Abstract
Background
A clinical informatics algorithm (CIA) was developed to systematically identify potential enrollees for a test-negative, case-control study to determine influenza vaccine effectiveness, to improve enrollment over manual records review. Further testing may enhance the CIA for increased efficiency.
Methods
The CIA generated a daily screening list by querying all medical record databases for patients admitted in the last 3 days, using specified terms and diagnosis codes located in admission notes, emergency department notes, chief complaint upon registration, or presence of a respiratory viral panel charge or laboratory result (RVP). Classification and regression tree analysis (CART) and multivariable logistic regression were used to refine the algorithm.
Results
Using manual records review, 204 patients (<4/day) were approached and 144 were eligible in the 2014–2015 season compared with 3531 (12/day) patients who were approached and 1136 who were eligible in the 2016–2017 season using a CIA. CART analysis identified RVP as the most important indicator from the CIA list for determining eligibility, identifying 65%–69% of the samples and predicting 1587 eligible patients. RVP was confirmed as the most significant predictor in regression analysis, with an odds ratio (OR) of 4.9 (95% confidence interval [CI], 4.0–6.0). Other significant factors were indicators in admission notes (OR, 2.3 [95% CI, 1.9–2.8]) and emergency department notes (OR, 1.8 [95% CI, 1.4–2.3]).
Conclusions
This study supports the benefits of a CIA to facilitate recruitment of eligible participants in clinical research over manual records review. Logistic regression and CART identified potential eligibility screening criteria reductions to improve the CIA’s efficiency.
Keywords: acute respiratory infection, influenza vaccination, respiratory viral panel
A clinical informatics algorithm (CIA) was developed to systematically identify potential enrollees for an influenza vaccine effectiveness study that improved enrollment efficiency over manual record review. Classification and Regression Tree analysis indicated modifications to further increase the CIA’s efficiency.
Recent influenza vaccine effectiveness (VE) studies have relied on the test-negative case-control study design to identify cases (positive for influenza) and controls (negative for influenza). Influenza can have a wide range of possible symptoms and complications. Influenza complications include pneumonia and exacerbations of asthma and chronic obstructive pulmonary disease (COPD) [1], as well as substantial numbers of cardiac complications including myocardial infarction [2–4] and exacerbations of congestive heart failure [5]. In addition, influenza can have neurologic [6] and inflammatory complications [7].
Thus, eligibility criteria for influenza studies must be broad enough to ensure that individuals across the spectrum of acute respiratory illness are identified. This broad scope of eligibility may result in a long list of individuals to approach and screen. Identification of cases from a broad spectrum of influenza-related complications and symptoms while reducing the risk for selection bias based on the most common hospital presentations of influenza such as pneumonia can be challenging and resource intensive. The availability of a streamlined approach to identify potentially eligible patients can save time and other resources and result in a more productive screening and enrollment process.
In the pilot phase of the Hospitalized Adults Influenza Vaccine Effectiveness Network (HAIVEN), manual review of the electronic health record (EHR) of potentially eligible participants culled from admissions lists was conducted. This time-intensive process was replaced by a computerized algorithm developed by the Pittsburgh HAIVEN team to increase the speed, thoroughness, and efficiency with which individuals were identified as potential participants by electronically searching the EHR. With the advent of centralized archiving of data sources, searches of medical records for the symptoms, signs, and International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM) diagnosis codes that represent influenza and its myriad complications are feasible. Given the breadth of the search criteria, we searched for further efficiencies. The purposes of this study are (1) to describe a clinical informatics algorithm (CIA) for identifying potential participants for an influenza VE study; (2) to describe the recursive partitioning analyses used to determine which search criteria provide a more precise screening algorithm; and (3) to suggest its broader applicability for recruitment in other research studies that recruit hospitalized patients, to track an influenza pandemic across all facilities within a health system, and for identification of patients at risk of a particular clinical outcome who may benefit from an intervention.
METHODS
The HAIVEN study is a Centers for Disease Control and Prevention (CDC)–funded, multicenter, test-negative case-control study to determine the influenza VE against hospitalization. Methods for the HAIVEN study have been previously published [8]. In brief, adults ≥18 years of age who have been hospitalized for <72 hours and who have a new-onset (≤10 days) acute respiratory illness (ARI) or newly worsening cough are eligible for enrollment. Newly admitted patients with any of a host of diagnoses or symptoms (see Appendix 1) as indicated in the EHR, are eligible for screening.
At the Pittsburgh site in the pilot year of the study (2014–2015), manual records review of the EHR was used to identify potentially eligible patients from a daily list of new admissions. For the 2015–2016 season, a CIA was developed to generate a daily screening list by querying the institution’s data repository (Medical Archival Retrieval System [MARS]) for each of the participating HAIVEN hospitals for patients admitted in the last 3 days. MARS is a repository for information forwarded from the health system’s electronic clinical, administrative, and financial databases that include Epic, Medipac, and others. MARS is indexed on every word in every report and will recover all reports for a given patient between specified dates [9]. MARS employs the Boolean search operators “and,” “or,” and “not,” which can be used in any combination. To permit more specific retrieval, MARS supports searches by forward and backward distance operators to control the proximity of the search terms. A search of ([er +1] diagnosis & flu) will require that the terms “diagnosis” and “flu” exist within 1 term of each other in a positive direction.
To begin the study, we retrieved 1 year of admission (ADM) reports and emergency department (ED) notes from several hospitals in the health system. We used these sets (1 for each hospital to account for variations in dictating patterns) to identify mentions of concepts in each report from the Unified Medical Language System (UMLS) Metathesaurus (version 2014AB) using the UMLS Terminology Service (https://uts.nlm.nih.gov/home.html). We matched the concepts to the disease list specified by the CDC and created a list of concepts for each disease, using the concept name as search terms. We also reviewed the notes for the relevant section headings to search for the terms. The health system uses a template-driven system to complete ADM and ED notes containing predefined section headings. The sections for “chief complaint” and “diagnosis” were the most relevant to our search and 14 terms were identified for the search list. To handle negation of the search terms, we used the NegEx algorithm [10] search terms, developed at our institution, which focuses on directly negated phrases such as “absence of” and “negative for” rather than pseudo-negated phrases such as “not certain if” and “not rule out.”
