Abstract
Targeted molecular imaging allows specific visualization and monitoring of tumors. Cancer-specific peptides have been developed for imaging and therapy. Peptides that specifically target cancer have several advantages including, ease of synthesis, low antigenicity, and enhanced diffusion into tissues. We developed the HVGGSSV peptide as a molecular targeting/imaging agent. HVGGSSV targets Tax interacting protein 1 (TIP1) which is a 14kDa PDZ domain-containing protein that is overexpressed in cancer. We docked HVGGSSV in silico using the three-dimensional structure of TIP1 and found the binding energy was −6.0 kCal/mol. The binding affinity of HVGGSSV to TIP1 protein was found to have a KD of 3.3 × 10−6 M using surface plasmon resonance. We conjugated a 40kDa PEG to HVGGSSV to enhance the circulation and evaluated the tumor binding in nude mice bearing heterotopic cervical (HT3), esophageal (OE33), pancreatic (BXPC3), lung (A549) and glioma (D54) tumors. NanoSPECT/CT imaging of the mice was performed 48 h and 72 h after injecting with 111Indium (111In) labeled PEG-HVGGSSV or PEG-control peptide. SPECT imaging revealed that 111In-PEG-HVGGSSV specifically bound to cervical, esophageal, pancreatic, lung and brain tumors. Post SPECT biodistribution data further validated tumor-specific binding. Overall, HVGGSSV peptide specifically binds to the major groove of the TIP1 protein surface. PEGylated-HVGGSSV could be used to target cancers that overexpress TIP1.
Keywords: Peptide, TIP1, molecular imaging, SPECT imaging
Introduction
Early and specific detection of cancer can improve cancer therapy and survival rates. Molecular imaging allows visualization and measurement of biological processes at the cellular level in living systems. Targeted molecular imaging utilizes agents that specifically identify and bind to their receptors without non-specific binding. This is a pre-requisite for cancer diagnosis and monitoring response to treatment. Cancer treatments require precise spatial identification of tumors. Molecular imaging using single-photon emission computed tomography (SPECT) or positron emission tomography (PET) are efficacious imaging modalities for identification of cancer-specific agents [1]. Several radiolabeled targeting agents, including small molecules, peptides, proteins as well as antibodies and antibody fragments, have been utilized in non-invasive imaging and specific detection of tumors. Targeting peptides offer several advantages over other imaging agents. Their small size allows enhanced diffusion, target accessibility and high sensitivity as imaging tracers.
Additionally, peptides have little antigenicity and are economical to synthesize [2]. Several peptides, such as somatostatin (SST) peptide, vasoactive intestinal peptide (VIP), Arg-Gly-Asp (RGD) peptide, and bombesin/gastrin-releasing peptide (BBN/GRP), have been successfully characterized for tumor receptor imaging [2–5]. Despite these advances, very few peptides have been approved by the FDA for cancer imaging and diagnosis. There is a need for developing novel cancer-specific targets and their peptide ligands, which could be designed as tracers for noninvasive imaging of cancers.
The HVGGSSV peptide was discovered by in vivo phage-display biopanning and was found to selectively bind to tumors that have been irradiated [6]. HVGGSSV peptide was further characterized and found to bind Tax interacting protein 1 (TIP1), which was overexpressed in cancers [7] We have previously demonstrated using noninvasive imaging that phage-displayed HVGGSSV successfully monitored response to tumor treatment [6]. We have also shown tumor-specific delivery of doxorubicin using liposome displaying HVGGSSV peptide [8]. TIP1 was initially identified as one of the binding partners of the T-cell leukemia viral oncoprotein Tax [9]. A single PSD-95/DlgA/ZO-1 (PDZ) domain (89 amino acids) is the only structural and functional unit in the small protein (total of 124 amino acids in human and mouse), distinguishing TIP-1 from other PDZ proteins [9, 10]. The TIP1 expression is elevated in various cancers including lung, breast, and gliomas. Radiation stress leads to induction of TIP1 within the cells, as well as the cell surface [7, 10]. Cell-surface TIP1 is detected on many types of cancer cells in vivo and is not usually present on normal cells [7, 10]. Expression of TIP1 on the surface of human cancer cells is associated with tumorigenesis, tumor progression, angiogenesis, and metastasis, thereby demonstrating its utility as an anticancer therapeutic target [11, 12].
In this study, we further developed and optimized the HVGGSSV peptide. We identified the binding site and affinity of HVGGSSV peptide to TIP1. We modified the peptide by PEGylation to increase circulation time in vivo. Using radioisotope labeling, we show here that HVGGSSV peptide specifically binds to various tumors and it can be potentially developed as an imaging agent for multiple cancers.
Materials and Methods
Cell lines
A549 cells are adenocarcinomas derived from human alveolar basal epithelial cells [13]. D54 cells were derived from a Grade IV glioblastoma multiforme [14]. Both A549 and D54 cells were cultured in DMEM/F12 media. The cell line OE33, also known as JROECL33, was established from the adenocarcinoma of the lower esophagus [15]. OE33 cells were cultured in RPMI 1640 media. HT3 cells were derived from a metastatic site of cervical cancer [16]. These cells were cultured in IMDM media. All media were supplemented with 10% fetal bovine serum, penicillin, and streptomycin. The cells were maintained at 37°C in a 5% CO2 incubator. D54 was a gift from Dr. Yancey Gillespie (University of Alabama at Birmingham). All other cell lines were from ATCC.
Peptide synthesis
DTPA-PEG-(KKK)-HVGGSSV (mPEG40K-carbonyl-Lys(CHX-A”-DTPA)-AEEAc-Lys(CHX-A”-DTPA)-AEEAc-Trp-Lys(CHX-A”-DTPA)-AEEAc-Gly-Ile-Arg-Leu-Arg-Gly-OH trifluoroacetate Salt) and DTPA-PEG-(KKK)-control (mPEG40K-carbonyl-Lys(CHX-A”-DTPA)-AEEAc-Lys(CHX-A”-DTPA)-AEEAc-Trp-Lys(CHX-A”-DTPA)-OH) containing 40 kDa PEG were synthesized by Bachem (USA). FITC-HVGGSSV and FITC scrambled peptide (HGVIGRV) were synthesized by China peptides (Shanghai, China) using standard solid-phase Fmoc chemistry. The peptides were purified to a minimum purity of 95% by high-performance liquid chromatography (HPLC) and were isolated by lyophilization.
In silico molecular docking studies
Docking of HVGGSSV to TIP1 was performed by using Autodock Vina software [17]. Peptide structures were prepared by using online software Probuilder (http://159.149.85.2/probuilder.htm). The output file that we generated was pdb2.2, secondary structure was alpha helix, phi -1350C, Psi- 1350C and omega 1800C. Peptide structure was minimized and hydrogens added by chimera software. The high-resolution crystal structure of TIP1 in complex with beta-catenin (PDB ID: 3DIW) was obtained from the RCSB protein data bank (http://www.pdb.org). The waters and ligand were removed from the original crystal structure. Protein structure was minimized, and hydrogen was added by chimera software. For minimization, the steepest descent step was 100 and step size 0.02Å. The incomplete side chains were replaced using Dunbrack rotamer library [18]. Energy-minimized TIP1 was docked with energy-minimized HVGGSSV using Autodock Vina software. The overall quality of the minimized model was evaluated to ensure the model quality by utilizing PROCHECK [19] for evaluation of Ramachandran plot quality. The structure of HVGGSSV in complex with TIP1 was generated by Pymol software (Schrodinger Inc.). Ligplot was used to identify the hydrogen bonds and to perform hydrophobic interaction analysis [20].
