Abstract
Multi-modal single-cell assays provide high-resolution snapshots of complex cell populations but are mostly limited to transcriptome plus an additional modality. Here, we describe Expanded CRISPR-compatible Cellular Indexing of Transcriptomes and Epitopes by sequencing (ECCITE-seq) for the high-throughput characterization of at least five modalities of information from each single cell. We demonstrate application of ECCITE-seq to multimodal CRISPR screens with robust direct sgRNA capture and to clonotype-aware multimodal phenotyping of cancer samples.
INTRODUCTION
High-throughput single cell RNA sequencing (scRNA-seq) has rapidly progressed from a tremendous technical achievement to a standard tool for phenotypic interpretation of complex biological systems. scRNA-seq has empowered researchers to deeply phenotype cells, enabling detection of rare cell populations and determination of developmental trajectories of distinct cell lineages. Recently, substantial progress has been made in combining readouts of other modalities with scRNA-seq in high throughput assays, including genome sequence1, chromatin accessibility2,3, methylation4–6, immunophenotype7,8 and synthetic markers of cell lineage9–12. Additionally, several approaches have recently been reported that allow detection of CRISPR-mediated perturbations along with the transcriptome of single cells using specialized vectors that link the expression of single guide RNAs (sgRNAs) to separate transcripts that can be captured by standard scRNA-seq methods13–16. Collectively, these methods enable the use of scRNA-seq as an unbiased readout of pooled CRISPR-based genetic screens, but all current methods suffer from limitations related to the need to determine the identity of the guide by a proxy polyadenylated transcript17–20.
Previously, we and others have layered detection of proteins on top of scRNA-seq to enable integration of robust and well-characterized protein markers with unbiased transcriptomes of single cells7,8. Our method, Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-seq) is compatible with oligo-dT based scRNA-seq approaches and enables simultaneous protein detection using DNA oligo-labeled antibodies against cell surface markers. Given that protein levels are typically much higher than corresponding mRNAs, detection of proteins via antibody-derived tags (hereafter called protein tags) is a more robust measure of gene expression. In a series of experiments, we demonstrated the value of multimodal analysis to reveal phenotypes that could not be discovered using scRNA-seq alone, as well as the use of CITE-seq for studies of post-transcriptional gene regulation at the single-cell level7.
Here, we extend the utility of CITE-seq and the related Cell Hashing method for multiplexing and doublet detection21 to 5’ capture-based scRNA-seq methods, exemplified by the 10× Genomics 5P / V(D)J system, allowing the detection of surface proteins together with the scRNA-seq and clonotype features currently offered by the 10× Genomics system22,23. Importantly, we further adapt the system to enable direct and robust capture of sgRNAs from existing guide libraries and commonly used vectors compatible with pooled cloning, for use in Perturb-seq / CRISPR-seq / CROP-seq type experiments. ECCITE-seq overcomes the limitations of existing systems for CRISPR screens with scRNA-seq readout, while also demonstrating the power of combining protein detection with scRNA-seq as a readout for CRISPR screens.
RESULTS
ECCITE-seq enables the detection of at least five modalities of cellular information from single cells
To enable profiling of protein markers together with V (D)J regions and transcriptomes, we modified our previously described CITE-seq method7. Oligos partially complementary to the gel bead-associated template switch oligos (TSO) in the 10× Genomics 5P / V(D)J kit were covalently conjugated to antibodies as described21,24 and used to label cells. Annealing and extension during the reverse transcription (RT) reaction associates the cell barcode and unique molecular identifier (UMI) from the gel bead oligo with the antibody tag in parallel with the addition of these sequences to the first strand cDNA copies of cellular mRNAs in the same droplet (Fig. 1a) (see methods). Separate detection of CITE-seq protein tags for differentially expressed proteins and cell hashtags for multiplexing is achieved using different amplification handles21. In contrast to commonly used 3’ tag scRNA-seq methods where cell barcode information is appended to transcripts through the use of barcoded RT oligos, the 10× Genomics 5P workflow appends the barcode via TSO, using a generic, soluble poly(dT) oligo to prime RT, opening up the possibility of adding custom RT primers to sequences of interest. sgRNAs have a structure that lends themselves to direct capture: the variable region that guides Cas9 to its target site is at the 5’ end while the 3’ end is an invariant scaffold25,26. We leveraged the scaffold as an annealing site for an additional RT primer, which after copying the variable guide sequence and template switching with the bead-derived TSO, acquires a cell barcode and UMI in parallel with other modalities (mRNA, protein tags, hashtags)(Fig. 1a). A mixture of human and mouse cells transduced with different sets of non-targeting sgRNAs was well resolved by transcriptome, surface protein and sgRNA content, demonstrating the specificity of this approach (Fig. 1b, Supplementary Fig. 1a, Supplementary Tables 1,2).
