Abstract
Understanding how plants respond to nitrogen in their environment is crucial for determining how they use it and how the nitrogen use affects other processes related to plant growth and development. Under nitrogen limitation the activity and affinity of uptake systems is increased in roots, and lateral root formation is regulated in order to adapt to low nitrogen levels and scavenge from the soil. Plants in the legume family can form associations with rhizobial nitrogen-fixing bacteria, and this association is tightly regulated by nitrogen levels. The effect of nitrogen on nodulation has been extensively investigated, but the effects of nodulation on plant nitrogen responses remain largely unclear. In this study, we integrated molecular and phenotypic data in the legume Medicago truncatula and determined that genes controlling nitrogen influx are differently expressed depending on whether plants are mock or rhizobia inoculated. We found that a functional autoregulation of nodulation pathway is required for roots to perceive, take up, and mobilize nitrogen as well as for normal root development. Our results together revealed that autoregulation of nodulation, root development, and the location of nitrogen are processes balanced by the whole plant system as part of a resource-partitioning mechanism.
Key words: Nitrogen transport, Medicago, root system architecture, transcriptomics, autoregulation of nodulation, rhizobia
Despite understanding the impact of nitrogen on nodulation, little is known about the effects of nodulation on nitrogen responses. By integrating molecular and phenotypic data, we show that the autoregulation of nodulation pathway is required for Medicago truncatula roots to perceive, take up, and mobilize nitrogen as part of resource-partitioning mechanism for functional root development.
Introduction
Legumes benefit from symbiotic association with soil nitrogen (N)-fixing rhizobia for N uptake. Nodulation relies on two closely coordinated processes: the infection process, including the colonization of the bacteria inside the host plant, and the organogenic process, in which the nodule tissue is formed to accommodate the bacteria (Madsen et al., 2010). Signal exchange for bacterial entry takes place between rhizobia that release nod factors (NF) and host plant roots that release flavonoids. NF perception by receptor-like kinases such as NFP in Medicago truncatula activates nodulation specific genes including DMI1, DMI2, and DMI3 and NSP2 (Amor et al., 2003) and downstream calcium signaling (Peiter et al., 2007). Nodulation signal transduction involved in initiating calcium spiking ultimately results in the activation of calcium/calmodulin-dependent protein kinase (CCaMK) (Tirichine et al., 2006) and subsequent physiological and morphological changes (Kosuta et al., 2008, Capoen et al., 2009) including root hair curling, infection thread formation, and root nodule primordial development (Oldroyd and Downie, 2008). These developmental processes are coordinated across different root cell types by a number of transcription factors including NIN, NSP1, NSP2, ERN1, and ERN2 (Kaló et al., 2005, Cerri et al., 2012; Vernié et al., 2015). This developmental molecular coordination also involves induction of early nodulin-like proteins including ENOD11, ENOD12, and RIP1 in the root epidermis, and induction of ENOD20 and ENOD40 in the root cortex and pericycle (Catoira et al., 2000). In the M. truncatula-Sinorhizobium meliloti symbiosis, host legume root cells undergo repeated rounds of genome duplication and increase in volume 80-fold, internalizing the rhizobia in a specialized compartment called “symbiosome”. Within this structure rhizobia differentiate to form N-fixing-specialized polyploid bacteroids unable to replicate (Maroti and Kondorosi, 2014). These bacteroids exchange biologically fixed N for an allocation of photosynthate from the host legume (Jones et al., 2007) with exchange at the symbiosome membrane through specific channels (Weaver et al., 1994).
Nitrogen responses in plants have been well studied, most commonly in the model plant Arabidopsis thaliana (reviewed in, e.g., O'Brien et al., 2016). Nitrogen responses in roots involve coordinated regulation of metabolic and cellular pathways that modulate N uptake and root system architecture (Walker et al., 2017). The nitrate transporter/sensor NRT1.1 plays a key role in modulating root responses in response to varying external N. These include repression of lateral root branching in response to deplete nitrate by diverting accumulating auxin from lateral root primordia (Bouguyon et al., 2015). In Medicago, nitrate transporters similarly play a key role in signaling as well as the distribution of internal N (nodulation-sourced) and external N (taken up from the environment) for growth and development (Pellizzaro et al., 2017). In addition, major intrinsic proteins, and more specifically those expressed specifically in nodules, nodule intrinsic proteins, appear to play an important role in the movement of symbiosome-sourced N in the form of ammonia (Benedito et al., 2008). This is one example of the many regulatory controls that exist to balance photosynthate payout with N payback in order to optimize whole plant growth, and the ability of the root system to develop lateral roots for other nutrient-harvesting purposes. Peptide and amino acid transporters also play important roles as part of regulation of cellular N metabolism during different stages of plant development (Miranda et al., 2003).
In legume species, high concentrations of N inhibit nodulation (van Noorden et al., 2016) and there are differing inhibitory effects depending on whether the source is nitrate or ammonium (Barbulova et al., 2007). These inhibitory effects can occur at a very early stage of NF signaling (Heidstra et al., 1997), and plant N status affects symbiotic competence (Omrane et al., 2009). Nitrogen status can also act later to modulate nodule functioning and activity. Repressing nodulation when the N status of the plant is sufficient involves a series of mobile signals. In soybean, Nitrogen-Induced CLE1 (NIC1), a CLE (CLV3/EMBRYO SURROUNDING REGION) peptide induced by nitrate, is involved in the local inhibition of nodulation (Reid et al., 2011). Grafting experiments show that NIC1 is perceived by a root-localized CLAVATA1-like Leucine-Rich Repeat Receptor-Like Kinase (LRR-RLK) called Nodulation Autoregulation Receptor Kinase (GmNARK) (Searle et al., 2003). In Lotus japonicus this gene is encoded by LjHAR1 (Hypernodulation Aberrant Root Formation 1) (Wopereis et al., 2000) and in M. truncatula by SUperNumary Nodules, MtSUNN (Penmetsa et al., 2003, Schnabel et al., 2005). These mutants display disruption in the autoregulation of nodulation (AON) pathway, which consists of at least two systemic regulatory circuits to control nodule numbers and activity (Kassaw et al., 2015). AON uses a feedback-suppression mechanism from a root-derived signal that is thought to move via the phloem (Oka-Kira and Kawaguchi, 2006). Along with SUNN, RDN1 (Root-Determined Nodulation) controls AON responses in M. truncatula, and CLE peptides are thought to be involved as both the local and systemic nodulation status signal (Mortier et al., 2012). In L. japonicus it was shown that Nitrate Unresponsive Symbiosis 1 (NRSYM1), a NIN-like gene, regulates nodule numbers and nodule development in response to nitrate levels (Nishida et al., 2018).
Hypernodulating mutants developing excess nodules escape autoregulation even in the presence of high levels of nitrate, indicating that nitrate exerts at least part of its effect via the autoregulatory pathway. The set of genes controlling AON also seem to affect nitrate perception and signaling; for example, nodulation in Lotus LjHAR1 mutants is not downregulated by high nitrate because the mutant is unable to recognize LjCLE-RS2 peptides (Okamoto et al., 2009). There are a number of genetic links between the AON and other N-responsive root development pathways, including lateral root development (Huault et al., 2014). In a similar regulatory mechanism another mobile small peptide, MtCEP1 (C-terminally Encoded Peptide 1) is recognized by an LRR-RLK, MtCRA2 (Compact Root Architecture 2) to antagonistically regulate nodulation and lateral root architecture (Mohd-Radzman et al., 2013).
Accumulating evidence suggests an N-status-dependent molecular dialog between long-distance signaling of AON pathway and nodulation. The effect of N on nodulation has been investigated in a variety of studies (reviewed in Nishida et al., 2018) but the effect of nodulation on N responses is much less well known. Following a systems biology-integrated approach, here we have used a combination of phenotypic data and transcriptomic analyses of wild type and the hypernodulating sunn-1 mutant to examine the interaction of rhizobial responses and N resources.
Results and Discussion
Analysis of Rhizobia and Nitrogen Responses in M. truncatula
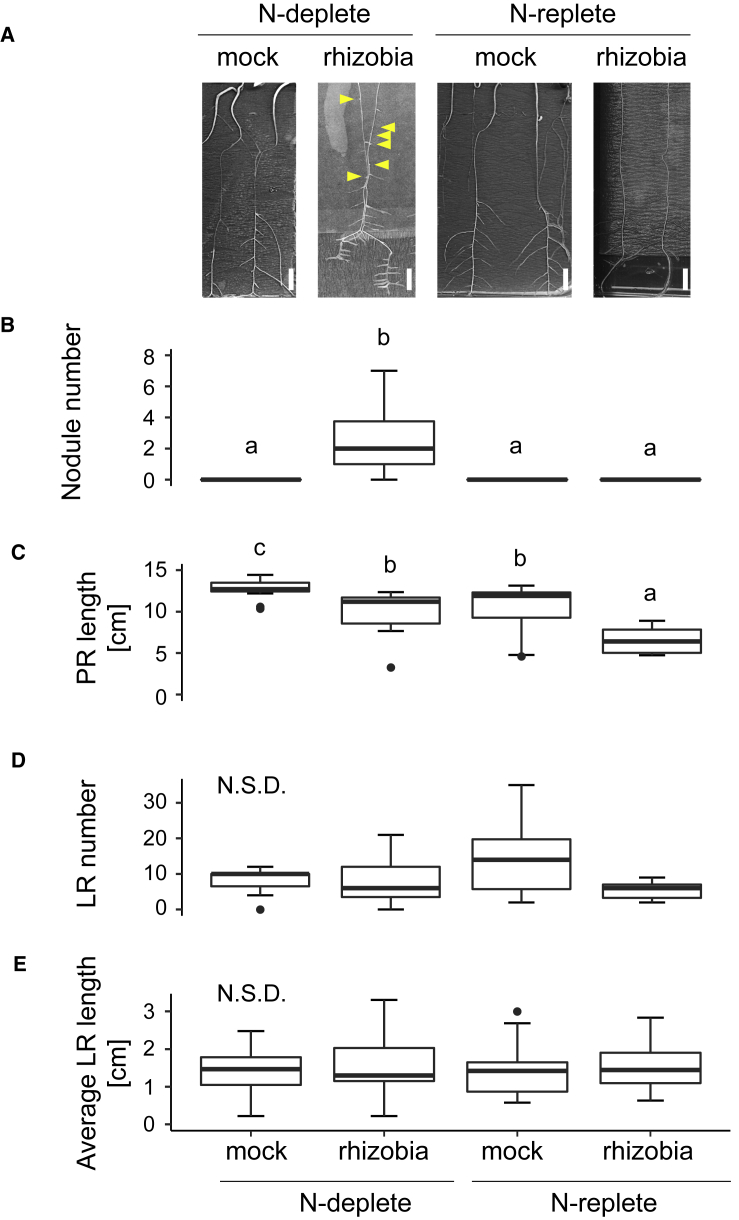
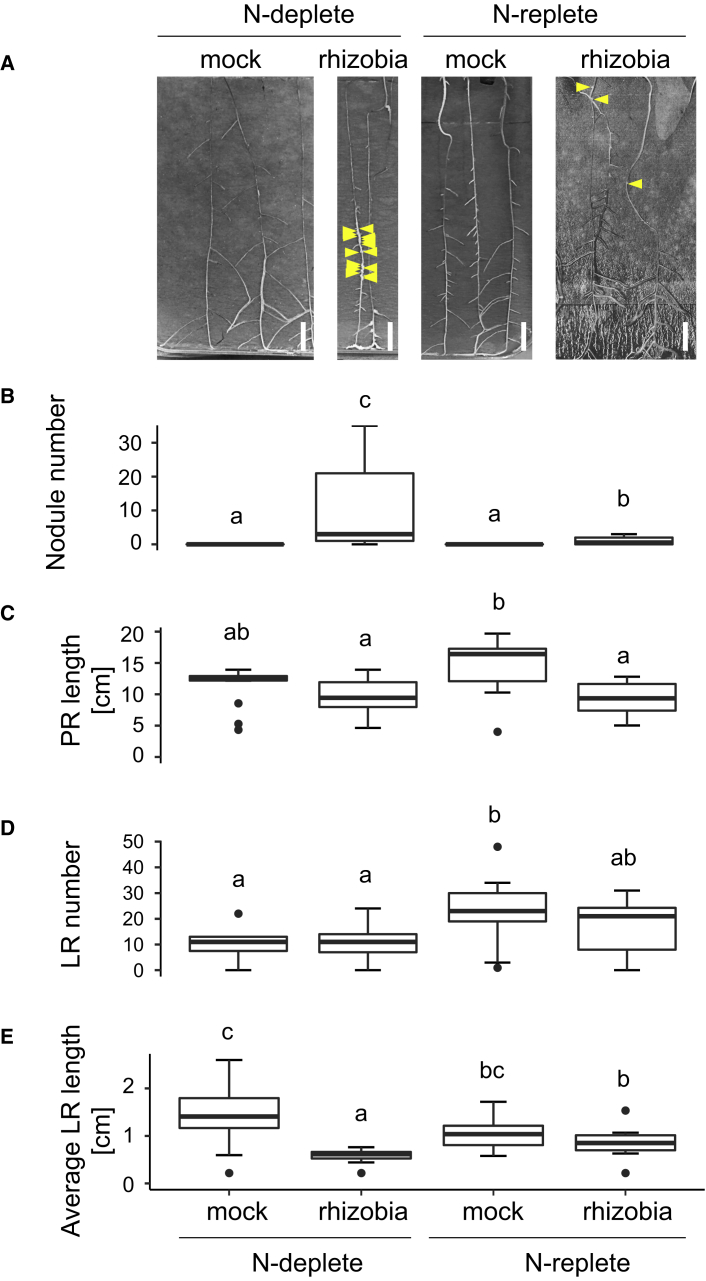
We measured root system architecture (RSA) in M. truncatula plants grown on deplete-N (0.1 mM NH4NO3) conditions treated with either S. meliloti (“rhizobia”) or mock for 14 days and then either treated with deplete N or replete N (5 mM NH4NO3) for 16 more days to study their individual and combinatorial effects on RSA (see Methods). Deplete and replete levels were chosen based on tests that showed inhibition of nodulation at levels higher than 2 mM NH4NO3 (Supplemental Table 1A), and in accordance with this we only saw nodulation in the N-deplete condition (Figure 1A and 1B; Supplemental Table 1B).
