Abstract
Background
Respiratory syncytial virus (RSV) infection in infants causes significant morbidity and is the strongest risk factor associated with asthma. Metabolites, which reflect the interactions between host cell and virus, provide an opportunity to identify the pathways that underlie severe infections and asthma development.
Objective
To study metabolic profile differences between infants with RSV infection, and human rhinovirus (HRV) infection, and healthy infants. To compare infant metabolic differences between children who do and do not wheeze.
Methods
In a term birth cohort, urine was collected while healthy and during acute viral respiratory infection with RSV and HRV. We used 1H-NMR to identify urinary metabolites. Multivariate and univariate statistics were used to discriminate metabolic profiles of infants with either RSV ARI, or HRV ARI, and healthy infants. Multivariable logistic regression was used to assess the association of urine metabolites with 1st-, 2nd-, and 3rd-year recurrent wheezing.
Results
Several metabolites in nicotinate and nicotinamide metabolism pathways were down-regulated in infants with RSV infection compared to healthy controls. There were no significant differences in metabolite profiles between infants with RSV infection and infants with HRV Infection. Alanine was strongly associated with reduced risk of 1st-year wheezing (OR 0.18[0.0, 0.46]) and 2nd-year wheezing (OR 0.31[0.13, 0.73]), while 2-hydroxyisobutyric acid was associated with increased 3rd-year wheezing (OR 5.02[1.49, 16.93]) only among the RSV infected subset.
Conclusion
The metabolites associated with infant RSV infection and recurrent-wheezing are indicative of viral takeover of the cellular machinery and resources to enhance virulence, replication, and subversion of the host immune-response, highlighting metabolic pathways important in the pathogenesis of RSV infection and wheeze development.
Keywords: Urine metabolomics, Respiratory syncytial virus, Recurrent wheezing, Infant
1. Background
RSV is the major cause of morbidity requiring hospitalization and the major contributor to infant deaths in children worldwide (Nair et al. 2010; Shi et al. 2017; Wright and Piedimonte 2011). Moreover, substantial and consistent evidence links infant RSV acute respiratory infection (ARI) with increased risk of recurrent wheezing and long-term respiratory morbidity, including asthma (Bacharier et al. 2012; Beigelman and Bacharier 2013; Jartti et al. 2009; Sigurs et al. 2010; Stein et al. 1999; Wu et al. 2008).
Despite the significant contribution of RSV to acute and chronic respiratory morbidity and mortality, no effective vaccine or anti-viral treatment is available and treatment is currently limited to supportive care (Hurwitz 2011; Simoes et al. 2015). Progress in understanding the pathogenesis of RSV and host response using in vivo experiments in small animal models have not always translated to humans (Mestas and Hughes 2004). This coupled with the physiological differences between adult and pediatric populations (Papin et al. 2013) and the ethical and technical difficulty of studying infants and children are major obstacles to advancing our understanding of RSV pathogenesis, and the mechanisms through which RSV contributes to recurrent wheezing and asthma development.
Recent developments in high-throughput molecular techniques provide an opportunity for understanding pathogenesis of RSV infection, but a biomarker that accurately and consistently predicts susceptibility to RSV infection, infection severity, and association with subsequent development of persistent wheezing and asthma has yet to be developed (Larkin and Hartert 2015; Openshaw 2013; Rosas-Salazar et al. 2015). Metabolic pathways, as a mirror of the interactions between host cell and virus (Pearce and Pearce 2013; Peeples and Levine 1980), provide an opportunity to understand the mechanisms that underlie severe infections and pathways through which RSV ARI contributes to the development of asthma.
The objective of this study is to identify metabolic pathways associated with RSV ARI. A secondary objective is to assess the association between infant urine metabolites and recurrent wheezing outcomes at 1st-year, 2nd-year, and 3rd-year of life. Urine-based biomarkers are especially appealing in infants, as urine collection is noninvasive, easy to collect, abundant, and comprehensive in metabolite composition. In addition, urine based biomarkers are most efficient in resource-constrained settings where blood drawing requires training and specialized equipment, which is especially important as 99% of RSV-bronchiolitis related mortality occurs in developing countries (Nair et al. 2010; Shi et al. 2017).
2. Methods
2.1. Ethics statement
The Institutional Review Board at Vanderbilt University Medical Center approved the cohort study protocol and informed consent documents. A parent of each infant provided written informed consent for participation in this study.
2.2. Study population
The study was conducted on a subsample of a birth cohort of healthy term infants enrolled in the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE) cohort. The infants were enrolled in the southeastern United States from 2012 to 2014. The cohort was specifically designed to capture each infant’s first RSV infection by enrolling infants born during June through December and facing their first winter virus season in the northern hemisphere. Survey questions about demographic characteristics, exposures, and health were administered in person and biospecimens were collected at enrollment. We conducted biweekly surveillance during winter virus season and infant in-person acute respiratory illness visits with assessment of illness severity, and biospecimen collection at each acute viral illness visit with respiratory virus identification by PCR, as has been previously described (Larkin et al. 2015). A total of 140 urine sample analyses were conducted: 70 urine samples from infants with RSV infection, 60 urine samples from healthy (non-infected) infants, and 10 urine samples from infants with HRV infection.
2.3. Urine collection
The urine samples were collected using an external bag, and were immediately transferred to a specimen cup and stored at 4 °C until processing by the laboratory. The urine was then aliquoted into 2 mL aliquots and stored at − 80 °C until shipment for NMR spectroscopy without interim freeze-thaw cycle.
2.4. Study design
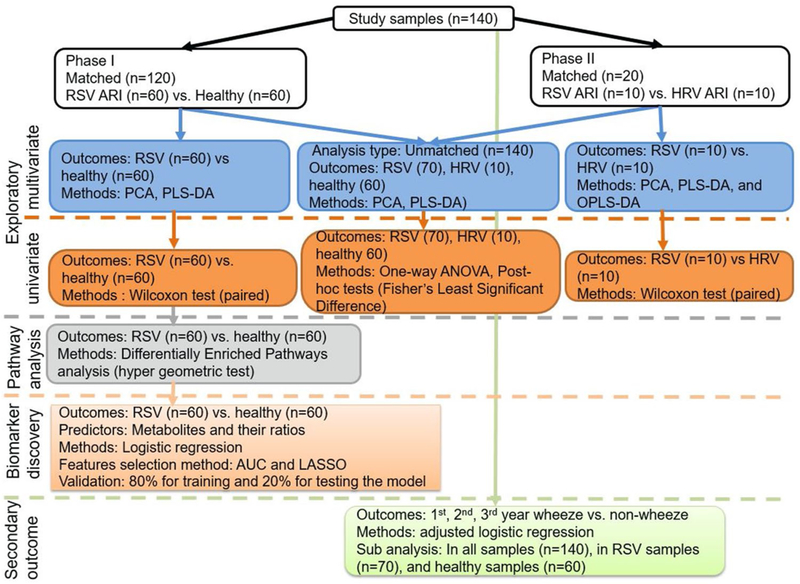
Analysis of a total of 140 urine samples was conducted in two phases (Fig. 1). In Phase 1, we matched 60 urine samples from infants with RSV ARI with 60 urine samples from healthy (no infection) infants. The samples were matched based on the infant age (in months) at urine collection, sex, race, and mode of feeding (exclusively breast fed, formula, or combination). In phase 2, we matched 10 urine samples from infants with RSV ARI with 10 urine samples from infants with HRV ARI to determine whether urine metabolites between healthy and RSV ARI samples were specific to RSV ARI or a global response to respiratory viral illness. In addition to matched analysis, we combined samples from phase 1 and 2 to compare unmatched samples for healthy controls (n = 60), RSV infection (n = 70) and HRV infection (n = 10).