Using the list of indicator terms and diagnoses, the algorithm created a search for word(s) or ICD-10 code(s) found in the ADM note, an ED note, or the chief complaint (CC) list; or indication of a respiratory viral panel charge or laboratory result (RVP). Each search was performed independently, and each patient could meet multiple criteria. The screening list contained all of the criteria from the algorithm that qualified each patient for the list.
In 2015–2016, the research assistants (RAs) used the algorithm-generated screening list to create an electronic spreadsheet of potentially eligible patients and from the spreadsheet proceeded to visit rooms to screen and enroll eligible patients. In 2016–2017, that process was automated such that the list from the CIA was automatically uploaded to a REDCap file that randomly assigned an order to each hospital unit for approaching listed patients each day (CIA-REDCap method).
After visiting a room, RAs recorded the disposition of each patient on the list. Once patients or their proxies had been approached, screened, and/or enrolled or had been on the list for >3 days (per HAIVEN exclusion criteria), they timed out and no longer appeared on the list. Patients were reapproached at least once a day until they timed out. Patients who were intubated or in the intensive care unit were not excluded, and patients transferred from other facilities were considered to be new admissions. A patient may not have been approached for a variety of reasons, including being out of the room, asleep, engaged with the clinical staff, or had been discharged or moved to a nonenrollment unit.
Hospitals
The study took place at 1 hospital in 2014–2015, a 750-bed, urban, quaternary care hospital (P); at 2 hospitals in 2015–2016, the aforementioned hospital and a 250-bed suburban, community hospital (SM); and in 2016–2017, a 520-bed urban, tertiary care hospital (SH) was added.
Enrollment surveys assessed demographics and self-reported vaccination status, which was subsequently verified using post–influenza season data searches from the EHR, state immunization registry, health insurance and employee health databases, and communication with medical offices and pharmacies. RVPs using the GenMark platform and/or research swabs using the CDC influenza reverse transcription polymerase chain reaction test confirmed the presence of an influenza infection, which was used to estimate VE. The sensitivity for both the CDC test [11] and the GenMark RVP [12] for influenza are high (>95%).
Data
Data from all patients who qualified and appeared on the daily screening lists in the 2016–2017 influenza season (11 November 2016–29 April 2017) were used for the primary analyses. Data for 2014–2015 and 2015–2016 were derived from daily admission lists from 1 December 2014 to 4 March 2015 and 13 December 2015 to 30 April 2016, respectively. Patients were only counted once per admission but could appear on multiple admissions so that analysis was performed at the admission level. This data set was combined with the approach log to determine the final disposition of each patient on the approach list. Because the research team only worked on designated days, some patients who appeared on the daily screening lists were never approached for screening. They represent the difference between identified patients and rooms visited.
Statistical Analysis
A 2-sample t test was used to test the differences between the mean number of patients approached per day across years.
The classification and regression tree (CART) method [13] was used to conduct recursive partitioning, a nonparametric statistical method for multivariable data. It uses a series of dichotomous splits, for example, presence or absence of symptoms or an ICD-10 code, to create a decision tree with the goal of correctly classifying members of the population, in this case, eligible patients or influenza cases. Each independent variable was examined and a split was made to maximize the sensitivity and specificity of the classification, resulting in a decision tree.
An impurity/purity measure, the Gini index was used for building the decision tree in CART. It was used to split off the largest category into a separate group, with the default split size set to enable growing the tree [13]. When the final tree was built, the tree was manually expanded or pruned to determine the lowest misclassification, highest clinical usefulness, and highest sensitivity, excluding the variables that did not further classify a substantial percentage of patients into the eligible or not eligible group or influenza case or control groups. Once a clinically meaningful structure of the decision tree on the CART evolved, pruning or expansion was discontinued.
To assess the model’s generalizability and to evaluate the overfitting of the model, a simple random sampling without replacement was used to split the sample into equally sized (50%/50%) development and validation samples. CART was applied first on the developmental sample then on the validation sample. Receiver operating characteristic (ROC) curves and the area under the curve were used to assess the performance of the CART model for the developmental and validation samples.
Three groups of patients were included in the CART analyses: (1) those on the CIA-generated daily screening list with whom an RA was able to speak directly or via a proxy; (2) a subset of these patients who were enrolled in the study; and (3) a subset of these enrolled patients who tested positive for influenza. In the primary CART analysis, the outcome variable was eligibility and the independent variables were RVP, ADM, ED, ICD-10 code, and CC. The study sample consisted of all the eligible patients who enrolled (n = 1136) and the ineligible patients (n = 1306). Secondary analyses used ICD-10 codes as the independent variables. To reduce the 545 ICD-10 codes to a manageable number, the frequency of ICD-10 codes across all patients was examined. ICD-10 codes were split into those occurring ≥10 times (n = 131) and those occurring <10 times (n = 414). In this CART analysis, 2 ICD-10 codes (Z86 and Z95) were excluded due to their unlikely relationship to influenza. The remaining 129 ICD-10 codes (select ICD-10 codes) as well as RVP, ADM, ED, and CC were included in the CART model.
A regression tree was also developed among enrolled patients only, using RVP, ADM, ED, CC, and ICD-10 code as the independent variables and influenza status—positive (n = 173 [17%]) or negative (n = 861 [83%])—as the outcome variable.