Figure 1.
A17 Root System Architecture Is Altered Differently upon N Treatment if Plants Are Inoculated with Rhizobia.
(A) Images of A17 seedlings that were mock or rhizobia inoculated and then grown in N-deplete (0.1 mM NH4NO3) or N-replete (5 mM NH4NO3) conditions. Scale bars, 1 cm.
(B–E) Root size and features were measured. (B) Number of nodules; (C) primary root (PR) length; (D) number of lateral roots (LRs); (E) average LR length. Data are presented as mean ± SD. Different letters denote statistically different groups for pairwise comparisons using Wilcoxon’s rank-sum test; n ≥ 11. P < 0.05; N.S.D., no significant difference.
See also Supplemental Table 1.
The primary root (PR) was significantly shorter when plants were inoculated with rhizobia, independently of the N treatment (Figure 1C), suggesting that investment in nodules is at the expense of RSA. Additionally, the shorter PR under rhizobia-inoculated conditions is also evident on replete N (Figure 1C), despite the fact that nodulation does not take place. This suggests a more complex regulatory effect at play, driven by external N availability. When comparing mock-treated plants it was observed that PR is shorter in replete N (Figure 1C), suggesting that PR growth might be driven by an N-scavenging response. Such an effect has previously been described in legumes (Mohd-Radzman et al., 2013) and maize (Gao et al., 2014). Moreover, rhizobia inoculation and N treatments seem to be additive in their effects on PR length, as PR in the rhizobia- and replete-N condition was shorter than in any other condition studied (Figure 1C). Between these conditions there was no significant difference in lateral root (LR) number or length (Figure 1 D and 1E; Supplemental Table 1B and 1C).
Rhizobia-Inoculated Plants Show a Different Nitrogen Response Compared with Mock-Inoculated Plants
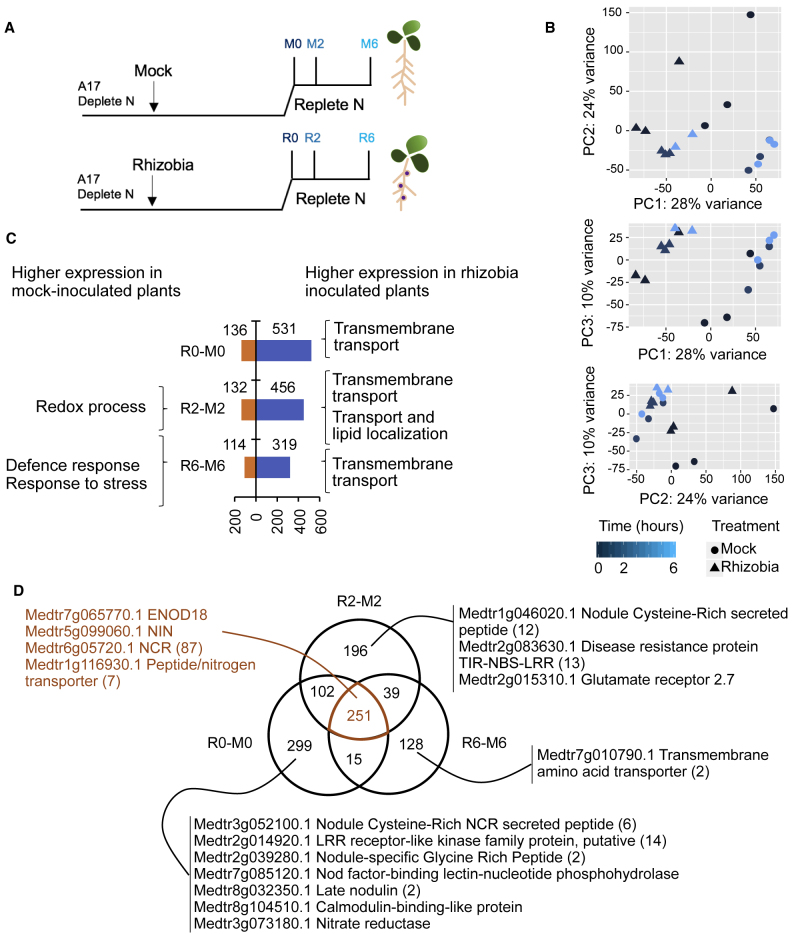
To investigate the early stages of the combinatorial effects of rhizobia and N responses on RSA, we performed the same experiments as for phenotyping by growing plants under N-deplete conditions and then transferring to N-replete conditions. Thus, we harvested roots at 0 h (at the moment of N addition), 2 h, and 6 h after the replete-N (5 mM NH4NO3) treatment (Figure 2A), carried out transcriptomic expression using microarrays (see Methods), and determined differentially expressed genes (DEGs) (Supplemental Table 2). Principal component analysis (PCA) was used to ask whether there were major sources of variation over the samples (Figure 2B). Earlier (0 h) time points in both rhizobia- and mock-inoculated samples had a greater degree of variation than later time points, and the variation over time was structured such that principal component 1 (PC1)/PC2 captured around 52% of the variation, PC1/PC3 38%, and PC2/PC3 34%. This time effect was also clear when plotting DEG heatmaps for mock- and rhizobia-inoculated experiments separately (Supplemental Figure 1). This analysis also showed that there was a greater change over time in the gene expression of samples that had been mock inoculated, suggesting that external N treatment has a greater impact in mock-inoculated plants.
Figure 2.
Nitrogen Responses in Rhizobia- and Mock-Inoculated Roots Suggests that A17 Seedlings Treated with Rhizobia Are More Responsive to External Nitrogen Than Mock-Inoculated Roots.
(A) Experimental design for transcriptomics experiment.
(B) Principal component analysis (PCA) reveals a greater difference between rhizobia- and mock-inoculated plants at time 0, and greater changes over time in the gene expression in mock-inoculated (M) than rhizobia-inoculated (R) roots.
(C) Number of DEGs in rhizobia versus mock at each time point and their associated gene ontology terms.
(D) Venn diagram representing the distribution of DEGs between rhizobia-inoculated and mock-inoculated roots over time after N treatment. Genes that are differentially regulated in rhizobia-inoculated and mock-inoculated roots; if other genes in the same family are also in the DEG list, the number is given in parentheses; n = 3 except for Rhizobia-6hNitrogen where n = 2.
See also Supplemental Tables 2 and 3.
When we compared rhizobia and mock-inoculated roots we found 1030 DEGs at at least one time point (Figure 2C). We then queried the lists of genes that change between these conditions (R0–M0, R2–M2, and R6–M6) to ask whether there were any gene ontology (GO) terms associated with genes regulated over time in each experiment. Genes related to defense, redox processes, and stress had a higher expression in mock-inoculated roots treated with replete N, suggesting that roots in these conditions have higher levels of stress than those inoculated with rhizobia. In contrast, genes that had higher expression in rhizobia-inoculated roots treated with replete N were associated with many GO terms related to the enriched term of “transport” (Figure 2C and Supplemental Table 3), which in the context of these experiments (rhizobia-inoculated plants with supplied external N treatment) could reflect alteration in N transport. Between R0 and M0, DE genes related to nodulation were found, including members of the nodule-specific small peptide (CCP/NCR) gene family and nodulins (Figure 2D). Genes that were expressed more highly in mock-inoculated roots included functions that could be part of N-scavenging responses including transmembrane amino acid transporter (Medtr5g023260) and Medtr2g097530, an IAA-amino acid hydrolase ILR1-like protein associated with auxin regulation and dormancy in Medicago (Du et al., 2017). There was a decrease in the number of nodulation-associated DE genes in rhizobia-inoculated roots over time (compared with mock) from 28 genes at 0 h to 26 genes at 2 h, and finally to three genes at 6 h of N treatment, suggesting that these regulated genes could be part of the N-repression of nodulation mechanism that has a greater impact over time (Figure 2D). These changes could be part of the regulation of nodulation system dynamics, whereby plants that have established a symbiotic relationship with rhizobia are more sensitive to the external N levels as one part of the mechanism to keep resource use for nodulation in balance with N demand.