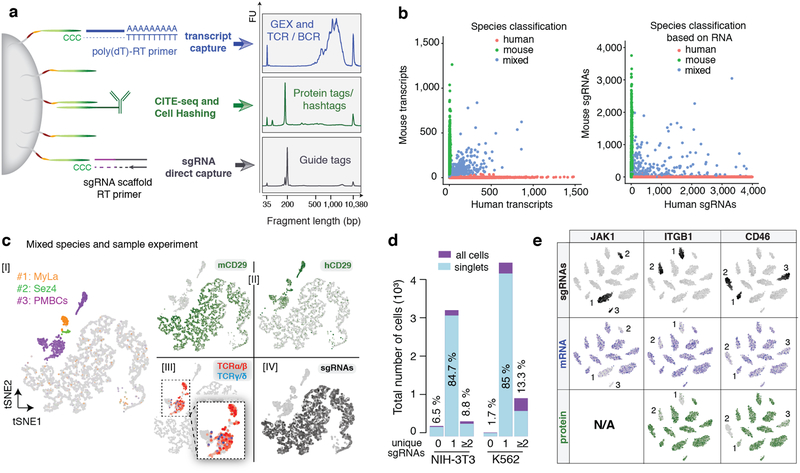
Figure 1:
ECCITE-seq allows simultaneous detection of transcriptome, proteins, clonotypes and CRISPR perturbations. a. Schematic overview of the multiple cellular modalities captured by ECCITE-seq. cDNAs derived from mRNA and sgRNA transcripts acquire cell barcodes and unique identifiers through a template switch reaction with the bead-bound oligo, whereas protein tags harbor sequences that allow direct annealing to the bead oligo. Reverse transcription and amplification yields products with distinct sizes that can be separated and amplified independently (right panel). b. Species-mixing experiment. Left: number of transcripts associated with each cell barcode (red, >90% human reads; green, >90% mouse reads; blue, >10% human and mouse (multiplet). Right: sgRNA reads associated with each cell barcode. Points are colored based on species classifications using transcripts. c. Transcriptome-based clustering of single-cell expression profiles of the mixed human and mouse sample, illustrating the 5 modalities of ECCITE-seq: Transcriptome, Cell Hashing [I], Protein [II], T cell antigen receptors (α/β: red, γ/δ: blue) [III] and sgRNAs [IV]. d. Number of cell barcodesbefore (purple) or after (blue) removal of cell doublets assigned zero, one or two unique guides per cell in two independent experiments and cell lines. e. K562 cells were clustered based on normalized and scaled sgRNA counts. Highlighted are counts for sgRNAs (black) targeting the indicated gene and counts for respective mRNA (blue) and protein tags (green) where applicable.
To illustrate the detection of six modalities (transcriptome, T cell receptor (TCR α/β and TCR γ/δ), surface protein, sample identity by hashtags, and sgRNA) in a single experiment (Supplementary Fig. 1b), we generated a cell mixture comprising human peripheral blood mononuclear cells (PBMCs), two human T cell lymphoma lines (MyLa and Sez4) and mouse NIH-3T3 cells that had been transduced with a library of non-targeting sgRNA-generating constructs (Fig.1c, Supplementary Table 2). Cell hashtags specific to human cells were used to distinguish the three human samples, and the hashtag distribution was consistent with transcriptome-based clustering (Fig.1c, [I]). CITE-seq antibodies directed against human or mouse CD29 label cells according to their species of origin [II], illustrating the ability of ECCITE-seq to detect differentially expressed proteins within a sample. Clonotypes for TCR α/β (following 10× protocol) and TCR γ/δ (custom adaptation, see methods) were detected in the PBMC and lymphoma cell clusters [III]. Finally, guide tags, derived directly from sgRNA molecules were specifically and robustly detected only in mouse cells [IV]. Importantly, the use of Cell Hashing together with sgRNA detection allowed us to distinguish between apparent “doublets” where cells have been infected with two viruses (n=325), from doublets resulting from co-encapsulation of two cells in the same droplet (n=65) (Fig. 1d). sgRNA capture was highly efficient, with sgRNAs detected in 93.5% of mouse cells (Fig.1d), in proportions consistent with genomic DNA-based detection from bulk cells (Supplementary Fig. 1c).