Surface Plasmon Resonance (SPR)
The affinity of HVGGSSV peptide to TIP1 protein was determined using the biosensor-based surface plasmon resonance (SPR) technique using an automatic apparatus BIAcore 2000 (GE Healthcare, Sweden) as described earlier [21]. The recombinant eukaryotic TIP1 protein (Prospec, USA) was immobilized by amine coupling on the CM5 sensor surface (ligand), and HVGGSSV peptide was used as the analyte. Experiments were performed at 25°C in HBS-EP buffer (GE healthcare). TIP1 protein was immobilized using surface preparation wizard for amine coupling. Briefly, equal volume (115 μl) of N-hydroxysuccinimide (NHS, 2.3 mg in 200 μl of water) and N-ethyl-N’ −3 (diethylamino propyl) carbodiimide (EDC, 15 mg in 200 μl of water) was mixed, and 75 μl of this solution was injected into the flow cell at the flow rate of 5 μl/min across the CM5 sensor chips to activate the carboxy methylated dextran surface for 15 min. TIP1 protein (50 μg/ml in 10 mM sodium acetate, pH 4.7) was injected at the flow rate of 5μl/min across the activated surface for 25 min. The residual NHS esters were inactivated with ethanolamine (50 μl) for 10 min. A blank reference surface was also prepared with the same procedure by activation with EDC/NHS and then inactivation with ethanolamine. The affinity of the interaction was determined from the level of binding at equilibrium as a function of the sample concentrations by BIA evaluation software 3.0. The rate constant KD was obtained by fitting the sensogram data after reference subtraction (data from the blank channel) using the BIA evaluation 3.0 software.
Flow cytometry
The surface expression of TIP1 on cancer cells A549, HT3, D54 and OE33 was evaluated using flow cytometer. A549, HT3, D54 and OE33 cells were irradiated (3Gy × 3) and harvested 48 h post-irradiation. Cells were incubated with anti TIP1 antibody (Abcam) for 1 h on ice followed by incubation for 1 h with anti-mouse Alexa488 labeled secondary antibody. Propidium iodide was added to the cells prior to acquisition for flow cytometric analysis for the exclusion of dead cells. Cells were analyzed using a MACSQuant Analyzer flow cytometer (Miltenyi Biotec) and the data were analyzed with FlowJo software (Tree Star Inc.).
Western immunoblot analysis
Total soluble protein from cells were isolated using M-PER mammalian protein extraction reagent (Thermo-Pierce, Rockford, IL, USA). Concentrations of cell lysates were determined by BCA assays, and equal amounts of protein were resolved by SDS-PAGE and transferred to PVDF membranes for immunoblotting. The blots were probed with anti TIP1 (Abcam) and GAPDH (Cell Signaling Technology) antibodies. Blots were imaged using ChemiDoc-MP Imaging System (Bio-Rad) and analyzed with Image Lab Software (Bio-Rad).
Synthesis of PEG-LS601 complex
PEG-LS601-PEG 5kD/10kDa. Methoxy-PEG-amine was dissolved in DMF and DIPEA was added. LS601-NHS ester was dissolved in DMF and added to the PEG solution. Synthesis of LS601-PEG40kDa was done following earlier published protocol [22]. Briefly, methoxy-PEG-amine with molecular weight 40kDa was dissolved in 0.1 M NaHCO3 buffer. LS601-NHS ester was dissolved in DMSO and added to the PEG solution. The reaction mixture of PEG-LS601 of 5, 10 and 40 kDa was left shaking at room temperature for 3 hours. The conjugate was purified on a Sephadex G-25 column and eluted with water. Fractions were evaluated by fluorescence anisotropy and SDS-PAGE. Fractions containing pure product were collected and lyophilized.
NIR imaging
Tumors were induced by injecting A549 (1 × 106) cells in the right hind limbs of nude mice. The tumors were irradiated with 3 fractions of 3 Gy or 0Gy (sham) over the course of 24 h. The tumor-bearing mice were then injected with Peg constructs via the tail vein. For optical imaging, the mice were anesthetized with 2% isoflurane. Mice were imaged using the Pearl NIR imaging system (Li-COR) under the following conditions: exposure time 30 s, ex/em. 685/700 and 785/810 nm. Images were processed with the Pearl Imaging System software (Li-COR) Background subtracted signal intensity was plotted using Graph Pad Prism software.
Cell uptake study
Cancer cells (A549, HT3, D54 and OE33) were grown on chamber slides (Millipore, USA) and incubated with 10μg/ml FITC-HVGGSSV or FITC-scrambled peptide for 2 h at 37°C in a CO2 incubator. The cells were then washed with PBS to remove unbound peptide and fixed with 4% paraformaldehyde at room temperature. The nuclei were stained with DAPI, and fluorescent images were captured using a Carl Zeiss microscope (USA).
Radiolabeling of peptides
The radiolabeling procedure was optimized by varying the pH, buffers, temperature, and amounts of 111InCl3 added per mg DTPA-PEG-compounds. DTPA-PEG-control and DTPA-PEG-HVGGSSV stock powders were dissolved in ammonium acetate buffer (0.1M) to obtain 5mg/ml solution. 111InCl3 (370MBq ml−1 in 0.5M HCl, pH 1.1–1.4) was obtained from Mallinckrodt Pharmaceuticals. Ammonium acetate (400μl of 0.5M) was added to the 111InCl3 stock solution (450μl) and carefully mixed; the final pH was between 5.5–5.8. The 111InCl3 was then added to the DTPA-PEG-control, and DTPA-PEGHVGGSSV at a ratio of 370:1 kBq:μg and the reaction mixture was incubated at 95°C with constant shaking for 1 h. The radiolabeling efficiency of the PEG peptides was determined using instant thin-layer chromatography, and labeled peptides of ≥95% purity were used for in vivo studies. Dynamic light scattering (DLS) studies were performed with the PEGylated peptides to confirm the absence of aggregates or changes in size (data not shown).
Serum Stability Studies
In vitro serum stability of the 111In radiolabeled complexes was performed to determine if the stability in the mouse serum. Ten μL (~100 μCi) of 111In labeled PEG-Control and PEG-HVGGSSV were added to 90 μL of mouse serum and incubated at 37 °C with agitation (300 rpm). Aliquots were removed at each time point (0.5, 1, 2, 4, 24, 48 and 72 h) and analyzed using Instant thin layer chromatography (ITLC) with 50 mM DTPA solution as the mobile phase. All reactions were conducted in triplicate and data plotted using GraphPad Prism software.
In vivo tumor models
All animal studies were performed in accordance with the guidelines of the IACUC and with protocols approved by the Washington University Division of Comparative Medicine. Tumor models were established in 6 to 8-week-old female athymic nude mice (obtained from Harlan Laboratories, USA) by sub-cutaneous injecting tumor cells (1–3 × 106) into both hind limbs for HT3 and OE33 heterotopic tumor models and the right hind limb only in the A549, BxPC3, and D54 heterotopic tumor models. The tumors were measured using calipers to a size of 1cm3 before using them for SPECT imaging. We used 1cm3 which is smaller than the maximum allowed size 2cm3 at which nonspecific enhanced permeability and retention (EPR) effect are more prominent. Prior to injection with radiolabeled peptide, the right hind limb in all tumor models was irradiated with 3 Gy IR, three times over 24 hours, using the RadSource RS2000 Biological Systems irradiator. The rest of the body (including the left hind limb tumor) was blocked from irradiation using lead plates.