Fig. 1.
Schematic representation of analysis workflow
2.5. Sample preparation
Urine samples were thawed, centrifuged (at 2000×gn for 5 min at 4 °C), filtered (at 12,000×gn for 60 min at 4 °C), and adjusted to pH 7.3 (± 0.05) using BTpH titration unit. The NMR sample was prepared combining 300 µL of urine filtrate with 300 µL of NMR buffer containing 100 mM phosphate buffer in D2O, pH 7.3, and 1.0 mM TMSP (3-Trimethylsilyl 2,2,3,3-d4 propionate). The final TMSP concentration was 0.5 mM in NMR sample.
2.6. Data acquisition and processing
We used a Bruker Avance II 600 MHz spectrometer to acquire one-dimensional (1D) 1H NMR spectra data, two-dimensional (2D) 1H–1H total correlation spectroscopy (TOCSY), and 2D 1H–13C heteronuclear single quantum coherence (HSQC) data. We subjected all free induction decays (FIDs) to an exponential line-broadening of 0.3 Hz. Upon Fourier transformation, we manually phased, baseline corrected, and referenced to the internal standard TMSP at 0.0 ppm for polar samples each spectrum using Topspin 3.5 software (Bruker Analytik, Rheinstetten, Germany). We assigned metabolites to the peak we found in the urine based on 1D 1H, 2D TOCSY, and 2D HSQC NMR experiments and by comparing the chemical shifts and spin–spin couplings with reference spectra found in databases, such as the Human Metabolome Database (HMDB) (Wishart et al. 2017), the biological magnetic resonance data bank (BMRB) (Ulrich et al. 2008), and Chenomx® NMR Suite profiling software (Chenomx Inc. version 8.1). The reference spectra database contains observed peak locations and ratios of heights of spectra from pure chemical compounds or their simulated counterparts, which was used to identify metabolites by matching the observed spectra to the reference spectra in the database. The area of the peaks from a metabolite is directly related to the abundance/quantity of the metabolite.
2.7. Data imputation, transformation, and scaling
We imputed missing metabolites using singular value decomposition (SVD) imputation method (Hastie et al. 1999; Stacklies et al. 2007) and normalized metabolite concentrations to urine creatinine levels. Creatinine concentrations (in millimolar, mM) for healthy control, RSV ARI, and HRV ARI are provided in Fig. E1. We transformed the data using generalized logarithm (glog) transformation method (with a constant of 1 added to all) and scaled using pareto scale before performing all analyses (van den Berg et al. 2006).
2.8. Statistical analysis
Continuous variables were described using medians and interquartile ranges (IQR, 25th and 75th) and categorical variables were summarized using frequencies and percent. We used principal component analysis (PCA) to identify patterns of the urine metabolites and for visual inspection of outliers, and none were detected.
2.8.1. Exploratory analysis: metabolites discriminate between RSV ARI, HRV ARI, and healthy controls
In exploratory analysis, we used Partial least square discriminant analysis (PLS-DA) (Barker and Rayens 2003) and its orthogonal extension (OPLS-DA) (Bylesjo et al. 2006; Trygg and Wold 2002; Wiklund et al. 2008) to discriminate between RSV ARI, HRV ARI and healthy control samples and to identify metabolites that are important for discrimination. To ensure model reliability PLS-DA models were internally validated using 10-fold cross validation. The predictive ability assessment (Q2) statistic was reported as a result of cross-validation to provide a qualitative measure of consistency between the predicted and original data. We used permutation (n = 2000) to test the significance of Q2 statistics (Westerhuis et al. 2008). A permutation test evaluates whether the specific classification of the individuals in the a priori comparison groups is significantly better than any other random classification in arbitrary groups by reshuffling the group label (Golland et al. 2005).
2.8.2. Exploratory analysis: statistically different metabolic profiles between RSV ARI, HRV ARI, and healthy controls
Although multivariate analysis helps us discriminate between groups (RSV ARI, HRV ARI and healthy control samples), it does not provide a profile of metabolites significantly different between these groups. Therefore, we used univariate statistics such as: Kruskal Wallis followed by Dunn’s post-hoc test for three factors comparisons (healthy control, RSV ARI, and HRV ARI), and Wilcoxon matched-pairs signed rank test to test for significant differences in metabolite profiles between two matched groups (healthy vs. RSV ARI, and HRV ARI vs. RSV ARI). To determine statistical significance, p values were evaluated at 0.05 after adjusting for multiple testing using Benjamini–Hochberg (BH) false discovery rate (FDR).
2.8.3. Differentially enriched metabolic pathways in infants with RSV infection compared to healthy infants
Further, pathway databases were queried with MetScape-3 (Karnovsky et al. 2012) and metaboanalyst (Xia and Wishart 2016) to identify metabolic pathways of significantly altered sets of metabolites in infants with RSV ARI versus healthy. We used the list of metabolites identified using univariate analyses that compared healthy controls versus RSV ARI to query the pathway databases. We used hyper-geometric test to determine the significantly enriched metabolic pathways.
2.8.4. RSV biomarker discovery and performance evaluation
To select biomarkers that distinguish between infants with RSV ARI and healthy controls, we evaluated models with metabolites and their ratios to each other (all possible pairwise ratios were calculated). We manually selected metabolites and their pairwise ratios (features) in the final model based on the prior knowledge about the metabolites, univariate prediction performance (using area under curve [AUC]) (Xia et al. 2015), and prediction performance (frequency of being select into the model using the least absolute shrinkage and selection operator [LASSO]) in multivariable logistic regression. During the selection of metabolites and their ratios using univariate statistics, p values were adjusted for multiple testing with FDR correction. Similarly, LASSO was applied in the context of sparse regression model including all metabolites and their ratios, where metabolites with non-zero regression coefficients from LASSO were considered to be “significant” in the sense that they are associated with the outcome (RSV vs. healthy). However, the “significance” criteria for LASSO should not be confused with significance for hypothesis testing (Xia et al. 2015). We randomly divided the sample into training (80%) and holdout (20%) sets. We built the model on the training set and tested the model on the holdout set of the data. The holdout data was not used during the model building process. The final model was evaluated based on AUC and a permutation test (n = 1000) conducted for accuracy of prediction and significance of AUC.
2.8.5. Analysis of association between infant urine metabolites and recurrent wheezing
For our secondary analysis we used multivariable logistic regression to estimate relative odds of urine metabolite unit increases with the 1st-, 2nd-, and 3rd-year recurrent wheezing outcomes adjusted for covariates: sex, race and ethnicity, maternal asthma, and second hand smoke exposure. The result of the p values from logistic regression were FDR corrected for multiple testing. A wheeze is a high-pitched, musical, adventitious lung sound produced by airflow through an abnormally narrowed or compressed airway(s) (Gong 1990). The outcome of recurrent wheeze was defined as three or more wheezing events in the last 12 months, or wheeze with use of asthma medications in the last 12 months based on parental report.
3. Results
3.1. Population characteristics
A total of 140 individual infant samples (60 healthy control, 70 RSV infection, 10 HRV infection) were analyzed for this study. The infants were 62.7% (89) female, 68.6% (97) white, 12.9% (18) black, and 11.4% (16) Hispanic (Table 1). The median age at time of urine collection was 120 days (IQR, 66–163) for all samples, 119 days (IQR = 66–158) for RSV ARI samples, and 104 days (IQR, 65–148) for healthy control samples.
