Abstract
We synthesized and screened a unique 46,656-member library composed of unnatural amino acids that revealed several hits against RRE IIB RNA. Among the hit peptides identified, peptide 4A5 was found to be selective against competitor RNAs and inhibited HIV-1 Rev-RRE RNA interaction in cell culture in a p24 ELISA assay. Biophysical characterization in a ribonuclease protection assay suggested that 4A5 bound to the stem-loop region in RRE IIB while SHAPE MaP probing with 234 nt RRE RNA indicated additional interaction with secondary Rev binding sites. Taken together, our investigation suggests that HIV replication is inhibited by 4A5 blocking binding of Rev and subsequent multimerization.
Graphical abstract

INTRODUCTION
Although highly active antiretroviral therapy (HAART) is effective at significantly reducing viral loads associated with HIV-1 infection, lifelong maintenance of viral suppression is required as there is no available cure.1,2 This is due to several factors, including evolution of resistance as well as persistent, latent viral loads post-treatment.2–4 Therefore, continued development of therapeutics to target HIV-1 is needed, ideally using strategies to inhibit mechanisms of viral replication less prone to mutation and drug resistance. One such strategy involves targeting conserved regions of the (+) RNA genome that are essential to promote the life cycle of the virus. RNA regulates many vital biological processes and its tertiary structure yields great potential as a target.5–7 Particular interest has been placed on targeting the Rev Response Element (RRE) RNA, a 351 nucleotide conserved region located in the env gene of the (+) strand genome.8, 9 Interaction of this RNA with its cognate ligand, Rev, promotes nuclear export of unspliced and singly spliced mRNAs complexed with nuclear export factors such as Ran-GTP, eIF-5A, and Crm-1.10, 11 This subsequently allows for both the translation of gag, pol, and env genes and packaging of the full length genome in budding virions.10–12 Therefore, disrupting the RRE-Rev interaction affords a viable strategy to inhibit HIV-1 replication. There has been a wide array of ligands designed to target RRE, including small molecules, heterocycles, aminoglycosides, antisense oligonucleotides, RRE decoys, peptide nucleic acids, metallopeptide complexes, cyclic peptides, α-helical peptidomimetics, and various bifunctional conjugates, but to date none has seen clinical success.13–19 This is in part due to the challenges inherent in targeting RNA that hinder the design of selective and potent ligands.20, 21 Several factors contribute to this difficulty: (i) the large surface area of RNAs available for binding interactions, (ii) limited permeability and stability of oligonucleotide-based RNA ligands, (iii) high binding affinities of cognate ligand:RNA interactions, and (iv) conformational dynamics inherent in RNA that make it difficult to both target the active conformation as well as obtain reliable secondary or tertiary structures for designing effective ligands.20, 22, 23
We surmised that branched peptides (BPs) could bind the tertiary structure of folded fragments of HIV-1 RNAs via multiple contact, allowing for increased surface area interactions that would improve both selectivity and affinity.24 An added benefit is that branched peptides are refractory to proteolysis compared to their linear counterparts.25 To this effect, our first BP library was developed to target a conserved HIV-1 RNA structure, the transactivation response element (TAR).24, 26 These BPs bound TAR in the sub-micromolar range, and were both non-toxic and cell permeable. We subsequently turned our attention to targeting the stem-loop IIB of HIV-1 RRE (RRE IIB), the high-affinity initial binding site for Rev.27 This second generation 46,656-member branched peptide boronic acid (BPBA) library exploited the use of two unnatural amino acids containing boronic acid to target RNA via formation of a reversible covalent bond between the empty p-orbital of boron and the 2ʹ-hydroxyl of the RNA.28, 29 One compound elicited a conformational change in the RNA structure upon binding and inhibited production of HIV p24 capsid protein in culture.30 As these BPs bound in the micromolar range, a second BPBA library consisting of 4,096 members was rapidly generated to improve binding affinity via incorporation of acridine, an intercalating moiety.31 We observed a significant improvement in binding affinity. However, selectivity towards RRE IIB was diminished, most likely due to lack of structural diversity in the library.
Herein, we disclose a unique peptide library that is composed of diverse chemical structures biased towards their propensity to interact with the tertiary fold of RNA. High-throughput screening generated several hit BPs that revealed sequence motifs that indicated preferred modes of binding. Inhibition of Rev-RRE RNA interaction was demonstrated in cell culture with one of the BPs showing much better inhibition than the others. Biochemical probing techniques identified binding of the BP to RRE IIB (primary Rev binding site) as well as the distant secondary binding site (IA). These studies suggest that the BP sterically blocks multimerization of Rev on the RRE and thus inhibit HIV mRNA nuclear export, a critical step in HIV-1 replication.
RESULTS AND DISCUSSION
Library Design and on-Bead High-Throughput Screening.
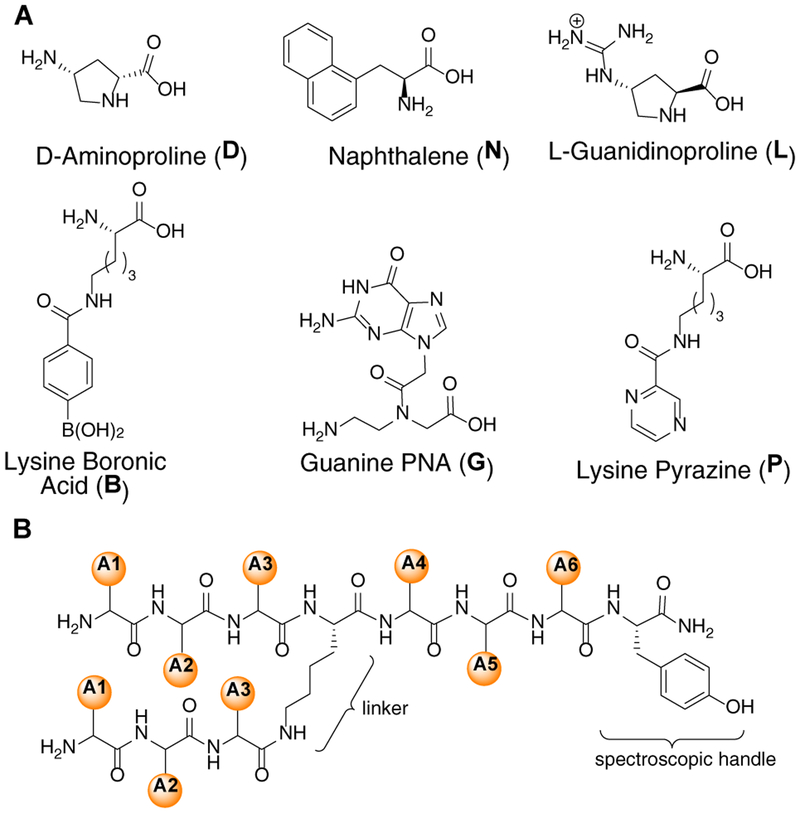
Our previous work with BP libraries revealed several guiding principles regarding opportunities for increasing affinity and selectivity towards the target RNA structure. Thus, we applied these principles using unnatural amino acids to facilitate a diversity of interactions with RRE IIB RNA (Figure 1A). For example, lysine boronic acid (B) was utilized to facilitate reversible covalent bonding between the Lewis acidic boron moiety and the RNA 2’-OH or Lewis basic nucleobases.28 L-guanidinoproline (L) and D-aminoproline (D) were selected as conformationally constrained and electrostatic mimetics of arginine and lysine. The incorporation of D-amino acids served to increase stereochemical diversity as well as to improve proteolytic stability of the peptides.32 Lysine pyrazine (P) and 1-Naphthylalanine (N) were incorporated to provide π-π stacking with nucleobases while lysine pyrazine can serve as a hydrogen bond donor/acceptor. Finally, the guanine peptide nucleic acid (G) was expected to initiate stacking and/or hydrogen bonding, and we envisioned that this PNA could also base pair with adenine and guanine.33 The branched peptide library was synthesized, screened, and sequenced using established protocols (see Supporting Information for details).24, 28, 34 The library was prepared such that there were three variable amino acid positions at both the N- and C-termini (A1–A3 and A4–A6, respectively) linked by a lysine core, and each variable position was composed of six unnatural amino acids. Tyrosine was incorporated at the C-terminus to quantify peptide yields spectroscopically. This yielded a one-bead- one-compound library of 46,656 unique sequences (Figure 1B).
