Keywords: nerve regeneration, novel microRNAs, miR-sc14, peripheral nerve injury, cell proliferation, cell migration, Schwann cells, fibroblast growth factor receptor 2, biological functions, peripheral nerve regeneration, regulatory mechanisms, neural regeneration
Abstract
MicroRNAs refer to a class of endogenous, short non-coding RNAs that mediate numerous biological functions. MicroRNAs regulate various physiological and pathological activities of peripheral nerves, including peripheral nerve repair and regeneration. Previously, using a rat sciatic nerve injury model, we identified many functionally annotated novel microRNAs, including miR-sc14. Here, we used real-time reverse transcription-polymerase chain reaction to examine miR-sc14 expression in rat sciatic nerve stumps. Our results show that miR-sc14 is noticeably altered following sciatic nerve injury, being up-regulated at 1 day and diminished at 7 days. EdU and transwell chamber assay results showed that miR-sc14 mimic promoted proliferation and migration of Schwann cells, while miR-sc14 inhibitor suppressed their proliferation and migration. Additionally, bioinformatic analysis examined potential target genes of miR-sc14, and found that fibroblast growth factor receptor 2 might be a potential target gene. Specifically, our results show changes of miR-sc14 expression in the sciatic nerve of rats at different time points after nerve injury. Appropriately, up-regulation of miR-sc14 promoted proliferation and migration of Schwann cells. Consequently, miR-sc14 may be an intervention target to promote repair of peripheral nerve injury. The study was approved by the Jiangsu Provincial Laboratory Animal Management Committee, China on March 4, 2015 (approval No. 20150304-004).
Chinese Library Classification No. R447; R363; R741
Introduction
Despite constituting more than 98% of total genomic sequence, non-coding DNAs used to be considered junk DNA. However, emerging studies show that non-coding DNAs play important biological functions, and indeed, emphasize the importance of non-coding DNAs (Wilusz et al., 2009; Pennisi, 2012; Gori et al., 2015). These non-coding DNAs are transcribed into many functional non-coding RNAs, including regulatory RNAs, ribosomal RNAs, and transfer RNAs. Among these non-coding regulatory RNAs, microRNAs (miRNAs) are of great research interest as they function as post-transcriptional regulators, negatively regulating up to 60% of human genes and mediating many physiological and pathological processes (Kloosterman and Plasterk, 2006; Taft et al., 2010; Dey et al., 2014).
miRNAs are a family of small single-strand non-coding RNAs of approximately 22 nucleotides in length, which directly binding to the 3′-untranslated region of target genes and negatively regulate them (Ambros, 2004; Bhattacharyya et al., 2006; Bartel, 2009; Fabian et al., 2010; Krol et al., 2010). miRNAs affect various tissues and organs in the body, including the nervous system (Fineberg et al., 2009; Yu et al., 2015; Ghibaudi et al., 2017). In the peripheral nervous system, miRNAs reportedly affect the phenotype of Schwann cells, the unique glial cells of the peripheral nervous system, and regulate their proliferation, migration, and myelination (Bremer et al., 2010; Pereira et al., 2010; Yun et al., 2010; Yu et al., 2012; Li et al., 2015; Yi et al., 2017b; Wang et al., 2018).
Previously obtained high-throughput sequencing data from our laboratory showed differential expression of numerous miRNAs in rat sciatic nerve stumps following peripheral nerve injury (Yu et al., 2011). Moreover, we identified a series of functionally annotated novel miRNAs (Li et al., 2011). Our subsequent studies examined the function of these novel miRNAs, including miR-sc3, miR-sc4, and miR-sc8, showing that they mediate phenotypic changes of Schwann cells (Gu et al., 2015; Yi et al., 2016; Qian et al., 2018). The functions of many other newly identified miRNAs remain largely unclear. Therefore in the present study, we focused on miR-sc14 by examining expression patterns of miR-sc14 in sciatic nerve stumps at different time points after nerve injury. Hence, we investigated the biological function of miR-sc14 on Schwann cells and its potential target genes.