Table 1.
Characteristics of the study population
| Demographic characteristic | Healthy n = 60 N (%) | RSV ARI n = 70 N (%) | HRV ARI n = 10 N (%) | Total n = 140 N (%) |
|---|---|---|---|---|
| Sex | ||||
| Female | 36 (60.7) | 44 (62.3) | 8 (80.0) | 88 (62.9) |
| Male | 24 (39.3) | 26 (37.7) | 2 (20.0) | 52 (37.1) |
| Race and ethnicity | ||||
| White | 38 (63.9) | 48 (68.1) | 10 (100.0) | 96 (68.6) |
| Black | 9 (14.7) | 9 (13.0) | 18 (12.9) | |
| Hispanic | 8 (13.1) | 8 (11.6) | 16 (11.4) | |
| Other/multiple | 5 (8.2) | 5 (7.3) | 10 (7.1) | |
| Recurrent wheezing (ages 0–1) | 18 (29.5) | 34 (49.3) | 4 (40.0) | 56 (40.0) |
| Recurrent wheezing (ages 1–2) | 13 (21.3) | 34 (49.3) | 5 (50.0) | 52 (37.1) |
| Recurrent wheezing (ages 2–3) | 10 (16.4) | 32 (46.4) | 3 (30.0) | 45 (32.1) |
| Exclusively formula fed at enrollment | 23 (37.7) | 28 (40.6) | 6 (60.0) | 57 (40.7) |
| Maternal smoking at enrollment | ||||
| Current | 12 (20.3) | 16 (26.2) | 1 (10.0) | 29 (23.3) |
| Former | 8 (13.6) | 7 (11.5) | 15 (12.5) | |
| Never | 39 (66.1) | 38 (62.3) | 9 (90.0) | 86 (64.2) |
| Maternal education level | ||||
| College | 32 (54.1) | 41 (60.9) | 7 (70.0) | 82 (58.5) |
| High school | 23 (37.7) | 21 (30.4) | 3 (30.0) | 47 (33.6) |
| < High school | 5 (8.2) | 6 (8.7) | 11 (7.9) | |
| Mean (SD) | Mean (SD) | Mean (SD) | ||
| Gestational age, weeks | 38.9 (1.2) | 38.9 (0.9) | 39.0 (1.1) | 38.9 (1.1) |
| Acute respiratory illness severity scorea | 4.4 (2.6) | 2.4 (1.1) | ||
RSV respiratory syncytial virus, HRV human rhinovirus
Respiratory severity score on scale to 0–12 with higher scores indicating more severe disease
3.2. Multivariate exploratory analysis of urine metabolomics data
Exploratory analysis of the combined and unmatched data (healthy control [n = 60], RSV ARI [n = 70], and HRV ARI [n = 10]) with PCA shows that 48.6% of the variation in the data was explained by five principal components. We did not observe a clustering pattern in the data. The first, second and third principal components represent 13.9%, 9.8%, and 9.2% of the variation in the data. PLS-DA analysis shown in the score plot shows slight separation between the groups (RSV vs. healthy, and HRV vs. healthy) (Fig. 2). The internal validation of the PLS-DA models using 10-fold cross validation shows the variation in the data can be represented by the first two components with prediction ability of Q2 = 0.18. The permutation analysis model performance (n = 2000) shows significant separation distance between groups (RSV vs. healthy, HRV vs. healthy, and RSV vs. HRV) groups (empirical p value < 0.002) and accuracy of predicting the groups in the training set (empirical p < 0.0035). The five most important metabolites that discriminated between healthy controls, RSV ARI, and HRV ARI were 1-methylnicotinamide, creatine, 4-deoxythreonic acid, citrate, 2-aminobutyrate, ordered as listed.
Fig. 2.
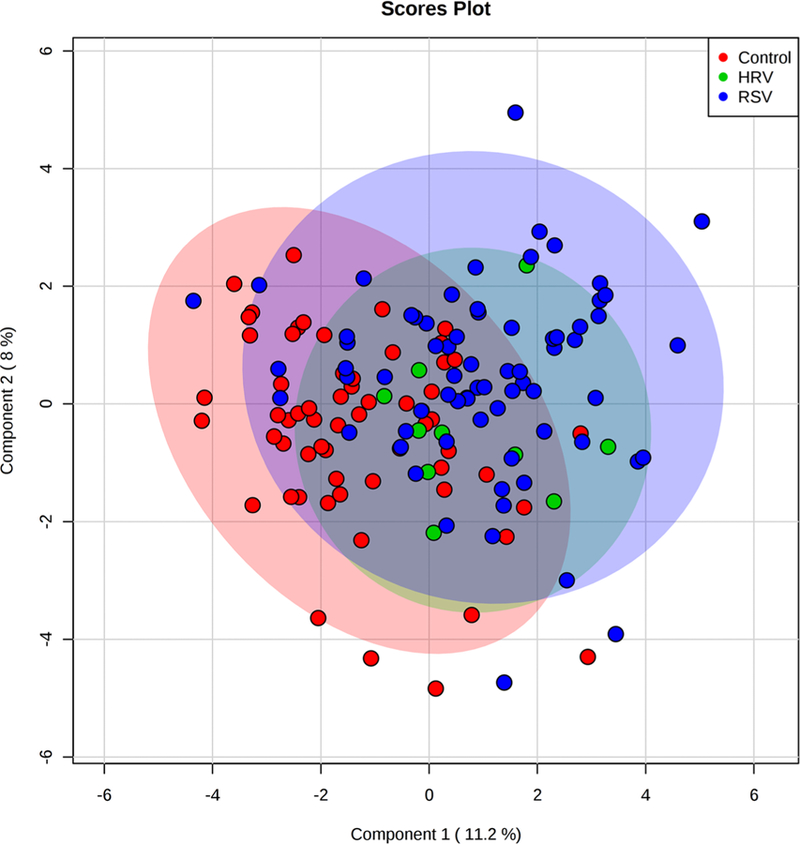
Partial least square discriminant analysis (PLS-DA) score plot showing the first two components of the urine sample projection based on the 31 metabolites identified in the unmatched data. The red, orange, and blue shaded ovals show the classification of the samples into healthy controls, human rhinovirus (HRV) ARI, and respiratory syncytial virus (RSV) ARI, respectively, based on the first two components of PLS-DA
For matched RSV ARI and healthy control urine samples, the internal validation of the PLS-DA models using 10-fold cross validation shows Q2 statistic (Q2 = 0.24) for two component based predictive ability. The permutation analysis (n = 2000) for model performance shows significant separation distance between RSV ARI and healthy control urine samples (empirical p value = 0.002) and accuracy of predicting the group membership during the training (empirical p value = 0.0005). 1-methylnicotinamide, creatine, citrate, 4-deoxythreonic acid, and 2-aminobutyrate, in the order listed, were the five most important metabolites in separating the healthy controls and RSV ARI groups.
For matched RSV and HRV urine samples, both 10-fold cross validation and permutation analysis of PLS-DA models, non-statistically-significant separation between the RSV and HRV urine samples or predictive accuracy of the groups was observed. OPLS-DA analysis for matched RSV and HRV ARI samples attributed 5.3% of the variation to the viruses, but the permutation analysis was not statistically significant (Q2 = 0.15, permutation p value = 0.38).