Nano CT/SPECT imaging and biodistribution studies
Mice (n=3 per group) were injected intravenously with approximately 500μCi 111In-DTPA-PEGHVGGSSV or PEG-control in a volume of 100 μl. Whole body SPECT images were obtained at 48, and 72 h post injection (p.i.) using a NanoSPECT/CT imager (Bioscan Inc., Washington, DC, USA) fitted with 2 mm pinhole collimators in helical scanning mode. Mice were placed in prone position and scanned under anesthesia (0.5 L/min 1.5% isoflurane in air). A 45-keV helical CT scan was performed first, and then the SPECT acquisition was performed at 24 projections and 60 s per projection, for a total scan time of about 45 min. Tomographic data were reconstructed iteratively with the manufacturer-supplied InVivoScope and HiSPECT software for CT and SPECT, respectively.
After scanning, 96 h p.i., the bio-distribution of the labeled peptide in various organs was determined. The animals were sacrificed, and organs of interest including blood were dissected/collected, weighed, and counted in a gamma counter along with a standard of the injected activity to allow calculation of the injected dose per organ (% ID/organ). The following calculations were used:
Autoradiography
After SPECT imaging the tumors from the mice were processed and sectioned. Muscle from the mouse leg was used as a control. The tumor and muscle sections were exposed to the phosphor imager plate for 16 h at negative 20° C. The plates were then scanned using phosphor imager plate scanner (Storm 840). Images were processed using ImageQuant 5.2 (Molecular Dynamics) software.
Statistical Analysis
Quantitative data are expressed as mean ± SD. Means were compared using the Student t-test. P values of less than 0.05 were considered statistically significant.
Results
HVGGSSV binds to the PDZ domain of TIP1
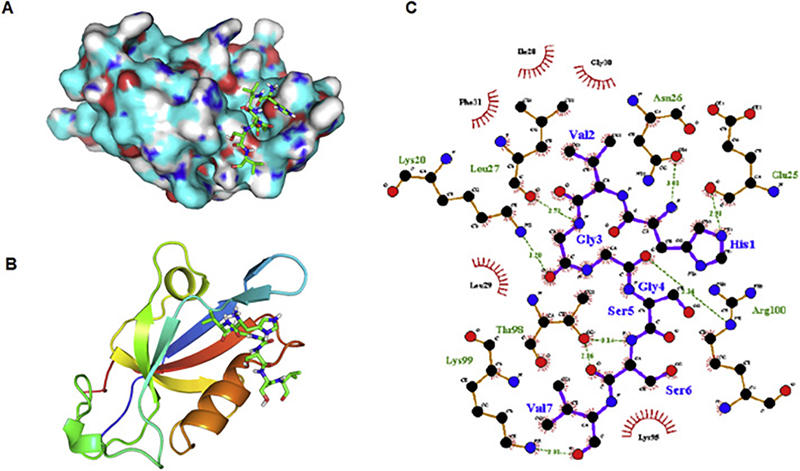
Energy-minimized TIP1 protein was docked with energy-minimized HVGGSSV peptide using Autodock Vina software [17]. The binding energy of HVGGSSV to TIP1 calculated by the software was −6.0 kCal/mol. We compared the binding energy of HVGGSSV to other potential peptides. We performed computational docking for the peptide sequences of iCAL36 (ANSRWPTSII), Kir2.3 (RRESI) and Glutaminase L (KENLESMV). The binding energies of these peptides to TIP1 were −6.1, −5.9 and −4.9 respectively (Supplementary table). Fig 1A shows HVGGSSV perfectly fitting in the major groove of the TIP1 protein surface. Fig 1B shows the peptide fits between the alpha helix and the anti-parallel beta sheets. Fig 1C shows the detailed interactions between the amino acids of the protein and the peptide. The TIP1 residues involved in interacting with HVGGSSV are Lys 20, Glu 25, Asn 26, Leu 27, Ile 28, Leu 29, Gly 30, Phe 31, Lys 95, Thr 98, Lys 99, Arg 100. The first Histidine of HVGGSSV forms a hydrogen bond with Glu 25 and Asn 26. The third and fourth glycine of HVGGSSV interacts with Leu 27 and Arg 100 respectively. The sixth serine of HVGGSSV interacts with Thr 98. The last valine of the peptide interacts with Lys 99. The overall interaction of the peptide with the protein is stabilized by hydrophobic interactions with Ile 28, Leu 29, Gly 30, Phe 31, Lys 95.
Figure 1.
Docking of HVGGSSV peptide into TIP1. (A) HVGGSSV (shown in stick representation) binding in the deep pocket on the TIP1 protein surface. (B) Cartoon representation of TIP1, showing binding of HVGGSSV (stick representation) between the α-helix and anti-parallel β-sheet. (C)Two-dimensional representation of interaction observed between TIP1 and HVGGSSV. Hydrogen bonds are identified by dashed lines, and hydrophobic interactions are shown as arcs. The figure was made by using LIGPLOT program.
Binding affinity of HVGGSSV peptide to TIP1
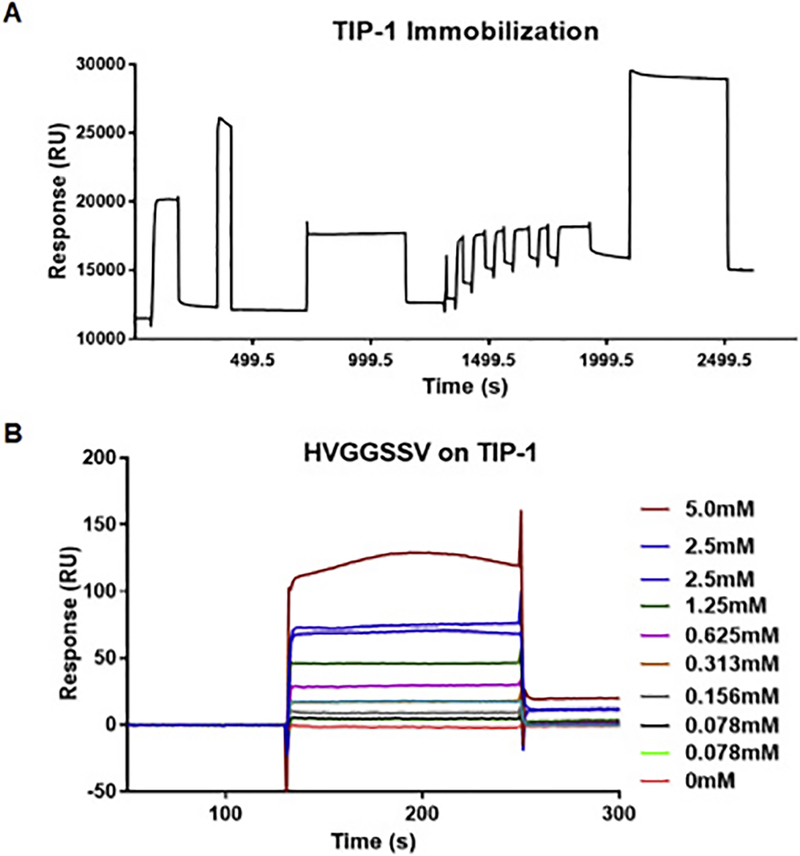
We used surface plasmon resonance technology on the BIAcore2000 instrument to determine the binding affinity of HVGGSSV peptide to TIP1 protein. TIP1 recombinant protein was immobilized on the surface of the CM5 sensor chip, and approximately 2911.2 resonance Units (RUs) of TIP1 protein were immobilized (Fig. 2A). The HVGGSSV peptide was dissolved in HBS-EP buffer at various concentrations (5, 2.5, 1.25, 0.625, 0.313, 0.156 and 0.078 mM) and passed over the immobilized TIP1 for 2 min (Fig. 2B). The rate constant Kd was obtained by fitting the sensogram data after reference subtraction (data from the blank channel) using the BIA evaluation 3.0 software. The change in RU with varying concentrations of peptide indicated the change in bound mass on the sensor surface with time, and the dissociation constant was found to be 3.3 × 10−6M. We also evaluated the binding of HVGGSSV peptide to TIP1 following pegylation with 40kDa PEG (PEG-HVGGSSV, Supplementary figure 1). We did not observe a significant difference in the binding of PEG-HVGGSSV vs. HVGGSSV to TIP1, indicating that PEGylation did not alter the binding affinity of HVGGSSV to TIP1 (Supplementary figure 1). The scrambled peptide did not show binding to the TIP1 protein (Supplementary figure 1),
Figure 2.