Materials and Methods
Animals
All experimental procedures involving animals were performed in accordance with Institutional Animal Care guidelines, and ethically approved by the Jiangsu Provincial Laboratory Animal Management Committee, China (approval No. 20150304-004) on March 4, 2015. In total, 30 specific-pathogen-free adult 2-month-old male Sprague-Dawley rats weighing 180–220 g were randomly separated into five groups (0, 1, 4, 7, and 14 days after nerve injury), with 6 rats in each group. Rats were subjected to surgical transection of sciatic nerves. Additionally, primary Schwann cells were collected from 60 specific-pathogen-free neonatal 1-day-old Sprague-Dawley rats of either sex. Both adult and neonatal rats were obtained from the Experimental Animal Center of Nantong University, China (animal license number: SCXK [Su] 2014-0001 and SYXK [Su] 2012-0031).
Real-time reverse transcription-polymerase chain reaction (RT-PCR)
Adult rat sciatic nerve injury was performed as previously described (Li et al., 2011). After anesthetization using an injection of mixed narcotics (85 mg/kg trichloroacetaldehyde monohydrate [RichJoint, Shanghai, China], 42 mg/kg magnesium sulfate [Xilong Scientific, Guangzhou, China], and 17 mg/kg sodium pentobarbital [Sigma, St. Louis, MO, USA]), rat sciatic nerve stumps were exposed to injury and 5 mm long proximal sciatic nerve stumps collected at 0, 1, 4, 7, and 14 days after injury. RNA samples were collected from nerve stumps and then reverse transcribed with Omniscript Reverse Transcription Kit (Qiagen, Valencia, CA, USA). Real-time RT-PCR was performed using QuantiNova SYBR Green PCR Kit (Qiagen) on a Stepone real-time PCR system (Applied Biosystems, Foster City, CA, USA). Abundance of miR-sc14 was determined using primer pairs ordered from Ribobio Company (Guangzhou, China), and normalized to U6. Primer pair sequences for mRNAs were: fibroblast growth factor receptor 2 (FGFR2) (forward) 5′-CGT ACT GGA CCA ACA CCG AA-3′ and (reverse) 5′-CCA GTG CTG GTT TCG TAC CT-3′; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (forward) 5′-CCT TCA TTG ACC TCA ACT ACA TG-3′ and (reverse) 5′-CTT CTC CAT GGT GAA GAC-3′. Expression levels of FGFR2 and CD44 were determined by the 2–ΔΔCt method, and normalized to GAPDH as the reference, as previously performed (Qian et al., 2018).
Schwann cell culture and transfection
The preparation method of sciatic nerve stumps from neonatal rats is the same as above. Sciatic nerve stumps of neonatal rats were used to isolate primary Schwann cells. Schwann cells were purified by removing fibroblasts, and then identified using the Schwann cell marker, anti-S100 (DAKO, Carpinteria, CA, USA), as previously described (Li et al., 2015). Schwann cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen), 1% penicillin and streptomycin (Invitrogen), 2 μM forskolin (Sigma), and 10 ng/mL human heregulin-β1 (Sigma) in a CO2 incubator. Prior to cell proliferation and migration assays, cultured Schwann cells were transfected with miR-sc14 mimic (Ribobio), non-targeting mimic control (Ribobio), miR-sc14 inhibitor (anti-miR-sc14; Ribobio), or non-targeting inhibitor control (Ribobio) using Lipofectamine RNAiMAX transfection reagent (Invitrogen).
Cell proliferation assay
The proliferation rate of Schwann cells was measured using the Cell-Light™ EdU DNA Cell Proliferation Kit (Ribobio), as previously described (Yi et al., 2016). Briefly, 96-well plates were coated with poly-L-lysine (Zhongshan Bio-tech Co., Beijing, China) and cells were seeded onto coated plates. EdU (50 μM; Ribibio) was added to cell cultures, and Schwann cells were cultured for an additional 24 hours prior to fixation with 4% paraformaldehyde (Xilong Scientific, Guangzhou, China) in phosphate buffered saline. Schwann cells were also stained with Hoechst 33342 (Solarbio, Beijing, China). Images were obtained using a DMR fluorescence microscope (Leica Microsystems, Bensheim, Germany). The proliferation rate of Schwann cells was calculated by dividing the number of EdU-positive cells by the number of total cells.