CRISPR screens with single cell multimodal readout
ECCITE-seq is designed to enable interrogation of single cell transcriptomes together with surface protein markers in the context of CRISPR screens. To illustrate this, we infected K562 cells with a CRISPR library comprising guides targeting genes encoding cell surface markers (CD29 and CD46), intracellular signaling molecules (JAK1 and p53), as well as two non-targeting controls (Supplementary Table 1). We leveraged the Cell Hashing feature to remove cell doublets and observed very high rates of guide capture (confident detection of guide sequences in 98.3% of cells), in proportions consistent with genomic DNA-based detection (Fig. 1d and Supplementary Fig. 1d). Clustering based on sgRNA counts of cells assigned to one guide revealed 13 distinct clusters, corresponding to the 13 guides in the experiment. Loss of expression of target genes at the level of mRNA and protein was readily apparent for ITGB1 (the gene encoding CD29 protein) and CD46 (Fig. 1e), and similarly apparent at the mRNA level for JAK1. TP53 transcript was poorly detected, consistent with K562 cells expressing a single allele of TP53 that is likely a substrate for nonsense-mediated decay27. In parallel, we performed scRNA-seq alone on the same aliquot of cells and confirmed no reduction in transcripts per cell (Supplementary Fig. 1e.), demonstrating no detrimental effect of capturing additional modalities on transcript capture.
Cellular perturbations measured at transcript and protein level by ECCITE-seq reveal important features to consider, exemplified by CD46: most cells have detectable levels of protein, which collapse in cells with detectable levels of sgRNAs directed against CD46, but not in cells with non-targeting sgRNAs (Supplementary Fig. 1f). mRNA reduction is also apparent in cells with targeting sgRNAs, albeit less notably. Many cells have undetectable levels of CD46 mRNA even in the absence of targeting guides, likely reflecting the high drop-out rates of scRNA-seq, and the increased sensitivity that comes with protein detection.
The low dropout of protein detection7,8 suggests that ECCITE-seq could be more sensitive in detecting expression phenotypes than scRNA-seq alone. To test this for single genes, we used clusters assigned to each given guide against the two non-targeting clusters and determined the p-value of detecting the expected gene expression change in randomly-sampled cells ranging from 10 to 100 per group. (Supplementary Fig. 1g). This analysis suggests that the number of cells needed to detect the direct consequence of a given perturbation is markedly reduced when using protein detection as a readout compared to mRNA, increasing the numbers of perturbations that can be assessed for a given number of cells. Additionally, as exemplified by CD46, the gene expression change triggered by 2 out of 3 sgRNAs (CD46.1 and CD46.3) was confidently detected only at the level of protein, even when considering all cells assigned to these sgRNAs. In practical terms, future applications of this technology will rely on detection of changes in gene expression signatures and it stands to reason that these signatures will be more robust with protein components.