Logistic regression analyses were used to further explore the relationships between the set of explanatory variables identified in CART (RVP, ADM, ICD-10) and demographic variables and the outcome variable, study eligibility. Independent variables were compared between eligible and ineligible patients using χ2 tests. All independent variables were included in the models and they were assumed to be fixed. The maximum-likelihood estimation with the Newton-Raphson algorithm was used to estimate the parameter effects in the model. We built the model using the stepwise selection method. Odds ratios (ORs) and 95% confidence intervals (CIs) were computed using the profile likelihood function of the parameter estimates.
Analyses were performed using CART software Salford Predictive Modeler version 8.2 (San Diego, California) and SAS version 9.4 (Cary, North Carolina). An alpha level of .05 was used to test for statistical significance.
RESULTS
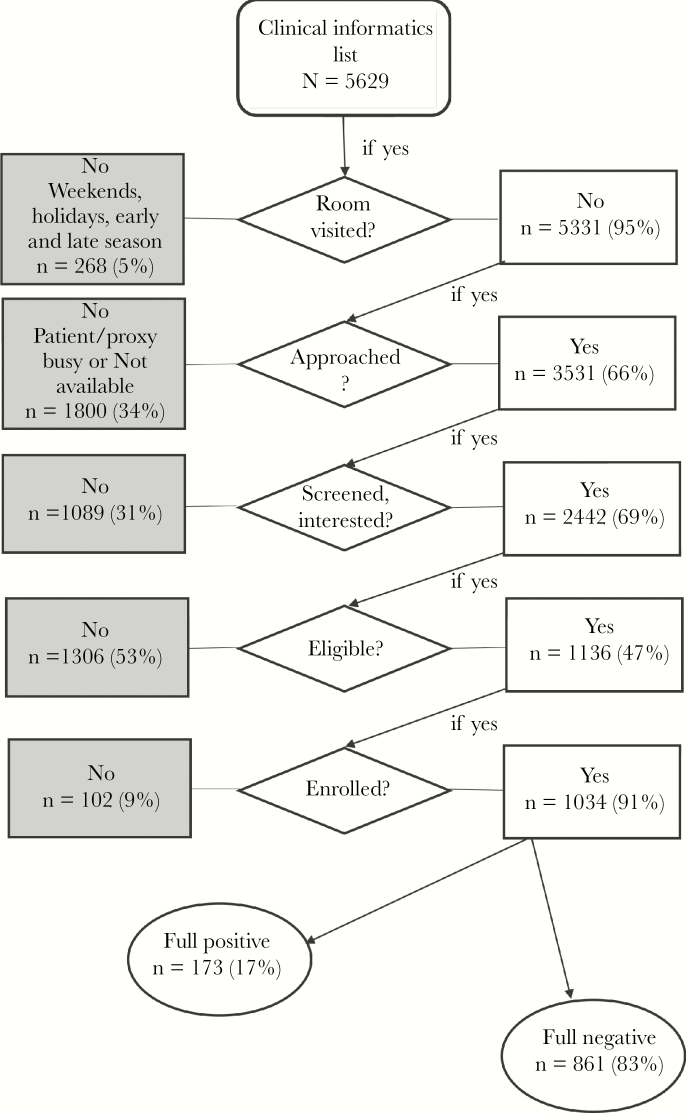
In 2014–2015, when 1 hospital was used as a recruiting site, EHR records were manually reviewed, and the enrollment season was 59 days, 210 records were identified with an average of 4 patients per hospital-day identified as potentially eligible admissions. In 2015–2016, when 2 hospitals were used as recruiting sites with the CIA, and the enrollment season was 201 hospital-days, 7332 records were identified, with an average of 36 patients per hospital-day identified as potentially eligible admissions. The following year, 2016–2017, with a longer enrollment period of 288 days across 3 hospitals and 5629 records identified using the CIA, 20 patients per day were identified as potentially eligible (Table 1). Ninety-five percent (5331) of those patients’ rooms were visited, with the remainder appearing on the list on days when RAs were not staffing the hospital for the study. Figure 1 shows the flow of patients from appearance on the CIA list to enrollment and influenza status. A total of 3531 patients were approached. However, 13 did not have CIA factors available for analysis and were not included. Of the remaining 3518 approached patients, 2442 agreed to be screened by the RA for eligibility. Of those, 1136 patients were eligible and 1034 patients were enrolled. Although percentages of patients visited, approached, eligible, and enrolled were higher for the manual review, the number of patients enrolled using this method was 20% of the number of enrollees from the CIA-REDCap method after adjusting for the number of hospital-days. The manual review resulted in 2 enrollees per day compared with 3.6 enrollees per day from the CIA-REDCap review, while accounting for its larger number of hospital-surveillance days.
Table 1.
Identification and Enrollment Process Over 3 Years
| Year 1 (Pilot) | Year 2 | Year 3 | Year 3 | |||
|---|---|---|---|---|---|---|
| 2014–2015 (P) | 2015–2016 (P + SM) | 2016–2017 (P + SM + SH) | 2016–2017 | |||
| Patients | Manual EHR Search of Those on Admissions List | CIA List as Spreadsheeta | CIA List Loaded to REDCap | Refinement of CIA Using CART | ||
| Patients identified for review, No. | 210 | 7332 | 5629 | ICD-10 codes J18, J44, R05, J20 | RVP, ED note, ADM note | RVP, ED note, ADM note, ICD-10 codes J18, J20 |
| Hospital-days of study surveillance, No. | 59 | 201 | 288 | |||
| Patients identified/ hospital-day, No. | 4 | 36 | 20 | |||
| Rooms visited, No. (%) | 204 (97) | 957 (13) | 5331 (95) | |||
| Patients approached, No. (%) | 204 (97) | 924 (13) | 3531b (63) | Projected Numbersc | ||
| Patients screened, No. (%) | 155 (74) | 711 (10) | 2442 (43) | 2442 | 2442 | 2442 |
| Patients eligible, No. (%) | 144 (69) | 549 (7) | 1136 (20) | 507 (21) | 1378 (56) | 1587 (65) |
| Patients enrolled, No. (enrolled/identified, %) | 126 (60) | 528 (7) | 1034 (18) | 461 (8) | 1254 (22) | 1444 (26) |
| Patients enrolled, No. (enrolled/eligible, %) | 126 (88) | 528 (96) | 1034 (91) | 461 (91) | 1254 (91) | 1444 (91) |
| Patients enrolled/hospital-days of surveillance, No. | 2.1 | 2.6 | 3.6 | 1.6 | 4.4 | 5.0 |
Abbreviations: ADM, admission; CART, classification and regression tree; CIA, clinical informatics algorithm; ED, emergency department; EHR, electronic health record; ICD-10, International Classification of Diseases, Tenth Revision; P, quaternary care hospital; RVP, respiratory viral panel; SH, tertiary care hospital; SM, community hospital.