3.3. Differential metabolomics profiles of infants with RSV ARI compared to healthy controls
Multivariate analysis was helpful in showing visual separation between groups (RSV ARI, HRV ARI, and healthy control) and providing the rank of metabolites important in group separation. We also wanted to identify metabolites that were significantly different between these groups using univariate statistics. Results from univariate statistical comparisons were used in downstream analysis, such as metabolic pathways differentially affected. We combined the two-phase sample data for unmatched non-parametric one-way ANOVA analysis (Kruskal Wallis test followed by Dunn’s test comparisons) with a total of 140 samples (10 HRV, 70 RSV, and 60 controls). We found 11 metabolites that were significantly different between RSV ARI, HRV ARI, and healthy control infant groups: 1-methylnicotinamide, 4-deoxythreonic acid, citrate, creatine, hypoxanthine, alanine, succinate, 3-hydroxyisovalerate, acetone, valine, and 2-aminobutyrate (Table 2 and Fig. E2). There was notable consistency in the results from univariate and PLS-DA analysis, as these metabolites were also among the top metabolites discriminating between RSV ARI, HRV ARI, and healthy control infant groups using PLS-DA analysis. The post-hoc analysis using Dunn’s test showed that 1-methylnicotinimide, succinate, 3-hydroxyisovalerate, acetone, alanine, and 2-aminobutyrate were significantly different only between healthy control infants and RSV ARI infants, while 4-deoxythreonic acid, citrate, creatine, and hypoxanthine were significantly different between both viruses (HRV ARI and RSV ARI) and healthy control infant urine samples. None of the metabolites were significantly different between HRV and healthy controls and between infants with RSV ARI and HRV ARI after adjusting FDR for multiple testing.
Table 2.
Comparison of urinary metabolites that discriminate between unmatched RSV acute respiratory infection (n = 70), HRV acute respiratory infection (n = 10), and healthy control (n = 60) infant urine samples using Kruskal Wallis test followed by Dunn’s post-hoc comparisons
| Metabolitesa | Chi-squared | Unadjusted p value | FDR adjusted p value | Dunn’s Post-hoc comparison |
|---|---|---|---|---|
| 1-Methylnicotinamide | 32.14 | 1.05 × 10−7 | 3.26 × 10−6 | Control − RSV |
| 4-deoxythreonic acid | 23.94 | 6.35 × 10−6 | 8.51 × 10−5 | Control − HRV; Control – RSV |
| Citrate | 23.41 | 8.23 × 10−6 | 8.51 × 10−5 | Control − HRV; Control − RSV |
| Creatine | 17.05 | 1.98 × 10−4 | 0.002 | Control − HRV; Control − RSV |
| Hypoxanthine | 15.93 | 3.48 × 10−4 | 0.002 | Control − HRV; Control − RSV |
| Alanine | 12.73 | 0.002 | 0.009 | Control − RSV |
| Succinate | 10.67 | 0.005 | 0.02 | Control − RSV |
| 3-Hydroxyisovalerate | 9.45 | 0.009 | 0.03 | Control − RSV |
| Acetone | 9.23 | 0.01 | 0.03 | Control − RSV |
| Valine | 8.59 | 0.01 | 0.04 | Control − HRV; Control − RSV |
| 2-Aminobutyrate | 8.43 | 0.02 | 0.04 | Control − RSV |
Significance was cut off at p value of 0.05 after adjusting for multiple testing using Benjamini–Hochberg false discovery rates (FDR) FDR false discovery rate, RSV respiratory syncytial virus, HRV human rhinovirus
Metabolites were normalized to creatinine
We next analyzed matched data comparing infants with RSV ARI with matched healthy controls (n = 120, 60 matched pairs). 1-methylnicotinamide, citrate, 4-deoxythreonic acid, 2-aminobutyrate, creatine, alanine, succinate, cis-aconitate, 3-acetone, hypoxanthine, tyrosine, 3-hydroxyisovalerate, and pantothenate were significantly different among the groups at p value < 0.05 with FDR correction (Table 3 and Fig. E3). While most of these metabolites were decreased in urine of infants with RSV ARI compared to healthy control infants, 2-aminobutyrate, acetone, cis-aconitate, and hypoxanthine were increased. The fold change (FC) for citrate, 1-methylnicotinamide, 4-deoxythreonic acid, creatine, 2-aminobutyrate, and succinate was greater than 1.5. The largest fold change of 2.03 was observed with acetone (FDR adjusted p value = 0.04 and AUC = 0.63). 1-methylnicotinamide and citrate were fairly good predictors of infants with RSV ARI, with AUC of 0.78 (CI 0.69, 0.86) and 0.77 ( CI 0.68, 0.84).
Table 3.
Comparison of urinary metabolites that discriminate between infants with RSV acute respiratory infection (n = 60) and matched healthy controls (n = 60) using Wilcoxon rank-sum test
| Metabolitesa | Fold change (RSV/healthy) | Wilcoxon rank-sum test, FDR adjusted p value | AUC |
|---|---|---|---|
| 1-Methylnicotinamide | 0.55 | 3.62 × 10−6 | 0.78 |
| Citrate | 0.65 | 3.62 × 10−6 | 0.77 |
| 4-Deoxythreonic acid | 0.59 | 0.0002 | 0.73 |
| 2-Aminobutyrate | 1.99 | 0.002 | 0.67 |
| Creatine | 0.52 | 0.003 | 0.68 |
| Alanine | 0.77 | 0.003 | 0.68 |
| Succinate | 0.59 | 0.003 | 0.67 |
| Cis-aconitate | 1.27 | 0.02 | 0.62 |
| Acetone | 2.03 | 0.04 | 0.63 |
| Hypoxanthine | 1.79 | 0.04 | 0.68 |
| Tyrosine | 0.79 | 0.04 | 0.63 |
| 3-Hydroxyisovalerate | 0.82 | 0.04 | 0.64 |
| Pantothenate | 0.85 | 0.04 | 0.62 |
Significance was cut off at p value of 0.05 after adjusting for multiple testing using Benjamini–Hochberg false discovery rates (FDR)
FDR false discovery rate, AUC area under the curve, RSV respiratory syncytial virus. Samples matched on age, sex, race and ethnicity, and feeding (breast vs. formula or combination)
Metabolites were normalized to creatinine
3.4. Metabolic pathways distinguishing infants with RSV ARI from healthy controls
We conducted pathway analysis of the set of significantly altered metabolites (in infants with RSV ARI compared to healthy controls) to understand the biological context of the significantly different metabolite profiles between healthy controls and acute RSV respiratory infection infant urine samples. We used the list of metabolites we identified with univariate statistical analysis that differentiated between healthy control and ARI RSV subjects. The significantly enriched pathways were determined with hyper-geometric test. As shown in Fig. 3 and Table 4, the following metabolic pathways were the most differentially affected during RSV ARI: nicotinate and nicotinamide metabolism; glycine, serine and threonine metabolism; selenoamino acid metabolism; alanine metabolism; glucose-alanine cycle; citric acid cycle; glutamate metabolism; arginine and proline metabolism; and catecholamine biosynthesis.
Fig. 3.
