Abstract
Receptors of the leukocyte receptor cluster (LRC) play a range of important functions in the human immune system. However, the evolution of the LRC remains poorly understood, even in m\ammals not to mention nonmammalian vertebrates. We conducted a comprehensive bioinformatics analysis of the LRC-related genes in the publicly available genomes of six species that represent eutherian, marsupial, and monotreme lineages of mammals. As a result, the LRCs of African elephant and armadillo were characterized, two new genes, IGSF1 and A1BG, were attributed to the LRC of eutherian mammals, the LRC gene content was substantially extended in the short-tailed opossum and Tasmanian devil and, finally, four LRC genes were identified in the platypus genome. These findings have for the first time provided a solid basis for inference of the LRC phylogeny across mammals. Our analysis suggests that the mammalian LRC family likely derived from two ancestral genes, which evolved in a lineage-specific manner by expansion/contraction, extensive exon shuffling, and sequence divergence. The striking structural and functional diversity of eutherian LRC molecules appears largely lineage specific. The only family member retained in all the three mammalian lineages is a collagen-binding receptor OSCAR. Strong sequence conservation of a transmembrane domain known to associate with FcRγ suggests an adaptive role of this domain subtype in the LRC evolution.
Keywords: marsupial, monotreme, OSCAR, IGSF1, A1BG, GPVI
Introduction
The leukocyte receptor cluster (LRC) is a family of structurally related genes for immunoregulatory receptors. Originally, the term LRC was introduced to emphasize the linkage of the genes encoding killer immunoglobulin-like receptors (KIRs), leukocyte Ig-like receptors (LILRs), and FcαR on human chromosome 19q13.4 (Wagtmann et al. 1997; Wende et al. 1999). Subsequently, it has been found that the region contains some other structurally related genes, such as NCR1, GPVI, LAIR1, LAIR2, and OSCAR (Meyaard et al. 1997; Sivori et al. 1997; Clemetson et al. 1999; Kim et al. 2002). Most recently, the LRC has been further extended by adding two more genes named VSTM1/SIRL1 and TARM1 (Steevels et al. 2010; Radjabova et al. 2015). These were found to be closely related to each other and linked to OSCAR.
Except for LAIR2, which is a secreted protein, all human LRC products are type I cell surface receptors with extracellular regions composed of 1–4 C2-type Ig-like domains. The functional properties of these proteins are strikingly diverse. Human KIRs play a crucial role in the regulation of NK cell activity by binding to MHC class I molecules on target cells (Bottino et al. 1995; Lanier and Phillips 1995). In humans, KIR and HLA class I genotypes are associated with various diseases ( reviewed by Parham et al. [2010] and Trowsdale et al. [2015]). The LILR function is less clear although interaction with classical and nonclassical MHC has been shown for some human and mouse (where they are designated PIRs) LILRs (Borges et al. 1997; Liang et al. 2002; Willcox et al. 2003; Takai 2005; Shiroishi et al. 2006). Additionally, a broad range of non-MHC ligands have been reported for human and mouse members of the LILR family (reviewed by Trowsdale et al. [2015]). NCR1 recognizes membrane-associated heparan sulfate proteoglycans (Hecht et al. 2009) and has been described as an influenza virus hemagglutinin receptor (Mandelboim et al. 2001). Human FcαR is a receptor for IgA (Maliszewski et al. 1990), whereas GPVI, LAIR1, LAIR2, and OSCAR bind to collagen (Moroi et al. 1996; Meyaard et al. 1997; Lebbink et al. 2008; Barrow et al. 2011).
According to their signaling properties, LRC receptors are generally subdivided into two major forms—inhibitory or activating. The inhibitory forms are expressed as monomers bearing the ITIM motifs in their cytoplasmic tails. The activating forms usually have short cytoplasmic regions and associate with ITAM-containing signaling transmembrane (TM) subunits, such as DAP12 (activating KIRs) or FcRγ (activating LILRs, NCR1, FcαR, GPVI, OSCAR, and TARM1), on the cell surface. The association is facilitated by the presence of aspartate residue in the TM regions of the signaling subunits and positively charged residue in the TMs of LRC molecules. Activating KIRs have lysine residue in their TMs. Except for this feature, TMs of activating and inhibitory KIRs are highly similar suggesting that the activating forms of KIRs have evolved from the inhibitory ones (Abi-Rached and Parham 2005). The TMs of all other activating LRC members contain the arginine residue embedded into the NxxR motif (fig. 1). Notably, this TM subtype is strongly conserved. It has been recognized in the LRC molecules from as distant species as amphibians and birds (Guselnikov et al. 2010).
Fig. 1.
—Alignment of amino acid sequences of the TM regions of human (Hs) FcRγ-associating LRC members and the predicted LRC members from Tasmanian devil (Sh), opossum (Md), and platypus (Oa). Attribution of opossum and platypus OSCAR, as well as devil GPVI is based on the results of phylogenetic analysis. Other opossum sequences are designated according to the gene position in the current version of the genome (fig. 6A). Identical and similar residues are shown by white letters on black background or black letters on gray background, respectively. Consensus (>50%) sequence is shown below. Asterisks indicate invariant residues.
Studies of the LRC and, in particular, KIR family significantly contributed to such important immunological concepts as “missing self-recognition” (Kärre et al. 1986) and “immunoregulation through a balance of positive and negative signals” (Ravetch and Lanier 2000). That is why it was of particular interest to understand the evolution of this group of molecules. Unexpectedly, the comparative studies revealed that, despite its proposed functional importance, the LRC is a highly diverse and rapidly evolving gene family. Simultaneously with the identification of KIRs, it has been found that, in mice, the function of MHC class I-specific recognition on NK cells is carried out by the Ly49 receptor family that belongs to the C-type lectin superfamily (Karlhofer et al. 1992). Subsequent studies have demonstrated that the expansion of the KIR family is specific for primates and many other mammals possess just one or a few KIR-like molecules (Kelley et al. 2005; Hammond et al. 2009). A similar lineage-specific evolution has been shown in the case of the LILR family (Hoelsbrekken et al. 2005; Hogan et al. 2012).
Investigation of the chicken and marsupial genomes revealed a dramatic expansion of the LRC-like genes in both lineages. However, members of the chicken and marsupial LRC families, designated CHIRs and MAIRs, respectively, showed only weak similarity to the LRC genes of eutherian mammals (Dennis et al. 2000; Nikolaidis et al. 2005; Viertlboeck et al. 2005; Laun et al. 2006; Belov et al. 2007; van der Kraan et al. 2013). The CHIR genes were found as a separate cluster on microchromosome 31. One more chicken gene for the LRC-related receptor designated ggFCR has been mapped to the region syntenic to human 20q13 (Viertlboeck, Schmitt, et al. 2009). Functional studies have demonstrated that ggFcR and some CHIRs are chicken IgY receptors (Viertlboeck et al. 2007; Viertlboeck, Schmitt, et al. 2009; Viertlboeck, Schweinsberg, et al. 2009). No LRC-like genes have been found in a monotreme platypus and in avians duck and zebra finch (Wong et al. 2009; Windau et al. 2013). The amphibian Xenopus tropicalis has been shown to possess four LRC-related genes all encoding activating receptors with the NxxR-motif-containing TMs (Guselnikov et al. 2010). The extracellular domains of Xenopus LRC proteins showed only a weak similarity to those of the chicken and human members of the family. In bony fishes, several gene families encoding paired Ig-like receptors expressed in lymphoid tissues have been described and suggested to play a role in NK cell mediated immunorecognition (reviewed by Yoder and Litman [2011] and Montgomery et al. [2011]). However, it remains uncertain if any of these genes are diverged counterparts of LRC or represent other subsets of IgSF.
Therefore, after two decades since the LRC definition, it is still unclear as to when this family might have arisen, what the function of the primordial LRC genes was, why they evolved so rapidly, and whether there is any functional activity shared by the LRC proteins from different vertebrates. To begin addressing these questions, we have analyzed LRC-like genes in the sequenced genomes of six mammalian species, representing placental mammals (humans, elephant, and armadillo), marsupials (short-tailed opossum and Tasmanian devil) and monotrems (platypus). We found that the eutherian LRC family, in addition to commonly recognized members, includes two new, IGSF1 and alpha-1-B glycoprotein (A1BG). This finding facilitated more accurate characterization of the LRC genes in marsupials and platypus and ultimately uncovered the structural relationships among the LRC members in the three mammalian lineages.
Materials and Methods
Genomic Analysis
In this study, the genomes of five mammalian species were searched: African elephant (Loxodontra africana), nine-banded armadillo (Dasypus novemcinctus), gray short-tailed opossum (Monodelphis domestica), Tasmanian devil (Sarcophilu sharrisii), and duck-billed platypus (Ornithorhynchus anatinus) (Mikkelsen et al. 2007; Warren et al. 2008; Murchison et al. 2012). The genome assembly versions were Loxafr3.0, Dasnov3.0, MonDom5, Devil_ref v7.0, and OANA5 according to the Ensembl designations (www.ensembl.org). Preliminary analysis showed the presence of numerous automatically generated models for the LRC-like genes in the annotations to these genomes. However, we had to disregard these models because of the known poor capacity of automatic gene prediction algorithms to recognize multiple structurally related genes. The species-specific pattern of gene and exon organization in such families, subdivision of the encoded proteins into various forms, such as secreted proteins, activating and inhibitory receptors, profoundly impede the automatic gene prediction. The most common output errors are gene truncation or fusing of two or more genes into a single one because of poor recognition of short exons coding for TMs or cytoplasmic tails. Furthermore, an automatic mode of homology attribution in the genome annotations is often misleading, as it relies solely on which protein in the protein database produced the highest matching BlastP score without careful structural considerations of such matching.