ECCITE-seq couples clonotype determination with immunophenotyping
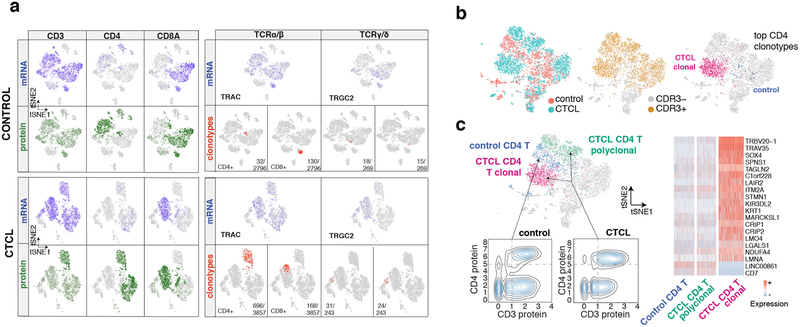
We next constructed a 49 marker panel of ECCITE-seq antibodies to deeply profile PBMCs from a healthy donor and a Cutaneous T Cell Lymphoma (CTCL) patient (Fig. 2, Supplementary Fig.2 and Supplementary Table 3) and prepared libraries for hashtags, ADTs, TCR α/β, TCR γ/δ and transcriptome. After hashtag demultiplexing to remove doublets, cells were clustered based on transcriptome (Fig. 2a and Supplementary Fig. 2). The majority of markers showed enrichmentat the level of both protein and RNA (not shown) in expected clusters, consistent with our previous 3’ CITE-seq results7. We additionally recovered TCR α/β and γ/δ clonotype information for both the control and CTCL samples. Select markers and clonotypes are shown in Fig. 2a and Supplementary Fig. 2. The control sample had 1,606 detected clonotypes from 2,796 barcodes, with the top CD4+ clonotype (defined by TRB CDR3 sequence: CASSTLQGKETQYF) accounting for ~1% of recovered clonotype-associated barcodes. In contrast, in the CTCL sample, clonal expansion was readily apparent with a single TRB CDR3 sequence (CSARFLRGGYNEQFF) present in 36% of cells for which we recovered clonotype information (1,390 out of 3,857 barcodes).
Figure 2:
ECCITE-seq couples clonotype determination with immunophenotyping. a. Transcriptome-based clustering of PBMCs from healthy donor (top) and CTCL patient (bottom) after removing cell doublets. Projected is the mRNA (blue) and protein (green) signal for CD3, CD4 and CD8, as well as the most abundant CD4+ or CD8+ TCR α/β or two most abundant TCR γ/δ clonotypes (red). b. Transcriptome-based clustering of the combined dataset after merging, depth normalization and cell alignment. Highlighted from left to right is sample identity, productive TCR rearrangement and most abundant CD4+ TCRα/β clonotype. c. Differentially expressed genes between CD3+ CD4+ T clusters of control and CTCL PBMCs, as defined by in silico gating based on CD3 and CD4 protein counts coupled with clonotype determination.
For further comparative analysis, cells from both samples were computationally merged28 and clustering based on either RNA or protein showed agreement in detecting most cell sub-populations and their gene-expression signatures (Supplementary Fig. 3). in silico gating based on CD3 and CD4 protein levels coupled with clonotypic information enabled differential gene expression analysis comparing monoclonal T cells with polyclonal T cells from both the patient and the healthy donor sample (Fig. 2b,c), revealing a distinct gene expression signature of the malignant CTCL cells, consistent with prior studies29, and illustrating the power of ECCITE-seq to combine immunophenotype, clonotype and transcriptome information.
DISCUSSION
The enhancements to the CITE-seq toolkit provided by ECCITE-seq enable detailed phenotypic and functional characterization of single cells. The recovery of clonotype information together with surface protein marker expression allowed fine separation of specific cell populations of interest, enabling careful determination of molecular phenotypes. Analogous to the use of TCR clonotype information in this study, we have recently used expressed mutations to define and further characterize clonal populations in scRNA-seq data-sets (Genotyping of Transcriptomes, GoT30), an approach that could readily be combined with ECCITE-seq. The method we describe is inherently customizable and we envisage additional oligo-tagged ligands, such as peptide-loaded MHC complexes for detecting specific TCRs, labeled antigens for detection of antigen specific B cells, or antibodies directed against intracellular proteins being added to future iterations of this system. The combination of Cell Hashing together with direct sgRNA capture will enhance perturbation screens with single cell readouts by allowing the analysis of greater numbers of cells for a given budget by allowing discrimination between single cells with multiple expressed sgRNAs and “doublets”, with 2 cells each with their own guide. The “super-loading” afforded by this knowledge will additionally drive down the per-cell cost of single cell CRISPR screens, which will also require less cells per guide to detect expression phenotypes that feature both protein and mRNA. The modular nature of ECCITE-seq allows the tailoring of readouts of such screens, potentially allowing the investigator to interrogate panels of transcripts and proteins of interest in response to their perturbations in addition to, or instead of, the transcriptome. This is in line with the high-dimensional phenotyping of multiple proteins in CRISPR-based pooled screens using Pro-Codes and CyTOF as readout31. While this method can more economically achieve precise quantification of intracellular and extracellular protein levels in millions of single cells, it cannot interrogate the single-cell transcriptome simultaneously, it lacks the scalability of DNA barcoding and requires sgRNA cloning in special constructs. ECCITE-seq is readily applicable to any sgRNA library with the Cas9 S. pyogenes scaffold sequence and, by allowing direct capture of sgRNA molecules, overcomes documented problems of barcode swapping events observed with Perturb-seq that have the potential to confound single cell perturbation screens17–20. Direct capture has the added benefit of capturing a highly abundant RNA polymerase III transcript, contributing to the observed high rates of guide recovery. While this work was under review, a pre-print describing two strategies for direct guide detection in the context of scRNA-seq was posted32. One of the described methods is conceptually similar to the method described here and was demonstrated to have superior rates of guide capture compared to the 3’-based approach. Direct and robust capture of sgRNAs will allow these related approaches to be further used for applications using multiple guides per cell enabling the targeting of multiple genes or genomic regions, either through engineered constructs33–35, high multiplicity of infection transductions36, the parallel use of different CRISPR systems for combining, for example, mutation and activation of selected genes37, or lineage tracing with multiple homing sgRNAs12. Our approach additionally provides a roadmap for targeted capture of specific RNA molecules including non-polyadenylated transcripts.