We compared changes over time between rhizobia and mock-inoculated roots and found 251 genes to be different at all time points; among these are key nodulation DEGs such as Nodule Inception NIN (Medtr5g099060) (Vernié et al., 2015) (as validated using qPCR; Supplemental Table 4), ENOD18 (Medtr7g065770) (Hohnjec et al., 2003), as well as a range of nodulins and NCRs (Figure 2D). We then compared the effect of N on mock-inoculated roots and found that 3126 genes change expression over the 6 h. We used a MapMan analysis (Thimm et al., 2004) to gain more detail into the processes that change and found that redox responses and biotic and abiotic stresses were more prevalent among genes that change in mock-inoculated roots, in accordance with the GO-term analysis described above (Figure 2C and Supplemental Figure 1A–1D). Finally, we compared the effect of N on rhizobia-inoculated roots and found that 857 genes change expression (Supplemental Figure 1E–1H and Supplemental Table 2). Despite the fact that rhizobia-inoculated roots have a much smaller sized response to N, the number of genes categorized as being involved in N transport and metabolism changing over time in the rhizobia-inoculated roots was similar to that in mock-inoculated roots (Supplemental Figure 1), thus the rhizobia N response seemed to relatively enriched. These results suggest that external N treatment affects both mock- and rhizobia-inoculated roots, showing interplay between N- and rhizobia-plant root responses.
Genes Controlling Nitrogen Transport Are Differently Regulated after Mock or Rhizobia Inoculation
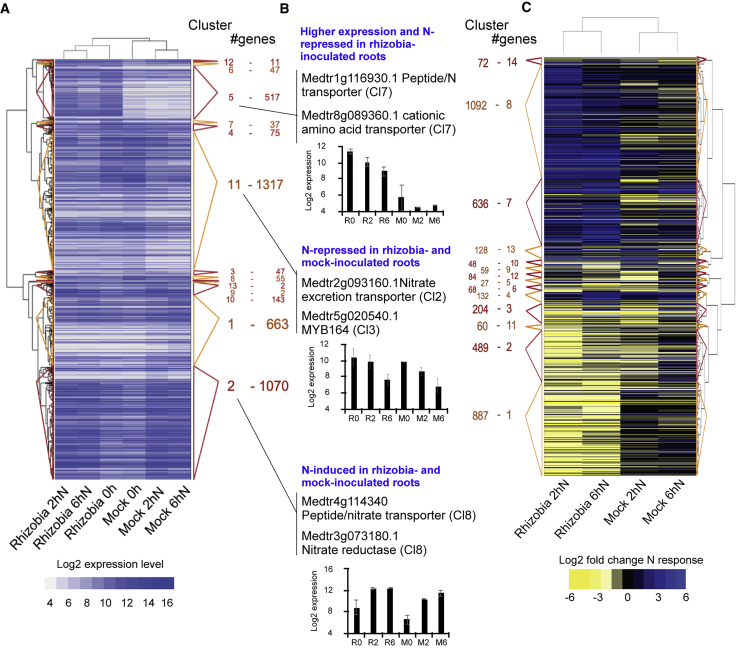
To understand the combined effect of N and rhizobial inoculation, and investigate the hypothesized reduction in nodulation-associated genes (Figure 2D), we used hierarchical clustering using silhouette plots to assess the expression patterns of all 3986 DEGs over all conditions (union of 3126 genes that are N-responsive over time in mock-inoculated roots, 857 genes that are N-responsive over time in rhizobia-inoculated roots, and 1030 genes that differ between mock- and rhizobia-inoculated roots) (Figure 3A and Supplemental Table 2). There were 13 clusters, with four predominant patterns (clusters 1, 2, 5, and 11) (Figure 2A). Genes in the largest cluster (cluster 11 with 1317 genes) were found to be N repressed in both rhizobia- and mock-inoculated roots, including the MYB transcription factor MYB164 (Figure 3B). This cluster also includes transport inhibitor response 1 (Medtr7g083610), an ortholog of A. thaliana AFB3, an auxin receptor involved in primary and LR growth inhibition in response to nitrate and a target of miR393 (Vidal et al., 2010) as well as lateral organ boundaries (LOB) domain protein Medtr1g095850, a member of the plant organ development key regulators (Xu et al., 2016). These genes could be involved in regulating root architecture in both nodulating and non-nodulating plants.
Figure 3.
Change in Gene Expression in Plant Roots Inoculated with Rhizobia and in Response to Nitrogen Treatment Shows that Regulation of Nitrogen Mobilization Genes Varies upon Nodulation Status.
(A and C) Heatmap and number of genes in each cluster for genes clustered over “raw” log2 values (A) and log2 fold N responses (C). Cluster sizes and membership is specified and also depicted by the colored triangles.
(B) Overview of main effect in clusters 2, 5, and 11, with representative genes (as referred to in text) and examples of the expression levels of representative genes. For each gene the cluster in the “response” data (C) is given in parentheses. n = 3 except for Rhizobia-6hNitrogen where n = 2.
See also Supplemental Tables 2 and 3.
Cluster 2 genes (1070 genes) were N-induced in both rhizobia-inoculated and mock-inoculated roots with greater N induction in mock-inoculated roots. The cluster includes range of N-response genes such as nitrate reductases and nitrate transporters (Figure 3B). Cluster 5 genes (517 genes) were much more highly expressed in rhizobial-inoculated roots than mock, and were N repressed. This cluster includes a range of N transporters and amino acid transporters (including Medtr8g089360, Figure 3B), which could be related to N-mobilization and N-metabolism rearrangement in plants undergoing nodulation. The changes in gene expression in these clusters indicate altered dynamics of N responses, potentially underlying the variation in root phenotypes under rhizobial inoculation compared with mock inoculation.
We also clustered the same genes by N response to directly compare the scale of N responses not dependent on the basal expression level (Figure 3C). Among the 14 N-response clusters there was enrichment of genes annotated with terms including nitrogen, transport, cysteine, redox, kinase, and jasmonate (Supplemental Tables 2, 3A, and 3B). The difference in N responses between rhizobia- and mock-inoculated roots suggested that N transport was activated in plants that had been inoculated with rhizobia.
Transcriptomic Analysis Shows that Altered Autoregulation of Nodulation Affects the Pathways Controlling Root Development, Nitrogen Perception, Uptake, and Transport
To investigate whether plants that were taking up fixed N in the root had altered N uptake, we used transcriptomic analysis of M. truncatula A17 compared with the SUPERNUMERARY NODULES (Medtr4g070970) sunn-1 mutant, since it has an extreme, hypernodulating phenotype. sunn-1 mutants continue to form nodules when A17 (wild-type) plant nodulation is repressed and thus sense that the N environment is perturbed (Jin et al., 2012). Previous work including use of split roots has shown that the altered N regulation occurs at both local and systemic levels (Jeudy et al., 2010).
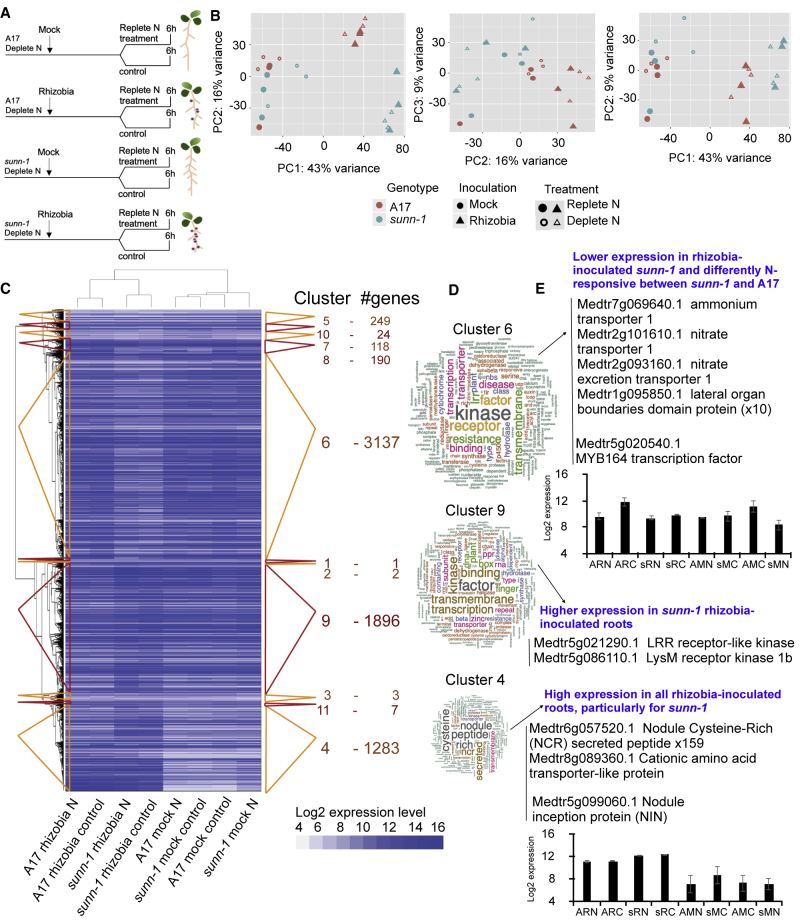
We used an experimental space with A17 and sunn-1 where we varied both N level and S. meliloti inoculation. We inoculated plants with S. meliloti (or mock inoculated) and then 14 days later carried out a 6-h replete-N (5 mM NH4NO3) treatment or left on deplete N (0.1mM NH4NO3, control) before harvesting roots (Figure 4A). PCA was used to identify major sources of variation over the samples (Figure 4B). PC1/PC2 captured 59% of the variation, reflecting that the greatest differences observed come from the mock-rhizobia comparisons in both genotypes. The differences between sunn-1 and A17 genotypes are greater in rhizobia-inoculated plants. In terms of N responses, there seemed to be a stronger impact in mock-inoculated sunn-1 plants. These results suggested a possible crosstalk between AON pathway and N transport in mock-inoculated plants.
Figure 4.
The sunn-1 Mutant Does Not Respond to the Combination of Rhizobia + N as Strongly as A17 and Could Be Transporting N Less Efficiently.
(A) Experimental design for transcriptomic experiment. Schematic indicating root architecture at point of N treatment for rhizobia-/mock-inoculated A17/sunn-1.
(B) PCA shows that samples are separated principally on genotype and to a greater extent by inoculation status.
(C) Clustering heatmap of the DEGs alongside cluster number and number of genes in the cluster. Cluster sizes and membership is depicted by the colored triangles.
(D) Wordclouds representing the most significant terms for the three largest clusters that together represent 91.4% of the total DEGs. Enriched terms were found to be: cluster 4, cysteine; cluster 6, kinase, transport, redox, calcium, nitrogen, and UDP.
(E) Examples of the expression levels of representative genes (as referred to in the text). n = 3.
See also Supplemental Tables 3 and 5.