Cell migration assay
The migration rate of Schwann cells was measured using a transwell-based assay, as previously described (Li et al., 2015). Briefly, the bottom surfaces of 6.5-mm transwell chambers with 8 mm pores (Costar, Cambridge, MA, USA) were coated with fibronectin. Schwann cells in Dulbecco’s modified Eagle’s medium were seeded onto the upper chamber, and the bottom chamber was filled with complete culture medium. After 24 hours of culture, Schwann cells remaining on the upper surface were removed by a cotton swab. Schwann cells that migrated to the lower surface were stained by crystal violet (Beyotime, Shanghai, China). Migration images were obtained using a DMR inverted microscope (Leica Microsystems). Next, 33% acetic acid was used to wash crystal violet labeled migrated Schwann cells, and absorbance of crystal violet (Bio-tek, Winooski, VT, USA) was measured at 570 nm to determine cell migration ability.
Bioinformatic analysis
FGFR2-related biological functions and interacting genes were identified from Ingenuity Pathways Knowledge Base using QIAGEN’s Ingenuity Pathway Analysis software program (Ingenuity Systems Inc., Redwood City, CA, USA). Briefly, Ingenuity Pathway Analysis software screened FGFR2-related genes and biological functions to form a network.
Statistical analysis
All data are expressed as the mean ± SEM. Student’s t-test or one-way analysis of variance was used to evaluate statistical differences using GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA, USA). A value of P < 0.05 was considered statistically significant.
Results
Expression levels of miR-sc14 after sciatic nerve injury
Real-time RT-PCR was used to examine temporal expression patterns of miR-sc14 in sciatic nerve stumps at 0, 1, 4, 7, and 14 days after sciatic nerve injury. Compared with the 0 day control, miR-sc14 was differentially expressed after nerve injury and showed a curved expression trend with a trough value at 7 days after sciatic nerve injury (Figure 1). These observations demonstrate that miR-sc14 is differentially expressed in sciatic nerve stumps after sciatic nerve injury.
Figure 1.
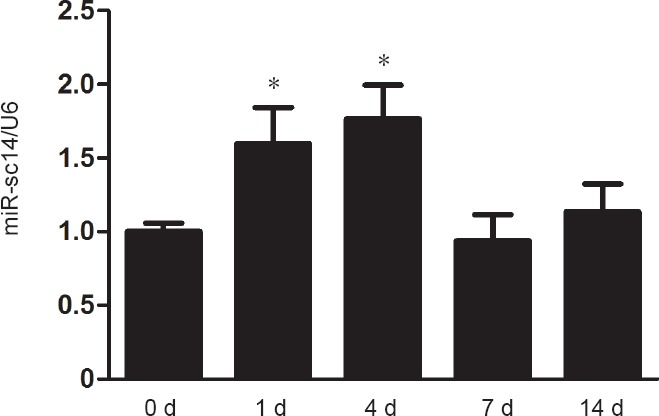
miR-sc14 is altered in sciatic nerve stumps after nerve injury.
Expression levels of miR-sc14 in sciatic nerve stumps at 0, 1, 4, 7, and 14 days after nerve injury were measured by real-time reverse transcription polymerase chain reaction. Abundance of miR-sc14 was normalized to U6, and relative levels of miR-sc14 at 1, 4, 7, and 14 days were compared with 0 day. Data are expressed as the mean ± SEM (n = 6). *P < 0.05, vs. 0 day (one-way analysis of variance). The experiment was conducted in triplicate. d: Day (s).
Effect of miR-sc14 on Schwann cell proliferation
Schwann cells are believed to be the main cell type in sciatic nerve stumps, and play significant roles during peripheral nerve regeneration (Palomo Irigoyen et al., 2018). Thus, we first investigated miR-sc14 effects on Schwann cells. Cultured primary Schwann cells were transfected with miR-sc14 mimic, miR-sc14 inhibitor, or corresponding non-targeting negative control to examine the effect of increased or decreased miR-sc14 on Schwann cell proliferation. Outcomes from the EdU cell proliferation assay showed that compared with Schwann cells transfected with non-targeting mimic control, Schwann cells transfected with miR-sc14 mimic exhibited an elevated proliferation rate (P = 0.0027). This suggests that increased expression of miR-sc14 increases the proliferation rate of Schwann cells (Figure 2). In contrast, Schwann cells transfected with miR-sc14 inhibitor showed a reduced proliferation rate (P = 0.0356), demonstrating that decreased expression of miR-sc14 suppresses the proliferation rate of Schwann cells (Figure 2). Taken together, these outcomes suggest that miR-sc14 promotes proliferation of Schwann cells.