The binding affinity of HVGGSSV peptide to TIP1 protein using surface plasmon resonance. (A) Sensogram for immobilization of TIP1 protein on the surface of the sensor chip via amine coupling method. (B) Overlay sensograms of the indicated concentrations of the HVGGSSV peptide that passed over the chip.
HVGGSSV peptide binds to cancer cells in vitro
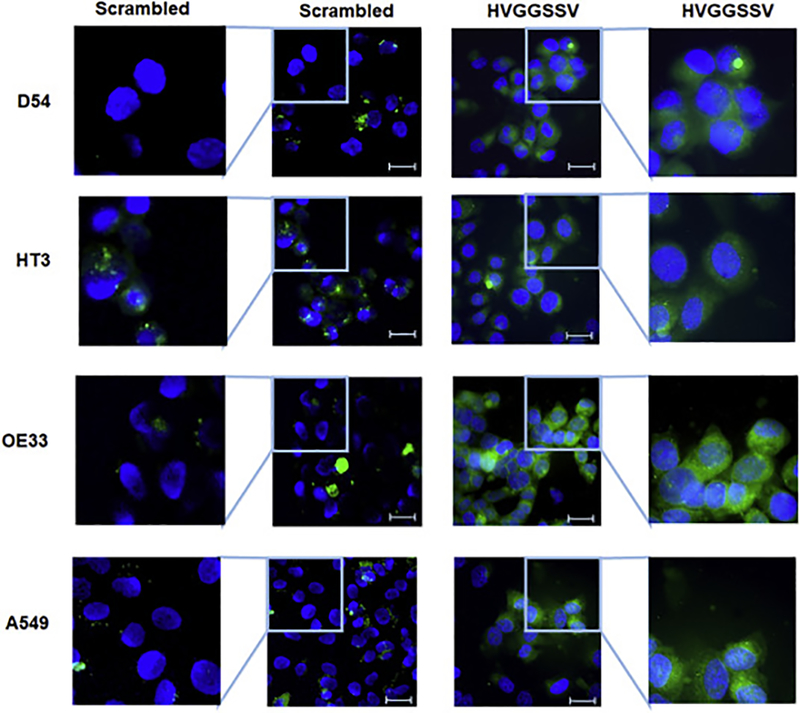
We evaluated the surface and whole cell expression of TIP1 in D54 (glioma), HT3 (cervical), OE33 (esophageal) and A549 (lung cancer) by flow cytometry and western immunoblot respectively (Supplementary Figure 2). We found cell surface expression of TIP1 in all the four cell lines which was further enhanced by radiation (Supplementary Figure 2A and B). We also found whole cell expression of TIP1 in D54, HT3, OE33 and A549, which also enhanced at 24h and 48h post radiation (Supplementary Figure 2C). Further, we evaluated the binding of HVGGSSV peptide to D54, HT3, OE33, and A549. Cancer cells D54, HT3, OE33, and A549 were incubated with HVGGSSV peptide, or scrambled peptide conjugated with FITC. We found uptake of HVGGSSV peptide in D54, HT3, OE33, and A549 and there was little or no uptake of the scrambled peptide (Fig. 3).
Figure 3.
HVGGSSV binds to cancer cells in vitro. Binding of FITC-conjugated scrambled and FITC-conjugated HVGGSSV peptide to various cancer cell lines as observed under fluorescent microscope (Resolution 200X). Scale bar = 10μm
Radiolabeling of PEG peptides
The size-dependent accumulation of macromolecules in tumors as a result of the EPR effect has been reported for various polymers, including PEG [23]. Recent studies [24] of PEGylated nanoparticles suggests that increasing the size of the nanoparticle can also result in increased tumor accumulation [25]. We observed a similar size-dependent accumulation of PEG (Supplementary Figure 3). The larger PEG40kDa-LS601 (MW~41 kDa) complex exhibited the most accumulation per injected dose when compared to either PEG10kDa-LS601 (MW~11 kDa) or PEG5kDa-LS601 (MW~6 kDa).
No appreciable signal was detected for free dye with a molecular weight less than 1 kDa. We selected PEG40kDa for our future radiolabeling studies with HVGGSSV. The radiolabeling of PEG peptides was optimized by varying the pH, buffer, temperature and amount of 111InCl3 added per mg DTPAPEG-compounds. For all in vivo studies, DTPA-PEG-control and DTPA-PEG-HVGGSSV peptide were labeled with 111Indium. The labeling efficiency was optimized by trying different temperatures, buffers, and pH as described earlier [26]. The best yield was obtained by carrying out all reactions under absolute metal-free condition. The optimal pH was 5.5, the optimal temperature was 95°C, and ammonium acetate buffer resulted in best labeling yield. Dynamic light scattering (DLS) studies were performed with the PEGylated peptides to confirm the absence of aggregates or changes in size (data not shown). DTPA-PEG-compounds were radiolabeled at a specific activity of 370MBq (10mCi) (greater than 95% radiolabeling efficiency) per mg compound. The ITLC chromatogram of 111In-DTPA-compounds is shown in supplementary figure 4.
HVGGSSV peptide binds specifically to tumors in vivo
To evaluate the efficacy of cancer-specific binding of the 111In-labeled PEG-HVGGSSV peptide, we noninvasively imaged cervical (HT3) and esophageal (OE33) heterotopic tumors using nanoSPECT/CT. Nude mice bearing HT3 and OE33 tumors in both the hind limbs were used (~1cm3). To evaluate the metabolic stability of the 111In labeled peptides, we performed serum stability assay in vitro. The 111In-labeled PEGHVGGSSV peptide was stable at all the time points (0.5, 1, 2, 4, 24, 48 and 72 h) tested (Supplementary Figure 5). The PEG-Control and PEG-HVGGSSV-111In complexes were observed to be 99.2 ± 0.6% and 93.5 ± 0.4% intact in serum at the 72-h time point.