We carried out gene expression analysis using microarrays as before (see Methods) (Supplemental Table 5) and identified a set of 6910 regulated genes. Hierarchical clustering was used to assess gene expression patterns (Figure 4B). There were 11 clusters, with three predominant patterns (clusters 4, 6, and 9) representing 91% of DEGs (Figure 4C). Cluster 4 (1283 DEG) contained genes that have increased expression in rhizobia-inoculated roots compared with mock, both in sunn-1 and A17; however, rhizobia induction is stronger in sunn-1 than in A17, independent of the N treatment. As would be expected, many genes in this rhizobia-enhanced cluster are involved in the nodulation pathway, e.g., NIN (Medtr5g099060, Figure 4E) (Vernié et al., 2015), MtNSP2 (Medtr3g072710) (Kaló et al., 2005) (both validated by qPCR), nodule-specific cysteine-rich peptides (159 genes), leghemoglobins (10 genes), late nodulins (25 genes), and glycine-rich proteins (19 genes). Cluster 4 also includes a number of genes involved in N metabolism and transport, including the amino acid transporter Medtr8g089360, also found to be strongly expressed in rhizobia-inoculated roots in the previous experiment (Figure 3B). This gene, as well as other N transporters (e.g., Medtr3g069420) are downregulated by N specifically in sunn-1 mock-inoculated plants, and could be part of the crosstalk between the AON pathway and N transport in mock-inoculated plants. The fact that the induction of these genes was stronger in sunn-1 than in wild-type A17 is likely part of the molecular alteration underlying the hypernodulation phenotype of sunn-1. As well as nodulation genes in cluster 4, there were calmodulin binding proteins (four genes) required for the recruitment of ubiquitous Ca2+ for endosymbiotic N fixation and cytochromes P450 (12 genes) that interact with calmodulin binding proteins to act as mediators in multiple catalytic pathways (Yamada et al., 1998, Li et al., 2012). We also found regulatory genes such as LRR receptor-like kinases (16 genes), MYB (11 genes), GRAS (4 genes), MADS-box transcription factors (3 genes), zinc finger proteins (19 genes), and members of the F-box protein family (17 genes) as well as transport genes including peptide transporters (10 genes) and peptide/nitrate transporters (6 genes) (Figure 4B and Supplemental Table 5).
Cluster 6 (3137 DEGs) was enriched for genes with annotation terms including kinase (276 genes), transport (142 genes), redox (26 genes), calcium (28 genes), nitrogen (35 genes), and UDP (37 genes) (Figure 4C and 4D; Supplemental Table 3A and 3C). In this cluster, A17 and sunn-1 exhibited opposite gene expression responses to replete-N treatment when rhizobia inoculated (Figure 4C), with upregulation in A17 but not in sunn-1. Interestingly, we found N-related genes in this cluster including nitrate transporters (4 genes), peptide/nitrate transporters (8 genes), nitrogen fixation proteins (2 genes), and ammonium transporters (2 genes out of 12 ammonium transporters in the whole genome). These findings suggested that N transport may be more efficient in A17 than in sunn-1 in the presence of rhizobia.
Within cluster 6, differentially N-responsive between sunn-1 and A17 there was Medtr7g092930, an ortholog of the squamosa promoter-binding-like protein SPL9, involved in LR development in A. thaliana (Yu et al., 2015). This cluster also contains 10 lateral organ boundaries (LOB-domain) genes. One of these LOB-domain genes, Medtr6g005080, is specifically downregulated in sunn-1 compared with A17 in rhizobia-inoculated roots, but only in N-deplete conditions, that typically have essential roles in integrating development in response to environmental changes (Xu et al., 2016) as well as two MYB transcription factors (including MYB164, Figure 4E). The altered N regulation of these genes suggests that LR growth might also be different in A17 compared with sunn-1. Cluster 9 (1896 DEGs) was more strongly expressed in sunn-1 with rhizobia inoculation than in any other experiment and independent of the N treatment. This cluster had many regulatory gene annotations, including kinase (95 genes), transmembrane (86 genes), and transcription (91 genes) (Figure 4C and 4D). Upregulation of the 35 LRR kinases and 11 LysMs in this cluster could be related to the altered perception of rhizobia in the sunn-1 mutant.
Overall, using transcriptomic analysis we found evidence that the SUNN and N-transport pathways are integrated. It has been previously hypothesized that SUNN perceives the N/carbon (C) ratio in the shoot and then sends a signal to the root to control nodule number (Jin et al., 2012). From our experiments, we hypothesized that SUNN is involved in root perception of external N levels and is responsible for uptake and transport of N to the shoot. To test this hypothesis, we analyzed the RSA phenotype of sunn-1 under different levels of N after mock or rhizobia inoculation and its putative involvement in N transport in an N-uptake assay followed by mineral analysis.
Nodule Numbers, Root Development, and Nitrogen Uptake Are Balanced Differently in A17 and sunn-1
We grew sunn-1 seedlings and inoculated them with S. meliloti (or carried out mock inoculation). Fourteen days later we treated them with deplete or replete levels of N (as used in the microarray experiments) for 16 days, then measured RSA (Figure 5 and Supplemental Table 6). As found previously, sunn-1 has significantly more nodules than A17, even on replete levels of N (5 mM NH4NO3) when wild type consistently shuts down nodulation (Figure 5A and 5B). We found that sunn-1 mutants had a significantly longer PR than A17 on replete N, either with or without rhizobia inoculation, although the PR was longer when mock inoculated (Figure 5B). On deplete N all plants had similar numbers of lateral roots but on replete N, sunn-1 mutants had significantly more LRs and greater lateral root density than A17 when rhizobia or mock inoculated (Figure 5D and Supplemental Table 6).
Figure 5.
sunn-1 Root System Architecture Changes after N Treatment, with Effects Dependent on Rhizobia-Inoculation Status.
(A) Images of sunn-1 seedlings that were mock or rhizobia inoculated and then grown in N-deplete or N-replete conditions. Scale bar, 1 cm.
(B–E) number of nodules (B), PR length (C), number of LRs (D), and average LR length (E); data are presented as mean ± SD. Different letters denote statistically different groups for pairwise comparisons using Wilcoxon’s rank-sum test, P < 0.05; n ≥ 11.
See Supplemental Table 6.
sunn-1 mutants that were rhizobia inoculated had shorter LRs than mock-inoculated plants only in deplete-N conditions (Figure 5E); in fact, sunn-1 LRs in these conditions are also significantly shorter than in A17 (Supplemental Figure 2 and Supplemental Table 7). Shorter LRs in sunn-1 when rhizobia inoculated could be explained by the regulation of LOB-domain genes (including Medtr6g005080 that is specifically downregulated in sunn-1 in these conditions) as described earlier (Figure 4C). These genes could be key in the regulation of LR length, integrating the internal and external N signals to mount an appropriate developmental response. Autoregulation mutants have previously been found to have nodulation-independent phenotypes, such as the increased LR density and a shorter root system in the ljhar1 mutant (Wopereis et al., 2000). In our experiments we observe that this LR phenotype is even more significant under rhizobia inoculation. These results suggest that the RSA phenotypes in AON mutants are also under the control of a long-distance signaling system. Auxin has been implicated in the shoot-to-root signaling regulating nodule and LR development (Jin et al., 2012), and we found “auxin” to be an enriched term in our regulated genes; DEGs related to auxin are mostly in clusters 6 and 9. The presence of auxin genes in cluster 6 (upregulated in A17 but not in sunn-1) is consistent with the phenotypic differences in A17 and sunn-1 in rhizobia + replete-N conditions (Figure 4C): shorter PR phenotype in A17 when compared with sunn-1 (Supplemental Figure 2 and Supplemental Table 7).
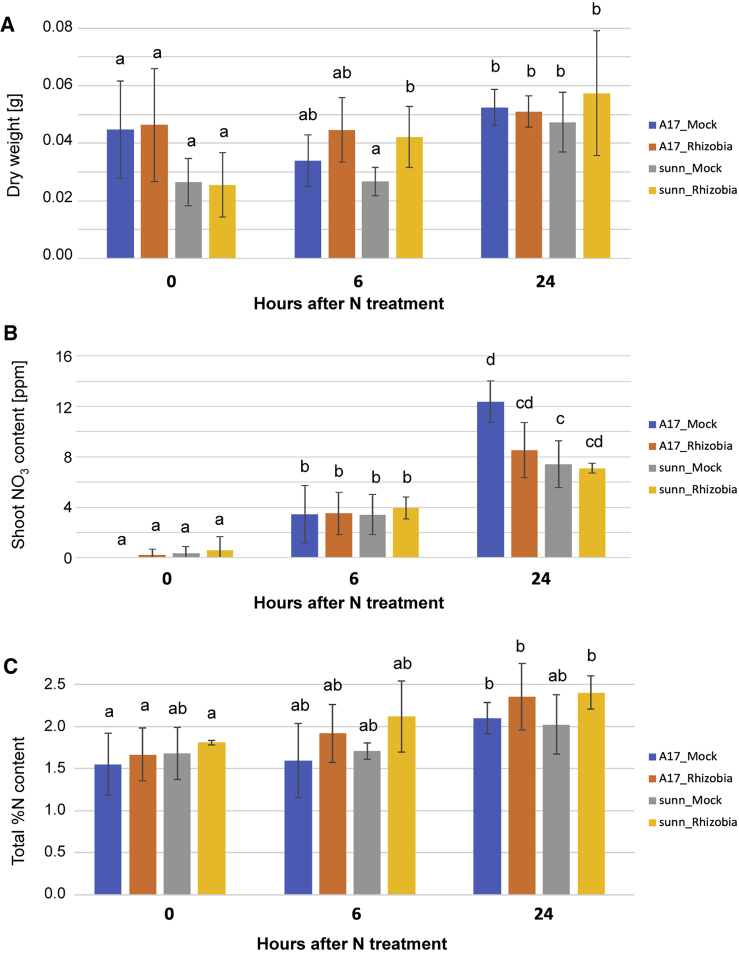
Based on the differing N transcriptome response of the sunn-1 hypernodulating mutant, we then asked whether sunn-1 mutants mobilized N to the shoot differentially, and if this affected whole plant size. We measured the shoot dry weight, free nitrate, and percentage of total N and total C shoot content of A17 and sunn-1 plants grown in perlite pots. As with the study for transcriptomics, we inoculated plants with S. meliloti (or mock inoculated) and then 14 days later carried out a replete-N (15mM NH4NO3) treatment before harvesting shoots after 6 and 24 h as well as at 0 h to assess N uptake. After 24 h of growth with N supply, dry weight of all shoots was increased (Figure 6A and Supplemental Table 8). sunn-1 rhizobia-inoculated plants appeared to increase in dry weight more quickly than all other plants (they are significantly different in dry weight at 6 h, but then similar at 24 h; Figure 6A and Supplemental Table 8).
Figure 6.
A17 Mock-Inoculated Plants Mobilize More N to Shoots Than sunn-1.
(A) Root and shoot fresh weight is larger in N-replete conditions but even more so when plants are rhizobia inoculated. This is not a rhizobia-alone effect as fresh weight is the same between mock- and rhizobia-inoculated conditions in N-deplete levels.
(B) Free NO3− content is higher in N-replete than N-deplete conditions and higher in the shoots of rhizobia-inoculated plants; free NO3− is similar in the roots of mock- and rhizobia-inoculated plants.
(C) Total N content is higher in N-replete than N-deplete conditions but is similar in mock- and rhizobia-inoculated plants.