Figure 1.
(A) Structures of unnatural amino acid. (B) Schematic of 46,656 membered branched peptide library.
The presence of diverse moieties on the bead surface would allow interaction of DY547-labeled RRE IIB with all possible modes of binding. Nonspecific binding was minimized by blocking the on- bead library with excess bovine serum albumin and competitor tRNA, followed by the introduction of DY547-labeled RRE IIB during the second incubation. Increased fluorescence of beads, which was monitored by confocal microscopy, suggested binding to the target RNA. A total of 90 BPs were consequently selected, photocleaved, and sequenced by MALDI-TOF MS/MS.34
Binding Affinities of Hit Branched Peptides to HIV-1 RRE RNA.
The 22 selected hits were synthesized by solid phase peptide synthesis (SPPS) and their dissociation constants (Kd values) were measured by using a 2-AP displacement assay (Table 1).35 Selection criteria of hits for further biophysical characterization included i) repeated detection during the screening process, ii) frequency of the peptide sequence, iii) presence of repeated N- or C-terminal motifs within the peptide sequence, or iv) presence of a diverse amino acid sequence.
Table 1.
Dissociation Constant of BPs
| Entry | BPs | Sequence | Kd (μM) | Theoretical Mass | Mass Observed |
|---|---|---|---|---|---|
| 1 | 4B3 | (DLL)2LGBY | 0.41 ± 0.05 | 1989.9 | 1989.7 |
| 2 | 13B8 | (LDL)2LGBY | 0.63 ± 0.03 | 1989.9 | 1989.3 |
| 3 | T1 | (LDN)2GGNY | 0.66 ± 0.02 | 2014.9 | 2015.5 |
| 4 | 11F6 | (DLD)2DLDY | 0.72 ± 0.03 | 1443.8 | 1443.7 |
| 5 | 4A5 | (GGD)2PGLY | 0.88 ± 0.02 | 2377.1 | 2376.7 |
| 6 | 10B10 | (LDD)2PGDY | 1.02 ± 0.07 | 1702.9 | 1702.4 |
| 7 | 16D9 | (PDL)2GLDY | 1.11 ± 0.05 | 1866.9 | 1867.6 |
| 8 | RA7 | (LDL)2PPGY | 1.12 ± 0.08 | 1908.9 | 1908.4 |
| 9 | 19B3 | (LDG)2GPBY | 1.18 ± 0.05 | 2343.0 | 2342.9 |
| 10 | RA8 | (LDP)2LLPY | 1.30 ± 0.03 | 1851.9 | 1851.4 |
| 11 | 7F8 | (LDD)2LPDY | 1.32 ± 0.06 | 1565.9 | 1566.2 |
| 12 | RA9 | (LDB)2NPLY | 1.50 ± 0.04 | 2215.0 | 2214.7 |
| 13 | 18D10 | (GDP)2LLPY | 1.64 ±0.03 | 2126.0 | 2125.7 |
| 14 | TT1 | (LDD)2GGDY | 1.76 ± 0.13 | 1759.9 | 1760.1 |
| 15 | 5B9 | (LDL)2DDBY | 2.57 ± 0.03 | 1767.9 | 1768.2 |
| 16 | 7F5 | (LDD)2PNDY | 2.98 ± 0.11 | 1068.9 | 1069.1 |
| 17 | 8B1 | (DBN)2PGDY | 3.18 ± 0.07 | 2353.0 | 2352.2 |
| 18 | 5E12 | (LDD)2BBLY | 4.46 ± 0.20 | 2007.9 | 2007.5 |
| 19 | 7C7 | (LBN)2PNLY | 4.96 ± 0.48 | 2385.1 | 2385.5 |
| 20 | 18D1 | (LBB)2PBNY | >10 | 3019.2 | 3019.3 |
| 21 | 6B3 | (GNG)2BBLY | >10 | 2809.1 | 2809.1 |
| 22 | 15B7 | (PPP)2PPPY | >10 | 2416.2 | 2416.5 |
In general, the library afforded BPs in the low micromolar range, but five demonstrated Kd values in the sub-micromolar range (entries 1−5). Sequence 4B3 ((DLL)2LGBY) was the best hit with a Kd value of 410 nM. L-Guanidinoproline (L) and D-aminoproline (D) were strongly preferred at the N terminus and were the most preferred amino acids in the library. This is not unexpected as the N-terminus of the arginine rich motif (ARM) of Rev interacts with the Rev-binding element (RBE) of RRE.36 However, a large number of positively charged residues in the BPs did not necessarily correlate with high binding affinity (compare entries 2 and 3 with 15 and 18) indicating that electrostatic interaction alone is insufficient for binding and suggests that alternative modes of binding coupled with sequence were responsible for some of the low Kds observed. Hit sequences with very few or no basic residues (entries 20–22) demonstrated the weakest binding affinities for RRE IIB, possibly reflecting limited electrostatic interactions. N was preferred at the N-terminus, in particular at A3, revealing a preference for stacking and aromatic interactions of the peptide with the RNA in this position. P and G were preferred at the C-terminus in positions A4 and A5. This suggests that aromatic functional groups capable of hydrogen bonding are crucial at these positions.
Gratifyingly, there were several sequence motifs present in the hit peptides that indicated sequence and structural requirements in demonstrating affinity for the RNA target. For example, at the C terminus, analysis of BPs containing the motif (LDD)2*PXDY (entries 6 and 16) revealed aromatic residues (G and N) at variable position A5. Substitution of guanine PNA with unnatural amino acid containing naphthalene (entry 16) produced a 3-fold decrease in binding affinity. We hypothesize that the preference for guanine PNA stems from possible aromatic stacking interaction as well as H- bonding engagement with the RNA. Other peptides such as 4B3 and 13B8 displayed good binding affinity and shared motifs at the N-terminus, (entries 1 and 2). These isomers differed in the location of D and L in positions A1 and A2 with only a 2-fold difference in Kd. As both amino acids are positively charged, the lack of amino acid preference in these positions suggests that electrostatic interaction is an important parameter within the motif (XXL)2*LGBY.
Additionally, comparable binding affinities were found with both RA8 and 18D10 (entries 10 and 13), which share the sequence (XDP)2*LLPY. Considering that variable position A1 contains either L or G and little change in binding affinity is observed, it is possible that the guanidine functional group in plays a similar role in both peptides. This may also hold true for two other BPs with similar binding affinities (entries 5 and 6), where the major difference in sequence involves substitution of G with L or D in variable positions A1 and A2, respectively. Collectively, these findings demonstrated that certain sequence motifs show a preference in RRE IIB binding and warranted further investigation to elucidate the binding interactions of these BPs with the RNA.
In vitro evaluation of BPs.