The approach for gene identification and prediction that was used in this series of articles has been described in our earlier publications (Fayngerts et al. 2007; Guselnikov et al. 2008, 2010, 2011). It is based on the identification of separate exons for the Ig-like and TM domains in the genomic sequences followed by feeding the groups of such exons into gene prediction pipelines. To minimize ambiguity, we did not consider exons for the signal peptides and intracellular regions unless their existence was supported by the EST data or by high similarity. The preliminary subdivision of the predicted receptors into activating or inhibitory was based on the presence or absence of the positively charged residues in their TMs.
Phylogenetic Analysis
Phylogenetic analysis was performed with the MEGA6 software (Tamura et al. 2013) using nucleotide sequences of individual exons or sequences encoding N-terminal Ig domain pairs. Alignment of the nucleotide sequences was guided by the amino acid sequence alignment generated by either Clustal or Muscle utilities of the MEGA6 software. For each data set, different variants of alignment including those corrected manually or with the certain part removed were tested in the phylogenetic analysis. Phylogenetic trees were constructed using the bootstrap tests of the Neighbor-Joining (NJ) method with p-distances (proportion of differences). The trees shown in this article were selected because of the topology that was the most tolerant to the variations in alignment and the tree generation settings. The corresponding alignments are available upon request. Nucleotide sequences were retrieved and analyzed using utilities at the NCBI (https://www.ncbi.nlm.nih.gov/, last accessed May 20, 2019) and Ensemble (http://www.ensembl.org, last accessed May 20, 2019) websites. Amino acid sequence alignments were shaded using the BoxShade program (https://embnet.vital-it.ch/software/BOX_form.html, last accessed May 20, 2019). GenScan program (Burge and Karlin 1997, http://hollywood.mit.edu/GENSCAN.html, last accessed May 20, 2019) was used for the automated gene structure prediction. The LRC-flanking non-LRC genes were generally identified according to the genomic annotations. However, in some cases, such annotations were verified by sequence comparisons at the NCBI website using BlastP.
Results
A1BG and IGSF1 Are LRC-Related Genes
In our previous studies, it was observed that the Ig-like domains of the frog and chicken LRC proteins reproducibly showed homology not only to known LRC members but also to the products of four mammalian genes that to our knowledge have never been considered in the phylogenetic analyses of LRC. These genes are VSTM1, TARM1, A1BG, and IGSF1. VSTM1 and TARM1 are the most recently identified members of the human LRC (Steevels et al. 2010; Radjabova et al. 2015). A1BG encodes alpha-1 B glycoprotein, a soluble component of mammalian blood plasma that is known for half a century (Schultze et al. 1963). The protein is composed of five Ig-like domains and has been shown to bind to CRISP-3, a small polypeptide that is present in exocrine secretions of neutrophilic granulocytes and that is believed to play a role in innate immunity (Udby et al. 2004). In the human genome, A1BG maps to 19q13.4 some 3.3 Mb away from GPVI (fig. 3A). Finally, IGSF1 codes for a large surface protein whose extracellular region consists of 12 Ig-like domains with a spacer between the fifth and the sixth domains (Frattini et al. 1998; Mazzarella et al. 1998). As a result of posttranslational cleavage, two TM forms of IgSF1 are produced encompassing domains 1–5 and 6–12, respectively (Robakis et al. 2008). The protein is mainly expressed in adult testes and nervous tissues. Its ligand is unknown. Loss-of-function IGSF1 mutations cause an X-linked syndrome of central hypothyroidism and testicular enlargement (Sun et al. 2012). Human IGSF1 is mapped to Xq25.
Fig. 3.
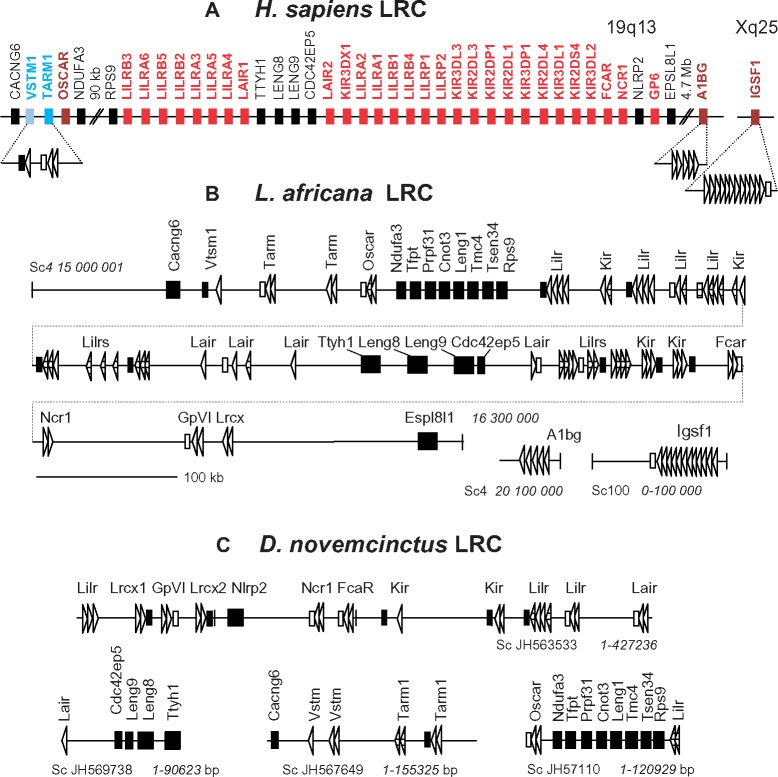
—Schematic representation of the LRC organization in the human (A), elephant (B), and armadillo (C) genomes. Italicized numbers show the position in a scaffold according to the current genome annotations at the Ensembl website. Exons for Ig-like domains are shown by triangles. The orientation of the exon-designating triangles corresponds to the transcription orientation. The attribution of the predicted genes is based on the results of the phylogenetic analysis (figs. 4 and 5). Non-LRC genes are shown by large black rectangles. Black bars show exons for TM regions without charged residues. Empty bars show exons for the NxxR-motif-containing TMs. Pattern-filled shapes show partially sequenced or aberrant exons.
To examine the relationships between these four genes/proteins and the previously described LRC members, individual Ig-like domains of VSTM1, TARM1, A1BG, and IGSF1 were compared against the known proteins by means of BlastP and PSI-BlastP analyses. All of the sequences were found to match known LRC proteins with scores much higher than those for other IgSF families (data not shown). The attribution of IGSF1 and A1BG domains to the LRC was supported by their 3D structures predicted using homology modeling (data not shown). In the case of TARM1 and IGSF1, an additional argument was that both of them contained the LRC-characteristic conserved TM region with the NxxR motif (fig. 1) that is known to facilitate an association with the FcRg subunit. Indeed, the ability of TARM1 to transmit signals through FcRγ has been recently demonstrated by Radjabova et al. (2015). As for IGSF1, this feature remains to be explored in functional studies.
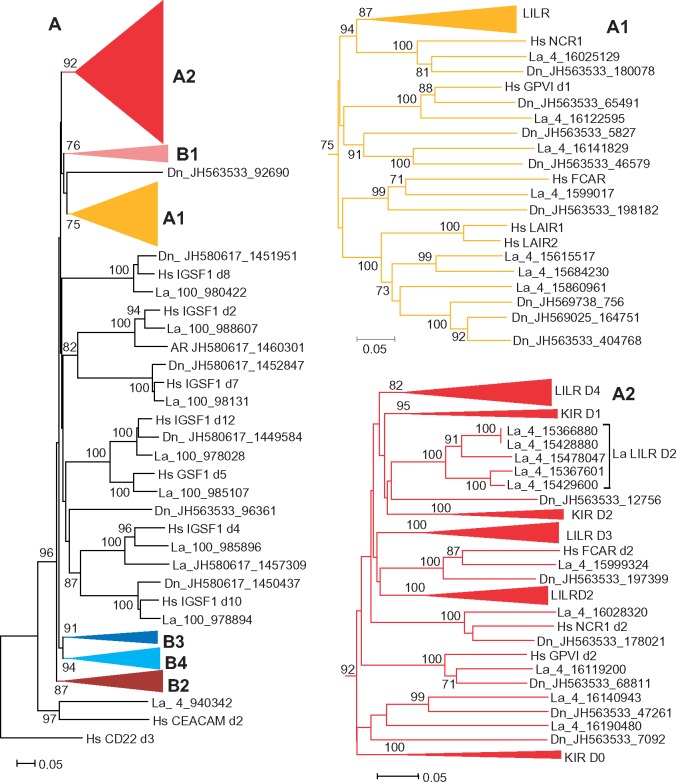
As for the next step, phylogenetic analysis of individual Ig-like domains was conducted using various algorithms available in the MEGA6 package. A representative NJ tree is shown in figure 2. In agreement with the results obtained previously (Hughes 2002; Nikolaidis et al. 2005), the domains of LILRs, KIRs, FcaR, NCR1, GPVI, and LAIR1/2 (herein referred to as the A group) fall into two main clusters supported by high bootstrap values. Here, we designate these clusters as A1 and A2. The A1 cluster includes the N-terminal domains (d1) of LILRs, NCRI, FcαR, GPVI, as well as the LAIR1 and LAIR2 domains. The A2 includes KIR domains, which are subdivided into three separate subtypes (KIR D0, D1, and D2), the remaining LILR domains with three subtypes designated here as LILR D2, D3, and D4 and, finally, the membrane-proximal domains (d2) of FcαR, GPVI, and NCR1.