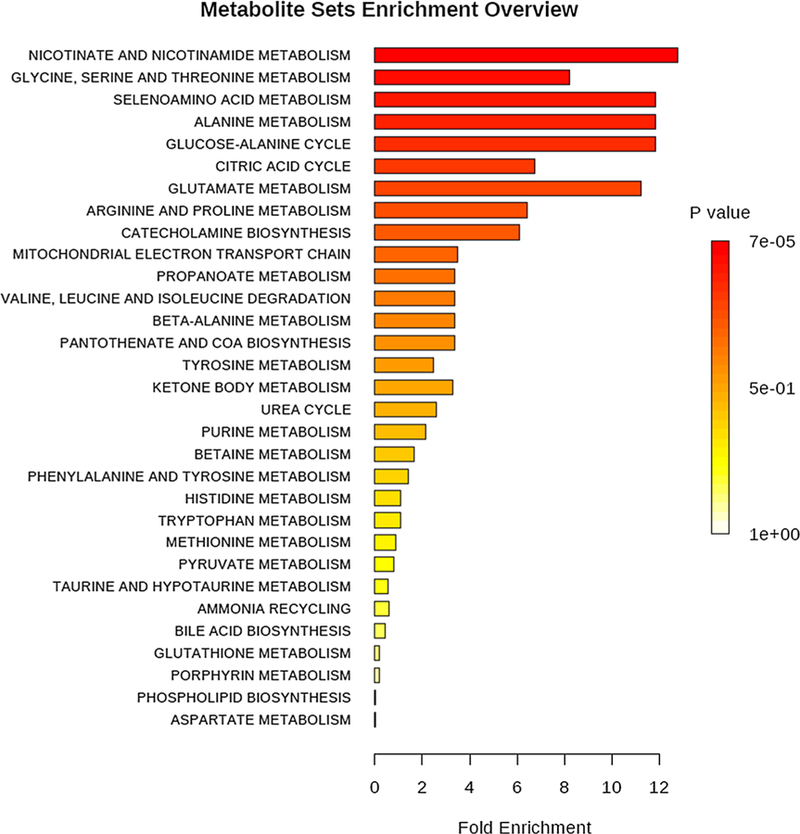
Differentially enriched metabolic pathways in infants with RSV acute respiratory infection compared to matched healthy control infants
Table 4.
Differentially enriched metabolic pathways in infants with RSV acute respiratory infection compared to matched healthy control infants using Wilcoxon rank-sum test and adjusting for multiple testing with Benjamini–Hochberg false discovery rate (FDR)
| Metabolitesa | Total compounds in the Pathway | Significantly altered metabolites | Q-statistic | FDR corrected p value |
|---|---|---|---|---|
| Nicotinate and nicotinamide metabolism | 13 | 2 | 9.74 | 6.94 × 10−5 |
| Glycine, serine and threonine metabolism | 26 | 3 | 6.26 | 0.003 |
| Selenoamino acid metabolism | 15 | 1 | 9.05 | 0.003 |
| Alanine metabolism | 6 | 1 | 9.05 | 0.003 |
| Glucose-alanine cycle | 12 | 1 | 9.05 | 0.003 |
| Citric acid cycle | 23 | 4 | 5.15 | 0.003 |
| Glutamate metabolism | 18 | 1 | 8.58 | 0.003 |
| Arginine and proline metabolism | 26 | 2 | 4.89 | 0.005 |
| Catecholamine biosynthesis | 5 | 1 | 4.67 | 0.04 |
Q-statistics is a global test statistic for quantitative metabolite enrichment analysis as an indicator of correlation with phenotype (RSV vs. healthy), FDR false discovery rate and RSV respiratory syncytial virus
Metabolites were normalized to creatinine
3.5. Biomarker discovery for RSV ARI
To identify biomarkers that distinguish between infants with RSV ARI and healthy controls, we evaluated models with metabolites and their ratio to each other (all possible ratios were calculated). With a total of 60 matched pairs, we randomly divided into 80% for training (n = 96, 48 pairs) and 20% for evaluation (n = 24, 12 pairs) holdout data. Holdout data was not used to build the model. We selected a model with two variables to distinguish between infants with RSV ARI and healthy controls: citrate/cis-aconitate ratio based on the highest univariate AUC performance (AUC = 0.84) and 1-methylnicotinamide/acetone (AUC = 0.79) which also had the highest frequency of being selected into multivariate logistic model using LASSO. The two variable model as classifier using logistic regression consistently yielded average AUC of 0.85 (95% CI 0.76–0.95) on 100-fold cross validation on training samples and AUC of 0.83 on the holdout samples (Fig. 4). The accuracy of the prediction of this model is 0.75 for training samples and 0.83 for holdout samples. The permutation test (n = 1000) for accuracy of prediction and AUC were statistically significant (empirical p value < 0.001). A p value < 0.05 means that given a randomly permuted outcome variable there is less than a 5% chance that a model of similar performance to the “true” non-permuted model will be produced (Bijlsma et al. 2006). This model is not modeling method dependent in that it yields similar performance results using SVM and PLS-DA. Moreover, as shown in Table 3, each of these metabolites was significantly different between infants with RSV ARI and matched healthy controls.
Fig. 4.
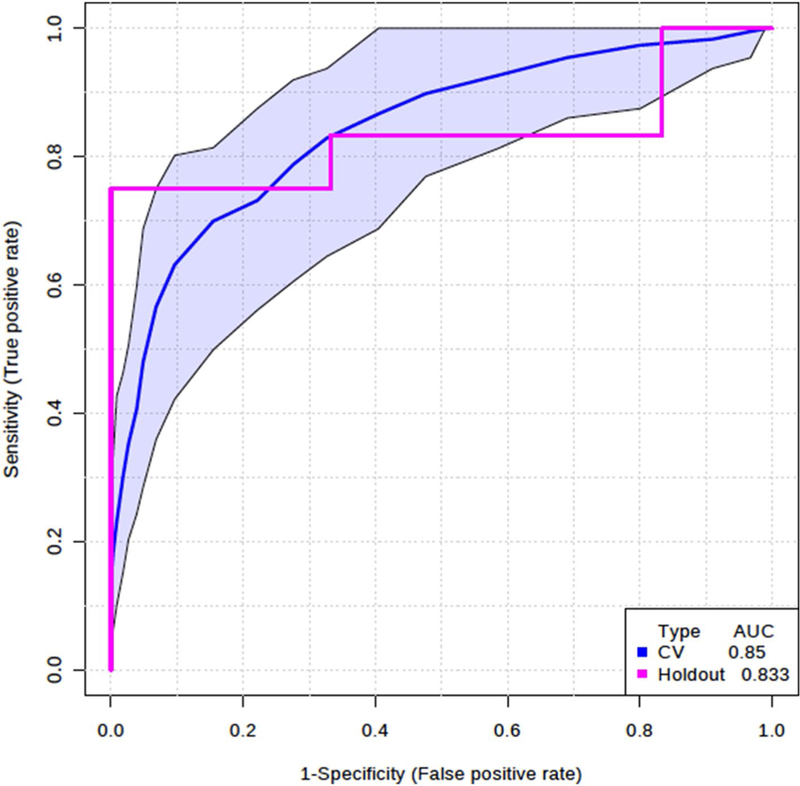
Model comprised of citrate/cis-aconitate and 1-methylnicotinamide/acetone ratios distinguishing infants with RSV acute respiratory infections from healthy controls
3.6. Differentiating RSV from other respiratory virus (HRV)
To see if the altered metabolites are specific to a virus, first we compared unmatched urine samples from infants with RSV ARI (n = 70) to urine samples from infants with HRV ARI (n = 10) using Wilcoxon signed-rank test. Glycine (p value = 0.03) and betaine (p value = 0.048) were significantly different between urine samples from infants with RSV and HRV ARI, however, none of the metabolites were significantly different after adjusting for FDR. The RSV ARI samples were combined from phase-1 (n = 60) and phase-2 (n = 10) to enhance the power, however, we should note that the sample sizes are unbalanced between the two comparison groups (RSV ARI vs. HRV ARI). As an additional conservative approach, we compared matched urine samples from infants with RSV ARI and HRV ARI (N = 20, 10 pairs). There were no significant differences in metabolite profiles even before FDR adjustment. See online supplement for details.