Figure 2.
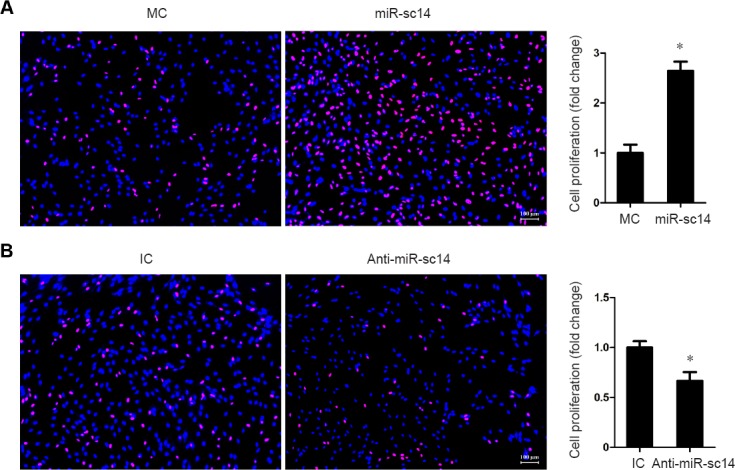
miR-sc14 reduces Schwann cell proliferation.
Representative images of EdU staining in Schwann cells transfected with miR-sc14 mimic (miR-sc14), MC and miR-sc14 inhibitor (anti-miR-sc14), or non-targeting IC. Red represents EdU staining and blue represents Hoechst 33342 staining. Scale bars: 100 μm. Schwann cell proliferation rate was calculated by dividing EdU-positive cells to total cells. The relative proliferation rate was normalized to control (MC or IC). Data are expressed as the mean ± SEM (Student’s t-test). *P < 0.05, vs. control. The experiment was conducted in triplicate. MC: Mimic control; IC: inhibitor control.
Effect of miR-sc14 on Schwann cell migration
Schwann cells transfected with miR-sc14 mimic or miR-sc14 inhibitor were further seeded onto transwell plates to study miR-sc14 effects on Schwann cell migration. Outcomes from the transwell-based migration assay demonstrated that compared with the non-targeting negative control, transfection with miR-sc14 mimic led to an increased migration rate (P = 0.0131), while transfection with miR-sc14 inhibitor led to a decreased migration rate (P = 0.0185) (Figure 3). These outcomes suggest that miR-sc14 also promotes Schwann cell migration.
Figure 3.
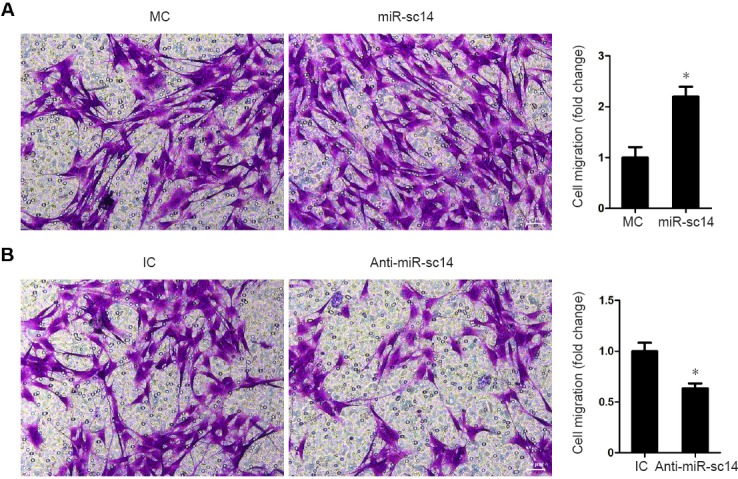
miR-sc14 reduces Schwann cell migration.