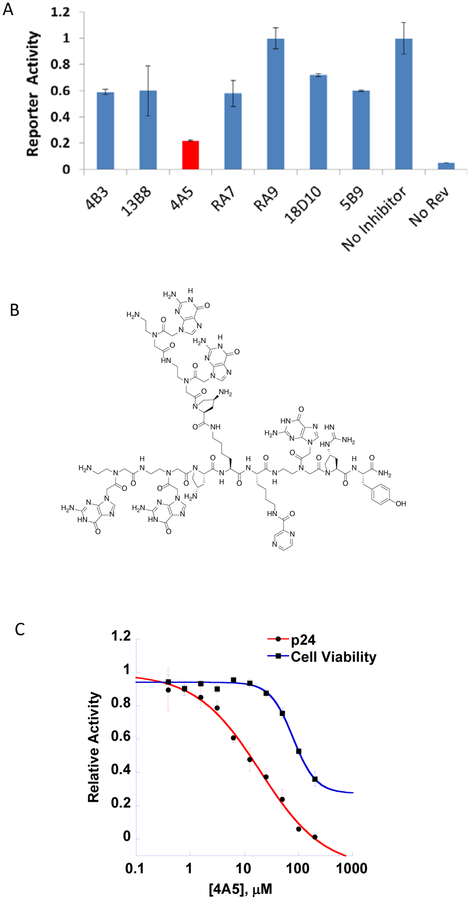
With sub-micromolar Kds observed for several of our BPs against RRE IIB, the BPs were tested in cell culture to determine their toxicity and ability to inhibit the primary RRE function, Rev-dependent nuclear export of intron-containing RNAs using a reporter assay system. In this assay, HEK 293T cells were transiently transfected with a Rev and CMV promoter-driven HIV-1 GagPol-RRE plasmid.37, 38 The expresion of HIV-1 Gag, present in an intron in the transcribed mRNA, requires successful Rev-RRE dependent nuclear export. This can be quantified by an ELISA for p24, a component of the Gag protein. We first tested the BPs at 70 μM that revealed seven BPs that displayed inhibition of p24 (>50%) (data not shown). Encouraged by these results, a subsequent screening was performed at 30 μM for 48 h to determine whether inhibition of p24 could be maintained at lower BP concentration over a longer period. Fortunately, after 48 h incubation, the Rev-RRE reporter activity in the presence of 4A5 was reduced by 80% (Figure 2A). To further characterize the effect of 4A5 (Figure 2B), a dose-dependent p24 inhibition assay was conducted in parallel with cell viability assay. As shown in Figure 2C, increased inhibition was observed as the concentration of 4A5 raised from 0.3 to 200 μM resulting in an IC50 value of 15.4 μM. Fortunately, the MTT assay indicated an CC50 of 135 μM, which suggests that the inhibition of p24 production is dependent on Rev-RRE interaction and not cellular toxicity. Because of the ability of 4A5 to inhibit Rev-RRE interaction, we further investigated its selectivity as well as determine its potential binding sites on the RRE RNA.
Figure 2.
A) HIV-1 Rev-RRE dependent nuclear export reporter activity assay in transiently transfected 293T cells measured as expression of HIV-1 p24 protein using ELISA. The assay was performed in the presence of 30 μM BPs. B) Structure of 4A5. C) Dose-response curves of HIV-1 Rev-RRE reporter activity and cell viability of HEK 293T cells with 4A5. Data points are mean ± standard deviation.
Selectivity of Hit BP 4A5 toward RRE IIB Tertiary Structure.
Selectivity was determined by a competition assay using 2-AP-labeled RRE IIB RNA in the presence of 10-fold excess unlabeled RNA variants: No Bulge RRE and TAR RNA as well as a homologous sequence RRE IIB DNA (see Supporting Information, Figure S3). The RNA variants probe potential binding sites in RRE and determines selectivity against a similar stem-loop bulge RNA (TAR) whereas the DNA counterpart determines selectivity regarding the three-dimensional structure. As shown in Table 2, 4A5 showed selectivity for RRE IIB in the presence of RRE DNA and RNA variants. These results were consistent from our previous work where, for example, we demonstrated that the loop and bulge in the RRE-IIB construct are essential for binding.29 For 4A5, an approximate 3-fold loss of binding affinity was observed when excess of No Buldge RRE was utilized suggesting the upper stem region of the RRE IIB arm might also contain the binding site for 4A5 (vide infra). Furthermore, the BP also showed less selectivity for RRE IIB in the presence of TAR RNA, with approximately a 4-fold decrease in binding affinity observed. Both TAR RNA and RRE IIB RNA share a similar stem-loop that contains a consensus sequence UGGG; this RNA sequence may play an important role in 4A5 binding.
Table 2.
Dissociation constant of 4A5 in presence of indicated RNAs.
| RNAs | Kd (μM) |
|---|---|
| Wild Type RRE IIB (without competitors) | 0.88 ± 0.02 |
| 10X RRE DNA | 0.99 ± 0.04 |
| 10X No Bulge RRE RNA | 2.04 ± 0.21 |
| 10X TAR RNA | 3.09 ± 0.10 |
Determination of BP:RNA binding sites by RNase protection assay.
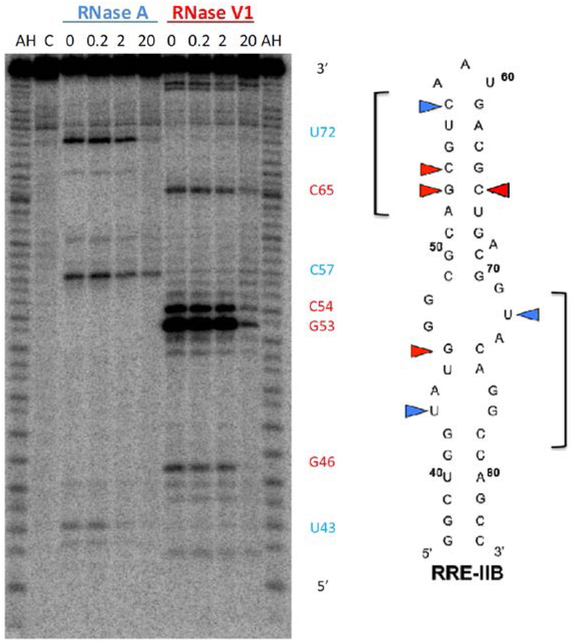
Binding sites of lead compound 4A5 on RRE IIB were investigated via footprinting assays. The RRE IIB RNA construct contains Stem Loop II, which is the primary Rev binding site of the RRE. Nucleotides outside RRE IIB that have been shown to maximally affect Rev-RRE function in previously reported Rev-RRE functional studies were also included.36, 38, 39 In this assay, increasing concentrations of 4A5 (0–20 μM) were incubated with 5ʹ−32P labeled RRE IIB, which was then subjected to enzymatic cleavage by RNase A, and VI (Figure 3). Protection of the RNA from enzymatic cleavage is indicative of potential contact points between the BP and RNA. For 4A5, the most noticeable cleavage was caused by RNase VI in the upper stem region of RRE IIB between bases A52 and G55, as this enzyme cleaves double-stranded RNA. 4A5 revealed similar binding sites along RRE IIB compared to previously characterized BPs from other 3.3.4 BP libraries.29 A concentration dependent protection from RNase VI in the upper stem region of the RNA was observed, as evidenced by the reduction of enzymatic cleavage seen at G46, G53, C54, and C65. This protection pattern further corroborated the decreasing binding affinity when No Bulge RRE was introduced as the competitor in the selectivity assay. Moreover, protection of RRE IIB from cleavage by RNase A, which cleaves the 3ʹ end of pyrimidines C and U, was also observed in the internal loop region of the RNA (U43 and U72). Interactions of the BP with this region of the RNA were not surprising, as this is also the region where Rev is shown to bind RRE IIB.10, 40 Overall, 4A5 interacts with both internal loop and upper stem / apical loop regions of RRE IIB, spanning a significant portion of the entire stem-loop structure.
Figure 3.
RNase protection assay of RRE IIB. The gel depicts the autoradiogram of alkaline hydrolysis (AH) and RNase protection experiments using RNases A, and V1 with increasing concentration of 4A5 (μM). Colored triangles highlight bases protected from cleavage by RNase A (blue) and RNase V1 (red).
SHAPE analysis of 4A5:RNA interactions.
The RRE is highly structured with likely long-range contacts and orchestrates cooperative binding of multiple Rev molecules on the RRE.41 Such long range interactions could influence the binding of 4A5 on SLII. To investigate this possibility, we expanded the 4A5 footprinting study to the much larger 234 nt RRE, the minimal sized functional RRE,42 using SHAPE-MaP (selective 2′ -hydroxyl acylation analyzed by primer extension and mutational profiling). SHAPE-MaP interrogates RNA flexibility using SHAPE chemistry (2’-hydroxyl nucleobase accessibility to an electrophile) and the ability of reverse transcriptase to record the conformational flexibility as mutations.43 The sites and frequency of these mutations are used to create a SHAPE reactivity profile. Nucleotides with significant decrease in reactivity in the presence of the BP are most likely bound by the BP. Free RNA and RNA incubated with various concentrations of 4A5 were treated with electrophile IM7, and reactivity differences were plotted as a function of nucleotide position. A cut-off of +/− 0.5 was arbitrarily chosen to identify nucleotides that were rendered more flexible or constrained in the presence of 4A5.