aEHR reviewed to rule out ineligible at P and approached only those with respiratory viral panel at SM.
bThirteen subjects did not have clinical informatics information available.
cCART analyses were run only on the subset of patients screened in year 3.
Figure 1.
Flow diagram for enrollment process starting from the clinical informatics algorithm–generated list.
CART Analysis
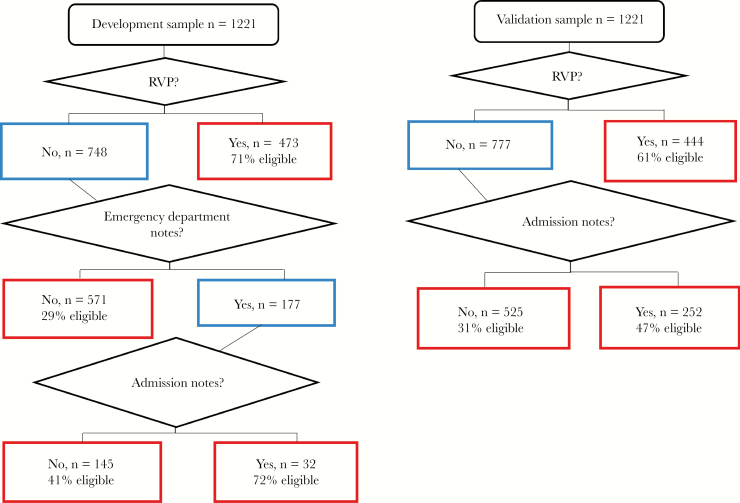
Because the CART analysis was conducted on both the group of approached and the subset of patients enrolled in 2016–2017, characteristics of the approached and enrolled patients are shown in Table 2. The sample of 2442 patients who agreed to be screened by the RA for eligibility was split into a developmental and a validation sample. Table 3 shows the characteristics of the 2 samples; they did not differ on demographic characteristics or indications on the clinical informatics list. The primary CART analysis, which used eligibility as the outcome variable and the indicators from the clinical informatics list as the independent variables, is shown in Figure 2. The most important indicator for eligibility is presence of an RVP, which identified 71% of the developmental sample and 61% of the validation sample. Adding ADM notes increased the likelihood of correctly identifying eligible patients in each sample, with ED notes increasing the likelihood further in the development sample.
Table 2.
Characteristics of Approached Patients and the Subset of Enrolled Patients
| Variables | Approached (n = 3518) | Enrolled (n = 1034) |
|---|---|---|
| Age, y, mean (SD) | 64 (17.5) | 62 (16.5) |
| Age group, y | ||
| 18–64 | 1581 (45) | 548 (53) |
| ≥65 | 1937 (55) | 486 (47) |
| Female sex (ref = male) | 1859 (53) | 599 (58) |
| Hospital | ||
| P | 1859 (53) | 460 (45) |
| SH | 443 (13) | 179 (17) |
| SM | 1216 (34) | 395 (38) |
| Eligible for screening indicated on: | ||
| ICD-10 | 1929 (55) | 476 (46) |
| Respiratory viral panel | 1278 (36) | 557 (54) |
| Admission note | 893 (25) | 301 (29) |
| Emergency department note | 544 (16) | 168 (17) |
| Chief complaints | 174 (5) | 44 (4) |
| Total indications (RVP, ADM, ED, ICD-10, CC) | ||
| 1 | 2500 (71) | 670 (65.4) |
| 2 | 770 (22) | 238 (23.0) |
| 3 | 217 (6) | 105 (10.3) |
| ≥4 | 31 (1) | 21 (2) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ADM, admission; ED, emergency department; ICD-10, International Classification of Diseases, Tenth Revision; P, quaternary care hospital; ref, reference category; RVP, respiratory viral panel; SD, standard deviation; SH, tertiary care hospital; SM, community hospital.
Table 3.
Characteristics of Development and Validation Samples for Classification and Regression Tree Analysis
| Characteristics | Development Sample (n = 1221) | Validation Sample (n = 1221) | P Value |
|---|---|---|---|
| Age, y, mean (SD) | 62.4 (17.7) | 63.5 (16.9) | .120 |
| Female sex | 667 (54.6) | 637 (52.2) | .224 |
| Age group, y | .544 | ||
| 18–64 | 598 (49.0) | 583 (47.8) | |
| ≥65 | 623 (51.0) | 638 (52.0) | |
| Identified on clinical informatics list by: | |||
| Respiratory viral panel | 473 (38.7) | 444 (36.4) | .226 |
| Admission note | 302 (24.7) | 313 (25.6) | .608 |
| Emergency department note | 200 (16.4) | 182 (14.9) | .316 |
| ICD-10 | 652 (53.4) | 677 (55.5) | .310 |
| Chief complaints | 72 (5.9) | 67 (5.5) | .662 |
| Eligible | 585 (47.9) | 551 (45.1) | .168 |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ICD-10, International Classification of Diseases, Tenth Revision; SD, standard deviation.
Figure 2.