Representative images of transwell migration in Schwann cells transfected with miR-sc14 mimic (miR-sc14), MC and miR-sc14 inhibitor (anti-miR-sc14), or non-targeting IC. Scale bars: 100 μm. The relative migration rate was obtained by counting the amount of migrated cells normalized to control (MC or IC). Data are expressed as the mean ± SEM. *P < 0.05, vs. control (Student’s t-test). The experiment was conducted in triplicate. MC: Mimic control; IC: inhibitor control.
Identification of candidate target genes of miR-sc14
Our previous study showed that FGFR2 and CD44 might be potential target genes of miR-sc14 (Li et al., 2011). Here, the relationship between miR-sc14 and these potential target genes was further studied. Binding relationships between the 3′-UTR of FGFR2 and miR-sc14 showed complete binding of FGFR2 to miR-sc14 (Figure 4A). Previously obtained microarray data were analyzed to demonstrate time-dependent expression profiles of FGFR2 in sciatic nerve stumps at 0, 1, 4, 7, and 14 days after nerve injury (Li et al., 2013). Heatmap showed that expression levels of FGFR2 were down-regulated at different times after nerve injury (Figure 4B). Real-time RT-PCR was also examined to validate the microarray data. Consistent with microarray outcomes, real-time RT-PCR demonstrated that FGFR2 exhibited an approximately negative relationship with miR-sc14 (Figure 4C). Similarly, binding relationships between the 3′-UTR of CD44 and miR-sc14 also showed complete binding of CD44 to miR-sc14. However, microarray data and real-time RT-PCR showed that CD44 exhibited an approximately positive relationship with miR-sc14 (data not shown). Taken together, and considering that miRNAs generally bind to the 3′-UTR of target genes and negatively regulate their expression, we believe that FGFR2, and not CD44, is the candidate target gene of miR-sc14.
Figure 4.
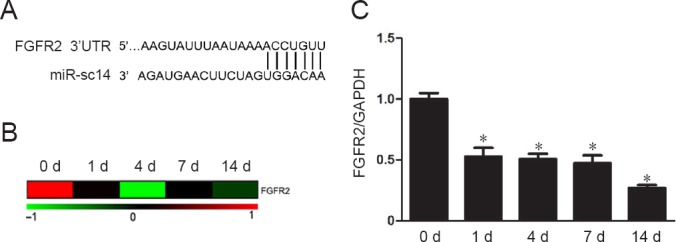
FGFR2 is a candidate target gene of miR-sc14.
(A) Predicted binding sites of miR-sc14 and its potential target gene, FGFR2. (B) Heatmap of FGFR2 mRNA expression in rat sciatic nerve stumps after nerve injury. Red represents up-regulation and green represents down-regulation. (C) Expression levels of FGFR2 in sciatic nerve stumps at 0, 1, 4, 7, and 14 days after nerve injury measured by real-time reverse transcription polymerase chain reaction. Abundance of FGFR2 was normalized to GAPDH, and relative levels of FGFR2 at 1, 4, 7, and 14 days were compared with 0 day. Data are expressed as the mean ± SEM. *P < 0.05, vs. 0 day (one-way analysis of variance). The experiment was conducted in triplicate. FGFR2: Fibroblast growth factor receptor 2; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; d: day(s).
Discussion
Peripheral nerve injury elicits disintegration of axons and myelin sheaths, degeneration of distal nerve stumps, reconstruction of the microenvironment, and subsequent regrowth of axons and regeneration of injured nerves (Siconolfi and Seeds, 2001; Zhang et al., 2016). Peripheral nerve injury and regeneration involve complicated cellular and molecular changes (Gu et al., 2011, 2014; Yi et al., 2015, 2017a; Yu et al., 2016). Gaining improved understanding of the dynamic gene expression changes after peripheral nerve injury may shed light on the underlying mechanisms of peripheral nerve regeneration and help identification of new therapeutic targets for peripheral nerve injury.