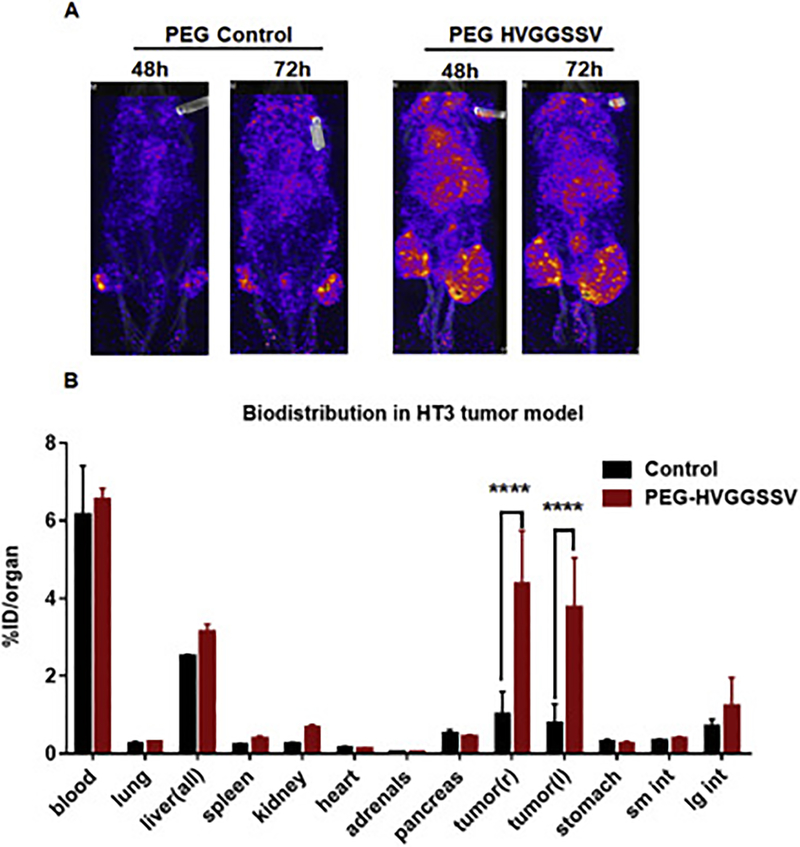
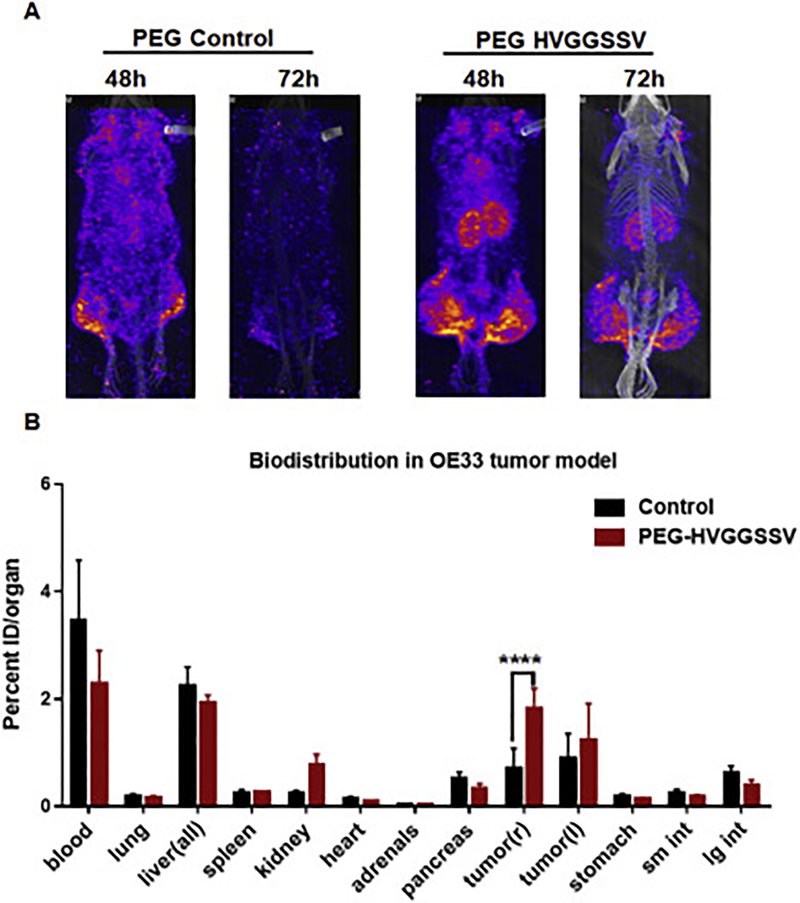
Since radiotherapy is the standard-of-care for treating local cancers including cervical and esophageal cancers, we irradiated the tumors to determine the specificity in binding cancer after radiation therapy. The tumor on the right hind limb was irradiated with 3 fractions of 3 Gy over the course of 24 h; the tumor on the left hind limb was used as unirradiated (sham) control. The mice were then injected with 500 μCi (10μCi/μg) of the radiolabeled peptide via the tail vein. In pilot studies, we performed SPECT imaging of the mice at 24, 48, 72 and 96h. At 24h we observed bright images with label throughout the mice body, and at 96 h the signal intensity was not sufficient to be detected by SPECT imager. We found optimal binding of PEG-HVGGSSV to tumors at 48h and 72h post-injection (Supplementary Figure 6) and hence used that time points for further studies. We found 111In-labeled PEG-DTPA-HVGGSSV peptide bound to both HT3 (Fig. 4A) and OE33 (Fig. 5A). We found very low or negligible binding of 111In-labeled PEG-DTPA-control in both HT3 (Fig. 4A) and OE33 (Fig. 5A) tumors. Irradiating the tumors (right hind limb) did not affect tumor binding of the radiolabeled HVGGSSV peptide to either HT3 OE33 tumors. Since we were not able to image the mice at 96h, we performed post-SPECT biodistribution at 96 h (Fig 4B and5B). The post-SPECT biodistribution data of the 111In labeled compounds in tumor-bearing mice are summarized in Fig. 4B and5B. The biodistribution results correlate with the SPECT imaging data. Tumor uptake of radiolabeled HVGGSSV was significantly higher (p<0.001) than control peptide. The biodistribution studies with radiolabeled peptides revealed the presence in the blood. Labeled peptide was also observed in the liver of both HT3 and OE33 tumor-bearing mice as these peptides were being cleared from the circulation. The levels of the labeled peptide in other organs were not significant. Further we evaluated the microscopic biodistribution of the peptide in the tumors. We sectioned the tumor (OE33) and exposed the tumor sections to phosphor imager plates. The muscle from the leg was used as control (Supplementary Figure 7). We found even distribution of 111In labeled HVGGSSV peptide in OE33 tumors and no peptide uptake on muscle.
Figure 4.
SPECT imaging and post-SPECT biodistribution with radiolabeled PEG-HVGGSSV and PEG-control peptide in nude mice with heterotopic cervical (HT3) tumors. The tumor on the right hind limb was irradiated with 3 doses of 3 Gy, and the left was sham control. Enhanced tumor binding of the PEGHVGGSSV peptide is observed in HT3 tumors at both 48 h and 72 h post injection.****p<0.001.
Figure 5.
SPECT imaging and post-SPECT biodistribution with radiolabeled PEG-HVGGSSV and PEG-control peptide in nude mice with heterotopic esophageal (OE33) tumors. The tumor on the right hind limb was irradiated with 3 doses of 3 Gy, and the left was sham control. Enhanced tumor binding of the PEG-HVGGSSV peptide is observed in OE33 tumors at both 48 h and 72 h post injection.****p<0.001.