MATERIALS AND METHODS
Antibody-oligo conjugates
Antibodies used for CITE-seq and Cell Hashing were obtained as purified, unconjugated reagents from BioLegend and were covalently and irreversibly conjugated to barcode oligos by iEDDA-click chemistry as previously described21,24. See Supplementary Tables 3, 4 and 5 for a list of antibodies, clones and barcodes used for ECCITE-seq.
Cell staining with barcoded antibodies
Cells were stained with barcoded antibodies as previously described for CITE-seq7 and Cell Hashing21. Briefly, approximately 1.5–2 million cells per sample were resuspended in 1× CITE-seq staining buffer (2% BSA, 0.01% Tween in PBS) and incubated for 10 min with Fc receptor block (TruStain FcX, BioLegend, USA) to block FC receptor-mediated binding. Subsequently, cells were incubated with mixtures of barcoded antibodies for 30 min at 4°C. Antibody concentrations were 1 μg per test, as recommended by the manufacturer (BioLegend, USA) for flow cytometry applications. For some highly expressed markers, tags can take up unacceptably high proportions of the protein-tag libraries. In these cases (determined empirically from prior experiments) we reduced the concentration of the oligo-tagged antibodies in the panel by diluting with un-tagged antibody. Oligo-labeled CD44 & CD45 were diluted 1:10 and therefore used at an effective concentration of 0.1μg per stain. After staining, cells were washed 3× by resuspension in PBS containing 2% BSA and 0.01% Tween, followed by centrifugation (300g 5 min at 4°C) and supernatant exchange. After the final wash, cells were resuspended in PBS and filtered through 40 μm cell strainers.
ECCITE-seq on 10× Genomics instrument
Stained and washed cells were loaded into 10× Genomics single cell V(D)J workflow and processed according to manufacturer’s instructions with the following modifications:
12 pmol of an RT-primer complementary to sgRNA scaffold sequences was spiked into the RT reaction (only when sgRNA capture was desired). gd_RT_v4: AGCAAGTGAGAAGCATCGTGTCAAAGCACCGACTCGGTGCCAC.
During the cDNA amplification step, 1 pmol of hashtag additive (GTGACTGGAGTTCAGACGTGTGCTC), 1 pmol of guide-tag additive (AGCAAGTGAGAAGCATCGTGTC) (only when sgRNA capture was desired) and 2 pmol of protein-tag additive primers (CCTTGGCACCCGAGAATTCC) were spiked into the cDNA amplification PCR.
Following PCR, 0.6X SPRI was used to separate the large cDNA fraction derived from cellular mRNAs (retained on beads) from the protein tag-, hashtag and guide tag-containing fraction (in supernatant). The cDNA fraction was processed according to the 10× Genomics Single Cell V(D)J protocol to generate the transcriptome library and the TCR α/β library. To amplify TCR γ/δ transcripts we implemented a strategy similar to TCR α/β approach from 10× Genomics with a two-step PCR: during target enrichment 1 we used SI-PCR (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTC) and a mix of R1_hTRDC (AGCTTGACAGCATTGTACTTCC) and R1_hTRGC (TGTGTCGTTAGTCTTCATGGTGTTCC), followed by target enrichment 2 with a generic P5 oligo (AATGATACGGCGACCACCGAGATCTACAC) and a mix of R2_hTRDC (TCCTTCACCAGACAAGCGAC) and R2_hTRGC (GATCCCAGAATCGTGTTGCTC). cDNA and TCR (α/β and γ/δ) enriched libraries were further processed according to the 10× Genomics Single Cell V(D)J protocol.