Differentially expressed miRNAs may play important roles in peripheral nerve injury and regeneration as miRNAs regulate many downstream genes and consequently a succession of biological activity. Previous sequencing data revealed novel miRNAs that were differentially expressed in sciatic nerve stumps after nerve injury. By performing real-time RT-PCR, we found that miR-sc14 was up-regulated in injured sciatic nerve stumps, especially at early time points after injury. In this study, we investigated the biological effect of miR-sc14 on Schwann cells via the EdU cell proliferation and transwell-based migration assays, and demonstrated that miR-sc14 plays a positive role on proliferation and migration of Schwann cells. Thus, following peripheral nerve injury, up-regulated miR-sc14 might contribute to peripheral nerve regeneration via its effect on Schwann cells.
Additionally, by combined use of bioinformatic tools, high throughput data, and real-time RT-PCR, potential target genes of miR-sc14 were identified, with FGFR2 revealed as a candidate downstream target gene of miR-sc14. FGFR2 is a tyrosine protein kinase that regulates development and regeneration of tissues and organs, including the nervous system. In the central nervous system, FGFR2 affects myelination of oligodendrocytes and regulates thickness of the myelin sheath (Messersmith et al., 2000; Furusho et al., 2012; Furusho et al., 2017). Besides oligodendrocytes, FGFR2 was also shown to be expressed in Schwann cells (Grothe et al., 2001). Further, it was demonstrated that conditionally inactivated FGFR1 and FGFR2 in Schwann cells of transgenic mice led to development of sensory axonal neuropathy (Furusho et al., 2009). Our study here showed that in contrast to miR-sc14, FGFR2 was down-regulated in rat sciatic nerve stumps at different time points after nerve injury. Therefore, dysregulated FGFR2, as a potential target of miR-sc14, might affect the phenotype of Schwann cells and mediate peripheral nerve regeneration. Furthermore, Ingenuity Pathway Analysis was also used to investigate biological functions and genes correlated with FGFR2. Ingenuity Pathway Analysis software screened genes directly linked with FGFR2, including fibroblast growth factor 6, fibroblast growth factor 19, fibroblast growth factor 21, fibroblast growth factor 22, klotho beta, centrosomal protein 295, transcription factor 7-like 2, and cAMP responsive element binding protein 1. Besides, these interacting genes, FGFR2 participation in biological functions associated with the nervous system was also determined. Correspondingly, a network of FGFR2-related genes and biological functions are demonstrated in Figure 5. In our future studies, dual luciferase assay and rescue experiments will be performed to determine whether miR-sc14 directly and negatively regulates FGFR.
Figure 5.
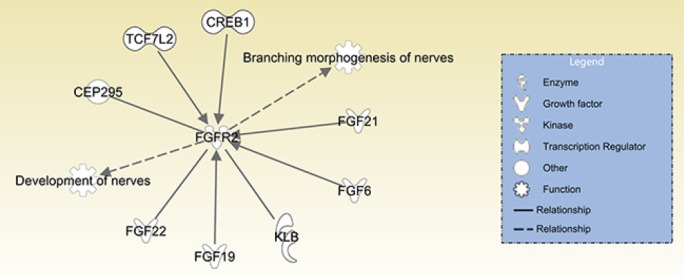
Ingenuity Pathway Analysis network of FGFR2-related genes and biological functions.
FGFR2 was directly linked with FGF6, FGF19, FGF21, FGF22, KLB, CEP295, TCF7L2, and CREB1. FGFR2 is associated with development of nerves and branching morphogenesis of nerves. FGF6: Fibroblast growth factor 6; FGF19: fibroblast growth factor 19; FGF21: fibroblast growth factor 21; FGF22: fibroblast growth factor 22; KLB: klotho beta; CEP295: centrosomal protein 295; TCF7L2: transcription factor 7 like 2; CREB1: cAMP responsive element binding protein 1; FGFR2: fibroblast growth factor receptor 2.
In summary, in this study, we investigated expression changes of the novel miRNA, miR-sc14, in rat sciatic nerve stumps at different time points after nerve injury. Accordingly, we found that mRNA expression levels of miR-sc14 are increased after peripheral nerve injury. Moreover, we further demonstrated that up-regulated miR-sc14 promoted proliferation and migration of Schwann cells. A candidate target gene of miR-sc14 was also revealed. Our current study enhances our understanding of the underlying mechanisms of peripheral nerve repair and regeneration, and may contribute to identification of novel therapeutic targets for peripheral nerve repair.
Footnotes
Conflicts of interest: None declared.