Development and validation samples for classification and regression tree analysis. The outcome was eligibility; independent variables were clinical informatics list indicators including respiratory viral panel (RVP), indicator word in admission notes, emergency department notes, or chief complaint or International Classification of Diseases, Tenth Revision code. Development sample receiver operating characteristic curve (ROC), 70%; sensitivity, 62%; specificity, 76%. Validation sample ROC, 65%; sensitivity, 71%; specificity, 54%. Boxes in red indicate terminal nodes.
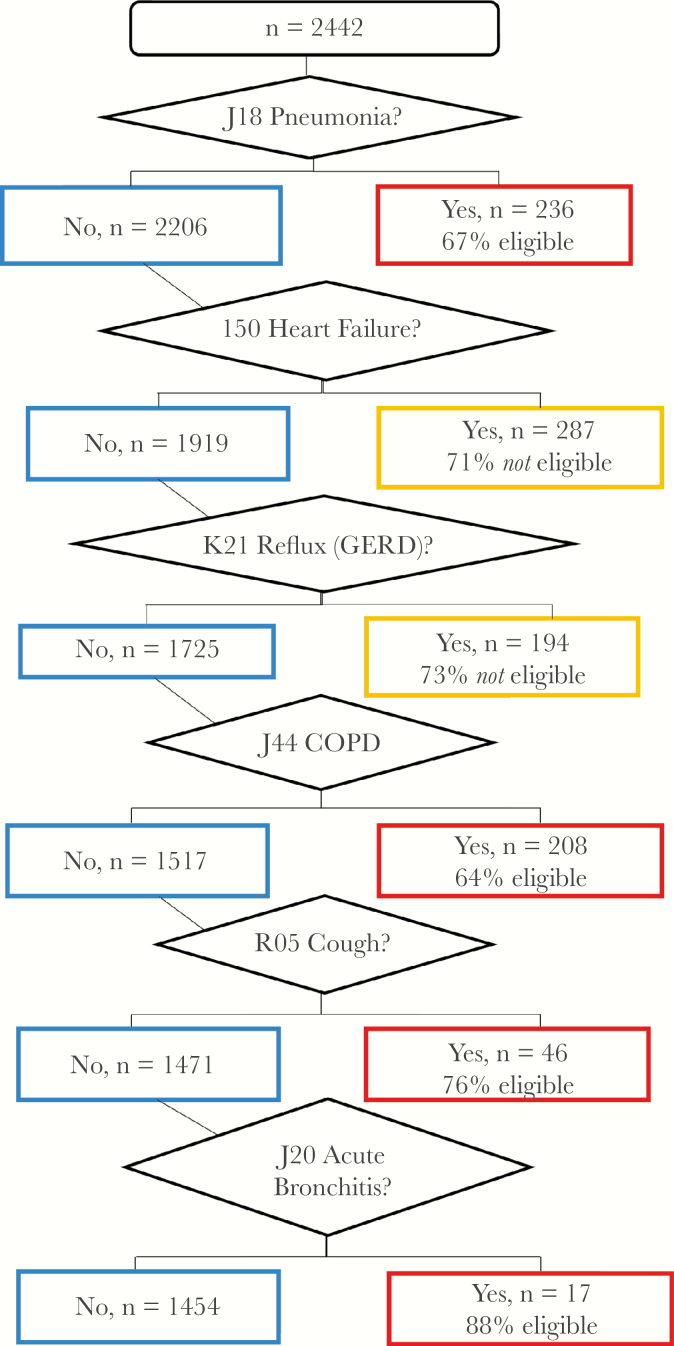
In a secondary CART analysis, the select ICD-10 codes were the independent variables with eligibility as the outcome variable. The regression tree is shown in Figure 3. Six conditions were identified in the CART, with 4 ICD-10 codes increasing the likelihood of eligibility (pneumonia, J18; COPD, J44; cough, R05; acute bronchitis, J20) and 2 ICD-10 codes decreasing the likelihood of eligibility (heart failure, I50; gastroesophageal reflux disease [GERD], K21). The ROC was 63%, sensitivity was 42%, and specificity was 73%.
Figure 3.
Classification and regression tree analysis for International Classification of Diseases, Tenth Revision (ICD-10) codes. The outcome was eligibility; independent variables were select ICD-10 codes (those that appeared in the clinical informatics list for ≥10 patients, n = 129). Receiver operating characteristic curve, 63%; sensitivity, 42%; specificity, 73%. Abbreviations: COPD, chronic obstructive pulmonary disease; GERD, gastroesophageal reflux disease.
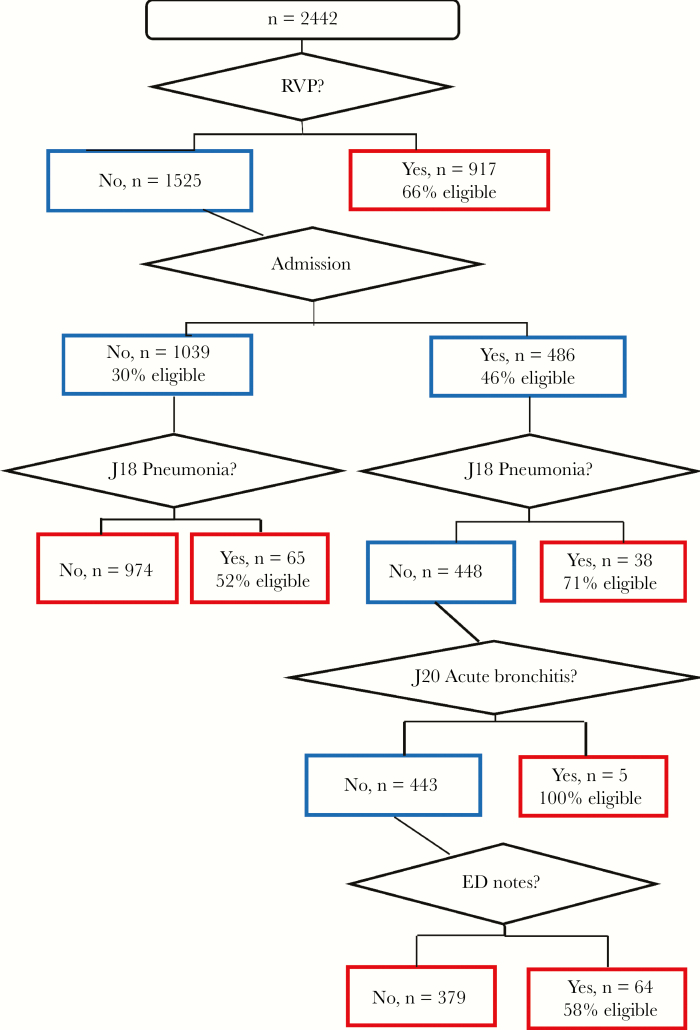
A subsequent secondary CART analysis used the clinical informatics list indicators and the select ICD-10 codes as the independent variables and eligibility as the outcome. The regression tree is shown in Figure 4. This tree contains RVP, ADM note, and ED note as well as pneumonia (J18) and acute bronchitis (J20). The ROC was 69%, sensitivity was 61%, and specificity was 71%.