3.7. Metabolite associations with clinical wheezing outcomes
For our secondary objective, we evaluated the effect of infant urine metabolites on the outcomes of 1st-, 2nd-, and 3rdyear recurrent wheezing using logistic regression adjusted for covariates including sex, race and ethnicity, maternal asthma, and second hand smoke exposure. Several metabolites were significantly associated with 1st-, 2nd-, and 3rdyear recurrent wheezing, however, only alanine remained significantly associated with reduced risk for 1st-year recurrent wheezing after FDR adjustment (FDR adjusted p value < 0.001). Alanine and tyrosine were consistently associated with reduced 1st- and 2nd-year recurrent wheezing although not significant after adjusting for multiple testing for second year. Among infants with RSV ARI, alanine, tyrosine, and 4-deoxythreonic acid were associated with 1st-year recurrent wheezing; acetate was associated with 2nd-year recurrent wheezing, and 2-hydroxyisobutyrate was associated with 3rd-year recurrent wheezing. However, except for the association between alanine and 1styear recurrent wheezing using combined sample (n = 140) analysis, none of the results were significant after adjusting for multiple testing. See online supplement for details.
4. Discussion
RSV infection in infants may cause life-threatening disease and is a strongly and consistently associated risk factor for the development of asthma. However, our understanding of the mechanisms through which RSV causes asthma is limited largely to animal models or human association studies. Thus our study aimed to fill gaps in our knowledge that may contribute to the development of targeted preventive or therapeutic interventions by: (1) determining metabolic profiles that distinguish infants with RSV ARI from healthy infants and from those with HRV ARI, and (2) assessing the association of urinary metabolites with childhood recurrent wheezing outcomes.
We find that urine metabolites clearly distinguish between infants with acute RSV ARI or HRV ARI respiratory viral infection from healthy controls. Not surprisingly, however, the metabolite profiles of infants with RSV ARI and HRV ARI were largely overlapping. Betaine was the only significantly different metabolite after FDR correction, that distinguished between RSV ARI (n = 70) and HRV ARI (n = 10). We did not find any significantly different metabolites in matched infants (n = 10 paired infants with RSV ARI and HRV ARI) also after FDR correction. The identified metabolite patterns seen during infant viral infection are consistent with and indicative of active viral infection (Delgado et al. 2012; Milner et al. 2014; Munger et al. 2006; Ritter et al. 2010; Sanchez and Lagunoff 2015; Yu et al. 2011), but were not specific to RSV ARI. The only other study comparing children with RSV, non-RSV virus, and bacterial infection also found similar metabolic profile differences (Adamko et al. 2016). For example, similar to our study, Adamko et al. found that betaine was elevated in children with RSV infection compared to both non-infected children and children with non-RSV infection. However the Adamko et al. study differs from ours in that the children were hospitalized, nonRSV virus comparators were diverse (adenovirus, influenza, and parainfluenza), and age ranges of children during RSV ARI were large with median age about 8 (range 6.2–9.9) months, whereas our study prospectively followed infants from near birth to first RSV ARI and the median age at ARI was about 3 (interquartile range, 2–5) months.
Nicotinate and nicotinamide metabolic pathways were specifically decreased in infants with RSV ARI compared to healthy controls. 1-Methylnicotinamide, the amide form of niacin or vitamin B3, is a precursor in the synthesis of NAD + and NADP +, which plays a role in numerous metabolic pathways including energy production, regulation of cellular redox, circadian rhythm, and longevity (Musfeld et al. 2001). Anti-inflammatory properties of 1-methylnicotinamide have also been recently described, including as a potential scavenger of reactive forms of oxygen, in particular superoxide radical anions and hydroxyl radicals, as well as reducers of adherence of pro-inflammatory cells and molecules to the surface of vascular endothelium (Gebicki et al. 2003). Thus, infants’ ability to fend off inflammation is likely decreased during RSV infection as infants with RSV acute respiratory infection have lower 1-methylnicotinamide compared to healthy infants.
In exploring urine metabolites as a biomarker for RSV, the model composed of citrate/cis-aconitate ratio and 1-methylnicotinamide/acetone ratio consistently yielded average AUC of 0.85 (95% CI 0.76–0.95) on 100-fold cross validation on training samples and AUC of 0.83 on the holdout samples. In citrate/cis-aconitate ratio, both metabolites are in the citric acid cycle pathway; cis-aconitate is an intermediate metabolite during conversion of citrate to isocitrate. In addition to energy production, citrate is used for production of the pro-inflammatory molecule prostaglandin E2 (PGE2) and oxaloacetate to make nicotinamide adenine dinucleotide phosphate (NADPH) needed for NO and ROS via Acetyl-CoA (Infantino et al. 2013, 2014). On the other hand, citrate is a substrate for production itaconate via cisaconitate, which acts as a negative regulator of inflammation by modulating the synthesis of the inflammatory mediators (Lampropoulou et al. 2016). Decreased levels of citrate and increased levels of cis-aconitate during RSV ARI compared to healthy controls may indicate that citrate is depleted to supply pro-inflammatory and anti-inflammatory molecule needs of immune cells to counter viral replication. Both metabolites in 1-methylnicotinamide/acetone ratio are precursors to NADP + production. As discussed above, 1-methylnicotinamide plays a role in several metabolic pathways including energy production. Acetone plays a role in energy supply to vital organs during metabolic catastrophe (Kalapos 1999). The exact relationship between 1-methylnicotinamide and acetone has not been described in the literature, however, it is possible that the increase in acetone and decrease in 1-methylnicotinamide levels in infants with RSV ARI compared to healthy infants are indicators of a cellular switch in energy production to survival mode and a tendency toward higher inflammatory metabolism due to stress from viral infection. Nonetheless, what is clear from the literature is that all metabolic markers discussed are involved in energy production and inflammation pathways critical during infections. This study is the first attempt to define metabolic biomarkers differentiating infants with RSV ARI and healthy infants. These promising results need next to be validated in a larger sample.
Lastly, we also identified metabolites associated with wheezing outcomes following RSV. Only the association of alanine with 1st-year recurrent wheezing remained significant after FDR correction at 0.05. Alanine has been previously identified as a metabolite discriminating healthy and chronic obstructive pulmonary disease in older adults (Wang et al. 2013). Alanine and tyrosine (a metabolite with neurotransmitter properties) metabolic pathways were the most consistently associated pathways with recurrent wheezing at 1- and 2-years following RSV infection. 2-hydroxyisobutyrate, which is mainly eliminated through exhaled air (Benson et al. 2001), was the strongest predictor of 3rd year recurrent wheezing after RSV infection. Previous studies show that 2-hydroxyisobutyrate and 4-deoxythreonic acid (which was associated with 1st-year wheezing following RSV) are organic acids subsequent to l-threonine and are elevated in urine of juvenile-onset (Type 1) diabetes mellitus patients (Kassel et al. 1986). While RSV ARI has long been associated with markedly increased risk of future asthma, the majority of infants with RSV ARI do not in fact develop asthma. Identifying metabolic pathways associated with increased wheezing and asthma risk enhances our understanding of the key pathways that may underlie this association.