Fig. 2.
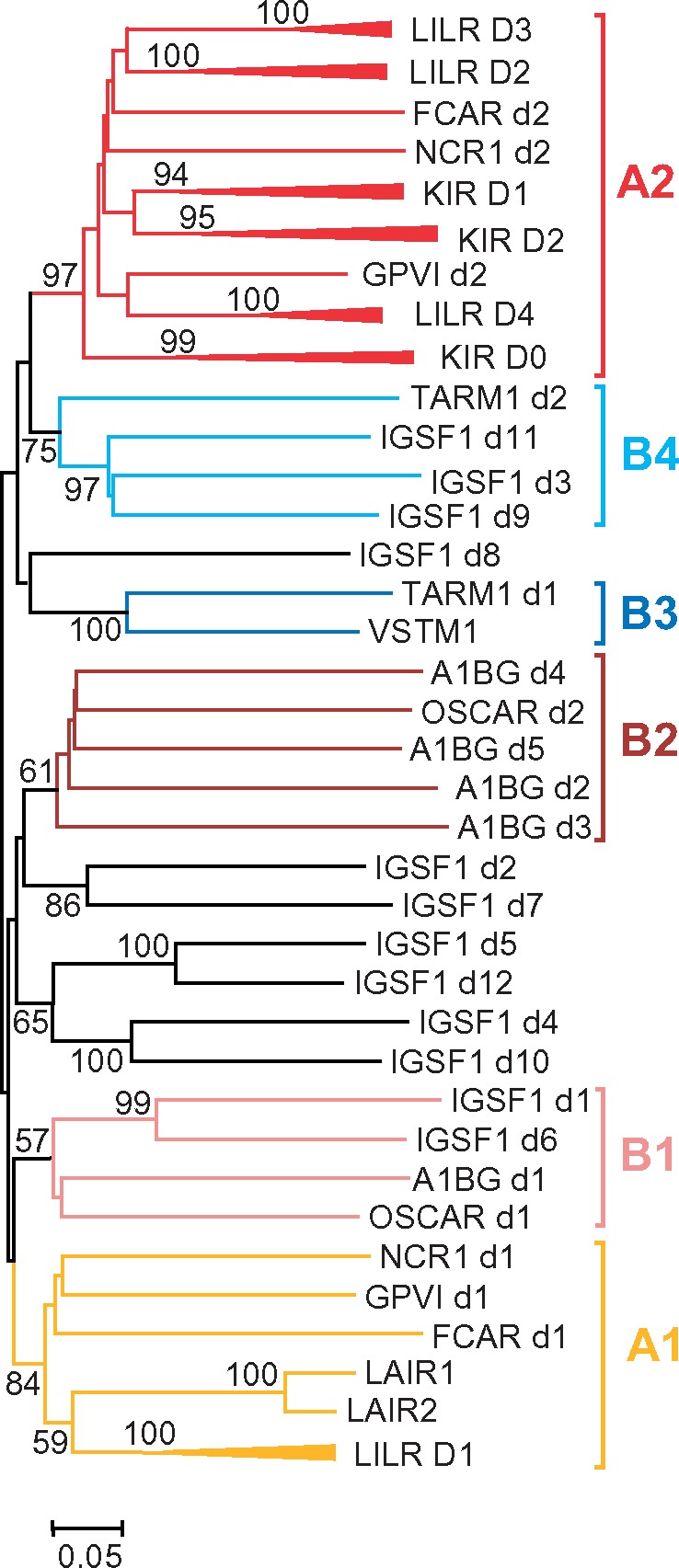
—Evolutionary relationships of human IGSF1 and A1BG with known LRC members. Unrooted NJ tree for nucleotide sequences encoding individual Ig-like domains is shown. Bootstrap values greater than 50% are shown at the nodes. Branch lengths depict evolutionary distance. The trees were generated using MEGA6 software with p-distances for nucleotide sequence sites and pair-wise deletion option. To reduce the size of the tree, the clusters for highly related Ig-like domains of various LILRs and KIRs were compressed and designated according to the domain subtype they represent (D1–D4 for LILRs and D0–D2 for KIRs).
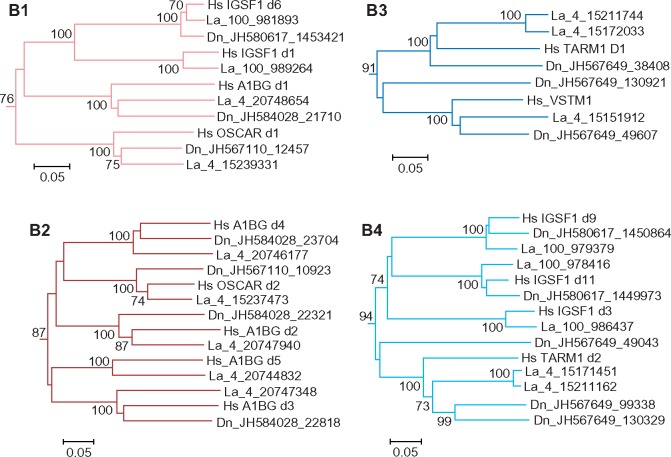
The relationships of OSCAR, IGSF1, A1BG, TARM1, and VSTM1 domains with each other and with those from the A group could only partially be resolved. High bootstrap values suggested close homology of the EC domain of VSTM1 to the N-terminal (D1) domain of TARM1 (cluster B3). The tree also showed high statistical support of close relationships of some IgSF1 domain to each other. Thus, the IGSF1 d1 domain is close to d6, d2 is close to d7, d3 to d9 and d11, d4 to d10, d5 to d12. This branching pattern suggests that IGSF1 might have originated by fusion of two highly similar ancestral genes. Noteworthy is that the D1 and D6 domains of IgSF1 fall into one clade with the N-terminal (d1) domains of A1BG and OSCAR (cluster B1). Closer relationship of A1BG and OSCAR was supported by clustering of the d2–d5 domains of A1BG with membrane-proximal (d2) domain of OSCAR (cluster B2). It might be inferred, therefore, that A1BG and OSCAR had a common origin and that A1BG evolved from an ancestral two-domain protein by loss of TM and quadruplication of its C-terminal domain. Finally, the membrane-proximal domain (d2) of TARM1 demonstrated closer relationships with IGSF1 d3, d9, and d11 (B4). Although bootstrap values for the B1, B2, and B4 clusters were not high, this branching pattern was reproduced at various tree generation settings. Altogether, these results support the attribution of IGSF1 and A1BG to the LRC and suggest their relatedness to OSCAR, TARM1, and VSTM1. To emphasize the relationships of these proteins and their difference from the group A members, we tentatively joined them into a distinct group B.
Similar LRC Content in Distant Species of Placental Mammals
The monophyletic origin of both the A1 and A2 clusters in this and the previous (Nikolaidis et al. 2005) analyses suggests that the EC regions of LILRs, KIRs, NCR1, FCAR, GPVI, and LAIR1/2 might have descended from a single ancestral protein composed of the A1 and A2 domain subtypes. We also hypothesize that the complex branching pattern formed by the individual domains of OSCAR, IGSF1, A1BG, TARM1, and VSTM1 may be explained by their origin from different parts of a common ancestral protein with the EC region composed of multiple highly diverged domains. To test these suggestions, we searched for the LRC genes in the genomes of African elephant (Loxodontra africana) and armadillo (Dasypus novemcinctus). These species represent the eutherian suborders Afrotheria and Xenarthra that are generally regarded as the most distant lineages of placental mammals (O’Leary et al. 2013).
The elephant genome analysis revealed 83 exons for LRC-like Ig-domains on the scaffolds 4 and 100. The schematic map of the exon arrangement is shown in figure 3B. A substantial fraction of exons (25) was suggested to be aberrant as they had stop codons, frameshifts, or disrupted splice-sites. However, sequencing errors cannot be excluded and, for that reason, we show the positions of all the exons. Additionally, 18 presumably functional exons for TM regions were revealed. Half of these encode conserved TMs with the NxxR motif. Yet, another half lack charged residues. Taking into account the positions of the TM exons, it may be assumed that the elephant LRC family includes at least 17 functional genes encoding cell surface receptors. There are seven putative genes in which TM exons were not detected. According to the phylogenetic analysis data (see below), one of them is the ortholog of human A1BG. As for the rest, it remains to be determined whether they are aberrant or encode secreted proteins or their TM exons for any reason have eluded identification. The latter possibility is quite probable because of multiple sequencing gaps in the examined version of the elephant genome.
The search in the armadillo genome revealed 115 exons for the LRC-like Ig-domains. Of these, 87 lacked frameshifts and stop codons. The exons were scattered on 21 scaffolds of varying length (4–2,649 kb) with 1–24 exons per scaffold. Figure 3C illustrates the exon organization on the four longest scaffolds. The predicted TM exons are also shown on this map. As in the elephant, TMs of the predicted armadillo LRC proteins fall into two types: with or without the NxxR motifs.