Data in (A) and (B) are presented as mean ± SD. Letters denote statistically different values: a–b, P < 0.05; a–c, P < 0.01.
See also Supplemental Table 8.
We measured shoot free NO3− content and found that it increased in all plants over the 24-h period (Figure 6B and Supplemental Table 8). This shows that transport from root to shoot occurs rapidly, within the first 6 h. However, by 24 h of N supply, free NO3− is higher in A17 mock-inoculated plants than sunn-1, suggesting that sunn-1 plants are less efficient at moving N to the shoots. This difference is not apparent for rhizobia-inoculated plants. We then measured total percent N content and found it increases from 0–6 h to 24 h in A17 (mock inoculated) but not in other conditions (Figure 6C and Supplemental Table 8). This again suggests that A17 mock-inoculated plants are more efficient at taking up NO3− and transporting it to the shoot. We also measured total percent C and found no significant changes between plants (Supplemental Table 8); regulation of photosynthesis upon higher N uptake could occur later than this 24-h time period.
Interactions between Nodulation and Nitrogen Regulatory Pathways
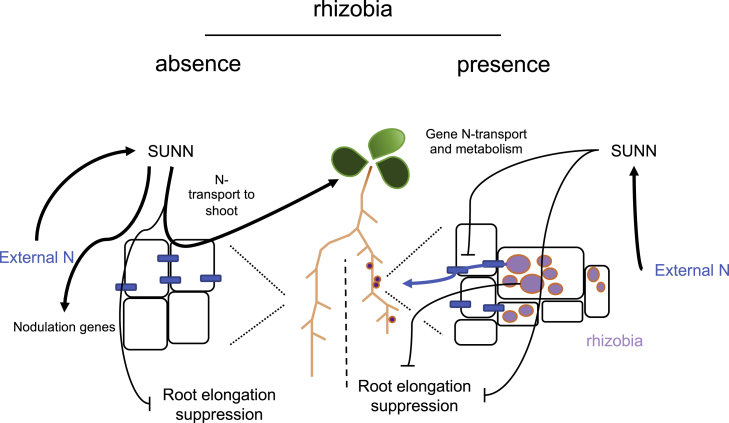
Put together, our transcriptomic and phenotypic analyses suggest that not only does external N treatment regulate the outcome of nodulation, but also that the AON signaling pathway regulates N uptake and metabolism in the absence of rhizobia. A key regulator of nodulation, NIN, was found not to respond to N in A17 wild type, but to be N repressed in sunn-1, only in mock conditions. A similar response pattern was found for many NCRs and N transporters. Most of these genes also seem to be more highly expressed in sunn-1 compared with A17 only in mock-inoculated and N-deplete conditions. This differential N responsiveness of sunn-1, implicating SUNN in control of N mobilization even when plants are not nodulating, is supported by our mineral analysis results. A17 appears to be able to transport more N to the shoot, compared with sunn-1, in mock conditions. Our new data enable us to propose a model to help elucidate the regulatory links between nodulation, root development, and plant nutritional status (Figure 7). Plants with a functional SUNN protein that are subject to rhizobia inoculation are able to perceive external levels of N and mobilize this to the shoot. N perception, uptake, and transport could thus be the first step required to trigger signaling that contributes to the AON.
Figure 7.
Model Showing the Interactions between Rhizobia and N Treatment on Medicago Root Architecture, and the Function of SUNN in Mediating These Interactions.
Nitrogen and N transport affects distal responses in leaves and mediates root architecture, including the balance between lateral and PR growth. SUNN was previously implicated in controlling the lateral–PR balance. With our new phenotypic and transcriptomic data, we hypothesize that the SUNN impact on root architecture (and ultimately the ability to nodulate) is partly due to playing a role in transport of N to the shoot. Plants with a functional SUNN protein are able to perceive external levels of N and mobilize this to the shoot more rapidly. Nitrogen perception, uptake, and transport could be the first step required to trigger signaling that contributes to the autoregulation of nodulation.
Methods
Plant Material and Growth
M. truncatula var. Jemalong A17 seeds were obtained from the Aberystwyth seed biobank, and the sunn-1 mutant (Schnabel et al., 2005) seeds were kindly provided by Giles Oldroyd (John Innes Center, Norwich, UK). Each experiment for transcriptomic analysis was carried out in triplicate; experiments for phenotypic analysis were carried out in triplicate with the images shown representative of the results. Seed was abrasion-scarified, bleach-surface sterilized, stratified for 24 h at 4°C, and sown onto plates containing 1.5% water-agar for 5 days. Six to seven germinated seedlings were grown on 0.8% agar-modified Fahräeus medium (MFM) plates, between the two layers of a plant growth pouch (CYG Growth Pouch, Mega International, Minneapolis, MN, USA) overlaid on MFM (Vincent, 1970). Medium contained 0.5 mM MgSO4·7H2O, 0.7 mM KH2PO4, 20 mM ferric citrate, 0.4 mM Na2HPO4·2H2O, 0.9 mM CaCl2 with 1 mg ml−1 each of MnSO4, CuSO4, ZnCl2, H3BO3, and Na2MoO4, NH4NO3 at the concentration required (deplete = 0.1 mM; replete = 5 mM), and adjusted to pH 6.5. 0.075 μM of the ethylene inhibitor (S)-trans-2-amino-4-(2-aminoethoxy)-3-butenoic acid hydrochloride (AVG) was added after autoclaving. Plates were sealed with microporous tape and roots were covered in black polythene covers (Bagman of Cantley, Lingwood, Norfolk, UK). Plates were grown in a Sanyo growth chamber (MLR-351H, Sanyo, E&E Europe, Loughborough, UK) vertically with a 16/8-h photoperiod at 50 μmol m−2 s−1 and constant 25°C.
For mineral analysis, M. truncatula A17 seeds were sterilized, stratified, and germinated as above, then transplanted into FP7 (7 × 7cm) pots filled with sterile perlite with a 2-cm layer of sterile vermiculite. Plants were growth in a glasshouse compartment with a 16/8-h photoperiod at 50 μmol m−2 s−1 and 24°C day/22°C night. Pots were watered with nutrient solution following a recipe modified from Broughton and Dilworth (1971) (1 mM KH2PO4, 1 mM CaCl2·2H2O, 75 μM FeNa EDTA, 1 mM MgSO4·7H2O, 0.25 mM K2SO4, 6 μM MnSO4·H2O, 1 μM ZnSO4·7H2O, 0.5 μM CuSO4·5H2O, 0.05 μM CoSO4·7H2O, 20 μM H3BO3, and 0.1 μM Na2MoO4·2H2O, adjusted to pH 6.2–6.4).
Plant Treatments
S. meliloti 1021 was grown in TY/Ca2+ medium plates at 28°C for 2 days. S. meliloti 1021 cultures were grown overnight in TY/Ca2+ liquid medium at 28°C with shaking at 220 rpm. Seedlings were grown vertically in the MFM plates or perlite pots for 4 days, then the seedlings were either inoculated with 2 ml of OD600 = 0.02 of S. meliloti 1021 in liquid MFM or mock inoculated with the same volume of modified liquid MFM. Treatments were performed by pipetting solutions directly onto the roots of seedlings. For N treatments in plates, 14 days post inoculation (dpi) roots were treated with 2 ml of liquid MFM supplemented with NH4NO3 at the concentration required, then placed on fresh solid MFM containing the same NH4NO3 concentration as the treatment. We confirmed rhizobia inoculation or N-treatment effect using qPCR of ENOD11, NRT2.1, NPL, NSP2, and NIN genes (Supplemental Table 4). Treated seedlings were kept in the incubator for 2–6 h, then roots were excised from the shoot and frozen in N2(l) for microarray experiments, or seedlings were kept for up to 16 more days for RSA analysis. For N treatments in perlite pots, at 14 dpi pots were watered with pot nutrient solution as above, containing 15 mM NH4NO3 as a replete-N treatment, after which samples were harvested at 0 (immediately before treatment), 6, and 24 h for mineral analysis.
RNA Analysis, qPCR, and Microarray Hybridization
RNA was extracted from roots using the Qiagen RNeasy Plant Mini Kit, following the manufacturer’s instructions. Total RNA samples were then treated with the TURBO DNA-free kit (Life Technologies). Lack of genomic DNA contamination was confirmed using bioanalyzer and PCR with primers Medtr3g091400_F: 5ʹ-TCATCTTCAACGCAGACCCC-3ʹ and Medtr3g091400_R: 5ʹ-ACTCACACTTACACGCGACA-3ʹ. cDNA was synthesized from RNA using the Ovation Pico WTA System (NuGEN Technologies, San Carlos, CA, USA) and purified using Qiagen Qiaquick PCR purification kit. dscDNA was labeled with Cy3 using the Nimblegen one-color DNA labeling kit. qPCR was performed on dscDNA using SYBR Green JumpStartTaq ReadyMix (Sigma-Aldrich, St Louis, MO) according to the manufacturer's instructions on an Mx3005P qPCR System (Agilent Technologies). Expression of five transcripts was tested for confirmation of microarray quantification results (for values and primer sequences see Supplemental Table 4). mRNA levels were normalized relative to Ubiquitin-Conjugating Enzyme E2 9 (UBC9) (Medtr7g116940) with primers as used in Kakar et al. (2008), UBC9F: 5ʹ-GGTTGATTGCTCTTCTCTCCCC-3ʹ and AAGTGATTGCTCGTCCAACCC, and quantified using standard curves generated for each primer pair. Cy3-labeled cDNA was quantified using NanoDrop, then hybridized using the Roche-Nimblegen (Roche Applied Science) single-color microarray hybridization kit to 12×135k Roche-Nimblegen probe arrays. This array is a custom design for the M. truncatula Mt3.5 genome with two to three unique 60-mer oligonucleotide probes. We only consider probes that map to the Mt4.0 genome version; this measures expression of 27 671 Mt4.0 genes and only these genes were analyzed in this work (see Gene Expression Omnibus GPL25305 array platform and data deposit GEO: GSE116789). Fluorescence intensity signals of the labeled cDNA excited at 532 nm wavelength were quantified using the Nimblegen MS 200 scanner set at 2-μm resolution. The data were collected as XYS files (raw probe level data by coordinates on the microarray).
Statistical Analysis of Transcriptomic Data
Microarray analysis was performed in R using custom scripts. We assessed microarray reproducibility and confirmed that housekeeping genes Medtr6g079920.1 (pentatricopeptide repeat protein), Medtr3g090960.1 (polypyrimidine tract-binding-like protein), and Medtr3g091400 (the ubiquitin-like phosphatidylinositol 3- and 4-kinase family protein) did not change expression. An annotation package associated with the array design was built by using the pdInfoBuilder package (Falcon and Carvalho, 2013) and the .ndf design file provided by the manufacturer. The quality of data was checked by generating box plots, smooth histograms, and heatmaps of Pearson correlation coefficients between all arrays of the raw data using the Oligo package (Carvalho and Irizarry, 2010). For background adjustment, quantile normalization, and summarization (to gene level by median polish), robust multi-array averaging (RMA) was performed with the Oligo package. To compare global expression levels between replicates in the same experiment, we used an R2 test; values were typically in the range of 0.90–0.95, with the lowest being 0.85.