An additional 1.4X reaction volume of SPRI beads was added to the protein-tag/hashtag/guide-tag fraction from step 3, to bring the ratio up to 2.0X. Beads were washed with 80% ethanol, eluted in water, and an additional round of 2.0X SPRI performed to remove excess single stranded oligonucleotides carried over from the cDNA amplification reaction. After final elution, separate PCR reactions were set up to generate the protein-tag library (SI-PCR and RPI-x primers), the hashtag library (SI-PCR and D7xx_s) and the guide-tag library (SI-PCR and Next_nst_x). The protein-tag and hashtag libraries were prepared as previously described21. Following the cDNA amplification, the sgRNA sequences are converted to an Illumina library by amplification with smRNA_nst_x (v3): CAAGCAGAAGACGGCATACGAGATxxxxxxxxGTGACTGGAGTTCCTTGGCACC CGAGAATTCCATTCTAGCTCTAAAAC or Next_nst_x (v4): CAAGCAGAAGACGGCATACGAGATxxxxxxxxGTCTCGTGGGCTCGGAGATGTG TATAAGAGACAGTATTTCTAGCTCTAAAAC together with the SI-PCR primer. “x” nucleotides indicate the sample index sequenced by the Illumina i7 index read. Prior to the final library PCR, sgRNA molecules can be further enriched by performing extra rounds of amplification with guide-tag additive and SI-PCR primers.
Libraries were pooled to desired quantities and sequenced on either an Illumina HiSeq 2500 (rapid run flowcell: recipe 26 cycles read 1, 8 cycles index, 39 cycles read 2), or on a NovaSeq 6000 (S2 flowcell: recipe 26 cycles read 1, 8 cycles index, 91 cycles read 2). Reads were trimmed as required for downstream processing. A detailed and regularly updated point-by-point protocol for CITE-seq, Cell Hashing, ECCITE-seq and future updates can be found at www.cite-seq.com and on Nature Protocol Exchange.
Cells
Patient and control samples were collected at New York University Langone Medical Center in accordance with protocols approved by the New York University School of Medicine Institutional Review Board and Bellevue Facility Research Review Committee (IRB#i15–01162). CTCL patients were diagnosed according to the WHO classification criteria. After written informed consent was obtained, peripheral blood samples were harvested. PBMCs were isolated from the blood of patients and healthy controls by gradient centrifugation using Ficoll-Paque™ PLUS (GE Healthcare) and Sepmate™−50 tubes (Stemcell). Buffy coat PBMCs were collected and washed twice with PBS 2% FBS and cryopreserved in freezing medium (40% Roswell Park Memorial Institute medium (RPMI) 1640, 50% FBS and 10% DMSO). Cryopreserved PBMCs were thawed for 1–2 minutes in a 37°C water bath, washed twice in warm PBS 2% FBS and resuspended in complete medium (RPMI 1640 supplemented with 10% FBS and 2mM L-Glut). Control and CTCL PBMCs were stained with a 49-antibody panel (Supplementary Table 3) and Cell Hashing antibodies (Supplementary Table 5), before loading into two separate 10× Genomics Chromium lanes.
The Sez4 cell line is derived from the blood of an SS patient38, and the MyLa 2059 line is derived from a plaque biopsy sample of an MF patient39. Sez4 cells were cultured in RPMI 1640 medium with 2mM L-glutamine, 1% Pen/Strep, 500 units/ml of rh IL-2 (Corning), and 10% human serum. MyLa 2059 cells were cultured in RPMI 1640 medium with 2mM L-glutamine, 1% Pen/Strep, and 10% fetal bovine serum. All cells were incubated at 37°C, 5% CO2 in a humidified incubator. The cells were cryopreserved in 90% FBS 10% DMSO and aliquots of 1-1.5 million cells were thawed on the day of the experiment. PBMCs were obtained cryopreserved from AllCells (USA) and used immediately after thawing. NIH-3T3 and HEK293FT cells expressing non-targeting sgRNAs were maintained according to standard procedures in Dulbecco’s Modified Eagle’s Medium (Thermo Fisher, USA) supplemented with 10% fetal bovine serum (Thermo Fisher, USA) and 1μg/ml puromycin, at 37°C with 5% CO2. K562 cells expressing targeting and non-targeting guides were maintained in RPMI supplemented with 10% fetal bovine serum and 1μg/ml puromycin, at 37°C with 5% CO2.