Figure 4.
Classification and regression tree analysis for International Classification of Diseases, Tenth Revision (ICD-10) codes, respiratory viral panel (RVP), and key terms or symptoms in admission (ADM) notes, emergency department (ED) notes, or chief complaint. The outcome was eligibility; independent variables were select ICD-10 codes (those that appeared in the clinical informatics list for ≥10 patients, n = 6), plus RVP plus key term or symptom in ADM notes, ED notes, or chief complaint.
The right 3 columns of Table 1 present the predicted improvements in percentages of eligible and enrolled patients that could be possible if the CIA were refined using CART. Adding select ICD-10 codes generally did not improve the percentages of eligible or enrolled patients, whereas using just RVP, ED notes, and ADM notes with or without ICD-10 codes J18 and J20 increased the projected eligible or enrolled patients over the manual and the CIA-REDCap methods.
Table 4 compares the characteristics of eligible and ineligible patients. Eligible patients were significantly younger, more often female, identified by presence of an RVP, key term in ADM notes, CC, or select ICD-10 code, and admission to 1 of the nonquaternary care hospitals (P ≤ .001 for all). Multivariable logistic regression analysis was conducted to map the distribution and significance of CART predictors for eligibility. In regression analysis, the most significant predictor for eligibility was RVP, with an OR of 5.0 (95% CI, 4.1–6.0) (Table 5). Other significant factors were having an indicator word in the ADM notes (OR, 2.3 [95% CI, 1.9–2.8]) or ED notes (OR, 1.9 [95% CI, 1.5–2.4]). Age group, sex, and hospital were added to the regression and results are shown in Appendix 2. Likelihood of eligibility was higher in the older age group ≥65 years (OR, 1.5 [95% CI, 1.3–1.8]) and also varied by site; that is, the likelihood of eligibility of those admitted to the quaternary care hospital (P) was 2–3 times less likely than for those admitted to the 2 lower-acuity hospitals.
Table 4.
Characteristics of Eligible and Ineligible Patients
| Characteristics | Not Eligible (n = 1306 [53.5%]) | Eligible (n = 1136 [46.5%]) |
P Value |
|---|---|---|---|
| Age, y, mean (SD) | 63.5 (17.8) | 62.2 (16.6) | .061 |
| Age group, y | .001 | ||
| 18–64 | 592 (45.3) | 589 (51.8) | |
| ≥65 | 714 (54.7) | 547 (48.2) | |
| Female sex | 653 (50.0) | 651 (57.3) | <.001 |
| Hospital | <.001 | ||
| P | 869 (66.5) | 531 (46.7) | |
| SH | 96 (7.4) | 184 (16.2) | |
| SM | 341 (26.1) | 421 (37.1) | |
| Identified on clinical informatics list by: | |||
| Respiratory viral panel | 310 (23.7) | 607 (53.4) | <.001 |
| Admission note | 285 (21.8) | 330 (29.1) | <.001 |
| Emergency department note | 197 (15.1) | 185 (16.3) | .415 |
| ICD-10 | 794 (60.8) | 535 (47.1) | <.001 |
| Chief complaints | 92 (7.0) | 47 (4.1) | .002 |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ICD-10, International Classification of Diseases, Tenth Revision; P, quaternary care hospital; SD, standard deviation; SH, tertiary care hospital; SM, community hospital.
Table 5.
Factors From Clinical Informatics List Independently Associated With Eligibility From Multivariate Logistic Regression Analyses
| Factor | Odds Ratio (95% CI) | P Value |
|---|---|---|
| Respiratory viral panel (ref = no) | 5.0 (4.1–6.0) | <.001 |
| Admission note (ref = no) | 2.3 (1.9–2.8) | <.001 |
| Emergency department note (ref = no) | 1.9 (1.5–2.4) | <.001 |
International Classification of Diseases, Tenth Revision codes and chief complaint factors were included in the model but were not significant.
Abbreviations: CI, confidence interval; ref, reference category.
A CART analysis was conducted to examine the ability of the CIA to predict presence of influenza among enrollees, using the CIA indicators of RVP, ADM, ED, and CC as independent variables. The single best predictor was a clinician ordering an RVP with a ROC of 64%, sensitivity of 77%, and specificity of 51% (regression tree not shown). In sensitivity analyses, all select ICD-10 codes were added to the model with no changes in the resulting ROC, sensitivity, or specificity.