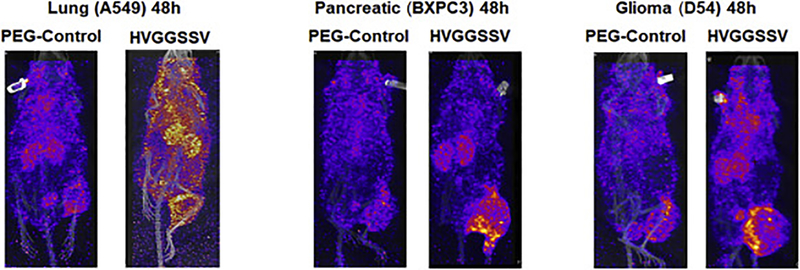
In addition, we also screened lung cancer (A549), pancreatic cancer (BxPC3) and glioblastoma (D54) tumor models for binding of radiolabeled PEG-HVGGSSV and PEG-control. SPECT/CT imaging at 48h showed enhanced binding of 111In-labeled PEG-DTPA-HVGGSSV in irradiated A549, BxPC3 and D54 tumors compared to 111In-labeled PEG-control (Fig. 6).
Figure 6.
SPECT imaging with radiolabeled PEG-HVGGSSV and PEG-control in nude mice with heterotopic lung (A549), pancreatic (BXPC3) and brain (D54) cancer. SPECT imaging (48 h post injection) in nude mice with heterotopic A549, BXPC3, D54 cancer. The tumor on the right hind limb was irradiated with 3 doses of 3 Gy. Enhanced tumor binding of the PEG-HVGGSSV peptide is observed in A549, BXPC3 and D54 tumors compared to the PEG-control.
Discussion
Imaging modalities including PET, CT, MRI, and SPECT have entered routine clinical use as structural imaging platforms [27, 28]. Over the last decade, molecular targeted imaging technologies have received growing attention as non-invasive methods to allow the visualization and monitoring of biological processes at a molecular level [27, 28]. These techniques make use of fluorescent or radioactive probes conjugated to a molecule that targets a cancer biomarker providing a signal at the target site. The targeting molecules being developed for tumor imaging include antibodies, peptides, small molecules, aptamers, etc. Peptides offer several advantages including rapid distribution, reduced or absent immunogenicity, ease and scalability of synthesis and affordable labeling [2, 29]. The first and most successful FDA approved peptide-based radiopharmaceutical is the somatostatin analog [30], 111In-DTPA-octreotide (111In-OctreoScan, 111In-pente-treotide) [2, 31, 32]. It is being used for SPECT imaging SST receptor-positive lesions, such as neuroendocrine tumors, mammary cancer and small cell lung cancer [30–32]. The successful clinical application of this radiopharmaceutical raised interest in the development of radiolabeled peptides to target other tumor-related peptide receptor systems. Recently, a similar PET imaging agent, 68Ga-DOTA-TATE (NETSPOT), was approved by the FDA and is being used widely for neuroendocrine tumors.
Currently, in vivo molecular imaging is limited by a shortage of biomarkers and probes with sound biological relevance. We have previously shown in tumor-bearing mice that the hexapeptide HVGGSSV demonstrated potential as a molecular imaging probe to distinguish tumors responding to ionizing radiation (IR) and/or tyrosine kinase inhibitor treatment from those of non-responding tumors [6]. We next discovered that Tax interacting protein 1 (TIP1) is the target of HVGGSSV peptide [7]. TIP1 is a radiation-inducible antigen that translocates to the plasma membrane in response to radiation [7]. In this study, we discovered the exact residues of TIP1 involved in binding with HVGGSSV peptide. We used computational docking and calculated the binding energy of HVGGSSV to TIP1 as −6.0 kCal/mol. HVGGSSV peptide fits between the alpha helix and the anti-parallel beta sheets of the major groove of TIP1. The TIP1 residues involved in interacting with HVGGSSV are Lys 20, Glu 25, Asn 26, Leu 27, Ile 28, Leu 29, Gly 30, Phe 31, Lys 95, Thr 98, Lys 99, Arg 100. We next used surface plasmon resonance (SPR) to evaluate the affinity of HVGGSSV to TIP1. SPR analysis showed that HVGGSSV had a Kd of 3.3 × 10−6M for TIP1. This high affinity can be attributed to the hydrogen bonds and the hydrophobic interactions between HVGGSSV and the interacting TIP1 residues that lead to stabilize the peptide binding to its target. [33, 34]. In cultured cancer cells, we found uptake of FITC-conjugated HVGGSSV peptide to glioma (D54), cervical (HT3), esophageal (OE33) and lung (A549) cells. The uptake of HVGGSSV in D54, HT3, OE33 and A549 is observed since these cancer cells overexpress TIP1.
Here, we evaluated the potential of HVGGSSV to be developed as a radiolabeled probe for cancer diagnosis. We used SPECT imaging which is highly sensitive and allows quantitative estimation of the amount of radioisotope localized to the sites of interest [27, 28]. Peptides, being small in size, are cleared quickly from the system. To overcome this, we conjugated the HVGGSSV peptide to a 40kDa PEG to enhance its circulation time and extravasation to the tumors. 40kDa PEG was chosen based on our NIR study with different sizes of PEG since we found the most accumulation of 40kDa in the mouse tumors. PEGylation has been frequently used and is clinically approved for various molecules used in the clinic [35]. PEGylation did not alter the binding affinity of HVGGSSV to the TIP1 protein.
For nano-SPECT imaging, PEG-HVGGSSV was radiolabeled with 111Indium by using DTPA as the chelator. The background signal was evaluated using 40kDa PEG construct having the chelator DTPA (PEG control). We performed pilot SPECT imaging studies at 24, 48, 72 and 96h post-injection of the radiolabeled peptides for various tumors. For the amount of injected radioactivity we found optimum images at 48h and 72h time points. These times were chosen for imaging cervical (HT3) and esophageal (OE33) heterotopic tumors in nude mice. PEG-HVGGSSV specifically bound to HT3 and OE33 tumors while little or no binding was observed in PEG-Control. This data indicated that PEG-HVGGSSV had specific binding to the HT3 and OE33 tumors most likely by binding to the TIP1 on the surface of the tumors. Since radiotherapy is the standard-of-care treatment for cervical and esophageal tumors, we had irradiated the right hindlimb in all tumor models to evaluate the effect of radiation on the binding of the PEG-HVGGSSV. No significant difference in the binding of 111In-PEG-HVGGSSV was observed between irradiated vs. non-irradiated tumors. This might be attributed to the abscopal effect of radiation which may have led to similar binding of the PEGylated peptide to the sham and irradiated tumor [33, 34]. We also observed binding of the FITC-labeled HVGGSSV to cancer cell line in vitro in the absence of radiation. TIP1 is expressed at a basal level on the surface of cancer cells. Thus binding of the 111In-PEG-HVGGSSV to the sham irradiated tumors may also be attributed to the basal level expression of TIP1 in the absence of radiation.
Post SPECT bio-distribution data further supported the SPECT imaging data. We observed higher binding efficacy of the radiolabeled PEG-HVGGSSV in the HT3 tumors compared to OE33 tumors. The higher binding in HT3 vs. OE33 may be attributed to the higher surface expression of TIP1 in HT3 compared to OE33 following IR as observed in our flow cytometry experiments. At 96 h, both the PEG-HVGGSSV and the PEG-control were observed in the blood and liver. This implies that this is a function of the PEG moiety and not the peptide. The higher blood and liver levels may be reduced by optimizing the size of the PEG moiety. We also evaluated the microscopic distribution of the radiolabeled peptides in the harvested tumor sections and muscle tissue (control). We found that the 111In-DTPA-PEG-HVGGSSV evenly distributed in the tumor. This supports the biodistribution signal in the tumor and suggests that this signal is not merely from the blood in the capillary space of the tumor. There was no radioactivity detected in the muscle.