Phylogenetic analysis of the individual exon sequences demonstrated that the LRC gene content in armadillo, elephant, and human is both conserved and heterogeneous. The inclusion of the elephant and armadillo sequences into phylogenetic analysis resulted in a tree topology very similar to that for the human LRC with higher bootstrap values for the clusters B1–B4 (figs. 4 and 5). The tree shows that elephant and armadillo possess orthologs of human FCAR, GPVI, NCR1, TARM1, VSTM1, OSCAR, A1BG, and IGSF1. Both species also possess the genes that are attributed to the KIR, LILR, and LAIR families. Elephant seems to have fewer LILR genes compared with humans. In contrast, the LILR family was expanded in the armadillo lineage. There are 10, 11, 11, and 11 exons coding for, correspondingly, the D1-, D2-, D3-, and D4-subtypes of LILR domains in the armadillo genome. The branching pattern observed is consistent with the species-specific expansion of the LAIR, KIR, and LILR families. For instance, the elephant sequences for LILR D2 subtype form a cluster separated from its human and armadillo counterparts (fig. 4). Both the elephant and armadillo genomes contain a GPVI-adjacent gene for the protein composed of the A1 and A2 domain subtypes (designated Lrcx in fig. 3), whose ortholog is absent in primates and rodents.
Fig. 4.
—Evolutionary relationships between the human (Hs), elephant (La), and armadillo (Dn) LRC molecules. (Left) NJ tree for individual LRC domains rooted with human CEACAM and CD22. To reduce the size of the tree, the clusters corresponding to certain domain subtypes were compressed and shown separately: A1 and A2 clusters are shown on the right, whereas B1–B4 are presented in figure 5. The trees were generated using MEGA6 software with p-distances for nucleotide sequence sites and pair-wise deletion option. The numbers on the tree nodes show values for the bootstrap tests after 500 replicates, the clusters for highly related Ig-like domains of various LILRs and KIRs were compressed and designated according to the domain subtype that they represent (D1–D4 for LILRs and D0–D2 for KIRs).
Fig. 5.
—Evolutionary relationships of the human (Hs), elephant (La), and armadillo (Dn) LRC molecules (continued from fig. 4). Uncompressed clusters B1–B4.
Although human VSTM1 has a single EC domain, its armadillo counterpart was predicted to have a two-domain EC region with d2 highly related to the TARM1 d2 (figs. 3 and 5). This fact further supports a common origin of the TARM1 and VSTM1 genes. Similarly to the human LRC tree, the d2 sequences of armadillo TARM1 and VSTM1 cluster with the d3, d9, and d11 domains of IGSF1 (fig. 5). Clustering of the N-terminal domains of OSCAR, IGSF1, and A1BG with each other and with IGSF1 d6 was also reproduced. Finally, the d2 domains of OSCAR cluster with the d2–d5 domains of A1BG (fig. 5). These results further justify grouping IGSF1, A1BG, OSCAR, TARM1, and VSTM1 into a distinct group B.
Taken together, the phylogenetic data show that the main set of LRC genes described previously in primates and rodents has emerged before the radiation of placental mammals. The same is true for the organization of the LRC at the chromosome level. Like in the human genome, elephant LRC is arranged into three main clusters separated by conserved groups of non-LRC genes. One such neighboring group includes Ttyh1, Leng8, Leng9, and Cdc24ep5, whereas the other includes Ndufa3, Tfpt, Prpf31, Cnot3, Leng1, Tmc4, Mboat7, Tsen34, and Rps9 (fig. 3A and B). The LRC-flanking genes are Cacng6 and Espl8l1. In the current version of the armadillo genome, the LRC genes are scattered across multiple scaffolds whose linkage with each other is unknown. However, the localization of the Cacng6, Cdc42ep5, Ttyh1, Ndufa3, Rps9, and Nlrp2 suggests a gene organization very similar to that in humans and elephant. In the human and mouse genomes, A1BG lays apart from the LRC on the same chromosome, whereas IGSF1 gene maps to the X chromosome. This arrangement seems to be conserved in the elephant and armadillo genomes. For instance, elephant A1bg maps roughly 4.7 Mb from GpVI. The elephant and armadillo Igsf1 are linked to the genes whose orthologs are localized on the X chromosome in humans (data not shown).
Expansion of the LRC in Marsupials
High similarity of the LRC gene content in placental mammals showed that the clues for the evolutionary history of the family should be sought in other mammalian lineages, such as marsupials and monotremes. LRC homologs of two marsupial species, the short-tailed opossum (Monodelphis domestica), and the Tasmanian devil (Sarcophilus sharrisii), have been described (Belov et al. 2007; van der Kraan et al. 2013). These authors have reported, correspondingly, 157 and 43 open reading frames (ORFs) for the LRC-like domains in these species’ genomes. The relationships of the marsupial LRC genes with the eutherian LILR or KIR families have not been resolved and, for this reason, the family has been designated as MAIR (Belov et al. 2007). With the finding that IGSF1, A1BG, TARM1, VSTM1, and OSCAR represent a distinct structural group of the eutherian LRC, it was of interest to reexamine the opossum and devil genomes to see if the new genomic and transcriptomic data may provide more accurate insight into the evolution of the mammalian LRC.
Our analysis of the opossum genome led to the identification of 233 exons for the LRC-like domains (fig. 6A and supplementary table 1, Supplementary Material online). Of these, 176 are clustered on the fourth chromosome according to the assembly annotation. Two exons map to the second chromosome and 55 to the regions with unknown (Un) chromosomal localization. One hundred and ninety-three exons appear to lack errors. The quantitative difference (193 vs. 157) in the present and previous findings is likely attributable to the different approaches used for the genome analysis. Belov et al. (2007) used a search with Markov’s models against LILRs and KIRs, and they considered only the matches with scores above the cutoff of 10. In this study, we selected LRC candidates based on their individual BlastP examination rather than on the matching scores. The accuracy of our approach was validated by the subsequent phylogenetic analysis. The attribution of the identified exons to the LRC was also supported by the structure of adjacent TM exons. The search revealed 74 TM exons (fig. 6A). Of them, 45 encoded TMs with the NxxR motif. Selected TM sequences are shown in figure 1. TM exons were not found in some of the predicted genes. Experimental examination is necessary to determine if these genes encode secreted proteins or their TM exons for any reason have eluded identification.
Fig. 6.
—Schematic representation of the LRC organization in the short-tailed opossum (A) and the platypus (B) genomes. Italicized numbers show position in a scaffold according to the current genome annotations at the Ensembl website. The colors of the opossum exons correspond to the color scheme of the phylogenetic tree in figure 7. The orientation of the exon-depicting triangles corresponds to the transcription orientation. Non-LRC genes are shown by large black rectangles. Black bars show exons for TM regions without charged residues. Empty bars show exons for the NxxR-motif-containing TMs.
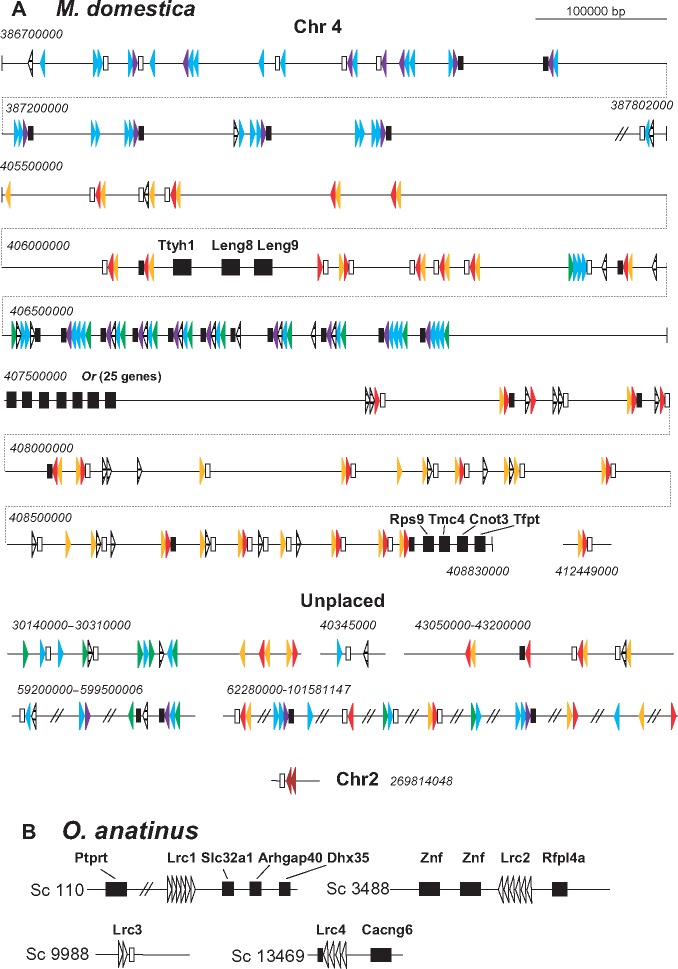
In opossum, the chromosomal region encompassing LRC genes is partially syntenic to the human 19q13.42 (figs. 3A and 6A). Two conserved gene groups, one consisting of Ttyh1, Leng8, Leng9 and the other of Rps9, Tsen34, Mboat7, Tmc4, Leng1, Cnot3, Prpf31, and Tfpt flank the 2.8-Mb region of the Chr4 containing 88 error-free LRC-related exons that may be organized into at least 39 genes judging by the presence of the TM exons. This large cluster is divided into two parts by a family of 26 genes encoding seven-span TM proteins homologous to olfactory receptors. A smaller cluster of 14 exons is present upstream of Ttyh1. It is flanked by a large group of genes encoding ZFN proteins. One more cluster of 39 exons is predicted further upstream at the distance of 17 Mb. This cluster is flanked upstream by a group of genes that map to the 19q13.3 region in the human genome (PPP5C, PTG1P, and GANG8) and downstream by the genes whose human orthologs map to 1p36 (C1ORF159, TTLL10, and TNFRSF18). A gene on the chromosome 2 is annotated as OSCAR-like by the sequencing consortium. Interestingly, it is located in the region syntenic to the human HLA.