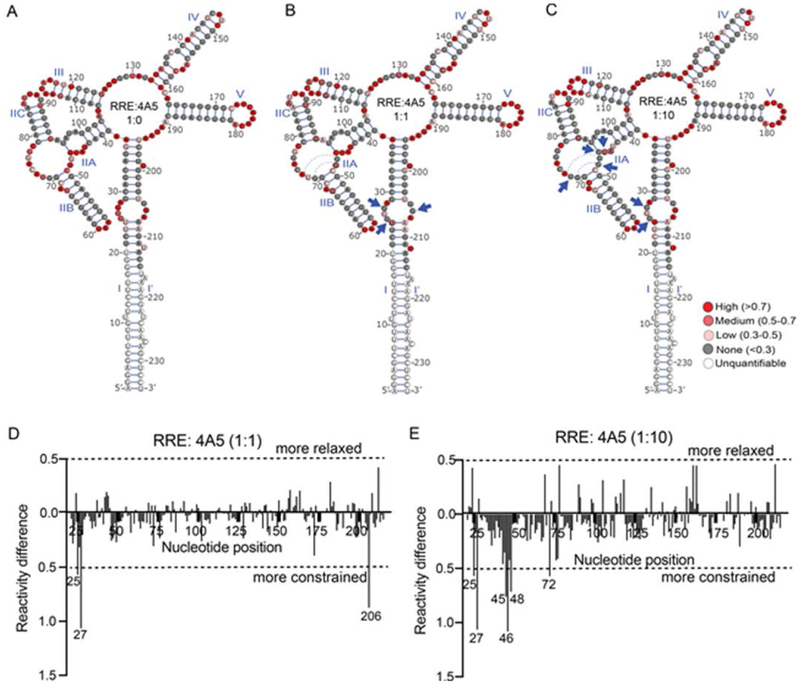
As shown in Figure 4, a good degree of protection was also observed in the secondary Rev binding site in the SLI region of the RRE. This protection is enhanced when the BP level is increased. At 1:1 RNA:4A5 ratio, decreased reactivity was observed at nucleotides 25, 27 and 206 suggesting that the peptide initially makes contacts on SLI. As these three nucleotides are part of a loop region that constitute the secondary Rev binding site, 4A5 may compete with Rev for binding at these positions. When RNA:4A5 ratio was increase to 1:10, the protection effect extended to nucleotides 45, 46, 48 and 72, which fall in the SLII region. These interactions between 4A5 and these nucleotides were also observed in the RNase protection assay, which provide evidence that direct and specific interactions between the RNA and peptide exists. Interestingly, the degree of modification at nucleotides 25 and 27 is not altered but the protection at nucleotide 206 is lost. We speculate that the binding of 4A5 to both the SLI and SLII regions induce local structural changes in SLI that renders nucleotide 206 inaccessible. However, nucleotides 46 and 48 that are protected in the presence of the BP have been previously shown to be essential for primary Rev binding. These two nucleotides make the two non-Watson-Crick base-pairs that widen the major groove of the RRE in that region by 5 Å allowing the helical Rev ARM to make specific contact with the RRE.36, 44 Therefore, the BP 4A5 might prevent Rev binding on the RRE directly by occupying the active Rev binding sites and/or allosterically by preventing the conformational change in the RRE required for Rev binding. Overall, since both the SLI and SLII are important Rev binding sites on the RRE,38, 45, 46 we hypothesize that the inhibitory effect of the BP on HIV replication is most likely due to the obstruction of Rev occupancy on these sites.
Figure 4.
SHAPE-MaP reactivity profiles of 234 nt RRE RNA in the presence of peptide 4A5. Upper panel:Reactivity values of the RRE superimposed on the 5 stem-loop structure of NL43 RRE (A) in the absence of 4A5, (B) in the presence of equal molar concentration of 4A5, and (C) in the presence of 10-fold molar excess of 4A5. The solid blue triangles point to nucleotides whose reactivity changes significantly in the presence of 4A5. Lower panel: Reactivity difference profile generated for (D) equimolar RRE:peptide ratio and (E) 1:10 RRE:peptide molar ratio by subtracting reactivity values of the RRE in the absence of the 4A5 from those in the presence of 4A5. Here +/− 0.5 difference is used as cut-off value to identify nucleotides that are more relaxed or more constrained in the presence of 4A5. Dotted blue arcs represent the non-canonical bonds shown to form upon Rev binding.
CONCLUSION
In summary, a BP library consisting of unnatural amino acids was developed to target HIV-1 RRE IIB RNA. These BPs bound RRE IIB in the sub-micromolar range, with several hit BPs demonstrating selectivity against various competitor nucleic acid structures. Structure-activity relationship studies revealed N-terminal, C-terminal, and overall sequence motifs that were important for the potent binding affinities observed. Among the BPs investigated, 4A5 displayed dose-dependent inhibition of HIV-1 p24 production in vitro in a Rev-RRE dependent manner. Biochemical investigations via RNase protection and SHAPE-MaP assays suggested binding of 4A5 in SLI and SLII regions of RRE, both of which are binding sites for Rev. The binding stoichiometry further indicates local RNA conformational changes upon peptide binding. Overall, the mode of action of 4A5 involves binding to RNA, inducing conformational changes, and competing for the Rev protein binding sites.
EXPERIMENTAL SECTION
1. General.
Unless otherwise noted, reagents and solvents were used as received from commercial suppliers. Mass spectra analyses were performed using Agilent 6200 LC-TOF-MS in ESI or APCI mode when appropriate. The purity of the compounds was analyzed by HPLC using an Agilent 1200 Series HPLC and Thermo TSQ Quantum mass spectrometry. All peptides used were >95% pure except for RA8 (see Supporting Information for details).
2. Synthesis of 3.3.4 branched peptide library with all unnatural amino acids.
Standard solid phase peptide synthesis techniques were used to generate the 3.3.4 library via the split and pool method using the previously described procedure.26 N-α-Fmoc protected L and D-amino acids, PyOxim (Novabiochem) and N,N-Diisopropylethylamine (DIEA, Aldrich) were used in coupling reactions. Fmoc-Tyr(tBu)-OH (Novabiochem), Fmoc-PNA-G(Bhoc)-OH (GPNA) (Link Technologies, Ltd.), and Fmoc-3-(1-naphthyl)-L-alanine (NAL1) (Chem Impex International Inc.) were purchased and used directly. The synthesis and full characterization of Fmoc-N-ε-(4- boronobenzoyl)-L-lysine (KBBA) and Fmoc-N-ε-(pyrazine-2-carboxamide)-L-lysine (KPYR) were performed in our lab. (2S,4R)-N-α- Fmoc-4-N,Nˊ-di-Boc-guanidinoproline (LPRO) and Fmoc-ANP-OH were synthesized as previously reported, and (2R,4R)-N-α-Fmoc-4-N-Bocaminoproline (DPRO) was synthesized using protocols previously reported for the (2S,4R) diastereomer.47, 48 Three copies of library were prepared simultaneously by using a three-fold excess of Tentagel Macrobead-NH2 resin (2.13 g, 0.51 mmol) (Peptides International). The resin was swollen in DCM (20 mL, 2 × 15 min) followed by DMF (20 mL, 15 min). The photocleavable linker Fmoc-ANP-OH (662 mg, 1.53 mmol) was first coupled to the resin in DMF for 3 h in the presence of PyOxim (806 mg, 1.53 mmol) and DIEA (530 μL, 3.06 mmol). After coupling, the resin was washed with DMF (2 × 20 mL, 1 min each), DCM (2 × 20 mL, 1 min each) and DMF (20 mL, 1 min). The same washing procedure was applied after every step. Then, 20% piperidine in DMF (20 mL, 2 × 10 min) was used for Fmoc deprotection. A Kaiser test was used after each coupling and deprotection step to confirm reaction completion. N-Fmoc amino acids (3 equiv.), PyOxim (3 equiv.), and DIEA (6 equiv.) were added to each reaction vessels in DMF and coupled for 30 min. Fmoc-Lys(Fmoc)-OH (Novabiochem) was used as a branching unit, and molar equivalencies of reagents were doubled in coupling reactions after installation of the branching unit. After Fmoc deprotection of the N-terminal amino acids, the resin was bubbled in a phenylboronic acid solution (0.2 g/mL, 1.6M) in DMF overnight to remove the pinacol groups of boron-containing side chains. Finally, the resin was treated with 95:2.5:2.5 TFA (Trifluoroacetic acid, Acros)/H2O/TIPS (Triisopropylsilane, Acros) (v/v/v) for 3 h. After deprotection, the resin was washed extensively with DMF, DCM, and MeOH before drying and storing at −20 °C.