The main strength of this study is that it was conducted on a population-based birth cohort of infants specifically designed to capture the first infant RSV ARI, and understand the metabolic patterns associated with RSV ARI. This design allowed for tremendous accuracy in phenotyping the infants. Because urine samples tested from healthy infants and from infants during RSV infection were matched on infant age, race and ethnicity, and sex, the variation in metabolite concentrations due to these factors were minimized. Despite the strengths in study design there are several limitations that must be considered. Even though our sample is well matched, there are genetic and environmental factors that we can neither measure nor control for, as well as residual confounding. Although this is a relatively large human metabolomics study, it is not adequately powered to discern the combined effects of metabolites, and the mediating effects of environmental exposures. The urine was collected only once a day, typically during the morning hours, which may not represent the variation in the urinary metabolome throughout the day, although morning is preferable if serial or 24-h collection is not possible, and infant diet is not as variable throughout the day as it is for older children and adults (Bernini et al. 2011). Lastly, even though our results are based on internal validation techniques we did not perform external validation on an independent sample.
5. Conclusions
We identified urinary metabolites that are involved in RSV ARI compared to healthy control infants and infants with HRV ARI. We also identified metabolites predictive of recurrent wheezing. We demonstrated that altered metabolic pathways provide insight into the pathogenesis of RSV ARI and development of recurrent wheezing in children, although the involvement of these pathways and predictive metabolite biomarkers has to be confirmed in larger sample sizes and replicated in independent samples. These findings together enhance our understanding of RSV pathogenesis and pathways dominant in asthma development and will hopefully lead to the establishment of novel urine based biomarkers for asthma that are easy to collect in future studies.
Supplementary Material
Acknowledgements
We thank the NMR-Based Metabolomics Core at Cincinnati Children’s Hospital Medical Center for advising on study design and processing our samples.
Funding This work was supported by the National Institute of Health U19AI95227, K24 AI 77930, R21HD087864, and T32HL087738. The funding agencies did not have any role in the study design, collection, analysis and interpretation of data, the writing of this report, or the decision to submit for publication. The funding was provided by National Institute of Allergy and Infectious Diseases.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11306–018-1431-z) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest Dr. Hartert has previously served as a consultant to Novavax and Regeneron. Dr. Kedir N Turi, Dr. Lindsey Romick-Rosendale, Tebeb Gebretsadik, Dr. Miki Watanabe, Dr. Steven Brunwasser, Dr. Larry J Anderson, Dr. Martin L Moore, Dr. Emma K Larkin, Dr. Ray Stokes Peebles and Dr. Tina V. Hartert have no conflicts of interest.
Ethical approval This study was approved by the ethical committee of Vanderbilt University Medical Center, Nashville TN.
Informed consent Informed written consent was obtained from the parent of each infant for study participation.
References
- Adamko DJ, Saude E, Bear M, Regush S, & Robinson JL (2016). Urine metabolomic profiling of children with respiratory tract infections in the emergency department: A pilot study. BMC Infectious Diseases, 16, 439 10.1186/s12879-016-1709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacharier LB, et al. (2012). Determinants of asthma after severe respiratory syncytial virus bronchiolitis. The Journal of Allergy and Clinical Immunology, 130, 91–100. 10.1016/j.jaci.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker M, & Rayens W (2003). Partial least squares for discrimination. Journal of Chemometrics, 17, 166–173. 10.1002/cem.785. [DOI] [Google Scholar]
- Beigelman A, & Bacharier LB (2013). The role of early life viral bronchiolitis in the inception of asthma. Current Opinion in Allergy and Clinical Immunology, 13, 211–216. 10.1097/ACI.0b013e32835eb6ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson JM, Barr EB, & Krone JR (2001). MTBE inhaled alone and in combination with gasoline vapor: Uptake, distribution, metabolism, and excretion in rats. Research Report Health Effects Institute, 102, 73–94; (discussion 95–109). [PubMed] [Google Scholar]
- Bernini P, Bertini I, Luchinat C, Nincheri P, Staderini S, & Turano P (2011). Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. Journal of Biomolecular NMR, 49, 231–243. 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- Bijlsma S, et al. (2006). Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Analytical Chemistry, 78, 567–574. 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- Boeke CE, et al. (2012). Gestational intake of methyl donors and global LINE-1 DNA methylation in maternal and cord blood Prospective results from a folate-replete population. Epigenetics, 7, 253–260. 10.4161/epi.7.3.19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylesjo M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, & Trygg J (2006). OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. Journal of Chemometrics, 20, 341–351. 10.1002/cem.1006. [DOI] [Google Scholar]
- Delgado T, Sanchez EL, Camarda R, & Lagunoff M (2012). Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLOS Pathogens, 8, e1002866 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Warskulat U, Hensel F, & Haussinger D (1998). Osmolyte strategy in human monocytes and macrophages: Involvement of p38(MAPK) in hyperosmotic induction of betaine and myoinositol transporters. Archives of Biochemistry and Biophysics, 354, 172–180. 10.1006/abbi.1998.0661. [DOI] [PubMed] [Google Scholar]
- Dominguez-Salas P, et al. (2014). Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nature Communications 10.1038/ncomms4746 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebicki J, Sysa-Jedrzejowska A, Adamus J, Wozniacka A, Rybak M, & Zielonka J (2003). 1-methylnicotinamide: A potent antiinflammatory agent of vitamin origin. Polish Journal of Pharmacology, 55, 109–112. [PubMed] [Google Scholar]
- Golland P, Grimson WE, Shenton ME, & Kikinis R (2005). Detection and analysis of statistical differences in anatomical shape. Medical Image Analysis, 9, 69–86. 10.1016/j.media.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong HJ (1990). Wheezing and Asthma. In: Walker HK, Hall WD, Hurst JW (eds) Clinical methods: The history, physical, and laboratory examinations Boston: Butterworths. [PubMed] [Google Scholar]
- Haggarty P, Hoad G, Campbell DM, Horgan GW, Piyathilake C, & McNeill G (2013). Folate in pregnancy and imprinted gene and repeat element methylation in the offspring. American Journal of Clinical Nutrition, 97, 94–99. 10.3945/ajcn.112.042572. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Sherlock G, Eisen M, Brown P, & Botstein D (1999). Imputing missing data for gene expression arrays Stanford: Stanford University Statistics Department Technical Report. [Google Scholar]
- Hurwitz JL (2011). Respiratory syncytial virus vaccine development. Expert Review of Vaccines, 10, 1415–1433. 10.1586/erv.11.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantino V, Iacobazzi V, Menga A, Avantaggiati ML, & Palmieri F (2014). A key role of the mitochondrial citrate carrier (SLC25A1) in TNF alpha- and IFN gamma-triggered inflammation. Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms, 1839, 1217–1225. 10.1016/j.bbagrm.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infantino V, Iacobazzi V, Palmieri F, & Menga A (2013). ATPcitrate lyase is essential for macrophage inflammatory response. Biochemical and Biophysical Research Communications, 440, 105–111. 10.1016/j.bbrc.2013.09.037. [DOI] [PubMed] [Google Scholar]
- Jartti T, et al. (2009). Systemic T-helper and T-regulatory cell type cytokine responses in rhinovirus vs. respiratory syncytial virus induced early wheezing: An observational study. Respiratory Research, 10, 85 10.1186/1465-9921-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalapos MP (1999). Possible physiological roles of acetone metabolism in humans. Medical Hypotheses, 53, 236–242. 10.1054/mehy.1998.0752. [DOI] [PubMed] [Google Scholar]