In addition to the cervical and esophageal cancer, we evaluated the binding of PEG-HVGGSSV in irradiated lung cancer (A549), pancreatic cancer (BXPC3) and glioblastoma (D54) models. Similar to the cervical and esophageal tumors, we found specific binding of PEG-HVGGSSV to lung cancer, pancreatic cancer, and glioblastoma compared to the PEG-control. Our future goal is to test this in orthotopic models. For orthotopic glioblastoma models we will compare various drug delivery methods such as sonication, high-intensity ultrasound etc to allow penetrance of this pegylated peptide to the brain.
Overall this study further supports that TIP1 is a suitable cell surface molecule and a molecular target for the development of diagnostics and therapeutics. PEG-HVGGSSV that targets TIP1 on the surface of the tumors is a potential peptide that will be developed further as a carrier for therapeutic agents. Further optimization of the size of PEG to improve long-term biodistribution for preclinical efficacy and safety testing will be performed before moving this peptide to clinical trials.
Supplementary Material
Supplementary Figure 1. Binding of HVGGSSV vs PEG-HVGGSSV to TIP1. Overlay sensograms showing the binding profile of HVGGSSV vs PEG-HVGGSSV to the immobilized TIP1 protein on CM5 sensor chip in BIAcore 2000 instrument.
Supplementary Figure 2. Expression of TIP1 in cancer cells following irradiation. Cancer cells D54 (glioma), HT3 (cervical), OE33 (esophageal) and A549 (lung cancer) were irradiated with 3 Gy 3 times or sham treated. Cells were analyzed for TIP1 surface expression using flowcytometry 48 h post-irradiation. Shown are the overlay histograms (A) and bar graphs (B) of cell surface staining in each treatment with SD of three samples. Western blot analysis of TIP1 in whole-cell lysates from D54 cells harvested at the indicated time-points after irradiation with 3Gy 3 times. The values represent the normalized densitometric output for each band divided by the corresponding loading control. ****p<0.0001.
Supplementary Figure 3. Size-dependent accumulation of PEG-NIR complexes in mice. Mice bearing hind-limb A549 tumors received either sham or 3Gy x3 irradiation. Following the treatment, PEG5kDa-LS601, PEG10kDa-LS601, PEG40kDa-LS601 was administered intravenously and accumulation of the probes in the tumor was observed with daily NIR imaging (* P < 0.05, **p<0.01).
Supplementary Figure 4. Instant thin layer chromatogram of 111In-DTPA-PEG-control (A) and 111In-DTPA-PEG-HVGGSSV (B). 111In-DTPA-PEG-HVGGSSV stays at the origin and 111In-DTPA moves with solvent front.
Supplementary Figure 5. In vitro serum stability assay for 111In-PEG-control and 111In-PEG-HVGGSSV. Both the compounds are stable until the end of the assay.
Supplementary Figure 6. SPECT imaging with radiolabeled PEG-HVGGSSV and PEG-control peptide in nude mice with heterotopic (A549 and OE33) tumors. The tumor on the hind limb was irradiated with 3 doses of 3 Gy. SPECT images were acquired at the indicated time points.
Supplementary Figure 7. Microdistribution of 111In PEG HVGGSSV in OE33 tumors. 200 micron tumor or muscle sections exposed to a phosphor imaging plate. Shown are the autoradiographs using phosphor imager plate scanner. The radiolabeled HVGGSSV peptide was found to be evenly distributed throughout the tumors.
Acknowledgments
This work was supported by National Cancer Institute grants 1R01CA140220–02, 5R01CA125757–06, 7R01CA112385–0 (D. Hallahan), Siteman Cancer Research Award, Elizabeth and James McDonnell III Endowment (D. Hallahan), American association for cancer research grant 12-60-26-HALL (D. Hallahan), The Barnes-Jewish Hospital Foundation (BJHF) and the Washington University Institute of Clinical and Translational Sciences (ICTS) grant (D. Hallahan), Department of Radiation Oncology Startup Funds (D. Thotala).
This work was performed with the support from the Siteman Cancer Center Small Animal Imaging Core. We thank Nicole Fettig and Amanda Klaas from the Mallinckrodt Institute of Radiology Pre-clinical PET/CT imaging facility for their help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosure:
None
Conflicts of interest:
All authors declare no conflicts of interest.
References
- [1].de Barros AB, Tsourkas A, Saboury B, Cardoso VN, Alavi A, Emerging role of radiolabeled nanoparticles as an effective diagnostic technique, EJNMMI Res, 2 (2012) 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schottelius M, Wester HJ, Molecular imaging targeting peptide receptors, Methods, 48 (2009) 161–177. [DOI] [PubMed] [Google Scholar]
- [3].Vomhof-DeKrey EE, Sandy AR, Failing JJ, Hermann RJ, Hoselton SA, Schuh JM, Weldon AJ, Payne KJ, Dorsam GP, Radical reversal of vasoactive intestinal peptide (VIP) receptors during early lymphopoiesis, Peptides, 32 (2011) 2058–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tweedle MF, Peptide-targeted diagnostics and radiotherapeutics, Accounts of chemical research, 42 (2009) 958–968. [DOI] [PubMed] [Google Scholar]
- [5].Laverman P, Sosabowski JK, Boerman OC, Oyen WJ, Radiolabelled peptides for oncological diagnosis, European journal of nuclear medicine and molecular imaging, 39 Suppl 1 (2012) S78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Han Z, Fu A, Wang H, Diaz R, Geng L, Onishko H, Hallahan DE, Noninvasive assessment of cancer response to therapy, Nat Med, 14 (2008) 343–349. [DOI] [PubMed] [Google Scholar]
- [7].Wang H, Yan H, Fu A, Han M, Hallahan D, Han Z, TIP-1 translocation onto the cell plasma membrane is a molecular biomarker of tumor response to ionizing radiation, PLoS One, 5 (2010) e12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lowery A, Onishko H, Hallahan DE, Han Z, Tumor-targeted delivery of liposome-encapsulated doxorubicin by use of a peptide that selectively binds to irradiated tumors, J Control Release, 150 (2011) 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P, The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins, Oncogene, 16 (1998) 643–654. [DOI] [PubMed] [Google Scholar]
- [10].Yan H, Kapoor V, Nguyen K, Akers WJ, Li H, Scott J, Laforest R, Rogers B, Thotala D, Hallahan D, Anti-tax interacting protein-1 (TIP-1) monoclonal antibody targets human cancers, Oncotarget, 7 (2016) 43352–43362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han M, Wang H, Zhang HT, Han Z, The PDZ protein TIP-1 facilitates cell migration and pulmonary metastasis of human invasive breast cancer cells in athymic mice, Biochem Biophys Res Commun, 422 (2012) 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang H, Han M, Whetsell W Jr., Wang J, Rich J, Hallahan D, Han Z, Tax-interacting protein 1 coordinates the spatiotemporal activation of Rho GTPases and regulates the infiltrative growth of human glioblastoma, Oncogene, 33 (2014) 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G, A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells, International journal of cancer. Journal international du cancer, 17 (1976) 62–70. [DOI] [PubMed] [Google Scholar]
