci 2017;58:128–36. 10.1167/iovs.16-20231 [DOI] [PubMed] [Google Scholar]
- 78. Carabotti M, Scirocco A, Maselli MA, et al. . The gut-brain axis: interaction between enteric microbiota, Central and enteric nervous systems. Ann Gastroenterol 2016;29. [PMC free article] [PubMed] [Google Scholar]
- 79. Shivaji S. We are not alone: a case for the human microbiome in extra intestinal diseases. Gut Pathog 2017;9 10.1186/s13099-017-0163-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. de Paiva CS, Jones DB, Stern ME, et al. . Altered mucosal microbiome diversity and disease severity in Sjögren syndrome. Sci Rep 2016;6 10.1038/srep23561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li J, Hao C, Ren L, et al. . Data mining of lung microbiota in cystic fibrosis patients. PLoS One 2016;11:e0164510 10.1371/journal.pone.0164510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lin P, Asquith M, Gruner H, et al. . The role of the gut microbiota in immune-mediated uveitis. Invest Ophthalmol Vis Sci 2015;56. [Google Scholar]
- 83. Nakamura YK, Metea C, Karstens L, et al. . Gut microbial alterations associated with protection from autoimmune uveitis. Invest Ophthalmol Vis Sci 2016;57:3747–58. 10.1167/iovs.16-19733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Horai R, Zárate-Bladés CR, Dillenburg-Pilla P, et al. . Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 2015;43:343–53. 10.1016/j.immuni.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. de Paula Freitas B, de Oliveira Dias JR, Prazeres J, et al. . Ocular findings in infants with microcephaly associated with presumed Zika virus congenital infection in Salvador, Brazil. JAMA Ophthalmol 2016;134:529–35. 10.1001/jamaophthalmol.2016.0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wen X, Hu X, Miao L, et al. . Epigenetics, microbiota, and intraocular inflammation: new paradigms of immune regulation in the eye. Prog Retin Eye Res 2018. [DOI] [PubMed] [Google Scholar]
- 87. Kaye SB, Rao PG, Smith G, et al. . Simplifying collection of corneal specimens in cases of suspected bacterial keratitis. J Clin Microbiol 2003;41:3192–7. 10.1128/JCM.41.7.3192-3197.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kaye S, Sueke H, Romano V, et al. . Impression membrane for the diagnosis of microbial keratitis. Br J Ophthalmol 2016;100:607–10. 10.1136/bjophthalmol-2015-307091 [DOI] [PubMed] [Google Scholar]
- 89. Graf EH, Simmon KE, Tardif KD, et al. . Unbiased detection of respiratory viruses by use of RNA sequencing-based Metagenomics: a systematic comparison to a commercial PCR panel. J Clin Microbiol 2016;54:1000–7. 10.1128/JCM.03060-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wilson MR, Shanbhag NM, Reid MJ, et al. . Diagnosing Balamuthia mandrillaris Encephalitis with metagenomic deep sequencing. Ann Neurol 2015;78:722–30. 10.1002/ana.24499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Huang Y, Yang B, Li W. Defining the normal core microbiome of conjunctival microbial communities. Clin Microbiol Infect 2016;22:643.e7–643.e12. 10.1016/j.cmi.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 92. Shigeyasu C, Yamada M, Aoki K, et al. . Metagenomic analysis for detecting Fusarium solani in a case of fungal keratitis. J Infect Chemother 2018. [DOI] [PubMed] [Google Scholar]
- 93. Brown JR, Bharucha T, Breuer J, et al. . Encephalitis diagnosis using metagenomics: application of next generation sequencing for undiagnosed cases. J Infect 2018;76:225–40. 10.1016/j.jinf.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]