Lentivirus production and transduction
The sgRNAs were individually synthesized (Integrated DNA T echnologies) and cloned into the lentiviral transfer vector LentiCRISPR v240 (Addgene Plasmid: 52961). Equal amounts of each sgRNA vector were mixed and packaged into lentiviral particles through transfection with packaging plasmids in HEK293FT cells, as previously described41.
For transduction of HEK293FT, the lentiviral guide pool consisted of 10 non-targeting human guides in one experiment and 10 non-targeting and 11 gene-targeting human guides in another experiment (Supplementary Table 1). For transduction of K562, the pool consisted of 2 non-targeting and 11 targeting human guides (Supplementary Table 1). For transduction of NIH-3T3, the pool consisted of 10 non-targeting mouse guides (Supplementary Table 2). NIH-3T3, HEK293FT and K562 cells were infected at MOI = 0.05 and selected and maintained in 1μg/ml puromycin. NIH-3T3 cells used in the proof-of-principle experiment were maintained in culture for several weeks, allowing drift in the representation of guides. Following transduction, K562 cells were stored in liquid nitrogen and were allowed to grow for 2 days before the ECCITE-seq run.
Single-cell data processing
Fastq files from the 10× libraries with four distinct barcodes were pooled together and processed using the cellranger count pipeline. Reads were aligned to the GRCh38 (human healthy and CTCL PBMC datasets) or hg19-mm10 concatenated reference (human-mouse experiment). For protein tag, hashtag and guide tag quantification, we used a previously developed tag quantification pipeline, available at https://github.com/Hoohm/CITE-seq-Count, run with default parameters (maximum Hamming distance of 1). For the TCR libraries, fastq files from the 10× libraries with four distinct barcodes were pooled together, processed using the cellranger vdj pipeline and reads were aligned to the GRCh38 reference genome.
Seurat
Normalization and downstream analysis of RNA data were performed using the Seurat R package (version 2.3.0)28 which enables the integrated processing of multi-modal single cell datasets. Protein tag, hashtag and guide tag raw counts were normalized using centered log ratio (CLR) transformation, where counts were divided by the geometric mean of the corresponding tag across cells, and log-transformed7. For demultiplexing based on hashtag or guide tag counts we used the HTODemux function within the Seurat package as described21. To calculate the significance in detecting the target gene expression change between the targeting guide clusters and the non-targeting clusters we used FindAllMarkers with maximum cell number ranging from 10 to 100, in 10 sampling iterations for each cell number. For the TCR libraries, productive clonotypes were filtered and their raw counts were inserted into the Seurat object under a new assay slot. Raw counts were normalized using centered log ratio (CLR) transformation and scaled. For comparison between the healthy donor and CTCL data, both Seurat objects were merged and depth-normalized when performing cell alignment (or batch normalization) using RunCCA with a default parameter of 30 canonical vectors28. The top 10 aligned components were used for visualization with t-SNE as well as clustering with modularity optimization. The top 20 genes upregulated in each cluster (FindAllMarkers) was used to label the cluster. For protein tag clustering, distance matrices of the combined object were computed before generating t-SNE plots.
Definition of CD4 T cells and Malignant clone
In analogous strategy to what is used for data visualization in flow cytometry, biaxial KDE plots were made using log(protein tag counts+1) of CD3 and CD4. Cells in both samples were gated at a threshold ≥ 4.5 (log scale) for CD4 protein tag counts and ≥ 1.0 (log scale) for CD3 protein tag counts, defining CD4+ T cells. CTCL Malignant cells were defined as CD4 T cells that possessed the most abundant TCRβ CDR3 amino acid sequence, CSARFLRGGYNEQFF, while CTCL CD4 polyclonal cells were CD4 T cells that did not possess this sequence.