3. On-bead screening assay.
DY547 labeled HIV-1 RRE-IIB RNA (5′-DY547- GGCUGGUAUGGGCGCAGCGUCAAUGACG CUGACGGUACAGGCCAGCC-3′) was purchased from Dharmacon and prepared according to the manufacturer’s protocol. To account for autofluorescence of Tentagel Macrobead-NH2 resins, control peptide (KKK)2*KKKY was incubated in 100 nM DY547 labeled HIV-1 RRE-IIB RNA for 3 hr in 1X phosphate buffer (10 mM potassium phosphate, 100 mM KCl, 1 mM MgCl2, 20 mM NaCl, pH 7.0). These beads were washed extensively and placed into a sterile 96-well plate (Nunc) and imaged by both a Zeiss Axiovert 200 fluorescent microscope under a rhodamine filter and a Zeiss LSM 510 microscope set to longpass 585. Fluorescence intensity of these RNA-incubated beads was compared with auto-fluorescence of the tentagel resins, and the detector sensitivity was adjusted for removal of auto-fluorescence. Library resins were placed into a 1.5 mL nonstick microfuge tube (Fisher) and incubated with various RNAs in phosphate buffer at a 600 μL final volume, with continual mixing by a Barnstead/Thermolyne Labquake rotisserie shaker. Beads were first treated with 1 mg/mL bovine serum albumin (BSA) (New England BioLabs), 2.5 mg/mL tRNA (Roche) (~1,000-fold molar excess to RRE-IIB RNA), 100 nM of Loop A/B/Bulge A Deleted RRE (5’- GGCTGGCAGCGTCATTGACGCTGCCAGCCCTATAGTGAGTCGTATTACAT-3’), 100 nM of LoopB/Bulge A Deleted RRE (5’-GGCTGGCCTGCAGCGTCATTGACGC TGCATACCAGCCCTATA GTGAGTCGTATTACAT-3’), and 100 nM of Stem B Deleted RRE (5’-GGCTGGCCTGTACCG TCACATTGTGCGCCCATACCAGCCCTATAGTGAGTCGTATTACAT-3’) for 5 h at room temperature to block nonspecific binding interactions. Subsequently, beads were washed 5 times with phosphate buffer (100 mM, pH = 7.4) and incubated with 100 nM DY547 labeled RRE-IIB RNA in phosphate buffer for 3 h at 4 °C. After the final incubation, beads were extensively washed with buffer and imaged under a confocal microscope in a 96-well plate using the previously optimized settings. Hit resins were isolated, washed by DMF, and photocleaved with clear non-stick 0.5 mL microfuge tubes in 20 μL of 1:1 MeOH: H2O (v/v) by irradiation at 365 nm with a 4W handheld UV lamp for 1 h. The supernatant was retained and subjected to MALDI-TOF analysis.
4. Peptide synthesis, purification and characterization.
Solid phase synthesis of branched peptides was achieved using N-α-Fmoc protected L- and D-amino acids (3 equiv.), Pyoxim (Novabiochem) (3 equiv.) in DMF as coupling reagent, and DIEA (Aldrich) (6 equiv.) on Rink amide MBHA resin (100–200 mesh) (Novabiochem) with 0.6 mmol/g loading. The Fmoc group was deprotected with 20% piperidine in DMF. Fmoc-Lys(Fmoc)-OH was used as a branching unit, and molar equivalencies of reagents were doubled in coupling reactions after installation of the branching unit. Solid phase synthesis was performed on a vacuum manifold (Qiagen) outfitted with 3-way Luer lock stopcocks (Sigma) in either Poly-Prep or Econo-Pac polypropylene columns (Bio-Rad). The resin was mixed in solution by bubbling argon during all coupling and washing steps. After Fmoc deprotection of the N-terminal amino acids, the resin was bubbled in a phenylboronic acid solution (0.2 g/mL, 1.6 M) in DMF overnight to remove the pinacol groups of boron-containing side chains. Finally, the resin was treated with 95:2.5:2.5 TFA (Trifluoroacetic acid, Acros)/H2O/TIPS (Triisopropylsilane, Acros) (v/v/v) for 3.5 h. The supernatant was dried under reduced pressure, and the crude peptide was triturated from cold diethyl ether. Peptides were purified using a Jupiter 4 μm Proteo 90 Å semiprep column (Phenomenex) using a solvent gradient composed of 0.1% TFA in Milli- Q water and HPLC grade acetonitrile. Peptide purity was determined using a Jupiter 4 μm Proteo 90 Å analytical column (Phenomenex), and peptide identity was confirmed by MALDI-TOF analysis. Peptide concentrations were measured in nuclease free water at 280 nm using their calculated extinction coefficient. The extinction coefficient for lysine pyrazine was experimentally determined (3374 M−1cm−1) by monitoring the absorbance of pyranoic acid in nuclease free water at 280 nm. Previously reported extinction coefficents were used for guanine PNA (7765 M−1cm−1) and the napthalene derivative (3374 M−1cm−1) at 280 nm in aqueous medium.49, 50
5. Transcription of HIV-1 RRE IIB RNA and RRE IIB variants.
Wild-type and mutant RRE-IIB RNA were transcribed in vitro by T7 polymerase using the Ribomax T7 Express System (Promega) following previously reported techniques.29, 51 The antisense template, sense complementary strand (5’- ATGTAATACGACTCACTATAGG-3’) and RRE-IIB reverse PCR primer (5’- GGCTGGCCTGTAC- 3’) were purchased from Integrated DNA Technologies. The antisense templates that were used are as follows: RRE IIB RNA 5’- GGCTGGCCTGTACCGTCAGCGTCATTGACGCTGCGCCCATACCAGCCCTATAGTGAGTCGTATT ACAT-3’; Loop A/B/Bulge A Deleted 5’- GGCTGGCAGCGTCATTGACGCTGCCAGCCCTATA GTGAGTCGTATTACAT-3’. HIV-1RRE-IIB Wild Type was PCR amplified using HotstarTaq DNA polymerase (Qiagen) followed by a clean-up procedure using a spin column kit (Qiagen). For preparation of all other sequences, the antisense DNA template was annealed with the sense DNA complementary strand in reaction buffer at 95 °C for 2 min then cooled on ice for 4 min. T7 transcription proceeded at 42 °C for 1.5 h. After transcription, DNA templates were degraded with DNase at 37 °C for 45 min and the RNA was purified by a 12% polyacrylamide gel containing 7.5 M urea. RNA of interest was excised from the gel and eluted overnight in 1x TBE buffer at 4 °C. The sample was desalted using a Sep-Pak syringe cartridge (Waters Corporation) and lyophilized. Purified RNA was stored as a pellet at −80 oC. RRE IIB DNA (5′- GGCTGGTATGGGCGCAGCGTCAATGACGCTGACGGTACAGGCCA GCC-3′) was purchased from Integrated DNA Technologies and stored at −20 °C.