The genome of the Tasmanian devil was found to contain 140 exons for the Ig-like LRC domains (supplementary table 2, Supplementary Material online). Of these, 126 lacked visible errors. In contrast to opossum, the current version of the Tasmanian devil’s genome assembly consists mainly of short scaffolds. Altogether we found 44 scaffolds ranging from 1.5 to 3.300 kb and containing 1–14 LRC-related exons (supplementary table 2, Supplementary Material online). It is quite likely that the actual size of the LRC in the Tasmanian devil’s genome is even larger. The scattering of the LRC exons on multiple scaffolds prevented accurate gene predictions. However, examination of the scaffold gene content demonstrated some signs of the conserved synteny. For instance, the scaffold GL849774 contains Ttyh1 and Leng8 together with seven predicted LRC genes. The scaffold GL850161, in addition to two LRC-like exons, contains a conserved group of genes that neighbor LRC in placental mammals: Rps9, Tsen34, Mboat7, Tmc4, Leng1, Cnot3, Prpf31, and Tfpt.
Phylogenetic analysis of individual exon sequences from the opossum and the Tasmanian devil LRC showed their close relationships and, on the other hand, species-specific expansion/contraction in certain LRC subsets. For a couple of opossum sequences, no closely related counterparts were identified in the devil genome. The same was also true for some devil sequences. Overall, the trees for marsupial LRC demonstrated the presence of five large clusters designated M1–M5 (fig. 7). Although bootstrap values for the M1 cluster were not high, the relatedness of the M1 sequences was supported by their position in the genome (fig. 6) and by subsequent phylogenetic analysis (fig. 8).
Fig. 7.
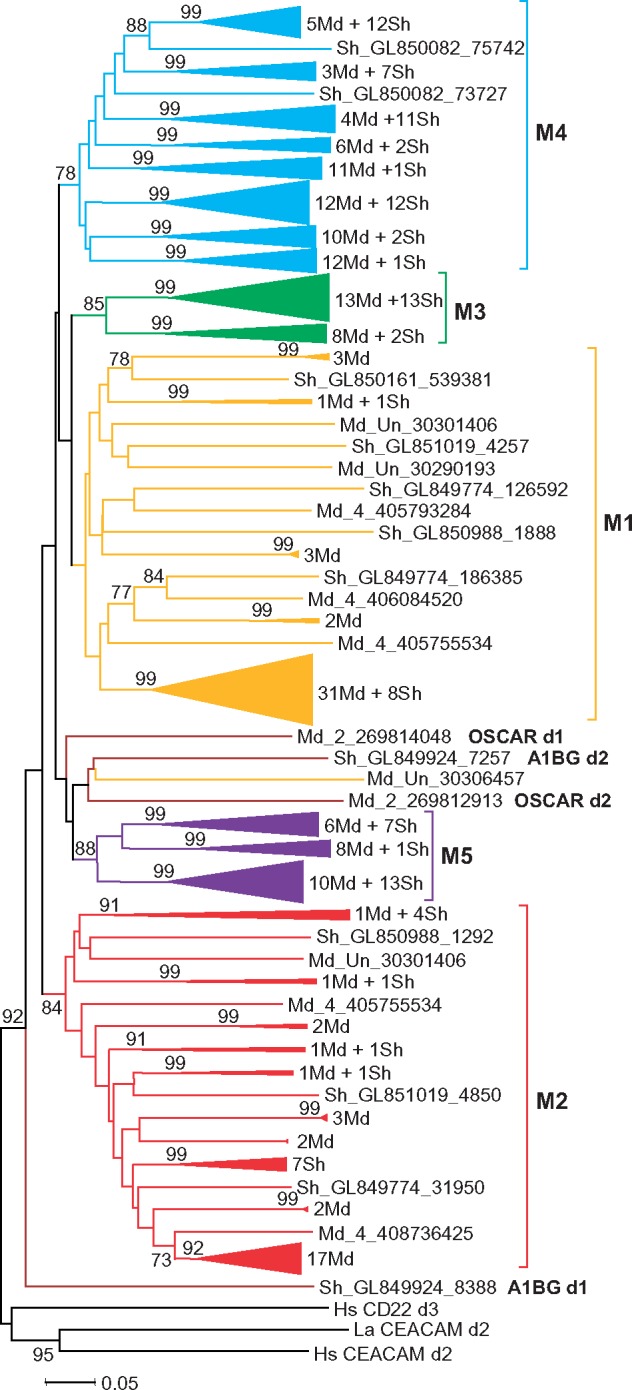
—Evolutionary relationships of the opossum (Md) and Tasmanian devil (Sh) LRC members. NJ tree of individual Ig-like domains rooted with human CEACAM and CD22 sequences. To reduce the size of the tree, the clusters with bootstrap values higher than 90% were compressed. A number of the opossum and devil sequences in each compressed cluster are shown at right.
Fig. 8.
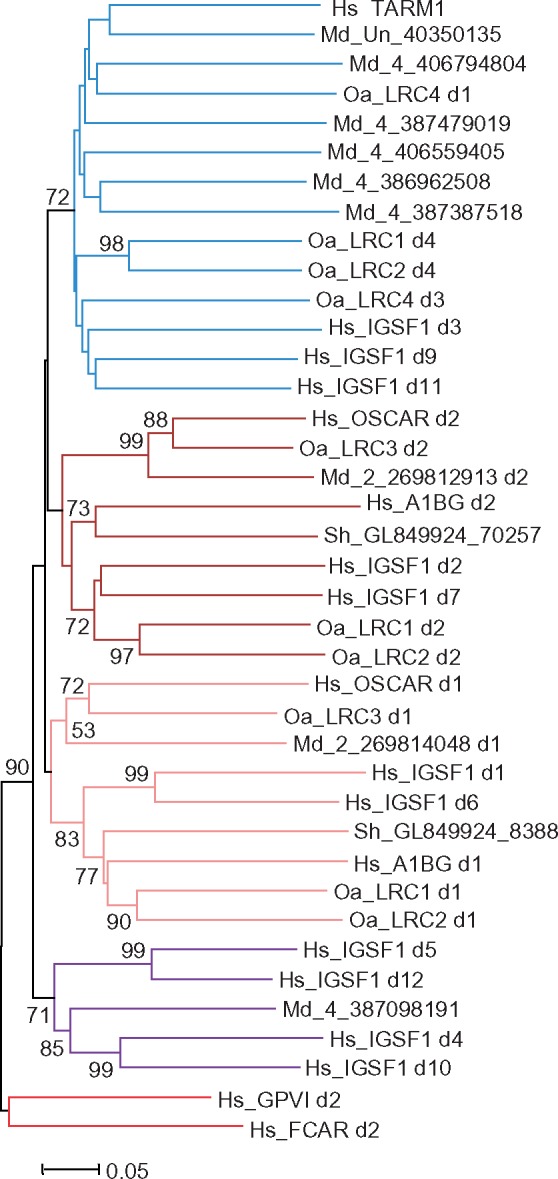
—Evolutionary relationships of the mammalian group B LRC members. NJ tree of a sample of individual Ig-like domains of the B group rooted with human group A sequences.
According to the results obtained, the vast majority of predicted opossum LRC-encoded proteins may be subdivided into two groups. The typical member of one of them has an EC region composed of two domains, of which the membrane-distal domain is attributed to the M1 and the membrane-proximal domain belongs to the M2 subtype. There are also genes in this group that encode proteins with a single-domain EC (either M1 or M2) or an EC region composed of two M1 domains. However, judging by the distribution (fig. 6) and attribution (not shown) of the pseudoexons, most of these might have derived from the genes encoding the M1M2 forms. The members of the second group have EC region composed of different combinations of the M3–M5 domain subtypes (fig. 6). Two more predicted marsupial LRC-like proteins cannot be attributed to any of these groups. One of these is encoded by the OSCAR-like gene on the opossum chromosome 2. Its counterpart is absent in the current version of the Tasmanian devil genome. Instead, the devil genome has two exons for a protein which is similar to the N-terminal part of A1BG (see below) and which is absent in the opossum genome.
Reduction of the LRC in Platypus
In their analysis of the platypus (Ornithorhynchus anatinus) genome, Wong et al. (2009) have revealed a large family of C-type lectin-like receptors. However, LRC homologs have not been found. The approach used in the present study made it possible to identify 18 LRC-like exons on the contigs 110 (1,790 kb), 3488 (53 kb), 9988 (23 kb), and 13469 (17 kb) (fig. 6B). The exon arrangement suggests the presence of four genes, designated here as Lrc1, Lrc2, Lrc3, and Lrc4 (fig. 6B). Lrc3 and Lrc4 encode cell surface receptors. LRC3 has TM containing the NxxR motif (fig. 1). TM of LRC4 lacks charged residues. Whether two other genes encode secreted proteins or their TM exons somehow eluded identification remains to be determined. The gene designated here as Lrc3 is described as OSCAR in the platypus genome annotation. Indeed, the D1 and D2 domains of the deduced protein share 46 and 67% identical residues with the D1 and D2 domains of human OSCAR, respectively.
Phylogenetic analysis of the platypus LRC single-exon sequences showed that LRC1 and LRC2 are most probably descendants of a common ancestor protein. Apart from the LRC1 d5, their individual domains are closely related (supplementary fig. 1, Supplementary Material online). LRC4 d1 and d3 appear to be closer related to d4 of LRC1 and LRC2. The relationships of the other sequences were poorly resolved. The synteny analysis revealed the presence Cacng6 gene linked with Lrc4 in the contig 13469. As mentioned above, Cacng6 is a neighbor of Vstm1 and Tarm1 in the genomes of placental mammals (fig. 3). The contig 110 contains Ptprt, Slc32a1, Arhgap40, and Dhx35 whose orthologs map to the 20q11-q13 region in the human genome. The contig 3488 contains three genes coding for proteins with zink finger (ZNF) and tripartite motifs in addition to Lrc2. One of these genes is most similar to the human Rfpl4a which is localized on 19q13.42. The contig 9988 appears to contain only Lrc3.