To explore the distribution of the variation, we performed PCA on the log2[signal intensity] data with average values for replicates. To characterize expression patterns and identify DEGs over time in mock or rhizobia-inoculated roots, we fitted a linear model for each gene across the series of arrays by a least-squares regression. To assess model fit, we used a t-statistic test and calculated P values by computing empirical Bayes statistics for differential expression using a significance cut-off of P < 0.05 and a fold change cut-off of 1.5 on log scale.
A linear model with moderated statistics (t-statistics and F-statistic) was fitted to the log2 normalized data with limma (Ritchie et al., 2015), adjusting P values with the false discovery rate algorithm (Benjamini and Hochberg, 1995). DEGs were defined as having an adjusted P value of <0.05 and fold change >1.5. DEGs were clustered using hierarchical clustering with an average linkage and Pearson correlation. Expression values for DEGs were averaged for each treatment, row normalized and clustered using hierarchical clustering with an average linkage and Pearson correlation using the clustergram function in MATLAB. Silhouette widths were plotted in MATLAB using the silhouette function for each hierarchical tree and used to determine where to cut the trees and define clusters, then the cluster patterns were visualized in MATLAB using the clustergram function. Annotation term enrichment was determined using Fisher’s Exact test with a P-value cut-off of <0.05. GO term annotations were obtained from AgriGO (http://systemsbiology.cau.edu.cn/agriGOv2/) for M. truncatula version Mt4. GO term overrepresentation was assessed using the topGO R package (Alexa and Rahnenfuhrer, 2018).
Identification of Regulation of Response Pathways
MapMan software (version 3.5.1R2) was used to visualize the time-dependent transcript profiles to N treatment. Experimental data files of mock (M2–M0, M6–M0) and rhizobia-inoculated root DEGs (R2–R0, R6–R0) were prepared containing the log2 fold change for each DEG. Mapping files to link the experimental data onto the Image Annotator was obtained from the MapMan store (https://mapman.gabipd.org/mapmanstore). The mapping files organize experimental data files for assignment into functional categories of BINs and sub-BINs that are defined in the SCAVENGER module (Thimm et al., 2004). Pathway analysis of all the major pathways (BINs) to visualize metabolic changes and cellular responses was performed by applying the built-in Wilcoxon rank-sum test with Benjamini–Hochberg correction (Thimm et al., 2004). For analysis of putative orthologs in M. truncatula of genes discovered in other species, we used a reciprocal best blast analysis using data from Ensembl plants (Bolser et al., 2017).
Microscopy Analysis
For visualization of nodule numbers, roots were viewed at 10× under differential interference contrast using an Olympus BX51 microscope.
Statistical Analysis of Phenotypic Data
Plant roots were scanned using a Scanjet G2710 flatbed scanner (Hewlett-Packard) at highest resolution, then phenotypic analysis of root architecture was performed using ImageJ (http://rsbweb.nih.gov/ij), measuring PR length, the number of LRs (LR num), and the length of every LR. From this the following were calculated: total LR length, average LR length (LR length ave), total LR plus PR length (PR + LR tot) and lateral root density. Phenotype data was analyzed using R with a Shapiro–Wilk test used to test data normality; a Bartlett test was used to test data variance. A pairwise Wilcox test was used to assess differences for significance using the Benjamini–Hochberg method. Data box plots were generated using the R package ggplot2.
Nitrate, Total Percent Nitrogen, and Total Percent Carbon Determination
Anion extraction was performed on 20 mg of freeze-dried ground plant material using 1.5 ml of milliQ water containing 20 mg of insoluble polyvinylpolypyrrolidone. Extraction was performed at 4°C for 60 min shaking at 200 rpm, then samples were heated up at 90°C for 15 min and centrifuged at 13 000 g and 4°C for 15 min. Clear supernatants were the samples used for analysis. Nitrate anion in samples was analyzed using a Thermo Scientific Dionex ICS-1100 Ion Chromatography System consisting of a guard column (IonPac AG14A), an analytical column (IonPac AS14A), a suppressor (Dionex ERS 500), and a column heater (Dionex ICS-1100 Column Heater). Samples were analyzed using a Na2CO3–NaHCO3 eluent at 1.4 ml/min, 28 mV (suppressor voltage), and a set temperature of 30°C (column heater). The total content of N was analyzed by total combustion using a LECO Trumac CN/N determinator. 0.5 g of freeze-dried ground samples were weighed into a ceramic boat, which was loaded into the furnace of the instrument set up at 1350°C.
Funding
M.L.G., S.O., and B.L. were funded by BBSRC grant BB/H109502/1, and M.L.G. was also funded by Defra grant WU0128. P.S. and M.L.G. were funded by BBSRC grant BB/P002145/1. J.H. was funded by BB/G014159/1. M.A. and A.J.P. were funded by BBSRC through the MIBTP. R.B.-P. was funded by a Warwick Chancellor’s International Scholarship. All microarray data have been deposited in GEO (series GEO: GSE116789), released upon publication.
Author Contributions
Conceptualization, M.L.G.; Methodology and Validation, M.L.G. and B.L.; Formal Analysis, S.O., J.H., B.L., S.V., B.L.R., C.S., A.J.P., and M.L.G.; Investigation, R.B.-P. and M.A.; Writing – Original Draft, M.L.G., B.L., M.A., and R.B.-P.; Writing – Review & Editing, M.L.G., B.L., A.J.P., S.O., and J.H.; Visualization, A.J.P. and B.L.; Supervision, M.L.G., J.H., P.S., and S.O.; Funding Acquisition, M.L.G.
Acknowledgments
The authors declare that there are no competing interests.
Published: April 3, 2019
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, SIBS, CAS.
Supplemental Information is available at Molecular Plant Online.
Supplemental Information
(A) Average (ave) and standard error (SE) for number of nodules of A17 M. truncatula seedlings grown over a range of concentrations of NH4NO3.
(B) Average (ave) and SE root trait values measured from 18-day-old A17 M. truncatula seedlings grown in the conditions specified.
(C) Adjusted P values from pairwise Wilcoxon rank-sum test with Holm's method for multiple testing; green shading indicates P value <0.05.
The table lists the Medicago microarray probes with their Medicago 3.5v3 and 4.0v1 gene IDs and annotation represented on the array (for design see GEO record GPL25305 for Nimblegen probe design) and their log2 scale expression values in all experiments as well as averaged (“Ave”) for each replicate set; see GEO: GSE116789 for complete raw and normalized individual replicate values. If a gene is significantly differentially expressed between samples the cluster number is listed (clustering on N-treatment responses and by “raw,” uncompared values). In comparisons between experiments for all genes with an Mt4.0 annotation the direction of regulation (“regulation” column, 0 = no change, −1 = downregulated, 1 = upregulated) and P value of significance (“Pval” column) are given. NA, not tested; n = 3 except for Rhizobia-6hNitrogen where n = 2.
(A) Terms enriched for differentially expressed genes (DEGs) whose expression changes over time in A17 or between A17 and sunn-1.
(B) Terms enriched in 14 clusters of DEG in the A17 time series data.
(C) Terms enriched in 11 clusters of DEG in the A17–sunn-1 experiment; red color = P value <0.05.
(A) Oligo sequences used for amplification of qPCR products shown in Supplemental Information and descriptions of each gene.
(B) Gene expression level from microarray hybridization.
(C) Gene expression level from qPCR; ND, not detectible.
The table lists the Medicago microarray probes with their Medicago 3.5v3 and 4.0v1 gene IDs and annotation represented on the array (for design see GEO record GPL25305 for Nimblegen probe design) and their log2 scale expression values in all experiments as well as averaged (“Ave”) for each replicate set; see GEO: GSE116789 for complete raw and normalized individual replicate values. If significantly differentially expressed between samples the cluster number is listed; NA, not tested.
(A) Average (ave) and SE for root trait values measured from 18-day-old sunn-1 M. truncatula seedlings grown in the conditions specified.
(B) Adjusted P values from pairwise Wilcoxon rank-sum test with Holm's method for multiple testing; green shading indicates P value <0.05.
Adjusted P values from pairwise Wilcoxon rank-sum test with Holm's method for multiple testing; green shading indicates P value <0.05.
(A) Dry weight and measurements of free nitrate, total % N, and total % C in A17 and sunn-1 M. truncatula shoots grown in the experiments designated; SD, standard deviation.
(B) Adjusted P values from t-tests comparing dry shoot weight or mineral analysis levels; green shading indicates P value <0.05.
References
- Alexa A., Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology R package. 2018 [Google Scholar]; Alexa A, Rahnenfuhrer J (2018). topGO: Enrichment Analysis for Gene Ontology R package.
- Amor B.B., Shaw S.L., Oldroyd G.E.D., Maillet F., Penmetsa R.V., Cook D., Long S.R., Denarie J., Gough C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313x.2003.01743.x. [DOI] [PubMed] [Google Scholar]; Amor, B.B., Shaw, S.L., Oldroyd, G.E.D., Maillet, F., Penmetsa, R.V., Cook, D., Long, S.R., Denarie, J., and Gough, C. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34:495-506. [DOI] [PubMed]
- Barbulova A., Rogato A., D'Apuzzo E., Omrane S., Chiurazzi M. Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol. Plant Microbe Interact. 2007;20:994–1003. doi: 10.1094/MPMI-20-8-0994. [DOI] [PubMed] [Google Scholar]; Barbulova, A., Rogato, A., D'Apuzzo, E., Omrane, S., and Chiurazzi, M. (2007). Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol. Plant Microbe Interact. 20:994-1003. [DOI] [PubMed]
- Benedito V.A., Torres-Jerez I., Murray J.D., Andriankaja A., Allen S., Kakar K., Wandrey M., Verdier J., Zuber H., Ott T. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]; Benedito, V.A., Torres-Jerez, I., Murray, J.D., Andriankaja, A., Allen, S., Kakar, K., Wandrey, M., Verdier, J., Zuber, H., Ott, T., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J 55:504-513. [DOI] [PubMed]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]; Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Series B 57:289-300.