6. Preparation of 32-P labeled RNA.
HIV-1 RRE IIB RNA was dephosphorylated with calf intestinal phosphatase (CIP) in NEBuffer 3 (New England Biolabs) according to the manufacturer’s protocol. The product was recovered by phenol extraction followed by ethanol precipitation. 10 pmol of dephosphorylated RNA was 5’ end-labeled with 20 pmol of [γ−32P] ATP (111 TBq mol−1) and 20 units of T4 polynucleotide kinase in 70 mM Tris•HCl, 10 mM MgCl2, and 5 mM dithiothreitol, pH 7.6. The mixture was incubated at 37 °C for 30 min, and then at room temperature for 20 min. The kinase was heat-inactivated at 65 °C for 10 min. The RNA was recovered by ethanol precipitation, and purity was confirmed by denaturing PAGE and autoradiography.
7. 2-Aminopurine (2-AP) assay.
All fluorescence spectra were measured on a Varian Cary Eclipse fluorescence spectrophotometer using a xenon flash lamp with a thermoelectrically controlled cell holder. The excitation slit width and the emission slit width was 10 nm. Excitation of the sample was performed at 310 nm and fluorescence spectra were collected from 340 nm to 450 nm. A quartz cell of 1 cm path length transparent on three sides (Starna Cells, Inc.) was used.
Dissociation constants for the branched peptides were determined by following the decrease in fluorescence at 372 nm of 2-AP labeled RRE IIB RNA (5’- CUGGUAUGGGCGCAGCGUCAA UGACGCUGACGG-2AP-ACAGGCCAGCC-3’, Integrated DNA Technologies) as a function of increase in branched peptide concentration.35 2-AP labeled RRE IIB was refolded by heating at 95 °C for 3 min and snap-cooling on ice. The concentration of RNA was fixed at 0.1 μM during titration and the peptide concentration was varied from 0–20 μM. Both peptides and RNA were prepared with 0.2 μm sterile-filtered 1X phosphate buffer (10 mM potassium phosphate, 100 mM KCl, 1 mM MgCl2, 20 mM NaCl, pH 7.0). Binding data were analyzed using a hill equation (Eq. 1) with Kaleidagraph (Synergy Software).52 In this equation, b and y are the fluorescence emission intensities of the RNA in the absence and presence of peptide; m is the fluorescence emission intensity of the RNA in the presence of an infinite drug concentration; R and x are the total concentrations of the RNA and peptide; K is the Kd value of the peptide binding to the RNA; n is the apparent cooperativity. Each experiment was performed at least in duplicate and error bars represent the standard deviation calculated over the replicates.
| Eq. 1 |
8. RRE functional activity reporter assay.
The ability of the peptides to inhibit Rev-RRE function was evaluated in cell culture using HEK 293T cells transiently transfected with a Rev-expressing plasmid and a CMV promoter-driven GagPol-RRE plasmid. HEK 293T cells in a 96-well plate were transfected with 1 ng of pCDNA4TO-3xFlag-Rev, 100 ng of pCMV-GagPol-RRE, 5 ng of a pCDNA4TO-firefly luciferase (to monitor transfection efficiency) and 20 ng of pBS carrier DNA using Polyjet transfection reagent (SignaGen). 5 hours after transfection, peptides were diluted to the desired concentration and added to the appropriate wells. Cells were lysed ~48 hours after transfection and transfection efficiency was monitored by measuring firefly luciferase activity. Rev- RRE reporter activity was quantified by measuring intracellular p24 levels using ELISA.
9. Cell viability assay (MTT).
Cell viability was monitored in parallel by an MTT assay starting with an identical set up as the reporter assay. For the MTT assay, ~48 hours after transfection, 10 μL of the MTT reagent (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide, Acros Organics), prepared at 5mg/mL in phosphate-buffered-saline, was added to each well. Following incubation at 37°C in an incubator for 4 hours, 100 μL of solubilization solution (10% sodium dodecyl sulphate, 0.01 N HCl) was added to the wells of the 96-well plate. After incubating overnight at 37°C in an incubator, the absorbance was read at 570 nm in a microplate reader.
10. Nuclease protection assays.
RNA was first refolded by heating a solution of 5’−32P-labeled RRE-IIB (10 nM) and excess unlabeled RRE-IIB (200 nM) at 95 °C for 3 min and snap-cooling on ice. Refolded RNA was incubated on ice for 4 h in a solution containing the BP and buffer composed of 10 mM Tris, pH 7, 100 mM KCl, and 10 mM MgCl2. RNase (Ambion) was added to the solution, which was further incubated on ice for 10 min (0.002 Units RNase V1), or 1 h (1 Unit RNase T1; 20 ng RNase A). Inactivation/precipitation buffer (Ambion) was added to halt digestion, and the RNA was precipitated with ethanol and collected by centrifugation at 13,200 rpm for 15 min. Precipitated RNA was redissolved in tracking dye and fractionated on a 12 % PAGE containing 7.5 M urea. The gel was dried at 80 °C for 1 h and imaged by autoradiography.
11. SHAPE analysis of 4A5.
RNA preparation:
234 nt NL4–3 5ʹ-RRE RNA was prepared by in vitro transcription using the MegaShortScript kit (Ambion/Life Technologies) per manufacturers’ recommendations. DNA template used in the transcription reaction was generated by PCR from a proviral pNL4–3 plasmid with high fidelity platinum Taq DNA polymerase (Invitrogen) using forward oligo 234RREf (5’ AGCGTACTTAATACGACTCACTATAGGGAGGAGCTTTGTTCCTTG) and reverse oligo 234RREr (5’ AGGAGCTGTTGATCCTTTAG). The forward primer was designed to introduce T7 promoter sequence at the 5’ end of the RRE. RNA was then treated with Turbo DNase I for 1 h at 37 °C, heated at 85 °C for 2 min and run on a denaturing gel (5% polyacrylamide (19:1), 1x TBE, 7 M urea) at constant temperature (45 °C, 30 W max). RRE RNA was excised, electroeluted at 200 V for 2 h at 4 °C, ethanol precipitated and stored at −20 °C in TE buffer (10 mM Tris, pH 7.6; 0.1 mM EDTA) prior to use.
RNA folding and modification:
5 pmoles of RNA, treated with renaturation buffer (final concentration: 10 mM Tris pH 8.0, 100 mM KCl, 0.1mM EDTA), in a volume of 5 μL was heated to 95°C for 2 mins and snap cooled on ice. Renatured RNA was incubated with RNA folding buffer (final concentration: 70 mM Tris pH 8.0, 180 mM KCl, 0.3 mM EDTA, 8 mM MgCl2, 5% of glycerol) in a total volume of 8 μL at 37°C for 15 min and cooled to 4°C. During this period 5 pmol/μL and 50 pmol/μL peptide dilutions were made in 1 X PBS. 1 μL of 1 X PBS or the above two peptide dilutions were then added to different tubes containing folded RNA to created peptide negative control reaction, 1:1, and 1:10 RNA:peptide ratios. Reactions were incubated at 37°C for 15 min and the RNA probed using 2.5 nM (final concentration) 1-methyk-7-nitroisatoic anhydride (1M7) in DMSO at 37°C for 5 min. 1M7 negative control reactions were generated as above with the exception that 1 μL of DMSO was added instead of 1M7. 1M7 (+) and (−) reactions were placed on ice and the denaturing control reaction was generated as previously described.