Phylogenetic Relationships of the Mammalian LRC Members
At the final step of this study, the relationships of the human, elephant, armadillo, opossum, devil, and platypus LRC sequences were inferred. The main goal of this analysis was to reveal if any domain subtypes are shared by eutherians, marsupials, and monotremes. Because of the very large size of marsupial LRC only a representative sample of the opossum and devil sequences was taken for analysis. For this purpose, the marsupial LRC tree was reduced by compression of clusters with a bootstrap support ≥95% (fig. 7). Forty-six sequences representing compressed clusters and separate branches were added to 18 platypus LRC sequences and to the above eutherian LRC data set, composed of the human, elephant, and armadillo sequences.
The trees generated at various software settings showed poor resolution in some parts and high/moderate support in others. Some sequences (IgSF1 d8, OaLRC1 d5, and OaLRC4 d2 and d4) reproducibly formed individual branches. The marsupial M3 subset, as well as the eutherian VSTM1 and TARM1 d1 sequences, was found to be lineage specific. However, the analysis also revealed the domain subtypes shared by the LRC molecules from all the three lineages. First of all, of importance was the clustering of the eutherian OSCAR sequences with those of the platypus LRC3 and with the opossum LRC gene from chromosome 2. In the case of OSCAR d2, this clustering was supported by high bootstrap values. Two exons from the Tasmanian devil contig GL849924 were found to encode the N-terminal domains of the marsupial counterpart of eutherian A1BG. However, no gene or gene fragments closely related to A1BG were found in the opossum genome. The d1 and d6 domains of IGSF1 demonstrated statistically supported relationships not only with d1 of A1BG but also with the d1 domains of platypus LRC1 and LRC2. The M4 subset of marsupial domains reproducibly fell into one clade together with the eutherian IGSF1 (d3, d9, d11) and TARM1 (d2), as well as with OaLRC1 (d4), OaLRC2 (d4) and OaLRC4 (d1, d3). Finally, the M5 subset of the marsupial LRC sequences showed closer relationships with d4, d5, d10, and d12 of eutherian IgSF1 (fig. 8). These results indicate that the group B of eutherian LRC genes might have emerged before radiation of the three mammalian lineages.
Of particular interest was the question whether platypus and marsupials possess receptors structurally related to the members of the “A” group of eutherian LRC, such as KIRs and LILRs. As indicated herein, this group most probably emerged by a series of duplications from a single ancestral protein with the A1- and A2-subtype domains. It remained, however, unclear as to which stage of the mammalian phylogeny these duplications might have occurred at. In the mammalian trees generated, the marsupial M2 subset showed reproducible clustering with the eutherian A2 sequences. Although the bootstrap values for this clustering were not high (55–65%), it was noteworthy that the marsupial M1 subset tended to associate with the eutherian A1 group sequences (not shown). As the M1 and M2 subsets characterize domains of a distinct group of marsupial LRC proteins (fig. 6), we suggested that this group is homologous to the eutherian group “A.”
To test this suggestion, we collected a data set of sequences encoding N-terminal domain pairs from 22 opossum and 5 devil predicted LRC genes (supplementary data set 1, Supplementary Material online). The choice was made on the basis of initial phylogenetic analysis to include sequences representing all the main branches of marsupial LRC (fig. 7). The data set was extended by addition of two-exon sequences for N-terminal parts of platypus LRC1, LRC2, LRC3, and LRC4 as well as of human and elephant FcαR, GPVI, NCR1, LILR1, TARM1, OSCAR, A1BG, and IgSF1. In the latter case, the sequence of a d6d7 pair of exons was added. Human CEACAM and CD22 sequences were used as an outgroup.
The representative NJ tree shows two main clusters (fig. 9). One of them includes the eutherian group “A” members (FcαR, GPVI, NCR1, and LILR1) and all the marsupial proteins with the M1M2 architecture. High statistical support suggests that the marsupial M1 domains are diverged counterparts of the eutherian A1, whereas the M2 is the counterpart of the A2. The results of this analysis also suggest that the devil gene from the GL851019 scaffold is an ortholog of eutherian GpVI in accordance with the notion by van der Kraan et al. (2013). Chromosomal localization of the marsupial genes belonging to the M1M2 subset is consistent with their closer relatedness to the eutherian A group. Like in the human and elephant LRC, these genes are linked to the Ttyh1, Leng9, and Rps9 genes (fig. 6).
Fig. 9.
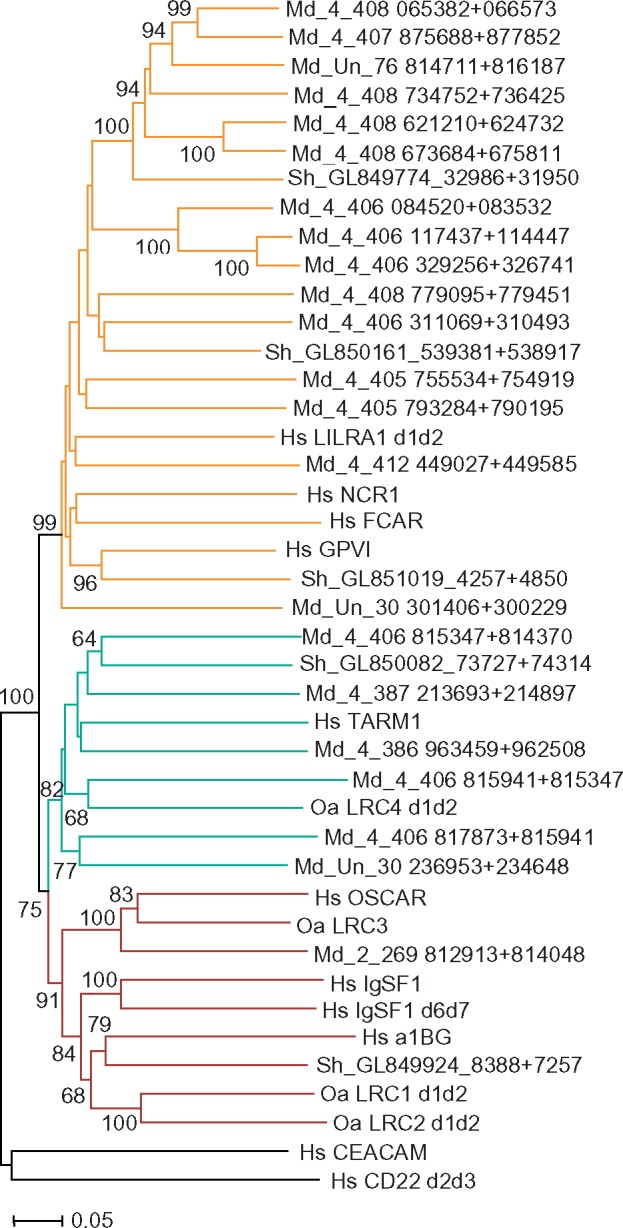
—Evolutionary relationships of the mammalian LRC members. NJ tree of N-terminal Ig-like domain pairs rooted with human CEACAM and CD22 sequences.
The remaining marsupial genes and all members of the platypus LRC fall into one cluster together with the group “B” eutherian sequences. This cluster is further subdivided into two main subclades. One of them includes eutherian TARM1, platypus LRC4 and all the marsupial proteins specified by the presence of M3–M5 domain subtypes. The second includes eutherian A1BG, IGSF1, and OSCAR together with the platypus LRC1, LRC2, LRC3, and the devil A1BG-like sequences. Eutherian OSCAR is closely related to the platypus LRC3/OSCAR and to the opossum OSCAR-like protein. Given that all these receptors have a TM region with the NxxR motif, they may be regarded as true orthologs. The reproducible clustering of the OSCAR, IGSF1, and A1BG in all the data sets analyzed strongly suggests their common origin.
Discussion
Despite a range of important functions served by mammalian LRC members, the evolutionary history of this gene family remains elusive. At the same time, genome annotations for various nonmammalian vertebrate species frequently contain gene names claiming attribution to the LRC. Although such relatedness may be true in some cases, the exact designations are usually misleading because they are generated in an automatic mode without taking into consideration the evolutionary plasticity of the family. The previous studies of the LRC evolution inferred a common origin of KIRs, LILRs, LAIRs FCAR, GPIV, and NCR1 (Hughes 2002; Nikolaidis et al. 2005). However, the relationships of these proteins with such eutherian LRC members as OSCAR and the recently described VSTM1 and TARM1 were unknown. Neither resolved were the relationships of human and rodent LRC members with their marsupial, chicken, and Xenopus counterparts. In this article, we present the results of the comprehensive bioinformatic analysis of the LRC gene structure and relationships in mammals as the first step in a study aimed at better understanding of the LRC evolution in jawed vertebrates.