- [14].Bigner SH, Bullard DE, Pegram CN, Wikstrand CJ, Bigner DD, Relationship of in vitro morphologic and growth characteristics of established human glioma-derived cell lines to their tumorigenicity in athymic nude mice, Journal of neuropathology and experimental neurology, 40 (1981) 390–409. [DOI] [PubMed] [Google Scholar]
- [15].Rockett JC, Larkin K, Darnton SJ, Morris AG, Matthews HR, Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization, British journal of cancer, 75 (1997) 258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fogh J, Wright WC, Loveless JD, Absence of HeLa cellF contamination in 169 cell lines derived from human tumors, Journal of the National Cancer Institute, 58 (1977) 209–214. [DOI] [PubMed] [Google Scholar]
- [17].Trott O, Olson AJ, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, Journal of computational chemistry, 31 (2010) 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dunbrack RL Jr., Rotamer libraries in the 21st century, Current opinion in structural biology, 12 (2002) 431–440. [DOI] [PubMed] [Google Scholar]
- [19].Hill CP, Anderson DH, Wesson L, DeGrado WF, Eisenberg D, Crystal structure of alpha 1: implications for protein design, Science, 249 (1990) 543–546. [DOI] [PubMed] [Google Scholar]
- [20].Wallace AC, Laskowski RA, Thornton JM, LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions, Protein engineering, 8 (1995) 127–134. [DOI] [PubMed] [Google Scholar]
- [21].Kapoor V, Singh AK, Dey S, Sharma SC, Das SN, Circulating cycloxygenase-2 in patients with tobacco-related intraoral squamous cell carcinoma and evaluation of its peptide inhibitors as potential antitumor agent, J Cancer Res Clin Oncol, 136 (2010) 1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gustafson TP, Cao Q, Achilefu S, Berezin MY, Defining a polymethine dye for fluorescence anisotropy applications in the near-infrared spectral range, Chemphyschem : a European journal of chemical physics and physical chemistry, 13 (2012) 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lukyanov AN, Gao Z, Mazzola L, Torchilin VP, Polyethylene glycol-diacyllipid micelles demonstrate increased acculumation in subcutaneous tumors in mice, Pharmaceutical research, 19 (2002) 1424–1429. [DOI] [PubMed] [Google Scholar]
- [24].Larsen EK, Nielsen T, Wittenborn T, Birkedal H, Vorup-Jensen T, Jakobsen MH, Ostergaard L, Horsman MR, Besenbacher F, Howard KA, Kjems J, Size-Dependent Accumulation of PEGylated Silane-Coated Magnetic Iron Oxide Nanoparticles in Murine Tumors, ACS nano, 3 (2009) 1947–1951. [DOI] [PubMed] [Google Scholar]
- [25].Schadlich A, Caysa H, Mueller T, Tenambergen F, Rose C, Gopferich A, Kuntsche J, Mader K, Tumor accumulation of NIR fluorescent PEG-PLA nanoparticles: impact of particle size and human xenograft tumor model, ACS nano, 5 (2011) 8710–8720. [DOI] [PubMed] [Google Scholar]
- [26].Kapoor V, Dadey DY, Nguyen K, Wildman SA, Hoye K, Khudanyan A, Bandara N, Rogers BE, Thotala D, Hallahan DE, Tumor-Specific Binding of Radiolabeled PEGylated GIRLRG Peptide: A Novel Agent for Targeting Cancers, Journal of nuclear medicine : official publication, Society of Nuclear Medicine, 57 (2016) 1991–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen ZY, Wang YX, Lin Y, Zhang JS, Yang F, Zhou QL, Liao YY, Advance of molecular imaging technology and targeted imaging agent in imaging and therapy, BioMed research international, 2014 (2014) 819324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lauber DT, Fulop A, Kovacs T, Szigeti K, Mathe D, Szijarto A, State of the art in vivo imaging techniques for laboratory animals, Laboratory animals, 51 (2017) 465–478. [DOI] [PubMed] [Google Scholar]
- [29].Staderini M, Megia-Fernandez A, Dhaliwal K, Bradley M, Peptides for optical medical imaging and steps towards therapy, Bioorganic & medicinal chemistry, 26 (2018) 2816–2826. [DOI] [PubMed] [Google Scholar]
- [30].Ambrosini V, Fani M, Fanti S, Forrer F, Maecke HR, Radiopeptide imaging and therapy in Europe, J Nucl Med, 52 Suppl 2 (2011) 42S–55S. [DOI] [PubMed] [Google Scholar]
- [31].Rufini V, Calcagni ML, Baum RP, Imaging of neuroendocrine tumors, Semin Nucl Med, 36 (2006) 228–247. [DOI] [PubMed] [Google Scholar]
- [32].Kaltsas GA, Papadogias D, Makras P, Grossman AB, Treatment of advanced neuroendocrine tumours with radiolabelled somatostatin analogues, Endocr Relat Cancer, 12 (2005) 683–699. [DOI] [PubMed] [Google Scholar]
- [33].Formenti SC, Demaria S, Systemic effects of local radiotherapy, Lancet Oncol, 10 (2009) 718–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Siva S, MacManus MP, Martin RF, Martin OA, Abscopal effects of radiation therapy: a clinical review for the radiobiologist, Cancer Lett, 356 (2015) 82–90. [DOI] [PubMed] [Google Scholar]
- [35].Bailon P, Won CY, PEG-modified biopharmaceuticals, Expert Opin Drug Deliv, 6 (2009) 1–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Binding of HVGGSSV vs PEG-HVGGSSV to TIP1. Overlay sensograms showing the binding profile of HVGGSSV vs PEG-HVGGSSV to the immobilized TIP1 protein on CM5 sensor chip in BIAcore 2000 instrument.
Supplementary Figure 2. Expression of TIP1 in cancer cells following irradiation. Cancer cells D54 (glioma), HT3 (cervical), OE33 (esophageal) and A549 (lung cancer) were irradiated with 3 Gy 3 times or sham treated. Cells were analyzed for TIP1 surface expression using flowcytometry 48 h post-irradiation. Shown are the overlay histograms (A) and bar graphs (B) of cell surface staining in each treatment with SD of three samples. Western blot analysis of TIP1 in whole-cell lysates from D54 cells harvested at the indicated time-points after irradiation with 3Gy 3 times. The values represent the normalized densitometric output for each band divided by the corresponding loading control. ****p<0.0001.
Supplementary Figure 3. Size-dependent accumulation of PEG-NIR complexes in mice. Mice bearing hind-limb A549 tumors received either sham or 3Gy x3 irradiation. Following the treatment, PEG5kDa-LS601, PEG10kDa-LS601, PEG40kDa-LS601 was administered intravenously and accumulation of the probes in the tumor was observed with daily NIR imaging (* P < 0.05, **p<0.01).
Supplementary Figure 4. Instant thin layer chromatogram of 111In-DTPA-PEG-control (A) and 111In-DTPA-PEG-HVGGSSV (B). 111In-DTPA-PEG-HVGGSSV stays at the origin and 111In-DTPA moves with solvent front.
Supplementary Figure 5. In vitro serum stability assay for 111In-PEG-control and 111In-PEG-HVGGSSV. Both the compounds are stable until the end of the assay.
Supplementary Figure 6. SPECT imaging with radiolabeled PEG-HVGGSSV and PEG-control peptide in nude mice with heterotopic (A549 and OE33) tumors. The tumor on the hind limb was irradiated with 3 doses of 3 Gy. SPECT images were acquired at the indicated time points.
Supplementary Figure 7. Microdistribution of 111In PEG HVGGSSV in OE33 tumors. 200 micron tumor or muscle sections exposed to a phosphor imaging plate. Shown are the autoradiographs using phosphor imager plate scanner. The radiolabeled HVGGSSV peptide was found to be evenly distributed throughout the tumors.