DISCUSSION
Recruitment of participants for research studies can be time consuming and resource intensive. As noted earlier, patients with ARIs report a wide range of possible symptoms. Therefore, recruitment for the HAIVEN study was based on presentation to the hospital with 1 or more signs, symptoms, diagnoses, and/or tests from a broad list. Using broad criteria for screening typically results in high sensitivity, but low specificity—that is, most actual cases would be included in the list of potentially eligible participants, but many who are included would not actually be eligible. Although there is a need to enroll negatives (controls) to satisfy requirements for the test-negative design, considerable time is required to screen out the ineligible individuals. Alternatively, a narrow approach with focused eligibility criteria would likely decrease sensitivity but increase specificity; that is, many cases may be missed, but fewer noncases would be included in the approach list. Despite expending less time identifying potential participants, the result may be a failure to reach recruitment goals.
When manual review was employed, 97% of identified patients were visited, compared with 95% when CIA was employed. But using the CIA-REDCap method, 1.5 times as many patients were enrolled per hospital-day compared with manual chart review. This difference is most likely a reflection of the amount of time required to manually review the EHR of each person on the new admissions list, suggesting that the CIA offers a more efficient system.
Using a CIA, we found that 49% (1106/2442) of individuals who were screened were ineligible for the study, suggesting that there is room for improvement in the methodology. The CART analysis revealed that for predicting eligibility, the clinical RVP was the single best indicator identified by the CIA. For hospitals without CIA capabilities, screening for those who had RVP ordered would simplify manual EHR searches. Adding a search for indicator words in the ADM or ED notes may be a useful addition as it added significantly to the ROC in the validation sample, but may be difficult to quickly identify in a manual search of the EHR. Using ICD codes alone may be a reasonable approach if the list is limited to heart failure and GERD for identification of likely ineligible patients, and pneumonia, COPD, cough, and acute bronchitis for identification of likely eligible patients. The use of ICD codes has limitations, which include the accuracy of coding by the treating physician and the timing of coding, which may occur only after hospital discharge. The most efficient and sensitive algorithm appears to include RVP, an indicator word in the ADM and ED notes, and the ICD-10 codes of J18* for pneumonia and J20* for acute bronchitis.
The most sensitive predictor for influenza positivity was a clinical RVP. Therefore, if a study’s eligibility criteria included the presence of influenza infection, an EHR search for RVP testing would be the most efficient choice.
Analysis of “big data” is a relatively new tool for using the combined resources of electronic health system medical records, molecular biology databases, administrative and insurance databases, national public health databases, and many other sources to predict health outcomes, improve service delivery, monitor epidemics, support timely clinical decision making, reduce healthcare costs, and facilitate research [14–16]. Specifically, clinical informatics has the potential to change the ways in which healthcare providers can determine which patients are candidates for specialized treatments and researchers can rapidly identify potentially eligible participants for study enrollment. Most research of clinical informatics has focused on the former use.
While the power of clinical informatics lies in the quality and quantity, depth, and breadth of the data, its usefulness lies in the precision of the outcomes with regard to a given study’s inclusion/exclusion criteria. Algorithms based on broad inclusion criteria should be tested for sensitivity and specificity using regression analyses such as CART as an enhancement to other multivariable regression analyses. By focusing searches on the most appropriate search terms, the algorithm can produce lists of potential participants that maximize the efficiency of the recruitment process.
Strengths and Limitations
Using a clinical informatics algorithm to generate an approach list for screening potential participants in an influenza VE study works well by systematically reviewing all patients. This method offers less chance for human errors and introduction of selection bias. The use of recursive partitioning and an algorithm that extends beyond ICD codes to include symptoms and signs and laboratory tests found in other parts of the EHR are strengths. A limitation of using a CIA is its time-intensive nature. However, once programmed, it runs automatically. Other limitations of this study are that these results are based primarily on 3 hospitals using the same EHR and should be replicated in other seasons and with a larger set of hospitals. Second, usefulness of the CIA has been demonstrated herein for acute respiratory illness only and exclusively for inpatients. CIAs should be tested in other recruitment settings and for other diseases or health conditions. It is possible that recruitment of individuals with diseases that are more or less common than ARI would not be as easily adapted to a CIA. This study assumes that research personnel are charged with screening and enrolling patients into the study and does not depend on clinical staff to perform these additional duties. Last, as mentioned above, use of ICD codes may be limited by the accuracy and timing of coding. In fact, we found that most ICD codes were not helpful for identifying potentially eligible patients.
In conclusion, this study supports the use of clinical informatics in preference to manual EHR review to facilitate recruitment of eligible participants in clinical research. Performing regression, especially CART analysis, offers the opportunity to hone a reasonably effective clinical informatics algorithm to further improve its efficiency.
APPENDIX 1. QUALIFYING SYMPTOMS/SYNDROMES FOR ACUTE RESPIRATORY ILLNESS
| ICD-10 Codes: Must Have Either ≥1 From Column A or ≥1 From Column B Top Plus Column B Bottom | ||||
|---|---|---|---|---|
| Column A: Beginning ≤10 d Ago | Column B | |||
| OR | Symptoms/syndromes | |||
| Influenza-like illness | J80 | Acute respiratory distress syndrome | ||
| J11.1 | Influenza-like illness | R50.9 | Fever | |
| J11.1 | Influenza-like disease | R09.81 | Nasal congestion | |
| J10.1 | Influenza | R09.89 | Chest congestion | |
| J06.9 | URI | R07.0 | Sore throat | |
| J06.9 | Viral URI | R68.83 | Chills | |
| R05 | Cough | R52 | Body aches | |
| J20.8 | Bronchitis | R53.83 | Fatigue | |
| Pneumonia | R06.03 | Respiratory distress | ||
| J18.9 | Pneumonia | R06.02 | Shortness of breath | |
| J15.9 | Bacterial pneumonia | R06.89 | Difficulty in breathing | |
| J18.9 | Community-acquired pneumonia | R06.00 | Dyspnea | |
| J18.9 | Healthcare-acquired pneumonia | A41.9 | Sepsis | |
| J69.0 | Aspiration pneumonia | E84.0 | Cystic fibrosis exacerbation | |
| J18.9 | Evaluate pneumonia | J98.8 | Respiratory medical, other | |
| J18.9 | Bibasilar pneumonia | I50.9 | Congestive heart failure | |
| Asthma and COPD | J84.112 | Idiopathic pulmonary fibrosis | ||
| J44.1 | COPD exacerbation | R41.82 | Altered mental status | |
| J45.901 | Asthma exacerbation |
AND New-onset, exacerbation, or change in ≥2 of the following symptoms with at least 1 respiratory symptom beginning ≤10 d ago: - Respiratory symptoms: cough, shortness of breath, nasal congestion, chest congestion, sore throat - Constitutional symptoms: fever/feverishness, chills, body aches, fatigue |
||
| J45.902 | Status asthmaticus | |||
| J45.901 | Asthmatic bronchitis | |||
Abbreviations: COPD, chronic obstructive pulmonary disease; ICD-10, International Classification of Diseases, Tenth Revision; URI, upper respiratory infection.