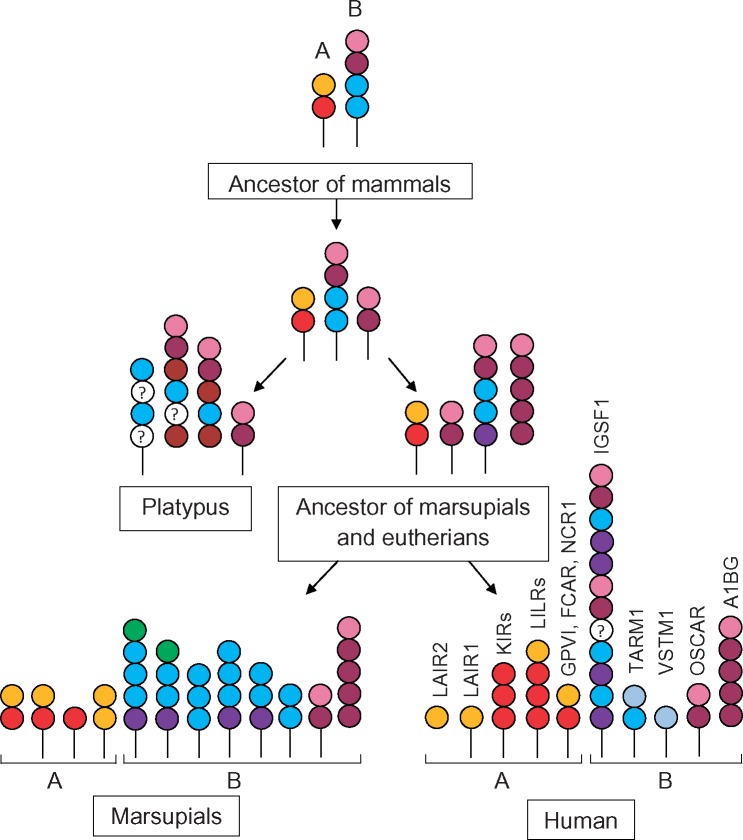
First of all, we showed that the previous descriptions of the family in eutherian mammals were incomplete as they did not consider such genes as Igsf1 and A1bg. These genes are not closely linked to other LRC genes in the human, armadillo, and elephant genomes. Nevertheless, their attribution to the LRC is unequivocally supported by sequence comparisons, 3D modeling and phylogenetic analysis. Actually, it was the inclusion of these genes into the study that made it possible to uncover the structural links among the eutherian, marsupial, and monotreme LRC. Second, we substantially revised the LRC gene content in marsupial species. Previous reports described 157 and 43 ORFs for LRC-like domains in the genomes of the short-tailed opossum (Belov et al. 2007) and the Tasmanian devil (van der Kraan et al. 2013), respectively. The approach used in this study enabled the identification of 199 and 123 error-free exons, respectively. The exhaustive characterization of the marsupial LRC as well as the identification of four platypus LRC genes for the first time provided a solid basis for the inferences of the LRC phylogeny in mammals.
Based on the phylogenetic analysis, the mammalian LRC members can be classified into two groups, each presumably derived from a single ancestor (figs. 9 and 10). The group A members are distinguished by the presence of the A1- and/or A2-like domain subtypes in their EC regions. In eutherians, both domain subtypes are present in LILRs, NCR1, GPVI, and FCAR. LAIRs encompass only A1 domains, and KIRs are composed of the A2 only. The marsupial A group includes receptors composed of the A1 and/or A2 counterparts (M1 and M2 domain subtypes, respectively, in fig. 7).
Fig. 10.
—A hypothetical scenario of the evolution LRC in mammals. A and B denote LRC classification into two groups. Domain subtypes distinguished by the phylogenetic analysis are shown by colored circles. The color scheme corresponds to the one used in the phylogenetic trees of individual domains (figs. 4, 7, and 8). Empty circles denote domain subtypes whose relationships were not resolved. In the marsupial lineage, only a fraction of predicted domain architectures is shown.
The B group is characterized by the presence of multiple domain subtypes. Some of them are strongly diverged and could not be classified reliably. However, of importance is the presence of at least three domain subtypes (B1, B2, and B4) shared in the domain architectures of B group members from different mammalian lineages. We suggest that the putative ancestor of the B group had a long EC region composed of several domain subtypes related to the B1-, B2-, and B4-like domains. The diverse architectures of the modern B group receptors may be explained by continuous exon shuffling and accumulation of multiple mutations in the domains less constrained by natural selection (fig. 10). One of the earliest events in the expansion of the B group was the emergence of OSCAR.
Both A and B groups evolved in a clear lineage-specific manner in monotremes, marsupials, and eutherians. The only member shared by all the three lineages is OSCAR. In the B group, platypus LRC1 and LRC2 are paralogs of marsupial and eutherian A1BG but domain architectures of these molecules are different in the three lineages. Platypus seems to have lost the A group genes (although assembly gaps in the current version of the genome cannot be excluded). Marsupials and eutherians share only GPVI in the A group. Given that OSCAR and GPVI are receptors for collagen (Clemetson et al. 1999; Barrow et al. 2011), it would be reasonable to suggest that mammalian LRC ancestors participated in collagen recognition. As to other functions known for eutherian LRC molecules, most of them might have emerged as a result of the eutherian-specific diversification of the family. For instance, we found no analogs of the lysine-containing KIR TM domain in species other than primates. This is in agreement with the proposed primate-specific emergence of KIRs as MHC class I-specific NK cell receptors (Parham et al. 2010).
Another highly probable neofunctionalization event in the LRC evolution is the emergence of Igsf1. The search in GenBank shows that numerous predicted genes from nonmammalian species are designated either as Igsf1 or Igsf1-like. However, our data indicate that Igsf1 is an eutherian-specific gene. It seems to have emerged by the fusion of two highly similar ancestor genes, which shared roots with the marsupial and eutherian A1bg as well as with the monotreme Lrc1 and Lrc2. Unlike other eutherian LRC members, IGSF1 is strongly conserved (75–80% identity at the amino acid level) and, most importantly, is predominantly expressed in the nervous and testicular tissues. According to the recent reports, Igsf1 mutations are implicated in central hypothyroidism (Sun et al. 2012; Joustra et al. 2016).
While attempting to uncover the exact function(s) of IGSF1, it would be important to keep in mind that its TM region belongs to the subtype known to transmit signals through the FcRγ subunit. Our data show that this particular TM domain is the most conserved structural element of the LRC family. Its characteristic arginine residue has been shown to be essential for functional association with FcRγ (Morton et al. 1995). However, there are other conserved features, such as the invariable Asp at the position −2 to the Arg, a consensus DYT motif at the N-terminus and rather unusual (for TMs) negatively charged residue(s) at the C-end (fig. 1). Interestingly, NxxR-containing TM subtype is also used by the Xenopus FCRL- and SLAM-like receptors (Guselnikov et al. 2008, 2011). Similarly to the marsupial LRC, the Xenopus FCRL and SLAM families are characterized by extraordinary size (at least 70–75 genes per family) and structural diversity. This striking parallelism argues in favor of advantageous role of the NxxR-TM in expansion and diversification of various receptor families.
The FcRγ subunit is known to associate with numerous cell surface molecules with highly diverged TM sequences. The exact reasons for NxxR-TM sequence conservation remain unclear. It is possible that this TM subtype has so far unknown functional properties affecting either surface distribution of the receptor, its mode of interaction with FcRγ or downstream signaling. Of relevance may be the fact that molecules containing this TM do not usually require FcRγ for surface expression and that, in certain circumstances, the FcRγ ITAM may fulfill the inhibitory function (Pasquier et al. 2005).
In summary, our study identifies IGSF1 and A1BG as members of eutherian LRC and redefines the LRC gene content in marsupial and monotreme genomes. The results of phylogenetic analysis show subdivision of the mammalian LRC into two groups and infer their origin from two ancestral genes. Rapid expansion/contraction accompanied by the extensive exon shuffling resulted in strikingly different LRC families in extant monotremes, marsupials, and eutherians. The only LRC member shared by the three lineages is OSCAR. The mode of the family evolution observed suggests that eutherian LRC functions may mostly have a lineage-specific origin. Of note is the strong conservation of a characteristic TM subtype known to interact with FcRγ. We suggest that the functional role of this particular TM sequence is underappreciated and it is worth further investigation.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgment
This work was supported by the Basic Scientific Research program (0310-2018-0012 to A.V.T.) and by the Russian Fund of Basic Research grant (17-04-01519-a to A.V.T.).