- Karnovsky A, et al. (2012). Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics, 28, 373–380. 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel DB, Martin M, Schall W, & Sweeley CC (1986). Urinary metabolites of l-threonine in type 1 diabetes determined by combined gas chromatography/chemical ionization mass spectrometry. Biomedical & Environmental Mass Spectrometry, 13, 535–540. [DOI] [PubMed] [Google Scholar]
- Lampropoulou V, et al. (2016). Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metabolism, 24, 158–166. 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin EK, et al. (2015). Objectives, design and enrollment results from the infant susceptibility to pulmonary infections and asthma following RSV Exposure Study (INSPIRE). BMC Pulmonary Medicine, 15, 45 10.1186/s12890-015-0040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin EK, & Hartert TV (2015). Genes associated with RSV lower respiratory tract infection and asthma: The application of genetic epidemiological methods to understand causality. Future Virology, 10, 883–897. 10.2217/fvl.15.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, & Hughes CC (2004). Of mice and not men: Differences between mouse and human immunology. Journal of Immunology, 172, 2731–2738. [DOI] [PubMed] [Google Scholar]
- Milner JJ, Wang J, Sheridan PA, Ebbels T, Beck MA, & Saric J (2014). 1H NMR-based profiling reveals differential immune-metabolic networks during influenza virus infection in obese mice. PLoS ONE, 9, e97238 10.1371/journal.pone.0097238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, & Rabinowitz JD (2006). Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathogens, 2, e132 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musfeld C, Biollaz J, Belaz N, Kesselring UW, & Decosterd LA (2001). Validation of an HPLC method for the determination of urinary and plasma levels of N-1-methylnicotinamide, an endogenous marker of renal cationic transport and plasma flow. Journal of Pharmaceutical and Biomedical Analysis, 24, 391–404. 10.1016/S0731-7085(00)00425-8. [DOI] [PubMed] [Google Scholar]
- Nair H, et al. (2010). Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. The Lancet, 375, 1545–1555. 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Openshaw PJ (2013). A gene expression signature for RSV: Clinical implications and limitations. PLoS Medicine, 10, e1001550 10.1371/journal.pmed.1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin JF, et al. (2013). Infant baboons infected with respiratory syncytial virus develop clinical and pathological changes that parallel those of human infants. American Journal of Physiology-Lung Cellular and Molecular Physiology, 304, L530–L539. 10.1152/ajplung.00173.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, & Pearce EJ (2013). Metabolic pathways in immune cell activation and quiescence. Immunity, 38, 633–643. 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeples M, & Levine S (1980). Metabolic requirements for the maturation of respiratory syncytial virus. Journal of General Virology, 50, 81–88. 10.1099/0022-1317-50-1-81. [DOI] [PubMed] [Google Scholar]
- Ritter JB, Wahl AS, Freund S, Genzel Y, & Reichl U (2010). Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC Systems Biology, 4, 61 10.1186/1752-0509-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Salazar C, et al. (2015). Urine club cell 16-kDa secretory protein and childhood wheezing illnesses after lower respiratory tract infections in infancy. Pediatric Allergy, Immunology, and Pulmonology, 28, 158–164. 10.1089/ped.2015.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez EL, & Lagunoff M (2015). Viral activation of cellular metabolism. Virology, 479–480, 609–618. 10.1016/j.virol.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, et al. (2017). Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. The Lancet, 390, 946–958. 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurs N, et al. (2010). Asthma and allergy patterns over 18 years after severe RSV bronchiolitis in the first year of life. Thorax, 65, 1045–1052. 10.1136/thx.2009.121582 . [DOI] [PubMed] [Google Scholar]
- Simoes EA, et al. (2015). Challenges and opportunities in developing respiratory syncytial virus therapeutics. The Journal of Infectious Diseases, 211(Suppl 1), S1–S20. 10.1093/infdis/jiu828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, & Selbig J (2007). pcaMethods–a bioconductor package providing PCA methods for incomplete data. Bioinformatics, 23, 1164–1167. 10.1093/bioinformatics/btm069 . [DOI] [PubMed] [Google Scholar]
- Stein RT, et al. (1999). Respiratory syncytial virus in early life and risk of wheeze and allergy by age 13 years. The Lancet, 354, 541–545. 10.1016/S0140-6736(98)10321-5. [DOI] [PubMed] [Google Scholar]
- Trygg J, & Wold S (2002). Orthogonal projections to latent structures (O-PLS). Journal of Chemometrics, 16, 119–128. 10.1002/cem.695. [DOI] [Google Scholar]
- Ulrich EL, et al. (2008). BioMagResBank. Nucleic Acids Research, 36, D402–D408. 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, & van der Werf MJ (2006). Centering, scaling, and transformations: Improving the biological information content of metabolomics data. Bmc Genomics 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevamurthy MK, Lever M, George PM, & Morison KR (2009). Betaine structure and the presence of hydroxyl groups alters the effects on DNA melting temperatures. Biopolymers, 91, 85–94. 10.1002/bip.21085 . [DOI] [PubMed] [Google Scholar]
- Veeranki SP, et al. (2014). Association of folic acid supplementation during pregnancy and infant bronchiolitis. American Journal of Epidemiology, 179, 938–946. 10.1093/aje/kwu019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, et al. (2013). Metabonomic profiling of serum and urine by (1)H NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PLoS ONE, 8, e65675 10.1371/journal.pone.0065675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerhuis JA, et al. (2008). Assessment of PLSDA cross validation. Metabolomics, 4, 81–89. 10.1007/s11306-007-0099-6. [DOI] [Google Scholar]
- Wettstein M, Weik C, Holneicher C, & Haussinger D (1998). Betaine as an osmolyte in rat liver: Metabolism and cell-to-cell interactions. Hepatology, 27, 787–793. 10.1002/hep.510270321. [DOI] [PubMed] [Google Scholar]
- Wiklund S, et al. (2008). Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Analytical Chemistry, 80, 115–122. 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. (2017). HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Research 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M, & Piedimonte G (2011). Respiratory syncytial virus prevention and therapy: Past, present, and future. Pediatric Pulmonology, 46, 324–347. 10.1002/ppul.21377 . [DOI] [PubMed] [Google Scholar]
- Wu P, et al. (2008). Evidence of a causal role of winter virus infection during infancy in early childhood asthma. American Journal of Respiratory and Critical Care Medicine, 178, 1123–1129. 10.1164/rccm.200804-579OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, & Wishart DS (2015). MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Research, 43, W251–W257. 10.1093/nar/gkv380 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, & Wishart DS (2016). Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Current Protocols in Bioinformatics, 55, 14 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- Yu Y, Clippinger AJ, & Alwine JC (2011). Viral effects on metabolism: Changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends in Microbiology, 19, 360–367. 10.1016/j.tim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, et al. (2015). Both the folate cycle and betaine-homocysteine methyltransferase contribute methyl groups for DNA methylation in mouse blastocysts. The FASEB Journal, 29, 1069–1079. 10.1096/fj.14-261131. [DOI] [PubMed] [Google Scholar]
- Zhao NN, Yang S, Hu Y, Dong HB, & Zhao RQ (2017). Maternal betaine supplementation in rats induces intergenerational changes in hepatic IGF-1 expression and DNA methylation. Molecular Nutrition & Food Research 10.1002/mnfr.201600940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.