APPENDIX 2. FACTORS INDEPENDENTLY ASSOCIATED WITH ELIGIBILITY FROM MULTIVARIATE LOGISTIC REGRESSION ANALYSES
| Factor | Odds Ratio (95% CI) | P Value |
|---|---|---|
| Respiratory viral panel (ref = no) | 4.9 (4.0–6.0) | <.001 |
| Admission note (ref = no) | 2.3 (1.8–2.8) | <.001 |
| Hospital (ref = P) | … | <.001 |
| SH | 3.2 (2.4–4.4) | |
| SM | 2.2 (1.8–2.7) | |
| Emergency department note (ref = no) | 1.8 (1.4–2.3) | <.001 |
| Age group 18–64 y (ref = ≥65 y) | 1.5 (1.3–1.8) | <.001 |
Individual International Classification of Diseases, Tenth Revision (ICD-10) codes were not used; rather, each factor was used as an indicator variable. Sex, ICD-10 code, and chief complaint were included in the model but were not significant factors.
Abbreviations: CI, confidence interval; P, quaternary care hospital; ref, reference category; SH, tertiary care hospital; SM, community hospital.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Centers for Disease Control and Prevention (CDC) or the National Institutes of Health (NIH).
Financial support. This work was supported by the CDC (grant number U01IP000969) and the NIH (grant number UL1TR001857).
Potential conflicts of interest. R. K. Z. has received research grants from Sanofi Pasteur, Pfizer, and Merck & Co. M. P. N. has received grant funding from Pfizer and Merck & Co. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Fiore AE, Bridges CB, Katz JM, Cox NJ. Inactivated influenza vaccines. In: Plotkin S, Orenstein W, Offit P, eds. Vaccines. 6th ed Philadelphia, PA: Elsevier Saunders; 2013:257–93. [Google Scholar]
- 2. Nguyen JL, Yang W, Ito K, et al. Seasonal influenza infections and cardiovascular disease mortality. JAMA Cardiol 2016; 1:274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ludwig A, Lucero-Obusan C, Schirmer P, et al. Acute cardiac injury events≤ 30 days after laboratory-confirmed influenza virus infection among US veterans, 2010–2012. BMC Cardiovasc Dis 2015; 15:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Estabragh ZR, Mamas MA. The cardiovascular manifestations of influenza: a systematic review. Int J Cardiol 2013; 167:2397–403. [DOI] [PubMed] [Google Scholar]
- 5. Nichol KL, Wuorenma J, von Sternberg T. Benefits of influenza vaccination for low-, intermediate-, and high-risk senior citizens. Arch Intern Med 1998; 158:1769–76. [DOI] [PubMed] [Google Scholar]
- 6. Studahl M. Influenza virus and CNS manifestations. J Clin Virol 2003; 28:225–32. [DOI] [PubMed] [Google Scholar]
- 7. Short KR, Kroeze EJBV, Fouchier RAM, Kuiken T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis 2014; 14:57–69. [DOI] [PubMed] [Google Scholar]
- 8. Balasubramani K, Saul S, Nowalk MP, et al. Does influenza vaccination status change physician ordering patterns for respiratory viral panels? Inspection for selection bias. Hum Vacc Immunother 2019; 15:91–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yount RJ, Vries JK, Councill CD. The medical archival system: an information retrieval system based on distributed parallel processing. Inf Process Manage 1991; 27:379–89. [Google Scholar]
- 10. Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 34:301–10. [DOI] [PubMed] [Google Scholar]
- 11. et al. Guidance for clinicians on the use of RT-PCR and other molecular assays for diagnosis of influenza virus infection. Available at: https://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm. Accessed December 18, 2018. [Google Scholar]
- 12. Popowitch EB, O’Neill SS, Miller MB. Comparison of four multiplex assays for the detection of respiratory viruses: Biofire FilmArray RP, Genmark eSensor RVP, Luminex xTAG RVPv1 and Luminex xTAG RVP FAST. J Clin Microbiol 2013; 51:1528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Breiman L, Friedman JH, Olshen RA, Stone CJ.. Classification and Regression Trees (Wadsworth Statistics/Probability). Boca Raton, FL: Chapman & Hall; 1984. [Google Scholar]
- 14. Alonso SG, de la Torre Díez I, Rodrigues JJPC, et al. A systematic review of techniques and sources of big data in the healthcare sector. J Med Syst 2017; 41:183. [DOI] [PubMed] [Google Scholar]
- 15. Luo J, Wu M, Gopukumar D, Zhao Y. Big data application in biomedical research and health care: a literature review. Biomed Inform Insights 2016; 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehta N, Pandit A. Concurrence of big data analytics and healthcare: a systematic review. Int J Med Inform 2018; 114:57–65. [DOI] [PubMed] [Google Scholar]