Literature Cited
- Abi-Rached L, Parham P.. 2005. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 201(8):1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow AD, et al. 2011. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J Clin Invest. 121(9):3505–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov K, et al. 2007. Characterization of the opossum immune genome provides insights into the evolution of the mammalian immune system. Genome Res. 17(7):982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges L, Hsu ML, Fanger N, Kubin M, Cosman D.. 1997. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 159(11):5192–5196. [PubMed] [Google Scholar]
- Bottino C, Vitale M, Pende D, Biassoni R, Moretta A.. 1995. Receptors for HLA class I molecules in human NK cells. Semin Immunol. 7(2):67–73. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S.. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268(1):78–94. [DOI] [PubMed] [Google Scholar]
- Clemetson JM, Polgar J, Magnenat E, Wells TN, Clemetson KJ.. 1999. The platelet collagen receptor glycoprotein VI is a member of the immunoglobulin superfamily closely related to FcalphaR and the natural killer receptors. J Biol Chem. 274(41):29019–29024. [DOI] [PubMed] [Google Scholar]
- Dennis G, Kubagawa H, Cooper MD.. 2000. Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc Natl Acad Sci U S A. 97(24):13245–13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayngerts SA, Najakshin AM, Taranin AV.. 2007. Species-specific evolution of the FcR family in endothermic vertebrates. Immunogenetics 59(6):493–506. [DOI] [PubMed] [Google Scholar]
- Frattini A, Faranda S, Redolfi E, Allavena P, Vezzoni P.. 1998. Identification and genomic organization of a gene coding for a new member of the cell adhesion molecule family mapping to Xq25. Gene 214(1-2):1–6. [DOI] [PubMed] [Google Scholar]
- Guselnikov SV, Laktionov PP, Najakshin AM, Baranov KO, Taranin AV.. 2011. Expansion and diversification of the signaling capabilities of the CD2/SLAM family in Xenopodinae amphibians. Immunogenetics 63(10):679–689. [DOI] [PubMed] [Google Scholar]
- Guselnikov SV, et al. 2008. The Xenopus FcR family demonstrates continually high diversification of paired receptors in vertebrate evolution. BMC Evol Biol. 8:148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guselnikov SV, et al. 2010. The amphibians Xenopus laevis and Silurana tropicalis possess a family of activating KIR-related Immunoglobulin-like receptors. Dev Comp Immunol. 34(3):308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P.. 2009. Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol. 182(6):3618–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht ML, et al. 2009. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res. 8(2):712–720. [DOI] [PubMed] [Google Scholar]
- Hoelsbrekken SE, Fossum S, Dissen E.. 2005. Molecular cloning of LILRC1 and LILRC2 in the mouse and the rat, two novel immunoglobulin-like receptors encoded by the leukocyte receptor gene complex. Immunogenetics 57(7):479–486. [DOI] [PubMed] [Google Scholar]
- Hogan L, et al. 2012. Characterisation of bovine leukocyte Ig-like receptors. PLoS One 7(4):e34291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. 2002. Evolution of the human killer cell inhibitory receptor family. Mol Phylogenet Evol. 25(2):330–340. [DOI] [PubMed] [Google Scholar]
- Joustra SD, et al. 2016. IGSF1 deficiency: lessons from an extensive case series and recommendations for clinical management. J Clin Endocrinol Metab. 101(4):1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlhofer FM, Ribaudo RK, Yokoyama WM.. 1992. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature 358(6381):66–70. [DOI] [PubMed] [Google Scholar]
- Kärre K, Ljunggren HG, Piontek G, Kiessling R.. 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319(6055):675–678. [DOI] [PubMed] [Google Scholar]
- Kelley J, Walter L, Trowsdale J.. 2005. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 1(2):129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Takami M, Rho J, Josien R, Choi Y.. 2002. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J Exp Med. 195(2):201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL, Phillips JH.. 1995. NK cell recognition of major histocompatibility complex class I molecules. Semin Immunol. 7(2):75–82. [DOI] [PubMed] [Google Scholar]
- Laun K, et al. 2006. The leukocyte receptor complex in chicken is characterized by massive expansion and diversification of immunoglobulin-like loci. PLoS Genet. 2(5):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbink RJ, et al. 2008. The soluble leukocyte-associated Ig-like receptor (LAIR)-2 antagonizes the collagen/LAIR-1 inhibitory immune interaction. J Immunol. 180(3):1662–1669. [DOI] [PubMed] [Google Scholar]
- Liang S, Baibakov B, Horuzsko A.. 2002. HLA-G inhibits the functions of murine dendritic cells via the PIR-B immune inhibitory receptor. Eur J Immunol. 32(9):2418–2426. [DOI] [PubMed] [Google Scholar]
- Maliszewski CR, March CJ, Schoenborn MA, Gimpel S, Shen L.. 1990. Expression cloning of a human Fc receptor for IgA. J Exp Med. 172(6):1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O, et al. 2001. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409(6823):1055–1060. [DOI] [PubMed] [Google Scholar]
- Mazzarella R, Pengue G, Jones J, Jones C, Schlessinger D.. 1998. Cloning and expression of an immunoglobulin superfamily gene (IGSF1) in Xq25. Genomics 48(2):157–162. [DOI] [PubMed] [Google Scholar]
- Meyaard L, et al. 1997. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity 7(2):283–290. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447(7141):167–177. [DOI] [PubMed] [Google Scholar]
- Montgomery BC, Cortes HD, Mewes-Ares J, Verheijen K, Stafford JL.. 2011. Teleost IgSF immunoregulatory receptors. Dev Comp Immunol. 35(12):1223–1237. [DOI] [PubMed] [Google Scholar]
- Moroi M, et al. 1996. Analysis of platelet adhesion to a collagen-coated surface under flow conditions: the involvement of glycoprotein VI in the platelet adhesion. Blood 88(6):2081–2092. [PubMed] [Google Scholar]
- Morton HC1, et al. 1995. Functional association between the human myeloid immunoglobulin A Fc receptor (CD89) and FcRγ chain. Molecular basis for CD89/FcR gamma chain association . J Biol Chem. 270:29781–29787. [DOI] [PubMed] [Google Scholar]
- Murchison EP, et al. 2012. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148(4):780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis N, Klein J, Nei M.. 2005. Origin and evolution of the Ig-like domains present in mammalian leukocyte receptors: insights from chicken, frog, and fish homologues. Immunogenetics 57(1-2):151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MA, et al. 2013. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339:662–667. [DOI] [PubMed] [Google Scholar]
- Parham P, et al. 2010. Primate-specific regulation of natural killer cells. J Med Primatol. 39(4):194–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier B, et al. 2005. Identification of FcαRI as an inhibitory receptor that controls inflammation: dual role of FcRγ ITAM. Immunity 22(1):31–42. [DOI] [PubMed] [Google Scholar]
- Radjabova V, et al. 2015. TARM1 is a novel leukocyte receptor complex-encoded ITAM receptor that costimulates proinflammatory cytokine secretion by macrophages and neutrophils. J Immunol. 195(7):3149–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravetch JV, Lanier LL.. 2000. Immune inhibitory receptors. Science 290(5489):84–89. [DOI] [PubMed] [Google Scholar]
- Robakis T, Bak B, Lin SH, Bernard DJ, Scheiffele P.. 2008. An internal signal sequence directs intramembrane proteolysis of a cellular immunoglobulin domain protein. J Biol Chem. 283(52):36369–36376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze HE, Heide K, Haupt H.. 1963. Isolation of an easily precipitable alpha1-glycoprotein of human serum. Nature 200:1103.. [DOI] [PubMed] [Google Scholar]
- Shiroishi M, et al. 2006. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc Natl Acad Sci U S A. 103(44):16412–16417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S, et al. 1997. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med. 186(7):1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steevels TA, Lebbink RJ, Westerlaken GH, Coffer PJ, Meyaard L.. 2010. Signal inhibitory receptor on leukocytes-1 is a novel functional inhibitory immune receptor expressed on human phagocytes. J Immunol. 184(9):4741–4748. [DOI] [PubMed] [Google Scholar]
- Sun Y, et al. 2012. Loss-of-function mutations in IGSF1 cause an X-linked syndrome of central hypothyroidism and testicular enlargement. Nat Genet. 44:1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T. 2005. A novel recognition system for MHC class I molecules constituted by PIR. Adv Immunol. 88:161–192. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J, Jones DC, Barrow AD, Traherne JA.. 2015. Surveillance of cell and tissue perturbation by receptors in the LRC. Immunol Rev. 267(1):117–136. [DOI] [PubMed] [Google Scholar]
- Udby L, et al. 2004. Cysteine-rich secretory protein 3 is a ligand of alpha1B-glycoprotein in human plasma. Biochemistry 43(40):12877–12886. [DOI] [PubMed] [Google Scholar]
- van der Kraan LE, Wong ES, Lo N, Ujvari B, Belov K.. 2013. Identification of natural killer cell receptor genes in the genome of the marsupial Tasmanian devil (Sarcophilus harrisii). Immunogenetics 65(1):25–35. [DOI] [PubMed] [Google Scholar]
- Viertlboeck BC, Schmitt R, et al. 2009. A novel activating chicken IgY FcR is related to leukocyte receptor complex (LRC) genes but is located on a chromosomal region distinct from the LRC and FcR gene clusters. J Immunol. 182(3):1533–1540. [DOI] [PubMed] [Google Scholar]
- Viertlboeck BC, Schweinsberg S, Schmitt R, Herberg FW, Göbel TW.. 2009. The chicken leukocyte receptor complex encodes a family of different affinity FcY receptors. J Immunol. 182(11):6985–6992. [DOI] [PubMed] [Google Scholar]
- Viertlboeck BC, et al. 2005. The chicken leukocyte receptor complex: a highly diverse multigene family encoding at least six structurally distinct receptor types. J Immunol. 175(1):385–393. [DOI] [PubMed] [Google Scholar]
- Viertlboeck BC, et al. 2007. The chicken leukocyte receptor complex encodes a primordial, activating, high-affinity IgY Fc receptor. Proc Natl Acad Sci U S A. 104(28):11718–11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long EO.. 1997. A new human gene complex encoding the killer cell inhibitory receptors and related monocyte/macrophage receptors. Curr Biol. 7(8):615–618. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453(7192):175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H, Colonna M, Ziegler A, Volz A.. 1999. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome 10(2):154–160. [DOI] [PubMed] [Google Scholar]
- Willcox BE, Thomas LM, Bjorkman PJ.. 2003. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 4(9):913–919. [DOI] [PubMed] [Google Scholar]
- Windau K, Viertlboeck BC, Göbel TW.. 2013. The Turkey Ig-like receptor family: identification, expression and function. PLoS One 8(3):e59577.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, et al. 2009. Identification of natural killer cell receptor clusters in the platypus genome reveals an expansion of C-type lectin genes. Immunogenetics 61(8):565–579. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Litman GW.. 2011. The phylogenetic origins of natural killer receptors and recognition: relationships, possibilities, and realities. Immunogenetics 63(3):123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.