- Bolser D.M., Staines D.M., Perry E., Kersey P.J. Ensembl Plants: integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol. Biol. 2017;1533:1–31. doi: 10.1007/978-1-4939-6658-5_1. [DOI] [PubMed] [Google Scholar]; Bolser, D.M., Staines, D.M., Perry, E., and Kersey, P.J. (2017). Ensembl Plants: integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol. Biol. 1533:1-31. [DOI] [PubMed]
- Bouguyon E., Brun F., Meynard D., Kubes M., Pervent M., Leran S., Lacombe B., Krouk G., Guiderdoni E., Zazimalova E. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants. 2015;1:15015. doi: 10.1038/nplants.2015.15. [DOI] [PubMed] [Google Scholar]; Bouguyon, E., Brun, F., Meynard, D., Kubes, M., Pervent, M., Leran, S., Lacombe, B., Krouk, G., Guiderdoni, E., Zazimalova, E., et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1:15015. [DOI] [PubMed]
- Broughton W.J., Dilworth M.J. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971;125:1075–1080. doi: 10.1042/bj1251075. [DOI] [PMC free article] [PubMed] [Google Scholar]; Broughton, W.J., and Dilworth, M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem J 125:1075-1080. [DOI] [PMC free article] [PubMed]
- Capoen W., Den Herder J., Sun J., Verplancke C., De Keyser A., De Rycke R., Goormachtig S., Oldroyd G., Holsters M. Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell. 2009;21:1526–1540. doi: 10.1105/tpc.109.066233. [DOI] [PMC free article] [PubMed] [Google Scholar]; Capoen, W., Den Herder, J., Sun, J., Verplancke, C., De Keyser, A., De Rycke, R., Goormachtig, S., Oldroyd, G., and Holsters, M. (2009). Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell 21:1526-1540. [DOI] [PMC free article] [PubMed]
- Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]; Carvalho, B.S., and Irizarry, R.A. (2010). A framework for oligonucleotide microarray preprocessing. Bioinformatics 26:2363-2367. [DOI] [PMC free article] [PubMed]
- Catoira R., Galera C., De Billy F. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell. 2000;12:1647–1666. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]; Catoira, R., Galera, C., and De Billy, F. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. The Plant Cell 12:1647-1666. [DOI] [PMC free article] [PubMed]
- Cerri M.R., Frances L., Laloum T., Auriac M.C., Niebel A., Oldroyd G.E., Barker D.G., Fournier J., de Carvalho-Niebel F. Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012;160:2155–2172. doi: 10.1104/pp.112.203190. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cerri, M.R., Frances, L., Laloum, T., Auriac, M.C., Niebel, A., Oldroyd, G.E., Barker, D.G., Fournier, J., and de Carvalho-Niebel, F. (2012). Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol 160:2155-2172. [DOI] [PMC free article] [PubMed]
- Du H., Shi Y., Li D., Fan W., Wang G., Wang C. Screening and identification of key genes regulating fall dormancy in alfalfa leaves. PLoS One. 2017;12:e0188964. doi: 10.1371/journal.pone.0188964. [DOI] [PMC free article] [PubMed] [Google Scholar]; Du, H., Shi, Y., Li, D., Fan, W., Wang, G., and Wang, C. (2017). Screening and identification of key genes regulating fall dormancy in alfalfa leaves. PloS one 12:e0188964. [DOI] [PMC free article] [PubMed]
- Falcon S., Carvalho B. pdInfoBuilder: Platform Design Information Package Builder. R package version 1.26. 2013 [Google Scholar]; Falcon, S., Carvalho, B. (2013). pdInfoBuilder: Platform Design Information Package Builder. R package version 1.26.
- Gao K., Chen F.-J., Yuan L.-X., Mi G.-H. Cell production and expansion in the primary root of maize in response to low-nitrogen stress. J. Integr. Ag. 2014;13:2508–2517. [Google Scholar]; Gao, K., Chen, F.-j., Yuan, L.-x., and Mi, G.-h. (2014). Cell production and expansion in the primary root of maize in response to low-nitrogen stress. J. Integr. Ag. 13:2508-2517.
- Heidstra R., Nilsen G., Martinez-Abarca F., van Kammen A., Bisseling T. Nod factor-induced expression of leghemoglobin to study the mechanism of NH4NO3 inhibition on root hair deformation. Mol. Plant Microbe Interact. 1997;10:215–220. doi: 10.1094/MPMI.1997.10.2.215. [DOI] [PubMed] [Google Scholar]; Heidstra, R., Nilsen, G., Martinez-Abarca, F., van Kammen, A., and Bisseling, T. (1997). Nod factor-induced expression of leghemoglobin to study the mechanism of NH4NO3 inhibition on root hair deformation. Mol. Plant Microbe Interact. 10:215-220. [DOI] [PubMed]
- Hohnjec N., Perlick A.M., Puhler A., Kuster H. The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol. Plant Microbe Interact. 2003;16:903–915. doi: 10.1094/MPMI.2003.16.10.903. [DOI] [PubMed] [Google Scholar]; Hohnjec, N., Perlick, A.M., Puhler, A., and Kuster, H. (2003). The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol. Plant Microbe Interact. 16:903-915. [DOI] [PubMed]
- Huault E., Laffont C., Wen J., Mysore K.S., Ratet P., Duc G., Frugier F. Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet. 2014;10:e1004891. doi: 10.1371/journal.pgen.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huault, E., Laffont, C., Wen, J., Mysore, K.S., Ratet, P., Duc, G., and Frugier, F. (2014). Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet 10, e1004891. [DOI] [PMC free article] [PubMed]
- Jeudy C., Ruffel S., Freixes S., Tillard P., Santoni A.L., Morel S., Journet E.P., Duc G., Gojon A., Lepetit M. Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses. New Phytol. 2010;185:817–828. doi: 10.1111/j.1469-8137.2009.03103.x. [DOI] [PubMed] [Google Scholar]; Jeudy, C., Ruffel, S., Freixes, S., Tillard, P., Santoni, A.L., Morel, S., Journet, E.P., Duc, G., Gojon, A., Lepetit, M., et al. (2010). Adaptation of Medicago truncatula to nitrogen limitation is modulated via local and systemic nodule developmental responses. New Phytol 185:817-828. [DOI] [PubMed]
- Jin J., Watt M., Mathesius U. The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiol. 2012;159:489–500. doi: 10.1104/pp.112.194993. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jin, J., Watt, M., and Mathesius, U. (2012). The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiol. 159:489-500. [DOI] [PMC free article] [PubMed]
- Jones K., Kobayashi H., Davies B. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jones, K., Kobayashi, H., and Davies, B. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nature Reviews 5:619-633. [DOI] [PMC free article] [PubMed]
- Kakar K., Wandrey M., Czechowski T., Gaertner T., Scheible W.R., Stitt M., Torres-Jerez I., Xiao Y., Redman J.C., Wu H.C. A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods. 2008;4:18. doi: 10.1186/1746-4811-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kakar, K., Wandrey, M., Czechowski, T., Gaertner, T., Scheible, W.R., Stitt, M., Torres-Jerez, I., Xiao, Y., Redman, J.C., Wu, H.C., et al. (2008). A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4:18. [DOI] [PMC free article] [PubMed]
- Kaló P., Gleason C., Edwards A., Marsh J., Mitra R.M., Hirsch S., Jakab J., Sims S., Long S.R., Rogers J. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]; Kalo, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R.M., Hirsch, S., Jakab, J., Sims, S., Long, S.R., Rogers, J., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308:1786-1789. [DOI] [PubMed]
- Kassaw T., Bridges W., Jr., Frugoli J. Multiple autoregulation of nodulation (AON) signals identified through split root analysis of Medicago truncatula sunn and rdn1 mutants. Plants. 2015;4:209–224. doi: 10.3390/plants4020209. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kassaw, T., Bridges, W., Jr., and Frugoli, J. (2015). Multiple autoregulation of nodulation (AON) signals identified through split root analysis of Medicago truncatula sunn and rdn1 mutants. Plants 4:209-224. [DOI] [PMC free article] [PubMed]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E.D. Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U S A. 2008;105:9823–9828. doi: 10.1073/pnas.0803499105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kosuta, S., Hazledine, S., Sun, J., Miwa, H., Morris, R.J., Downie, J.A., and Oldroyd, G.E.D. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105:9823-9828. [DOI] [PMC free article] [PubMed]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 2012;8:e1002767. doi: 10.1371/journal.pgen.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, G., Meng, X., Wang, R., Mao, G., Han, L., Liu, Y., and Zhang, S. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8:e1002767. [DOI] [PMC free article] [PubMed]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Madsen, L.H., Tirichine, L., Jurkiewicz, A., Sullivan, J.T., Heckmann, A.B., Bek, A.S., Ronson, C.W., James, E.K., and Stougaard, J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1:10. [DOI] [PMC free article] [PubMed]
- Maroti G., Kondorosi E. Nitrogen-fixing rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front. Microbiol. 2014;5:326. doi: 10.3389/fmicb.2014.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]; Maroti, G., and Kondorosi, E. (2014). Nitrogen-fixing rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front. Micro. 5:326. [DOI] [PMC free article] [PubMed]
- Miranda M., Borisjuk L., Tewes A., Dietrich D., Rentsch D., Weber H., Wobus U. Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol. 2003;132:1950–1960. doi: 10.1104/pp.103.024422. [DOI] [PMC free article] [PubMed] [Google Scholar]; Miranda, M., Borisjuk, L., Tewes, A., Dietrich, D., Rentsch, D., Weber, H., and Wobus, U. (2003). Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiol 132:1950-1960. [DOI] [PMC free article] [PubMed]
- Mohd-Radzman N.A., Djordjevic M.A., Imin N. Nitrogen modulation of legume root architecture signaling pathways involves phytohormones and small regulatory molecules. Front. Plant Sci. 2013;4:385. doi: 10.3389/fpls.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mohd-Radzman, N.A., Djordjevic, M.A., and Imin, N. (2013). Nitrogen modulation of legume root architecture signaling pathways involves phytohormones and small regulatory molecules. Front. Plant Sci. 4:385. [DOI] [PMC free article] [PubMed]
- Mortier V., De Wever E., Vuylsteke M., Holsters M., Goormachtig S. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 2012;70:367–376. doi: 10.1111/j.1365-313X.2011.04881.x. [DOI] [PubMed] [Google Scholar]; Mortier, V., De Wever, E., Vuylsteke, M., Holsters, M., and Goormachtig, S. (2012). Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 70:367-376. [DOI] [PubMed]
- Nishida H., Tanaka S., Handa Y., Ito M., Sakamoto Y., Matsunaga S., Betsuyaku S., Miura K., Soyano T., Kawaguchi M. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat. Commun. 2018;9:499. doi: 10.1038/s41467-018-02831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nishida, H., Tanaka, S., Handa, Y., Ito, M., Sakamoto, Y., Matsunaga, S., Betsuyaku, S., Miura, K., Soyano, T., Kawaguchi, M., and Suzaki, T. (2018) A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat Commun. 9:499. [DOI] [PMC free article] [PubMed]
- O'Brien J.A., Vega A., Bouguyon E., Krouk G., Gojon A., Coruzzi G., Gutierrez R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant. 2016;9:837–856. doi: 10.1016/j.molp.2016.05.004. [DOI] [PubMed] [Google Scholar]; O'Brien, J.A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., and Gutierrez, R.A. (2016). Nitrate transport, sensing, and responses in plants. Mol Plant 9:837-856. [DOI] [PubMed]
- Oka-Kira E., Kawaguchi M. Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 2006;9:496–502. doi: 10.1016/j.pbi.2006.07.012. [DOI] [PubMed] [Google Scholar]; Oka-Kira, E., and Kawaguchi, M. (2006). Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 9:496-502. [DOI] [PubMed]
- Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., Kawaguchi M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Phys. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]; Okamoto, S., Ohnishi, E., Sato, S., Takahashi, H., Nakazono, M., Tabata, S., and Kawaguchi, M. (2009). Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Phys. 50:67. [DOI] [PubMed]
- Oldroyd G.E., Downie J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]; Oldroyd, G.E., and Downie, J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59:519-546. [DOI] [PubMed]
- Omrane S., Ferrarini A., D'Apuzzo E., Rogato A., Delledonne M., Chiurazzi M. Symbiotic competence in Lotus japonicus is affected by plant nitrogen status: transcriptomic identification of genes affected by a new signalling pathway. New Phytol. 2009;183:380–394. doi: 10.1111/j.1469-8137.2009.02873.x. [DOI] [PubMed] [Google Scholar]; Omrane, S., Ferrarini, A., D'Apuzzo, E., Rogato, A., Delledonne, M., and Chiurazzi, M. (2009). Symbiotic competence in Lotus japonicus is affected by plant nitrogen status: transcriptomic identification of genes affected by a new signalling pathway. New Phytol. 183:380-394. [DOI] [PubMed]
- Peiter E., Sun J., Heckmann A.B., Venkateshwaran M., Riely B.K., Otegui M.S., Edwards A., Freshour G., Hahn M.G., Cook D.R. The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 2007;145:192–203. doi: 10.1104/pp.107.097261. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peiter, E., Sun, J., Heckmann, A.B., Venkateshwaran, M., Riely, B.K., Otegui, M.S., Edwards, A., Freshour, G., Hahn, M.G., Cook, D.R., et al. (2007). The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol 145:192-203. [DOI] [PMC free article] [PubMed]
- Pellizzaro A., Alibert B., Planchet E., Limami A.M., Morere-Le Paven M.C. Nitrate transporters: an overview in legumes. Planta. 2017;246:585–595. doi: 10.1007/s00425-017-2724-6. [DOI] [PubMed] [Google Scholar]; Pellizzaro, A., Alibert, B., Planchet, E., Limami, A.M., and Morere-Le Paven, M.C. (2017). Nitrate transporters: an overview in legumes. Planta 246:585-595. [DOI] [PubMed]
- Penmetsa R.V., Frugoli J.A., Smith L.S., Long S.R., Cook D.R. Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 2003;131:998–1008. doi: 10.1104/pp.015677. [DOI] [PMC free article] [PubMed] [Google Scholar]; Penmetsa, R.V., Frugoli, J.A., Smith, L.S., Long, S.R., and Cook, D.R. (2003). Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131:998-1008. [DOI] [PMC free article] [PubMed]
- Reid D.E., Ferguson B.J., Gresshoff P.M. Inoculation-and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 2011;24:606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]; Reid, D.E., Ferguson, B.J., and Gresshoff, P.M. (2011). Inoculation-and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 24:606-618. [DOI] [PubMed]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ritchie, M.E., Phipson, B., Wu, D., Hu, Y., Law, C.W., Shi, W., and Smyth, G.K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed]
- Schnabel E., Journet E.P., de Carvalho-Niebel F., Duc G., Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]; Schnabel, E., Journet, E.P., de Carvalho-Niebel, F., Duc, G., and Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58:809-822. [DOI] [PubMed]
- Searle I.R., Men A.E., Laniya T.S., Buzas D.M., Iturbe-Ormaetxe I., Carroll B.J., Gresshoff P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]; Searle, I.R., Men, A.E., Laniya, T.S., Buzas, D.M., Iturbe-Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299:109-112. [DOI] [PubMed]
- Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]; Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37:914-939. [DOI] [PubMed]
- Tirichine L., Imaizumi-Anraku H., Yoshida S., Murakami Y., Madsen L.H., Miwa H., Nakagawa T., Sandal N., Albrektsen A.S., Kawaguchi M. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]; Tirichine, L., Imaizumi-Anraku, H., Yoshida, S., Murakami, Y., Madsen, L.H., Miwa, H., Nakagawa, T., Sandal, N., Albrektsen, A.S., Kawaguchi, M., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441:1153-1156. [DOI] [PubMed]
- van Noorden G.E., Verbeek R., Dinh Q.D., Jin J., Green A., Ng J.L., Mathesius U. Molecular signals controlling the inhibition of nodulation by nitrate in Medicago truncatula. Int. J. Mol. Sci. 2016;17:E1060. doi: 10.3390/ijms17071060. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Noorden, G.E., Verbeek, R., Dinh, Q.D., Jin, J., Green, A., Ng, J.L., and Mathesius, U. (2016). Molecular Signals Controlling the Inhibition of Nodulation by Nitrate in Medicago truncatula. Int. J. Mol. Sci. 17:1060. [DOI] [PMC free article] [PubMed]
- Vernié T., Kim J., Frances L., Ding Y., Sun J., Guan D., Niebel A., Gifford M., de Carvalho-Niebel F., Oldroyd G.E.D. The NIN transcription factor coordinates diverse nodulation programmes in different tissues of the Medicago truncatula root. Plant Cell. 2015;27:3410–3424. doi: 10.1105/tpc.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vernie, T., Kim, J., Frances, L., Ding, Y., Sun, J., Guan, D., Niebel, A., Gifford, M., de Carvalho-Niebel, F., Oldroyd, GED. (2015). The NIN transcription factor coordinates diverse nodulation programmes in different tissues of the Medicago truncatula root. Plant Cell 27:3410-3424. [DOI] [PMC free article] [PubMed]
- Vidal E.A., Araus V., Lu C., Parry G., Green P.J., Coruzzi G.M., Gutierrez R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vidal, E.A., Araus, V., Lu, C., Parry, G., Green, P.J., Coruzzi, G.M., and Gutierrez, R.A. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107:4477-4482. [DOI] [PMC free article] [PubMed]
- Vincent J.M. Blackwell Scientific Press; Oxford: 1970. A Manual for the Practical Study of Root-Nodule Bacteria. [Google Scholar]; Vincent, J.M. (1970). A manual for the practical study of root-nodule bacteria. Oxford: Blackwell Scientific Press.
- Walker L., Boddington C., Jenkins D., Wang Y., Gronlund J.T., Hulsmans J., Kumar S., Patel D., Moore J.D., Carter A. Changes in gene expression in space and time orchestrate environmentally mediated shaping of root architecture. Plant Cell. 2017;29:2393–2412. doi: 10.1105/tpc.16.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]; Walker, L., Boddington, C., Jenkins, D., Wang, Y., Gronlund, J.T., Hulsmans, J., Kumar, S., Patel, D., Moore, J.D., Carter, A., et al. (2017). Changes in gene expression in space and time orchestrate environmentally mediated shaping of root architecture. Plant Cell 29:2393-2412. [DOI] [PMC free article] [PubMed]
- Weaver C.D., Shomer N.H., Louis C.F., Roberts D.M. Nodulin 26, a nodule-specific symbiosome membrane protein from soybean, is an ion channel. J. Biol. Chem. 1994;269:17858–17862. [PubMed] [Google Scholar]; Weaver, C.D., Shomer, N.H., Louis, C.F., and Roberts, D.M. (1994). Nodulin 26, a nodule-specific symbiosome membrane protein from soybean, is an ion channel. J Biol Chem 269:17858-17862. [PubMed]
- Wopereis J., Pajuelo E., Dazzo F.B., Jiang Q., Gresshoff P.M., De Bruijn F.J., Stougaard J., Szczyglowski K. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 2000;23:97–114. doi: 10.1046/j.1365-313x.2000.00799.x. [DOI] [PubMed] [Google Scholar]; Wopereis, J., Pajuelo, E., Dazzo, F.B., Jiang, Q., Gresshoff, P.M., De Bruijn, F.J., Stougaard, J., and Szczyglowski, K. (2000). Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J 23:97-114. [DOI] [PubMed]
- Xu C., Luo F., Hochholdinger F. LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 2016;21:159–167. doi: 10.1016/j.tplants.2015.10.010. [DOI] [PubMed] [Google Scholar]; Xu, C., Luo, F., and Hochholdinger, F. (2016). LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci 21:159-167. [DOI] [PubMed]
- Yamada T., Imaishi H., Oka A., Ohkawa H. Molecular cloning and expression in Saccharomyces cerevisiae of tobacco NADPH-cytochrome P450 oxidoreductase cDNA. Biosci. Biotechnol. Biochem. 1998;62:1403–1411. doi: 10.1271/bbb.62.1403. [DOI] [PubMed] [Google Scholar]; Yamada, T., Imaishi, H., Oka, A., and Ohkawa, H. (1998). Molecular cloning and expression in Saccharomyces cerevisiae of tobacco NADPH-cytochrome P450 oxidoreductase cDNA. Biosci. Biotechnol. Biochem. 62:1403-1411. [DOI] [PubMed]
- Yu N., Niu Q.W., Ng K.H., Chua N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015;83:673–685. doi: 10.1111/tpj.12919. [DOI] [PubMed] [Google Scholar]; Yu, N., Niu, Q.W., Ng, K.H., and Chua, N.H. (2015). The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83:673-685. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Average (ave) and standard error (SE) for number of nodules of A17 M. truncatula seedlings grown over a range of concentrations of NH4NO3.
(B) Average (ave) and SE root trait values measured from 18-day-old A17 M. truncatula seedlings grown in the conditions specified.
(C) Adjusted P values from pairwise Wilcoxon rank-sum test with Holm's method for multiple testing; green shading indicates P value <0.05.
The table lists the Medicago microarray probes with their Medicago 3.5v3 and 4.0v1 gene IDs and annotation represented on the array (for design see GEO record GPL25305 for Nimblegen probe design) and their log2 scale expression values in all experiments as well as averaged (“Ave”) for each replicate set; see GEO: GSE116789 for complete raw and normalized individual replicate values. If a gene is significantly differentially expressed between samples the cluster number is listed (clustering on N-treatment responses and by “raw,” uncompared values). In comparisons between experiments for all genes with an Mt4.0 annotation the direction of regulation (“regulation” column, 0 = no change, −1 = downregulated, 1 = upregulated) and P value of significance (“Pval” column) are given. NA, not tested; n = 3 except for Rhizobia-6hNitrogen where n = 2.
(A) Terms enriched for differentially expressed genes (DEGs) whose expression changes over time in A17 or between A17 and sunn-1.
(B) Terms enriched in 14 clusters of DEG in the A17 time series data.
(C) Terms enriched in 11 clusters of DEG in the A17–sunn-1 experiment; red color = P value <0.05.
(A) Oligo sequences used for amplification of qPCR products shown in Supplemental Information and descriptions of each gene.
(B) Gene expression level from microarray hybridization.
(C) Gene expression level from qPCR; ND, not detectible.
The table lists the Medicago microarray probes with their Medicago 3.5v3 and 4.0v1 gene IDs and annotation represented on the array (for design see GEO record GPL25305 for Nimblegen probe design) and their log2 scale expression values in all experiments as well as averaged (“Ave”) for each replicate set; see GEO: GSE116789 for complete raw and normalized individual replicate values. If significantly differentially expressed between samples the cluster number is listed; NA, not tested.
(A) Average (ave) and SE for root trait values measured from 18-day-old sunn-1 M. truncatula seedlings grown in the conditions specified.
(B) Adjusted P values from pairwise Wilcoxon rank-sum test with Holm's method for multiple testing; green shading indicates P value <0.05.
Adjusted P values from pairwise Wilcoxon rank-sum test with Holm's method for multiple testing; green shading indicates P value <0.05.
(A) Dry weight and measurements of free nitrate, total % N, and total % C in A17 and sunn-1 M. truncatula shoots grown in the experiments designated; SD, standard deviation.
(B) Adjusted P values from t-tests comparing dry shoot weight or mineral analysis levels; green shading indicates P value <0.05.