Mutational profiling:
SHAPE-MaP experiments were largely performed as previously described.43 Approximately 0.5 pmoles of purified RNA was used per reaction to generate cDNA, by mutational profiling (MaP) reverse transcription using RRE specific oligo 234-RRE (5’ AGGAGCTGTTGATCCTTTAG) as primers. RNA was hydrolyzed by adding 1 μL 2 N NaOH to each reaction and the solution neutralized by adding 1 μL 2N HCl. cDNA was purified over Sephadex G50 spin columns. The entirety of each cDNA was used as template in a 100 μl PCR (PCR1) reaction (1.1 μL each of 50 pmoles of forward oligo and reverse oligo, 2 μL of 10mM dNTPs, 20 μL of 5X Q5 reaction buffer, 1 μL of hot Start High-Fidelity DNA polymerase). Cycling conditions comprised: 98°C for 30 sec, 15 cycles of [98°C for 10 sec, 50°C for 30 sec, 72°C for 30 sec], 72°C for 2 mins. to generate ds DNA with half of the illumina adapters on each ends. The resulting PCR product was gel purified using a gel purification kit (Qiagen) and the entire product was used in a subsequent PCR reactions that added the remaining half of the Illumina adapters with appropriate indices. The PCR2 reaction and cycling conditions were same as those for PCR1. The resulting sequencing amplicon library fractionated run on a 2% agarose gel and the amplicons recovered and purified by electro- elution at room temperature for 2 hours followed by ethanol precipitation. Each library was quantitated by real time PCR using KAPA Universal Library Quantitation kit (cat# KK4824) per manufacturer’s protocol. Sequencing libraries were pooled and mixed with 20% phiX and sequenced using a Mid Output Kit (300-cycles) on a MiniSeq sequencer following the manufacturer’s protocol to generate 2 X 150 paired-end reads. SHAPE reactivity profiles were created by aligning reads to the 234-nt NL4–3 RRE sequence using ShapeMapper (v1.2, http://chem.unc.edu/rna/software.html) with default settings. Reactivity values corresponding to PCR1 primer binding regions (nt 1− nt 17 and nt 219 − nt 234) were excluded and the remainder renormalized to generate the final reactivity profiles.
Supplementary Material
ACKNOWLEDGEMENT
We thank K. Ray and R. Helm in the VT mass spectrometry incubator for their assistance with MALDI-TOF analysis, D. Tebit and D. Rekosh for the initial in vitro screening of the BPs, and K. Decourcy at the Fralin Life Science-Institute for her assistance with confocal microscopy. This work was supported by the National Institutes of Health [RO1 GM093834]. Funding for open access charge: National Institutes of Health.
ABBREVIATION USED
- 2-AP
2-aminopurine
- ARM
arginine rich motif
- BPs
Branched peptides
- BPBA
branched peptide boronic acid
- CMV
cytomegalovirus
- ELISA
Enzyme-linked immunosorbent assay
- HAART
highly active antiretroviral therapy
- HEK
Human embryonic kidney
- MTT
3-(4,5-Dimethylthiazol-2-yl)- 2,5-Diphenyltetrazolium Bromide
- PNA
Peptide nucleic acids
- RBE
Rev-binding element
- RRE
Rev-Response Element
- SHAPE-MaP
selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling- mutational profiling
- SL
Stem Loop
- SPPS
Solid phase peptide synthesis
- TAR
Transactivation response element
Footnotes
The authors declare no competing financial interest
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: General Procedure and characterization data for all the BPs and binding curve for 2-AP binding assay. (PDF)
REFERENCES
- 1.Arts EJ; Hazuda DJ HIV-1 Antiretroviral Drug Therapy. Cold Spring Harb. Perspect. Med.2012, 2, a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richman DD; Margolis DM; Delaney M; Greene WC; Hazuda D; Pomerantz RJ The Challenge of Finding a Cure for HIV Infection. Science 2009, 323, 1304–1307. [DOI] [PubMed] [Google Scholar]
- 3.Iyidogan P; Anderson KS Current Perspectives on HIV-1 Antiretroviral Drug Resistance. Viruses-Basel 2014, 6, 4095–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun TW; Fauci AS Latent Reservoirs of HIV: Obstacles to the Eradication of Virus. Proc. Natl. Acad. Sci. U S A 1999, 96, 10958–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Draper DE Protein-RNA Recognition. Annu. Rev. Biochem. 1995, 64, 593–620. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder R; Barta A; Semrad K Strategies for RNA Folding and Assembly. Nat. Rev. Mol. Cell Biol. 2004, 5, 908–919. [DOI] [PubMed] [Google Scholar]
- 7.Zaman GJ; Michiels PJ; van Boeckel CA Targeting RNA: New Opportunities to Address Drugless Targets. Drug Discov. Today 2003, 8, 297–306. [DOI] [PubMed] [Google Scholar]
- 8.Mann DA; Mikaelian I; Zemmel RW; Green SM; Lowe AD; Kimura T; Singh M; Butler PJ; Gait MJ; Karn J A Molecular Rheostat. Co-operative Rev Binding to Stem I of the Rev- Response Element Modulates Human Immunodeficiency Virus Type-1 Late Gene Expression. J. Mol. Biol. 1994, 241, 193–207. [DOI] [PubMed] [Google Scholar]
- 9.Van Ryk DI; Venkatesan S Real-time kinetics of HIV-1 Rev-Rev Response Element Interactions. Definition of Minimal Binding Sites on RNA and Protein and Stoichiometric Analysis. J. Biol. Chem. 1999, 274, 17452–17463. [DOI] [PubMed] [Google Scholar]
- 10.Kjems J; Brown M; Chang DD; Sharp PA Structural Analysis of the Interaction Between the Human Immunodeficiency Virus Rev Protein and the Rev Response Element. Proc. Natl. Acad. Sci. U S A 1991, 88, 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollard VW; Malim MH The HIV-1 Rev Protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [DOI] [PubMed] [Google Scholar]
- 12.Malim MH; Tiley LS; McCarn DF; Rusche JR; Hauber J; Cullen BR HIV-1 Structural Gene Expression Requires Binding of the Rev Trans-Activator to its RNA Target Sequence. Cell 1990, 60, 675–683. [DOI] [PubMed] [Google Scholar]
- 13.Hyun S; Lee KH; Yu J A Strategy for the Design of Selective RNA Binding Agents. Preparation and RRE RNA Binding Affinities of a Neomycin-Peptide Nucleic Acid Heteroconjugate Library. Bioorg. Med. Chem. Lett. 2006, 16, 4757–4759. [DOI] [PubMed] [Google Scholar]
- 14.Jin Y; Cowan JA Targeted Cleavage of HIV Rev Response Element RNA by Metallopeptide Complexes. J. Am. Chem. Soc. 2006, 128, 410–411. [DOI] [PubMed] [Google Scholar]
- 15.Lee SJ; Hyun S; Kieft JS; Yu J An Approach to the Construction of Tailor-Made Amphiphilic Peptides That Strongly and Selectively Bind to Hairpin RNA Targets. J. Am. Chem. Soc. 2009, 131, 2224–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li K; Davis TM; Bailly C; Kumar A; Boykin DW; Wilson WD A Heterocyclic Inhibitor of the Rev−RRE Complex Binds to RRE as a Dimer. Biochemistry 2001, 40, 1150–1158. [DOI] [PubMed] [Google Scholar]
- 17.Symensma TL; Baskerville S; Yan A; Ellington AD Polyvalent Rev Decoys Act as Artificial Rev-Responsive Elements. J. Virol. 1999, 73, 4341–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mei H-Y; Cui M; Heldsinger A; Lemrow SM; Loo JA; Sannes-Lowery KA; Sharmeen L; Czarnik AW Inhibitors of Protein−RNA Complexation That Target the RNA: Specific Recognition of Human Immunodeficiency Virus Type 1 TAR RNA by Small Organic Molecules. Biochemistry 1998, 37, 14204–14212. [DOI] [PubMed] [Google Scholar]
- 19.Wang S; Huber PW; Cui M; Czarnik AW; Mei H-Y Binding of Neomycin to the TAR Element of HIV-1 RNA Induces Dissociation of Tat Protein by an Allosteric Mechanism. Biochemistry 1998, 37, 5549–5557. [DOI] [PubMed] [Google Scholar]