Single-cell differential analysis
Comparisons were done using Wilcoxon rank sum test (FindMarkers) between “CTCL Malignant” and “CTCL CD4 polyclonal” as well as between “CTCL Malignant” and “control CD4 Normal”. Significant genes were defined using q-value < 0.05 and |avg_log2FC| > 1.0. All Ribosomal Protein (ΛRP[SL][:digit:]) genes as well as Y, X-escapee and X-variable genes were removed from the differentially expressed list. Heatmaps were made using the union of both sets of significant genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Odum and Kaltoft for the kind gift of cell lines. Patient samples were obtained with the help of Michal Bar Natan Zommer and Jo-Ann Latkowski. We thank Lu Yang, William Stephenson, Suma Jaini, and Kunal Pandit for helpful discussions. We thank Brian Fritz from 10× Genomics for providing kits for development of 5P compatible CITE-seq reagents, and Bertrand Yeung and Kit Nazor from BioLegend for providing some of the unconjugated antibodies used in this study.
COMPETING INTERESTS
MS and PS are listed as co-inventors on a patent application related to this work (US provisional patent application 62/515–180).
Footnotes
DATA AVAILIBILITY
Data has been deposited in the Gene Expression Omnibus (GEO) with accession code pending
REFERENCES
- 1.Macaulay IC et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nature Methods 12, 519–522 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Lake BB et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol 17, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark SJ et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Comms 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angermueller C et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature Methods 13, 229–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson VM et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol 35, 936–939 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Spanjaard B et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalhor R et al. Developmental barcoding of whole mouse via homing CRISPR. Science 361, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaitin DA et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Adamson B et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nature Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill AJ et al. On the design of CRISPR-based single-cell molecular screens. Nature Methods (2018). doi: 10.1038/nmeth.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feldman D, Singh A, Garrity AJ, bioRxiv PB 2018. Lentiviral co-packaging mitigates the effects of intermolecular recombination and multiple integrations in pooled genetic screens. biorxiv.org doi: 10.1101/262121 [DOI] [Google Scholar]
- 19.Xie S et al. Frequent sgRNA-barcode Recombination in Single-cell Perturbation Assays. biorxiv.org doi: 10.1101/255638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegde M, Strand C, Hanna RE, bioRxiv JD 2018. Uncoupling of sgRNAs from their associated barcodes during PCR amplification of combinatorial CRISPR screens. biorxiv.org doi: 10.1101/254334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeckius M et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neal JT et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 175, 1972–1988.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Buggenum JAGL et al. A covalent and cleavable antibody-DNA conjugation strategy for sensitive protein detection via immuno-PCR. 6, 22675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law JC, Ritke MK, Yalowich JC, Leder GH & Ferrell RE Mutational inactivation of the p53 gene in the human erythroid leukemic K562 cell line. Leuk. Res 17, 1045–1050 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fanok MH et al. Role of Dysregulated Cytokine Signaling and Bacterial Triggers in the Pathogenesis of Cutaneous T-Cell Lymphoma. J. Invest. Dermatol 138, 1116–1125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam AS et al. High throughput droplet single-cell Genotyping of Transcriptomes (GoT) reveals the cell identity dependency of the impact of somatic mutations. biorxiv.org doi: 10.1101/444687 [DOI] [Google Scholar]
- 31.Wroblewska A et al. Protein Barcodes Enable High-Dimensional Single-Cell CRISPR Screens. Cell 175, 1141–1155.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Replogle JM et al. Direct capture of CRISPR guides enables scalable, multiplexed, and multi-omic Perturb-seq. 1–26 (2018). doi: 10.1101/503367 [DOI] [Google Scholar]
- 33.Du D et al. Genetic interaction mapping in mammalian cells using CRISPR interference. Nature Methods 14, 577–580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han K et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 35, 463–474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen JP et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nature Methods 14, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasperini M et al. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 1–34 (2018). doi: 10.1016/j.cell.2018.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Najm FJ et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat Biotechnol 36, 179–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrams JT et al. A clonal CD4-positive T-cell line established from the blood of a patient with Sézary syndrome. J. Invest. Dermatol 96, 31–37 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Kaltoft K et al. Establishment of two continuous T-cell strains from a single plaque of a patient with mycosis fungoides. In Vitro Cell. Dev. Biol 28A, 161–167 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel SJ et al. Identification of essential genes for cancer immunotherapy. Nature 548, 537–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.