Financial support: This study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions, China. Funder had no involvement in the study design; data collection, analysis, and interpretation; paper writing; or decision to submit the paper for publication.
Institutional review board statement: The study was approved by the Jiangsu Provincial Laboratory Animal Management Committee, China on March 4, 2015 (approval No. 20150304-004).
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Huiyin Tu, Zhengzhou University, China.
Funding: This study was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions of China.
P-Reviewer: Tu H; C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: James R, Hindle A, Qiu Y, Song LP; T-Editor: Liu XL
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 4.Bremer J, O’Connor T, Tiberi C, Rehrauer H, Weis J, Aguzzi A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS One. 2010;5:e12450. doi: 10.1371/journal.pone.0012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5:e944014. doi: 10.4161/21541272.2014.944014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 7.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Furusho M, Ishii A, Bansal R. Signaling by FGF receptor 2, not FGF receptor 1, regulates myelin thickness through activation of ERK1/2-MAPK, which promotes mTORC1 activity in an Akt-independent manner. J Neurosci. 2017;37:2931–2946. doi: 10.1523/JNEUROSCI.3316-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furusho M, Dupree JL, Bryant M, Bansal R. Disruption of fibroblast growth factor receptor signaling in nonmyelinating Schwann cells causes sensory axonal neuropathy and impairment of thermal pain sensitivity. J Neurosci. 2009;29:1608–1614. doi: 10.1523/JNEUROSCI.5615-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furusho M, Dupree JL, Nave KA, Bansal R. Fibroblast growth factor receptor signaling in oligodendrocytes regulates myelin sheath thickness. J Neurosci. 2012;32:6631–6641. doi: 10.1523/JNEUROSCI.6005-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghibaudi M, Boido M, Vercelli A. Functional integration of complex miRNA networks in central and peripheral lesion and axonal regeneration. Prog Neurobiol. 2017;158:69–93. doi: 10.1016/j.pneurobio.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Gori M, Trombetta M, Santini D, Rainer A. Tissue engineering and microRNAs: future perspectives in regenerative medicine. Expert Opin Biol Ther. 2015;15:1601–1622. doi: 10.1517/14712598.2015.1071349. [DOI] [PubMed] [Google Scholar]
- 13.Grothe C, Meisinger C, Claus P. In vivo expression and localization of the fibroblast growth factor system in the intact and lesioned rat peripheral nerve and spinal ganglia. J Comp Neurol. 2001;434:342–357. doi: 10.1002/cne.1181. [DOI] [PubMed] [Google Scholar]
- 14.Gu X, Ding F, Williams DF. Neural tissue engineering options for peripheral nerve regeneration. Biomaterials. 2014;35:6143–6156. doi: 10.1016/j.biomaterials.2014.04.064. [DOI] [PubMed] [Google Scholar]
- 15.Gu X, Ding F, Yang Y, Liu J. Construction of tissue engineered nerve grafts and their application in peripheral nerve regeneration. Prog Neurobiol. 2011;93:204–230. doi: 10.1016/j.pneurobio.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, Chen C, Yi S, Wang S, Gong L, Liu J, Gu X, Zhao Q, Li S. miR-sc8 inhibits Schwann cell proliferation and migration by targeting Egfr. PLoS One. 2015;10:e0145185. doi: 10.1371/journal.pone.0145185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Yu B, Wang Y, Yao D, Zhang Z, Gu X. Identification and functional annotation of novel microRNAs in the proximal sciatic nerve after sciatic nerve transection. Sci China Life Sci. 2011;54:806–812. doi: 10.1007/s11427-011-4213-7. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Liu Q, Wang Y, Gu Y, Liu D, Wang C, Ding G, Chen J, Liu J, Gu X. Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One. 2013;8:e57000. doi: 10.1371/journal.pone.0057000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, Wang X, Gu Y, Chen C, Wang Y, Liu J, Hu W, Yu B, Wang Y, Ding F, Liu Y, Gu X. Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. Mol Ther. 2015;23:423–433. doi: 10.1038/mt.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messersmith DJ, Murtie JC, Le TQ, Frost EE, Armstrong RC. Fibroblast growth factor 2 (FGF2) and FGF receptor expression in an experimental demyelinating disease with extensive remyelination. J Neurosci Res. 2000;62:241–256. doi: 10.1002/1097-4547(20001015)62:2<241::AID-JNR9>3.0.CO;2-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomo Irigoyen M, Tamayo Caro M, Perez Andres E, Barreira Manrique A, Varela Rey M, Woodhoo A. Isolation and purification of primary rodent Schwann cells. Methods Mol Biol. 2018;1791:81–93. doi: 10.1007/978-1-4939-7862-5_7. [DOI] [PubMed] [Google Scholar]
- 24.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159, 1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 25.Pereira JA, Baumann R, Norrmen C, Somandin C, Miehe M, Jacob C, Luhmann T, Hall-Bozic H, Mantei N, Meijer D, Suter U. Dicer in Schwann cells is required for myelination and axonal integrity. J Neurosci. 2010;30:6763–6775. doi: 10.1523/JNEUROSCI.0801-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian T, Wang X, Wang Y, Wang P, Liu Q, Liu J, Yi S. Novel miR-sc4 regulates the proliferation and migration of Schwann cells by targeting Cdk5r1. Mol Cell Biochem. 2018;447:209–215. doi: 10.1007/s11010-018-3305-0. [DOI] [PubMed] [Google Scholar]
- 27.Siconolfi LB, Seeds NW. Induction of the plasminogen activator system accompanies peripheral nerve regeneration after sciatic nerve crush. J Neurosci. 2001;21:4336–4347. doi: 10.1523/JNEUROSCI.21-12-04336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, He J, Wang S, Wang X, Liu Q, Peng W, Qian T. miR-3075 inhibited the migration of Schwann cells by targeting Cntn2. Neurochem Res. 2018;43:1879–1886. doi: 10.1007/s11064-018-2605-9. [DOI] [PubMed] [Google Scholar]
- 30.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yi S, Tang X, Yu J, Liu J, Ding F, Gu X. Microarray and qPCR analyses of Wallerian degeneration in rat sciatic nerves. Front Cell Neurosci. 2017a;11:22. doi: 10.3389/fncel.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi S, Wang QH, Zhao LL, Qin J, Wang YX, Yu B, Zhou SL. miR-30c promotes Schwann cell remyelination following peripheral nerve injury. Neural Regen Res. 2017b;12:1708–1715. doi: 10.4103/1673-5374.217351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi S, Zhang H, Gong L, Wu J, Zha G, Zhou S, Gu X, Yu B. Deep sequencing and bioinformatic analysis of lesioned sciatic nerves after crush injury. PLoS One. 2015;10:e0143491. doi: 10.1371/journal.pone.0143491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi S, Wang S, Zhao Q, Yao C, Gu Y, Liu J, Gu X, Li S. miR-sc3, a novel microRNA, promotes Schwann cell proliferation and migration by targeting Astn1. Cell Transplant. 2016;25:973–982. doi: 10.3727/096368916X690520. [DOI] [PubMed] [Google Scholar]
- 35.Yu B, Zhou S, Yi S, Gu X. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Prog Neurobiol. 2015;134:122–139. doi: 10.1016/j.pneurobio.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Yu B, Zhou S, Wang Y, Ding G, Ding F, Gu X. Profile of microRNAs following rat sciatic nerve injury by deep sequencing: implication for mechanisms of nerve regeneration. PLoS One. 2011;6:e24612. doi: 10.1371/journal.pone.0024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu J, Gu X, Yi S. Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: insights into Wallerian degeneration. Front Cell Neurosci. 2016;10:274. doi: 10.3389/fncel.2016.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun B, Anderegg A, Menichella D, Wrabetz L, Feltri ML, Awatramani R. MicroRNA-deficient Schwann cells display congenital hypomyelination. J Neurosci. 2010;30:7722–7728. doi: 10.1523/JNEUROSCI.0876-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Yu B, Gu Y, Zhou S, Qian T, Wang Y, Ding G, Ding F, Gu X. Fibroblast-derived tenascin-C promotes Schwann cell migration through beta1-integrin dependent pathway during peripheral nerve regeneration. Glia. 2016;64:374–385. doi: 10.1002/glia.22934. [DOI] [PubMed] [Google Scholar]