- 20.Lu J; Kadakkuzha BM; Zhao L; Fan M; Qi X; Xia T Dynamic Ensemble View of the Conformational Landscape of HIV-1 TAR RNA and Allosteric Recognition. Biochemistry 2011, 50, 5042–5057. [DOI] [PubMed] [Google Scholar]
- 21.Morgan BS; Forte JE; Culver RN; Zhang Y; Hargrove AE Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA‐Targeted Bioactive Ligands. Angew.Chem. Int. Ed. 2017, 56, 13498–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidson BL; McCray PB Jr. Current Prospects for RNA Interference-based Therapies. Nat. Rev. Genet. 2011, 12, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherpa C; Rausch JW; Le Grice SF; Hammarskjold ML; Rekosh D The HIV-1 Rev Response Element (RRE) Adopts Alternative Conformations that Promote Different Rates of Virus Replication. Nucleic. Acids. Res. 2015, 43, 4676–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryson DI; Zhang W; McLendon PM; Reineke TM; Santos WL Toward Targeting RNA Structure: Branched Peptides as Cell-Permeable Ligands to TAR RNA. ACS Chem. Biol. 2012, 7, 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bracci L; Falciani C; Lelli B; Lozzi L; Runci Y; Pini A; De Montis MG; Tagliamonte A; Neri P Synthetic Peptides in the Form of Dendrimers Become Resistant to Protease Activity. J. Biol. Chem. 2003, 278, 46590–46595. [DOI] [PubMed] [Google Scholar]
- 26.Bryson DI; Zhang W; Ray WK; Santos WL Screening of a Branched Peptide Library with HIV-1 TAR RNA. Mol. Biosyst. 2009, 5, 1070–1073. [DOI] [PubMed] [Google Scholar]
- 27.Cook KS; Fisk GJ; Hauber J; Usman N; Daly TJ; Rusche JR Characterization of HIV- 1 REV Protein: Binding Stoichiometry and Minimal RNA Substrate. Nucleic Acids Res. 1991, 19, 1577–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W; Bryson DI; Crumpton JB; Wynn J; Santos WL Branched Peptide Boronic Acids (BPBAs): a Novel Mode of Binding towards RNA. Chem. Commun. 2013, 49, 2436–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W; Bryson DI; Crumpton JB; Wynn J; Santos WL Targeting folded RNA: a Branched Peptide Boronic Acid that Binds to a Large Surface Area of HIV-1 RRE RNA. Org. Biomol. Chem. 2013, 11, 6263–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wynn JE; Zhang W; Tebit DM; Gray LR; Hammarskjold M-L; Rekosh D; Santos WL Characterization and in vitro Activity of a Branched Peptide Boronic Acid that Interacts with HIV-1 RRE RNA. Bioorg. Med. Chem. 2016, 24, 3947–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wynn JE; Zhang W; Tebit DM; Gray LR; Hammarskjold ML; Rekosh D; Santos WL Effect of Intercalator and Lewis Acid-Base Branched Peptide Complex Formation: Boosting Affinity towards HIV-1 RRE RNA. Medchemcomm. 2016, 7, 1436–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong SY; Oh JE; Lee KH Effect of D-amino Acid Substitution on the Stability, the Secondary Structure, and the Activity of Membrane-Active Peptide. Biochem. Pharmacol. 1999, 58, 1775–1780. [DOI] [PubMed] [Google Scholar]
- 33.Datta B; Bier ME; Roy S; Armitage BA Quadruplex Formation by a Guanine-Rich PNA Oligomer. J. Am. Chem. Soc. 2005, 127, 4199–4207. [DOI] [PubMed] [Google Scholar]
- 34.Crumpton JB; Zhang W; L., S. W. Facile Analysis and Sequencing of Linear and Branched Peptide Boronic Acids by MALDI Mass Spectrometry. Anal. Chem. 2011, 83, 3548–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacourciere KA; Stivers JT; Marino JP Mechanism of Neomycin and Rev Peptide Binding to the Rev Responsive Element of HIV-1 as Determined by Fluorescence and NMR Spectroscopy. Biochemistry 2000, 39, 5630–5641. [DOI] [PubMed] [Google Scholar]
- 36.Battiste JL; Mao H; Rao NS; Tan R; Muhandiram DR; Kay LE; Frankel AD; Williamson JR Alpha Helix-RNA Major Groove Recognition in an HIV-1 Rev Peptide-RRE RNA Complex. Science 1996, 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 37.Srinivasakumar N; Chazal N; Helga-Maria C; Prasad S; Hammarskjold ML; Rekosh D The Effect of Viral Regulatory Protein Expression on Gene Delivery by Human Immunodeficiency Virus Type 1 Vectors Produced in Stable Packaging Cell Lines. J. Virol. 1997, 71, 5841–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayaraman B; Crosby DC; Homer C; Ribeiro I; Mavor D; Frankel AD RNA-Directed Remodeling of the HIV-1 Protein Rev Orchestrates Assembly of the Rev-Rev Response Element Complex. Elife 2014, 3, e04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel DP; Zapp ML; Green MR; Szostak JW HIV-1 Rev Regulation Involves Recognition of Non-Watson-Crick Base Pairs in Viral RNA. Cell 1991, 67, 529–536. [DOI] [PubMed] [Google Scholar]
- 40.Kjems J; Calnan BJ; Frankel AD; Sharp PA Specific Binding of a Basic Peptide from HIV- 1 Rev. EMBO J. 1992, 11, 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai Y; Tambe A; Zhou K; Doudna JA RNA-guided Assembly of Rev-RRE Nuclear Export Complexes. ELife 2014, 3, e03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malim MH; Hauber J; Le S-Y; Maizel JV; Cullen BR The HIV-1 Rev Trans-Activator Acts Through a Structured Target Sequence to Activate Nuclear Export of Unspliced Viral mRNA. Nature 1989, 338, 254–257. [DOI] [PubMed] [Google Scholar]
- 43.Smola MJ; Rice GM; Busan S; Siegfried NA; Weeks KM Selective 2’-Hydroxyl Acylation Analyzed by Primer Extension and Mutational Profiling (SHAPE-MaP) for Direct, Versatile and Accurate RNA Structure Analysis. Nat. Protoc. 2015, 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ippolito JA; Steitz TA The Structure of the HIV-1 RRE High Affinity Rev Binding Site at 1.6 Å Resolution J. Mol. Biol. 2000, 295, 711–717. [DOI] [PubMed] [Google Scholar]
- 45.Daugherty MD; D’Orso I; Frankel AD A Solution to Limited Genomic Capacity: Using Adaptable Binding Surfaces to Assemble the Functional HIV Rev Oligomer on RNA. Mol. Cell 31, 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jayaraman B; Mavor D; Gross JD; Frankel AD Thermodynamics of Rev–RNA Interactions in HIV-1 Rev–RRE Assembly. Biochemistry 2015, 54, 6545–6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamaki M; Han G; Hruby VJ Practical and Efficient Synthesis of Orthogonally Protected Constrained 4-Guanidinoprolines. J. Org. Chem. 2001, 66, 1038–1042. [DOI] [PubMed] [Google Scholar]
- 48.Tamamura H; Ojida A; Ogawa T; Tsutsumi H; Masuno H; Nakashima H; Yamamoto N; Hamachi I; Fujii N Identification of a New Class of Low Molecular Weight Antagonists Against the Chemokine Receptor CXCR4 Having the Dipicolylamine-zinc(II) Complex Structure. J. Med. Chem. 2006, 49, 3412–3415. [DOI] [PubMed] [Google Scholar]
- 49.Mallik B; Katragadda M; Spruce LA; Carafides C; Tsokos CG; Morikis D; Lambris JD Design and NMR Characterization of Active Analogues of Compstatin Containing Non-Natural Amino Acids. J. Med. Chem. 2005, 48, 274–286. [DOI] [PubMed] [Google Scholar]
- 50.Smith SJ; Rittinger K Preparation of GTPases for Structural and Biophysical Analysis. Methods Mol. Biol. 2002, 189, 13–24. [DOI] [PubMed] [Google Scholar]
- 51.Milligan JF; Groebe DR; Witherell GW; Uhlenbeck OC Oligoribonucleotide Synthesis Using T7 RNA Polymerase and Synthetic DNA Templates. Nucleic Acids Res. 1987, 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pagano JM; Clingman CC; Ryder SP Quantitative Approaches to Monitor Protein-Nucleic Acid Interactions Using Fluorescent Probes. RNA 2011, 17, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.